Key Points
Question
Does a benign genotype that is common among African American individuals and lowers white blood cell (WBC) counts contribute to unnecessary bone marrow biopsies?
Findings
In this genetic association study, among 399 African American individuals who underwent a bone marrow biopsy as part of routine clinical care at 3 medical centers, the rs2814778-CC genotype was highly prevalent when the biopsy was performed to evaluate isolated low WBC counts. In the absence of another cell type abnormality, these biopsies very rarely identified a hematologic abnormality.
Meaning
These findings suggest that genotyping for rs2814778-CC may avoid unnecessary bone marrow biopsies related to isolated low WBC counts among African American patients.
Abstract
Importance
Up to two-thirds of African American individuals carry the benign rs2814778-CC genotype that lowers total white blood cell (WBC) count.
Objective
To examine whether the rs2814778-CC genotype is associated with an increased likelihood of receiving a bone marrow biopsy (BMB) for an isolated low WBC count.
Design, Setting, and Participants
This retrospective genetic association study assessed African American patients younger than 90 years who underwent a BMB at Vanderbilt University Medical Center, Mount Sinai Health System, or Children’s Hospital of Philadelphia from January 1, 1998, to December 31, 2020.
Exposure
The rs2814778-CC genotype.
Main Outcomes and Measures
The proportion of individuals with the CC genotype who underwent BMB for an isolated low WBC count and had a normal biopsy result compared with the proportion of individuals with the CC genotype who underwent BMB for other indications and had a normal biopsy result.
Results
Among 399 individuals who underwent a BMB (mean [SD] age, 41.8 [22.5] years, 234 [59%] female), 277 (69%) had the CC genotype. A total of 35 patients (9%) had clinical histories of isolated low WBC counts, and 364 (91%) had other histories. Of those with a clinical history of isolated low WBC count, 34 of 35 (97%) had the CC genotype vs 243 of 364 (67%) of those without a low WBC count history. Among those with the CC genotype, 33 of 34 (97%) had normal results for biopsies performed for isolated low WBC counts compared with 134 of 243 individuals (55%) with biopsies performed for other histories (P < .001).
Conclusions and Relevance
In this genetic association study, among patients of African American race who had a BMB with a clinical history of isolated low WBC counts, the rs2814778-CC genotype was highly prevalent, and 97% of these BMBs identified no hematologic abnormality. Accounting for the rs2814778-CC genotype in clinical decision-making could avoid unnecessary BMB procedures.
This genetic association study examines whether the rs2814778-CC genotype is associated with an increased likelihood of receiving a bone marrow biopsy for an isolated low white blood cell count.
Introduction
The observation of lower mean white blood cell (WBC) counts among individuals of African ancestry compared with those of European ancestry is well established.1,2,3,4 The clinical finding of low WBC counts among healthy individuals of African ancestry is variably designated benign ethnic neutropenia and is not associated with an increased risk of infection.5,6,7 This phenomenon is attributed, in large part, to homozygosity for the rs2814778-C variant in the promoter of ACKR1 (OMIM 613665) gene. Loss of expression of ACKR1 on cell surfaces may lead to relative neutropenia attributable to sequestration of neutrophils in peripheral tissues, including the spleen.8 This variant is common (allele frequency, 0.96) among populations in sub-Saharan Africa but uncommon (allele frequency, 0.006) among White populations of European ancestry.9,10,11,12,13 Approximately 63% of African American individuals, who are of mixed European and African ancestries, have the CC genotype and would be anticipated to have benign ethnic neutropenia.9
Because reference ranges of normal values for WBC counts do not account for genotype, persons with the rs2814778-CC genotype are more likely to have a WBC count that falls below the reference range. Although many practitioners are aware of benign ethnic neutropenia, genotyping for the rs2814778-C variant is not routine, and the extent to which this genetic diagnosis is clinically recognized is unknown. We used data from 3 large, unselected clinical biobank populations of African American patients to ascertain whether those undergoing bone marrow biopsy (BMB) with a clinical history of isolated low WBC counts were more likely to have the rs2814778-CC genotype than those undergoing BMB for other indications and how likely the biopsies were to identify a hematologic abnormality.
Methods
Study Population
Individuals were identified at 3 institutions with DNA biobanks linked to electronic health record (EHR) resources. All participants provided written informed consent. For minors at Children’s Hospital of Philadelphia (CHOP), written informed consent was acquired from a parent or guardian with verbal assent from the minor. All data were deidentified. This study was evaluated and approved by the institutional review boards of Vanderbilt University Medical Center (VUMC), Icahn School of Medicine at Mount Sinai (ISMMS), and CHOP. The study followed the Strengthening the Reporting of Genetic Association Studies (STREGA) reporting guideline.
The cohort incorporated individuals from all sites and comprised African American patients (as specified by each site as described below) with prior genotyping of the rs2814778 (NC_000001.11:g.159204893T>C) variant or, at VUMC, DNA available for analysis and a BMB report. Ungenotyped individuals at VUMC with a WBC count abnormality noted on their BMB report underwent single-nucleotide variation genotyping.
The VUMC BioVU resource is constructed from discarded blood samples collected from individuals who had provided consent and linked to deidentified EHRs.14 Individuals participating in these analyses had an EHR-reported race that included the term Black and did not fall within the White European genetic ancestry cluster–based HapMap reference populations. Some individuals had previously undergone single-nucleotide variation genotyping using the Illumina Infinium MEGAEX platform or the Illumina Infinium Exome BeadChip, version 1.1 (Illumina Inc). Of 68 ungenotyped individuals with a BMB and a WBC abnormality, 48 had available DNA and were genotyped using the MEGAEX platform. Quality control analyses were performed by the Vanderbilt Technologies for Advanced Genomics Analysis and Research Design core, as previously described.15,16 Quality control and data processing used PLINK, version 1.90β3.42.17 Individuals missing genotype data for the rs2814778 variant were excluded.
The ISMMS BioMe Biobank is an ancestrally diverse, EHR-linked biobank ascertained through the Mount Sinai Health System in New York City. Genotyping of BioMe participants was performed in collaboration with the Regeneron Genetics Center. Analyses were restricted to genotyped African American/African participants, using self-reported race/ethnicity data ascertained from BioMe enrollment questionnaires, as previously described.18 The rs2814778 variant was directly genotyped across the BioMe Biobank as part of the global screening array (635 623 sites and 32 595 individuals in total). Individual-level quality control was performed, which resulted in the exclusion of 890 individuals in total who exhibited significant deviances in genome-wide heterozygosity (±6 SDs from the population mean), elevated missingness (>5% missingness across the array), or discordance between genetic and EHR recorded sex or were intentional genetic duplicates.
The Center for Applied Genomics at CHOP was founded in 2006 and incorporates biobanking and genotyping and sequencing facilities. Data from EHRs are available for all CHOP patients, and DNA samples are banked in a biorepository.19 Individuals 0 to 21 years of age are recruited during inpatient and outpatient visits, including emergency, ambulatory, surgical, general, and specialty pediatric practices. Analyses were restricted to participants with self-reported Black or African American race. All samples were genotyped on Illumina Infinium Beadchips with genome-wide coverage of at least 500 000 single-nucleotide variants. The rs2814778 variant genotype was imputed using the Minimac3 imputation software with the Haplotype Reference Consortium, version r1.1 imputation reference panel.
BMB Identification and Review
The BMB reports were identified by searching for the keywords bone marrow or hematopathology among electronic pathology reports, using the first report if individuals had multiple reports. Reports were reviewed by physicians blinded to genotype (N.S.A.H., M.S.E., and J.D.M.). First, a reviewing physician excluded biopsies performed for follow-up of a previously diagnosed or treated hematologic condition (eg, lymphoma) or cancer or staging of an established diagnosis (eg, sarcoidosis). The same physician extracted the clinical history provided to the pathologist (eg, high or low WBC, high or low levels of other hematologic cell types, and pancytopenia) from the biopsy report from a REDCap database. These data were categorized into 2 groups: isolated low WBC count or other. Included biopsy reports were then reviewed directly by or in consultation with a clinical hematologist (S.C.B.) to categorize the overall pathological findings (normal BMB result vs clinically significant hematologic abnormalities in any cell line).
Statistical Analysis
rs2814778 genotypes were coded as CC vs CT and TT (binary). Baseline characteristics of the study population were stratified by genotype, including the frequencies of the specified clinical histories and biopsy results. We fit a multivariable logistic regression model to examine the association of biopsy result (normal vs other) with biopsy indication (isolated low WBC count vs other) adjusted for age, site, and sex and using a 2-sided P < .05 significance level likelihood ratio test. This analysis was restricted to the subset of 277 patients with the CC genotype, given the small number of individuals with CT/TT genotype and isolated low WBC counts.
Results
Clinical and genetic data for a total of 399 individuals (mean [SD] age, 41.8 [22.5] years, 234 [59%] female) from the 3 academic medical centers who had undergone a BMB to establish a new hematological diagnosis were available for study (Figure, A-C). An isolated low WBC count was noted in the clinical history for 35 biopsies (9%). A total of 364 biopsies (91%) were performed for other clinical histories.
Figure. Selection of the Bone Marrow Biopsy Population.
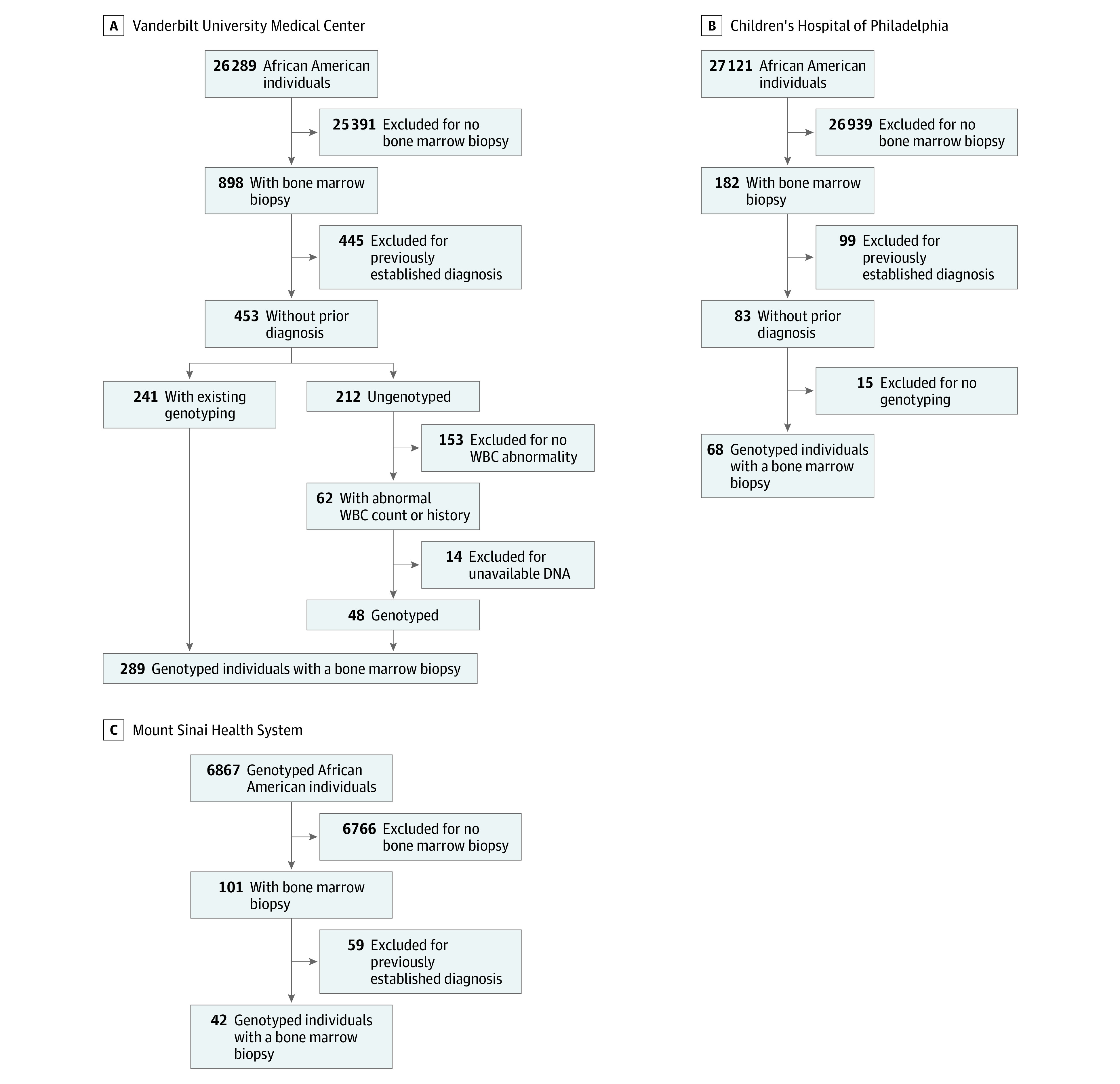
The initial populations were drawn from African American individuals identified through nonselected institutional DNA biobanks. WBC indicates white blood cell.
The CC genotype was far more common, occurring in 34 of 35 (97%) of those with a clinical history of isolated low WBC count vs 243 of 364 (67%) among those with other clinical histories (Table). The numbers for additional clinical histories are given in eTable 1 in the Supplement. The median WBC count at the time of biopsy among carriers of the CC genotype and with isolated low WBC count was 2.9/μL (interquartile range, 2.4-3.9/μL) (to convert to ×109/L, multiply by 0.001). Higher CC genotype representation among those with a clinical history of isolated low WBC counts was consistent when data were stratified by clinical site (eTable 2 in the Supplement) and by adult vs pediatric participants (eTable 3 in the Supplement).
Table. Characteristics of Individuals Receiving a Bone Marrow Biopsy by Genotypea.
| Characteristic | CC genotype (n = 277) | CT/TT genotype (n = 122) |
|---|---|---|
| Female | 163 (58.8) | 71 (58.2) |
| Male | 114 (41.2) | 51 (41.8) |
| Age, mean (SD), y | 42.2 (22.2) | 40.9 (23.1) |
| Clinical history | ||
| Isolated low WBC count | 34 (12.3) | 1 (0.8) |
| Other | 243 (87.7) | 121 (99.2) |
| Biopsy result | ||
| Normal | 167 (60.3) | 65 (53.3) |
| Abnormal | 110 (39.7) | 57 (46.7) |
Abbreviation: WBC, white blood cell.
Data are presented as number (percentage) of individuals unless otherwise indicated. The percentages represent the percentage of individuals within the genotype category.
Because so few people with the low WBC count indication had the CT/TT genotype (a total of 1 with isolated low WBC count), we restricted further analyses to those with the CC genotype. Among 34 individuals with the CC genotype and a clinical history of isolated low WBC counts, 33 (97%) had a biopsy that did not identify bone marrow abnormalities and 1 had an abnormal biopsy result (P < .001). In contrast, 134 of 243 individuals (55%) with the CC genotype and other clinical histories had a normal biopsy result and 109 had abnormal biopsy results (P < .001 from logistic regression). Similar trends are seen when results are stratified by clinical site (eTable 4 in the Supplement). Normal biopsy rates for other clinical histories are presented in eTable 5 in the Supplement. Review of the clinical record for the 1 individual with the abnormal biopsy and a low WBC history revealed that the biopsy was performed to follow-up findings of isolated lymphopenia and blast cells with Auer rods on a peripheral blood smear. Consistently, the biopsy identified acute myeloid leukemia.
Discussion
In this genetic association study, among African American individuals who underwent BMB, differences were found in the indications for the procedure by genotype groups. Namely, those with the rs2814778-CC genotype more often had a BMB because of isolated low WBC counts. Furthermore, individuals with this genotype were more likely to have a normal BMB result. For those with the rs2814778-CC genotype and an isolated low WBC indication for the procedure, nearly all the BMBs had normal results and may not have been necessary.
Genetic variation accounts for a significant portion of the variability in the levels of some clinical biomarkers, such as WBC counts. Reference ranges for biomarker measurements are generally defined within restricted populations, without consideration of this variation. When genetic variation is confounded with race, these inaccurate reference ranges contribute to health disparities among underrepresented populations, even when the differences in observed values are clinically recognized. These analyses highlight the potential costs and consequences of misclassifying individuals carrying benign genetic variation. BMB is an invasive and relatively expensive procedure pursued to identify the cause of hematologic findings, such as neutropenia. When the indication for biopsy was isolated low WBC count, no pathological findings were observed in 97% of the biopsies. Thus, among this group, there was substantial cost and risk with minimal to no added benefit. This finding was consistent across 3 academic medical centers and across adult and pediatric subgroups.
The costs attributable to this benign variant may be substantial and fall disproportionately on those of African ancestry. We can only estimate the burden of this testing for individuals with the rs2814778-CC genotype. According to the publicly available list of charges for services and procedures at VUMC,20 a BMB is associated with charges of at least $6000. The 34 BMBs among those with the CC genotype and isolated low WBC count as the indication would lead to charges of $204 000.
One proposed solution to address misclassification of outlying biomarker values is to define race-based reference ranges.21 However, self-reported race is an inaccurate marker of persons carrying the rs2814778-CC genotype. Although African ancestry is known to be associated with lower WBC counts, only 69% of African American persons in our study carried the rs2814778-CC genotype, so race-based clinical inferences about expected WBC counts would be expected to be incorrect approximately one-third of the time. Accurate clinical interpretation of a low WBC count is only possible by knowing the individual’s genotype. As the US population becomes increasingly admixed,18,22 reliance on self-identified race will become increasingly inaccurate.
Another potential solution is additional education for practitioners. The study data indicate that hematologists continue to include BMB in the workup of isolated low WBC counts in some African American patients. In current clinical practice, WBC variation attributable to this genotype is not easily distinguished from WBC variation attributable to disease. An alternative approach supported by this study is to define genotype-guided reference ranges.
This study focused on a single variant with known effects to assess for health care overuse attributable to ancestry-associated benign genetic variation. Given the substantial impact of mendelian and complex genetics on clinical biomarkers and long-standing reliance on data from individuals of European ancestry to define reference ranges, the potential contribution of heritable genetic variation to health disparities may be substantial. The moniker for the blood disorder benign ethnic neutropenia provides insight into an important feature of the problem: population-based norms often represent genetic variation common to European populations.23 In theory and practice, the moniker benign ethnic neutrophilia could be equally ascribed to White European ancestries. It is crucial to develop evidence-based approaches to define the scope of this problem, which is largely unexplored, and to enact appropriate solutions.
Limitations
This study has limitations. Although the data from this study came from 3 tertiary care medical centers, the results may not be generalizable to nonbiobank populations or other communities. The phenotype data were derived from EHRs, which reflect only the care obtained within the 3 care systems and do not include encounters from outside facilities. These data are retrospective, and causality cannot be assessed. However, genotypes by definition preceded all clinical activities, and those performing manually completed data extraction (eg, review of BMB data) were blinded to genotype at all sites. Data pertaining to the actual charges generated by BMBs and the associated clinical visits are not available for analysis to determine the financial cost. The generally high rate of normal biopsy results (and low rate of adverse pathological findings) for the subset of individuals included in the study limited the power to detect differences and to make definitive recommendations as to when biopsies are indicated vs inappropriate. Larger studies are warranted to provide recommendations with a high level of confidence to guide the care of patients of non-European ancestry.
Also, study of a single genetically defined trait that predominantly affects individuals of African ancestry fails to address the complexity of genomic biology. This study also does not explore larger but important issues, such as underrepresentation of many populations as stakeholders in our health care system, social determinants of health as mediators of health outcomes, and the effects of systemic racism on health and health care. Indeed, the use of the term African American throughout this article is an inadequate descriptor for those included in the study because the study cohort included diverse populations of individuals who received care at tertiary care medical centers based in Nashville, Tennessee, New York City, New York, and Philadelphia, Pennsylvania, and may not self-identify specifically as African American. The goal in presenting these data is to provide an example of when use of genetic data may have a positive impact to reduce a single contributor to health disparity. We hope these data also further the conversation of health disparities, recognizing that the terms for these discussions are evolving.
Conclusions
Among African American participants in in this study, 69% had the rs2814778-CC genotype, previously associated with lower WBC counts. This genotype was more frequent among those who underwent a BMB for an isolated low WBC count than among those who underwent a biopsy for another indication; most biopsies for an isolated low WBC count identified no hematologic abnormality. Accounting for the rs2814778-CC genotype in clinical decision-making may avoid unnecessary BMBs, which would be particularly beneficial for African American individuals, in whom the risk genotype is common.
eTable 1. Summary of Clinical Histories for Subjects Receiving a Bone Marrow Biopsy
eTable 2. Characteristics of Subjects Receiving a Bone Marrow Biopsy by Site
eTable 3. Characteristics of Subjects Receiving a Bone Marrow Biopsy by Age
eTable 4. Frequency of Normal Biopsy for Isolated Low WBC for Participants by Genotype and Site
eTable 5. Frequency of Normal Bone Marrow Biopsy Result, by Clinical History and Genotype
References
- 1.Bain B, Seed M, Godsland I. Normal values for peripheral blood white cell counts in women of four different ethnic origins. J Clin Pathol. 1984;37(2):188-193. doi: 10.1136/jcp.37.2.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brain P, Buckle GC, Jamieson M. Haematological differences in three population groups. S Afr Med J. 1979;55(16):635-636. [PubMed] [Google Scholar]
- 3.Shaper AG, Lewis P. Genetic neutropenia in people of African origin. Lancet. 1971;2(7732):1021-1023. doi: 10.1016/S0140-6736(71)90335-7 [DOI] [PubMed] [Google Scholar]
- 4.Broun GO Jr, Herbig FK, Hamilton JR. Leukopenia in Negroes. N Engl J Med. 1966;275(25):1410-1413. doi: 10.1056/NEJM196612222752504 [DOI] [PubMed] [Google Scholar]
- 5.Ortiz MV, Meier ER, Hsieh MM. Identification and clinical characterization of children with benign ethnic neutropenia. J Pediatr Hematol Oncol. 2016;38(3):e140-e143. doi: 10.1097/MPH.0000000000000528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakhotia R, Aggarwal A, Link ME, Rodgers GP, Hsieh MM. Natural history of benign ethnic neutropenia in individuals of African ancestry. Blood Cells Mol Dis. 2019;77:12-16. doi: 10.1016/j.bcmd.2019.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Atallah-Yunes SA, Ready A, Newburger PE. Benign ethnic neutropenia. Blood Rev. 2019;37:100586. doi: 10.1016/j.blre.2019.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rappoport N, Simon AJ, Amariglio N, Rechavi G. The Duffy antigen receptor for chemokines, ACKR1—‘Jeanne DARC’ of benign neutropenia. Br J Haematol. 2019;184(4):497-507. doi: 10.1111/bjh.15730 [DOI] [PubMed] [Google Scholar]
- 9.Nalls MA, Wilson JG, Patterson NJ, et al. Admixture mapping of white cell count: genetic locus responsible for lower white blood cell count in the Health ABC and Jackson Heart Studies. Am J Hum Genet. 2008;82(1):81-87. doi: 10.1016/j.ajhg.2007.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10(2):224-228. doi: 10.1038/ng0695-224 [DOI] [PubMed] [Google Scholar]
- 11.Reich D, Nalls MA, Kao WHL, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5(1):e1000360. doi: 10.1371/journal.pgen.1000360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Charles BA, Hsieh MM, Adeyemo AA, et al. Analyses of genome wide association data, cytokines, and gene expression in African-Americans with benign ethnic neutropenia. PLoS One. 2018;13(3):e0194400. doi: 10.1371/journal.pone.0194400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.1000 Genomes Project Consortium, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;526(7571):68-74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84(3):362-369. doi: 10.1038/clpt.2008.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo Y, He J, Zhao S, et al. Illumina human exome genotyping array clustering and quality control. Nat Protoc. 2014;9(11):2643-2662. doi: 10.1038/nprot.2014.174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mosley JD, Witte JS, Larkin EK, et al. Identifying genetically driven clinical phenotypes using linear mixed models. Nat Commun. 2016;7:11433. doi: 10.1038/ncomms11433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559-575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Belbin GM, Cullina S, Wenric S, et al. ; CBIPM Genomics Team; Regeneron Genetics Center . Toward a fine-scale population health monitoring system. Cell. 2021;184(8):2068-2083.e11. doi: 10.1016/j.cell.2021.03.034 [DOI] [PubMed] [Google Scholar]
- 19.Connolly JJ, Glessner JT, Li D, Sleiman PMA, Hakonarson H. The Center for Applied Genomics at the Children’s Hospital of Philadelphia—pediatric perspectives on genomic medicine. The Journal of Precision Medicine. Accessed May 18, 2021. https://www.thejournalofprecisionmedicine.com/the-journal-of-precision-medicine/the-center-for-applied-genomics-at-the-childrens-hospital-of-philadelphia-pediatric-perspectives-on-genomic-medicine
- 20.Estimates and charges. Vanderbilt Health Nashville, TN. Accessed May 15, 2020. https://www.vanderbilthealth.com/information/estimates-and-charges
- 21.Rappoport N, Paik H, Oskotsky B, et al. Comparing ethnicity-specific reference intervals for clinical laboratory tests from EHR data. J Appl Lab Med. 2018;3(3):366-377. doi: 10.1373/jalm.2018.026492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathias RA, Taub MA, Gignoux CR, et al. ; CAAPA . A continuum of admixture in the Western Hemisphere revealed by the African Diaspora genome. Nat Commun. 2016;7:12522. doi: 10.1038/ncomms12522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merz LE, Achebe M. When non-Whiteness becomes a condition. Blood. 2021;137(1):13-15. doi: 10.1182/blood.2020008600 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. Summary of Clinical Histories for Subjects Receiving a Bone Marrow Biopsy
eTable 2. Characteristics of Subjects Receiving a Bone Marrow Biopsy by Site
eTable 3. Characteristics of Subjects Receiving a Bone Marrow Biopsy by Age
eTable 4. Frequency of Normal Biopsy for Isolated Low WBC for Participants by Genotype and Site
eTable 5. Frequency of Normal Bone Marrow Biopsy Result, by Clinical History and Genotype


