Abstract
Campylobacter jejuni is the leading cause of food poisoning in the United States and Europe. The exterior cell surface of C. jejuni is coated with a capsular polysaccharide (CPS) that is essential for the maintenance and integrity of the bacterial cell wall and evasion of the host immune response. The identity and sequences of the monosaccharide components of the CPS are quite variable and dependent on the specific strain of C. jejuni. It is currently thought that the immediate precursor for the multiple variations found in the heptose moieties of the C. jejuni CPS is GDP-d-glycero-α-d-manno-heptose. In C. jejuni NCTC 11168 the heptose moiety is d-glycero-l-gluco-heptose. It has previously been shown that Cj1427 catalyzes the oxidation of GDP-d-glycero-α-d-manno-heptose to GDP-d-glycero-4-keto-α-d-lyxo-heptose using α-ketoglutarate as a co-substrate. Cj1430 was now demonstrated to catalyze the double epimerization of this product at C3 and C5 to form GDP-d-glycero-4-keto-β-l-xylo-heptose. Cj1428 subsequently catalyzes the stereospecific reduction of this GDP-linked heptose by NADPH to form GDP-d-glycero-β-l-gluco-heptose. The three-dimensional crystal structure of Cj1430 was determined to a resolution of 1.85 Å in the presence of bound GDP-d-glycero-β-l-gluco-heptose, a product analog. The structure shows that it belongs to the cupin superfamily. The three-dimensional crystal structure of Cj1428 was solved in the presence of NADPH to a resolution of 1.50 Å. Its fold places it into the short chain dehydrogenase/reductase superfamily. Typically, members in this family display a characteristic signature sequence of YXXXK, with the conserved tyrosine serving a key role in catalysis. In Cj1428, this residue is a phenylalanine.
Graphical Abstract
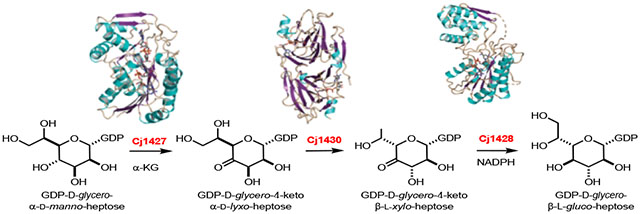
INTRODUCTION
Campylobacter jejuni is the leading cause of food poisoning in the United States and Europe.1 The pathogenicity of this organism, commonly found in chickens and cattle, is facilitated by its ability to evade the host immune response, high adaptability, and growing antibiotic resistance.2 Several carbohydrate-based structures are appended to the cell surface of C. jejuni including capsular polysaccharides (CPS) and lipooligosaccharides (LOS). The CPS and LOS are essential for the maintenance and integrity of the bacterial cell wall and the evasion of the host immune response upon infection.3 The repeating carbohydrate found in the CPS of C. jejuni NCTC 11168 is presented in Scheme 1.
Scheme 1:
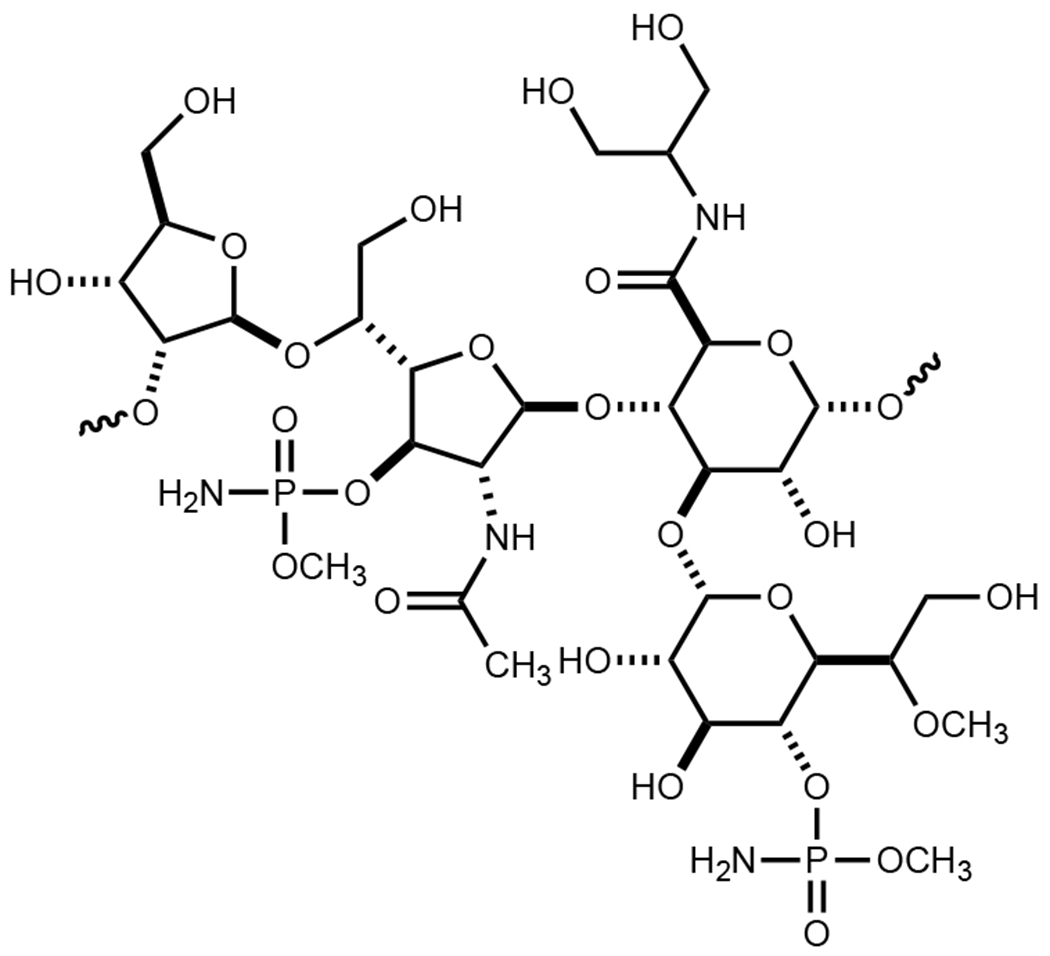
Structure of the repeating carbohydrate sequence in the CPS from C. jejuni NCTC 11168.
The chemical compositions of at least 12 CPS structures have been reported from various strains of C. jejuni.4 In the CPS of C. jejuni 11168 one of the four repeating carbohydrates has been identified as d-glycero-l-gluco-heptose.5 In other strains of C. jejuni 13 different structural variations of a heptose moiety have been identified. It is currently thought that all of the structurally distinct heptose sugars originate from GDP-d-glycero-α-d-manno-heptose (1). The proposed transformation from this heptose to GDP-d-glycero-β-l-gluco-heptose (4) is illustrated in Scheme 2.6, 7 This biosynthetic pathway was based, in part, on demonstrated catalytic activities of Cj1430 and Cj1428 with surrogate substrates that lacked the hydroxyl group of the physiological substate at C6.7 However, we recently demonstrated that Cj1427 can effectively oxidize C4 of GDP-d-glycero-α-d-manno-heptose (1) in the presence of α-ketoglutarate (α-KG) to form GDP-d-glycero-4-keto-α-d-lyxo-heptose (2).6, 8
Scheme 2:
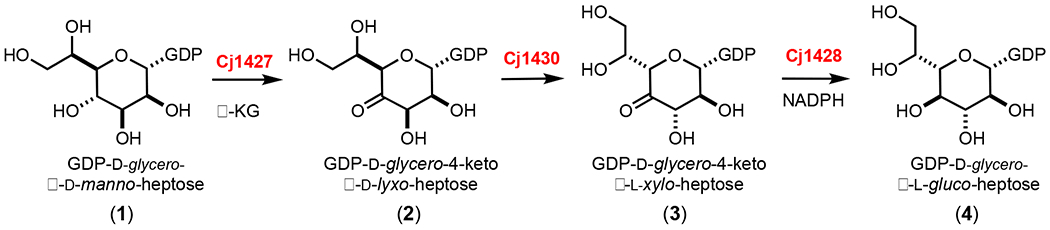
Proposed biosynthetic pathway for the formation of d-glycero-l-gluco-heptose in C. jejuni 11168.
In this report we demonstrate, for the first time, that GDP-d-glycero-β-l-gluco-heptose (4) can be synthesized from GDP-d-glycero-α-d-manno-heptose (1) from the combined activities of Cj1427, Cj1430, and Cj1428 in the presence of α-KG and NADPH. The kinetic constants for the catalytic activities of Cj1430 and Cj1428 have been measured, and the structures of these two enzymes have been determined in the presence of bound ligands.
MATERIALS and METHODS
Materials.
All materials and chemicals used were obtained from Sigma-Aldrich, GE Healthcare Bio-Sciences, or Carbosynth, unless otherwise stated. α-Ketoglutarate was purchased from AK Scientific.
Cloning of the Genes for Cj1427, Cj1428, Cj1430, and Cjj1430.
The cloning of the genes for Cj1427 (Uniprot entry: Q0P8I7), Cj1428 (Uniprot entry: Q0P8I6), and Cj1430 (Uniprot entry: Q0P8I4) from C. jejuni NCTC 11168 (HS:2) has been reported previously.6 All three clones were made with an N-terminal hexa-histidine tag in a pET30a+ expression vector between the BamHI and XhoI restriction sites. The codon-optimized plasmid for the expression of Cjj1430 (Uniprot entry: Q29VV4) from C. jejuni 81-176 (HS:23/36) was ordered from Twist Biosciences in a pET28 vector containing an N-terminal hexa-histidine tag.
Expression and Purification of Cj1427, Cj1428, Cj1430, and Cjj1430.
The methods for purification of Cj1427, Cj1428, and Cj1430 have previously been described.6 The protein encoded by the gene for Cjj1430 was prepared in the same manner. All four constructs were used to transform Escherichia coli BL21 (DE3) competent cells. Single colonies were used to create starter cultures that contained kanamycin (50 μg/mL). The starter culture was used to inoculate 1.0 L of lysogeny broth. The cultures were allowed to grow at 37 °C until an OD600 of 0.6-0.8 was reached, and expression was induced by the addition of 1.0 mM β-thiogalactoside (IPTG). The cultures were grown at 22 °C for 18 h and then harvested by centrifugation. The resulting pellets were flash frozen in liquid nitrogen and stored at −80 °C for future use or resuspended and purified immediately after harvest. The pellets were resuspended in 50 mM HEPES, 250 mM KCl, pH 8.5, and lysed by sonication. Cellular debris was removed by centrifugation, and the supernatant was loaded onto a prepacked 5-mL HisTrap column (GE Healthcare) and eluted with a linear gradient of 50 mM HEPES, 250 mM KCl, and 300 mM imidazole, pH 8.5. The protein was concentrated using a spin filter, aliquoted, and stored at −80 °C.
UV-vis Spectra of Cj1428.
UV-vis traces (λ = 300 - 400 nm) were collected in a 96-well NucC plate using a SpectraMax340 UV-visible plate reader. The spectra collected of Cj1428 (80 μM) included the purified enzyme, heat-denatured protein (95 °C for 60 s), and enzyme with added α-KG (2.0 mM) for 10 min, then heat-denatured. Precipitated protein from the heat-denaturation samples was removed using a PALL-Omega 10K centrifugal filtration column.
Cofactor Identification and Occupancy of Cj1428.
Cofactors bound to Cj1428 were determined by FPLC anion exchange chromatography. The FPLC methodology used was outlined previously.8 Briefly, concentrated Cj1428 (1.0 mM) was diluted with water to a final concentration of 400 μM and then heat-denatured (95 °C for 60 s) to liberate bound cofactor. Precipitated protein was removed using a PALL-Omega 10K centrifugal filtration device. The resulting flow-through was injected onto a BioRad FPLC system equipped with a 1.0 mL Resource Q Column (GE Healthcare). The column was washed with water, and the products were eluted with a linear gradient of 500 mM ammonium bicarbonate. The elution of product was monitored by changes in absorbance at 255 nm and quantified by integration of relative peak area. To the sample, a known amount of NADH (143 μM) was added and used as an internal standard to quantify the amount of NADP(H) observed in the sample to determine percent occupancy of the cofactor bound to the enzyme and relative ratios. A control spectrum was collected using known concentrations of NAD+ and NADH (16.2 nmoles) and NADP+ and NADPH (31.5 nmoles).
Determination of Kinetic Constants for Cj1428.
The kinetic constants for Cj1428 were determined by monitoring the oxidation of NADPH to NADP+ at 340 nm using a SpectraMax340 UV-visible plate reader. The substrate for Cj1428 (3) was generated by incubating GDP-d-glycero-α-d-manno-heptose (3.0 mM) and α-KG (10 mM) with Cj1427 (10 μM) and Cj1430 (10 μM) in 100 mM HEPES, pH 7.5, for 1 h. Cj1427 and Cj1430 were removed from the reaction mixture using a 10 kDa molecular weight cutoff filter (Pall Corporation). Total substrate used for the kinetic assayed varied between 25 and 540 μM. The assay for the determination of the kinetic constants was conducted in HEPES, pH 7.5, at 30 °C using 300 μM NADPH and 300 nM Cj1428. The apparent values of kcat, and kcat/Km were determined by fitting the initial velocity data to eq. 1 using GraFit 5, where v is the initial velocity of the reaction, E is the enzyme concentration, kcat is the turnover number, and Km is the Michaelis constant.
| (1) |
Determination of Kinetic Constants for Cj1430.
The kinetic constants for Cj1430 were determined using a coupled enzyme assay with Cj1428 by monitoring the oxidation of NADPH to NADP+ at 340 nm. GDP-d-glycero-4-keto-α-d-lyxo-heptose (2) was generated by incubation of GDP-d-glycero-α-d-manno-heptose (1, 3.0 mM) and α-KG (10 mM) with Cj1427 (10 μM) in 100 mM HEPES, pH 7.5, for 1 h. Cj1427 was removed using a 10 kDa molecular weight cutoff filter (Pall Corporation). The concentration of GDP-d-glycero-4-keto-α-d-lyxo-heptose (2) was determined by incubation with Cj1430 and Cj1428 with NADPH. For determination of the kinetic constants, the concentration of GDP-d-glycero-4-keto-α-d-lyxo-heptose (2) was varied between 7.5 μM and 1260 μM. The assay for the determination of the kinetic constants was conducted in HEPES, pH 7.5, at 30 °C using 300 μM NADPH, 200 nM Cj1430, and 10 μM Cj1428.
Synthesis of GDP-d-glycero-β-l-gluco-heptose (4).
A 1.5-mL reaction containing 25 mM GDP-d-glycero-α-d-manno-heptose (1), 30 mM α-KG, 1.0 mM NADPH, and 50 mM acetaldehyde was incubated with Cj1427 (15 μM), Cj1428 (15 μM), Cj1430 (15 μM), and aldehyde dehydrogenase (20 units/mL) in 150 mM HEPES/KOH, pH 8.0, for 18 h. Following the 18 h incubation period, the reaction mixture was loaded onto a prepacked 5-mL HiTrap Q HP column (GE Healthcare). The column was washed with water and then eluted using a linear gradient (0-20%) of 500 mM NH4HCO3, pH 8.0, over 20 column volumes. Fractions of 2-mL were collected and dried under vacuum. The resulting samples were resuspended in D2O and analyzed by 1H NMR spectroscopy.
Structural Analysis of Cj1428 and Cj1430.
The genes encoding Cj1428 and Cj1430 were placed into pET28t3g, a modified pET28b(+) vector (Novagen), which leads to proteins with an N-terminal polyhistidine tag as previously described, 9 and into pET31b(+) to give a version of the protein with a non-cleavable C-terminal polyhistidine tag. The plasmids were utilized to transform Rosetta2(DE3) E. coli cells (Novagen). Cultures were grown in lysogeny broth supplemented with kanamycin and chloramphenicol for pET28t3g constructs (both at 50 mg/L concentration) or ampicillin and chloramphenicol for the pET31b(+) constructs (100 mg/L and 50 mg/L concentrations, respectively) at 37 °C with shaking until an optical density of 0.8 was reached at 600 nm. The flasks were cooled in an ice bath, and the cells were induced with 1 mM IPTG and allowed to express protein at 21 °C for 24 h.
The cells were harvested by centrifugation and frozen as pellets in liquid nitrogen. These pellets were subsequently disrupted by sonication on ice in a lysis buffer composed of 50 mM sodium phosphate, 20 mM imidazole, 10% glycerol, and 300 mM NaCl (pH 8.0). The lysates were cleared by centrifugation, and the proteins were purified at 4 °C utilizing Prometheus™ Ni-NTA agarose (Prometheus Protein Biology Products) according to the manufacturer’s instructions. All buffers were adjusted to pH 8.0 and contained 50 mM sodium phosphate, 300 mM NaCl, and imidazole concentrations of 20 mM for the wash buffer and 300 mM for the elution buffer. The protein constructs with the N-terminal polyhistidine tag were divided into two pools, one where the tag was left in place, and a second that was treated with TEV protease to remove the tag. The TEV protease and remaining tagged proteins for these pools were removed by passage over Ni-NTA agarose. All purified proteins were dialyzed against 10 mM Tris-HCl (pH 8.0) and 200 mM NaCl. The Cj1428 proteins were concentrated to ~25 mg/mL based on an extinction coefficient of 0.98 (mg/mL)−1cm−1, whereas the Cj1430 proteins were concentrated to ~20 mg/mL based on an extinction coefficient of 0.63 (mg/mL)−1 cm−1.
Crystallization conditions were screened using a laboratory-developed 144 condition sparse matrix crystallization screen. Cj1428 constructs were tested in the presence of 2 mM NADPH and 5 mM GDP. Diffraction quality crystals were subsequently grown by hanging drop vapor diffusion at room temperature from 16-20% poly(ethylene glycol) 5000 and 150 mM MgCl2 buffered with 100 mM MOPS (pH 7.0) using the protein that had the tag removed. They belonged to the monoclinic space group P21 with unit cell dimensions of a = 57.4 Å, b = 131.3 Å, c = 58.9 Å, and β = 105.8°. The asymmetric unit contained one dimer. For X-ray data collection, the crystals were transferred to a cryo-protectant solution composed of 26% poly(ethylene glycol) 5000, 300 mM NaCl, 150 mM MgCl2, 18% ethylene glycol, 5 mM NADPH, and 100 mM MOPS (pH 7.0).
The Cj1430 constructs were screened in the presence 5 mM GDP. Multiple crystallization conditions were identified using all three of the protein constructs, all of which had some degree of twinning. It was possible to collect an initial data set on a crystal of the C-terminally tagged enzyme grown from 18-20% poly(ethylene glycol) 5000 and 100 mM MgCl2 in the presence of 5 mM GDP at pH 8.0 (100 mM HEPPS). The crystal belonged to the triclinic space group P1 with unit cell dimensions of a = 40.6 Å, b = 77.7 Å, c = 130.1 Å, α = 87.3°, β = 86.6°, and γ = 84.9°. The asymmetric unit contained four dimers. For X-ray data collection, the crystals were transferred to a cryo-protectant solution composed of 27% poly(ethylene glycol) 5000, 300 mM NaCl, 100 mM MgCl2, 18% ethylene glycol, 5 mM GDP, and 100 mM HEPPS (pH 8.0).
X-ray data for Cj1428 were collected at the Advanced Photon Source, Structural Biology Center (Beamline 19-BM) and processed with HKL3000.10 All X-ray data sets for Cj1430 were collected using a BRUKER D8-VENTURE sealed tube system equipped with HELIOS optics and a PHOTON II detector. These X-ray data sets were processed with SAINT and scaled with SADABS (Bruker AXS). Relevant X-ray data collection statistics are listed in Table 1.
Table 1.
X-ray Data Collection Statistics and Model Refinement Statistics
| Cj1428 NADPH | Cj1430 GDP | Cj1430 GDP-d-glycero-β-l-gluco-heptose | |
|---|---|---|---|
| Resolution limits (Å) | 50.0-1.50 (1.53 – 1.50)b | 50.0-2.10 (2.20 – 2.10)b | 50.0-1.85 (1.95 – 1.85)b |
| Number of independent reflections | 129208 (6167) | 69127 (8583) | 99769 (13932) |
| Completeness (%) | 96.9 (92.9) | 97.3 (93.0) | 97.7 (93.9) |
| Redundancy | 3.9 (2.8) | 5.2 (2.6) | 4.4 (2.6) |
| avg I/avg σ(I) | 35.5 (3.8) | 9.7 (2.4) | 10.3 (2.5) |
| Rsym (%)a | 6.9 (25.3) | 8.8 (39.0) | 8.3 (38.9) |
| cR-factor (overall)%/no. reflections | 16.4/129208 | 20.5/69127 | 19.2/99769 |
| R-factor (working)%/no. reflections | 16.3/122651 | 20.2/65719 | 19.0/94557 |
| R-factor (free)%/no. reflections | 18.3/6557 | 26.4/3408 | 23.3/5212 |
| number of protein atoms | 5472 | 8768 | 8865 |
| number of heteroatoms | 759 | 822 | 1079 |
| average B values | |||
| protein atoms (Å2) | 23.8 | 28.8 | 17.7 |
| ligand (Å2) | 15.8 | 29.7 | 11.8 |
| solvent (Å2) | 31.0 | 27.7 | 22.2 |
| weighted RMS deviations from ideality | |||
| bond lengths (Å) | 0.010 | 0.007 | 0.009 |
| bond angles (deg) | 1.65 | 1.60 | 1.60 |
| planar groups (Å) | 0.008 | 0.007 | 0.008 |
| Ramachandran regions (%)d | |||
| most favored | 98.7 | 96.0 | 97.5 |
| additionally allowed | 1.3 | 3.4 | 2.5 |
| generously allowed | 0.0 | 0.6 | 0.0 |
| PDB entry | 7M13 | 7M14 | 7M15 |
Rsym = (∑|I - Ī|/ ∑ I) x 100.
Statistics for the highest resolution bin.
R-factor = (∑|FO - FC| / ∑|FO|) x 100 where FO is the observed structure-factor amplitude and FC is the calculated structure-factor amplitude.
Distribution of Ramachandran angles according to PROCHECK.18
Cj1428 was solved by molecular replacement with Buccaneer11 which utilized PDB entry 1FXS as a search model.12 Cj4130 was solved via molecular replacement with Buccaneer using PDB entry 3RYK as a search model.13 For both Cj1428 and Cj1430, the Buccaneer software created partial models that were subsequently completed with iterative cycles of manual model-building with COOT 14, 15 and refinement with REFMAC.16
Refinement of the Cj1430 model plateaued at an R-factor of about 26% with many disordered loops. The packing of the dimers in the unit cell was examined to determine if there were any surface mutations that could be engineered to disrupt this crystalline packing and potentially lead to new crystal forms with better diffraction properties. Two contacts points were identified that both contained a pair of amino acids: K128 and E129 and E139 and E141. Using the Stratagene QuikChange method of mutagenesis, the K128A/E129A and E139A/E141A site-directed mutant proteins were prepared. Growth and purification of the pET28t3g and pET31b(+) constructs were performed as described above. The mutant proteins were all concentrated to ~25 mg/mL. Crystallization screens were initially conducted in the presence of 10 mM GDP. Well-ordered crystals of the K128A/E129A variant with the N-terminal tag removed were grown using 22-27% poly(ethylene glycol) 5000 at pH 7.0 (100 mM MOPS). They belonged to the space group C2 with unit cell dimensions of a = 134.3 Å, b = 83.7 Å, c = 110.0 Å and β = 94.7°. The asymmetric unit contained three dimers. For X-ray data collection, the crystals were transferred to a cryo-protectant solution composed of 32% poly(ethylene glycol) 5000, 300 mM NaCl, 15% ethylene glycol, 10 mM GDP, and 100 mM MOPS (pH 7.0). The structure was solved by molecular replacement using PHASER17 and the partially refined structure of the wild-type protein functioning as the search model. Substitution of GDP-d-glycero-β-l-gluco-heptose (4) for GDP in the crystallization trials led to a similar unit cell (space group C2 with unit cell dimensions of a = 136.4 Å, b = 81.6 Å, c = 109.4 Å and β = 94.4°), but the unit cell changes were significant enough that this structure also had to be solved using PHASER. X-ray data collection and model refinement statistics are presented in Table 1 for both of these structures.
RESULTS
Isolation and Characterization of Cj1428.
Cj1428 is a structural homologue of Cj1427 with an overall sequence identity of 24%. Previously, we showed that Cj1427 was functionally able to catalyze the conversion of GDP-d-glycero-α-d-manno-heptose (1) to GDP-d-glycero-4-keto-α-d-lyxo-heptose (2).6 We showed that the isolated Cj1427 contained a tightly bound NADH cofactor with an occupancy of 85%.6, 8 Additionally, we showed that Cj1427 does not exchange the bound cofactor during catalytic turnover, but required an α-keto acid co-substrate, preferably α-ketoglutarate, to re-oxidize the bound NADH to NAD+ such that Cj1427 could catalyze the oxidation of GDP-d-glycero-α-d-manno-heptose (1) to GDP-d-glycero-4-keto-α-d-lyxo-heptose (2).
Freshly purified Cj1428 exhibits properties similar to that of the purified Cj1427. The UV-vis spectrum of Cj1428 shows significant absorbance from 300 to 400 nm with a maximum at ~358 nm (Figure 1, red trace). Heat denaturation of Cj1428 liberated the bound cofactor from the protein, and after removal of the precipitated protein, the absorbance spectrum is shifted to a maximum at 340 nm (Figure 1, blue trace). This was expected if the absorbance of the purified Cj1428 was due to NADH or NADPH that is red-shifted when bound to the protein. It was concluded that Cj1428 purifies with a reduced cofactor as described previously for the Cj1427 homologue.6, 8 However, unlike Cj1427, incubation with α-KG did not diminish the absorbance at 358 nm and thus the bound cofactor remained reduced (Figure 1, green trace).
Figure 1.
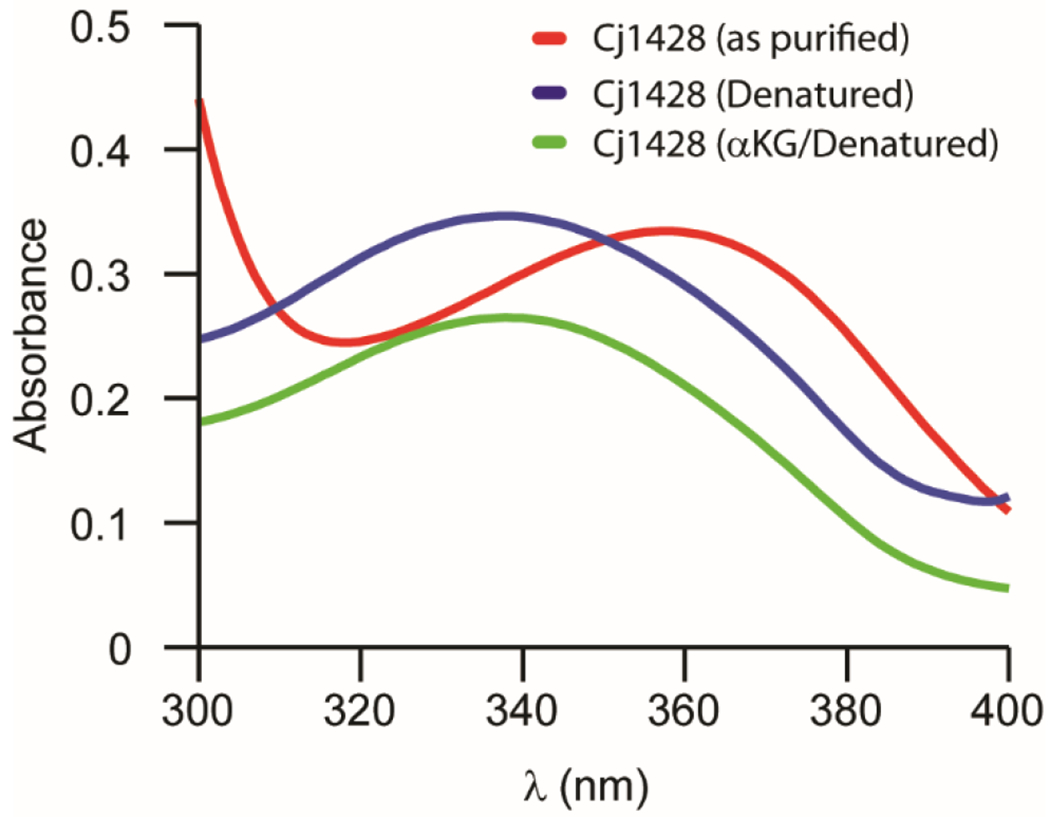
Absorbance spectra of Cj1428 (80 μM) after purification (red), heat-denaturation (blue), and the addition of 2.0 mM α-ketoglutarate (green). Spectra were collected in 50 mM HEPES/KCl buffer, pH 7.4 at 30 °C.
To determine the identity and occupancy of the bound cofactor, an FPLC method was employed, which can chromatographically separate NAD+, NADH, NADP+, and NADPH from one another.8 To quantify the amount of bound cofactor in the purified Cj1428, an internal standard of NADH (143 μM) was added to the heat-denatured sample and peak integration was used to quantify the amount of reduced cofactor liberated from Cj1428. The liberated cofactor was shown to be NADPH. Based on the integration with respect to a calibrated amount of NADPH, Cj1428 was isolated with an occupancy of 78%.
Isolation of GDP-d-glycero-β-l-gluco-heptose (4).
We have previously demonstrated that Cj1427 from C. jejuni NCTC 11168 catalyzes the oxidation of GDP-d-glycero-d-manno-heptose (1) at C4 to form GDP-d-glycero-4-keto-α-d-lyxo-heptose (2) in the presence of added α-KG via a reaction mechanism that involves the transient oxidation/reduction of a tightly-bound NADH in the active site.6, 8 In C. jejuni NCTC 11168 the only heptose sugar in the capsular polysaccharide is d-glycero-l-gluco-heptose. It has previously been proposed that the combined activities of Cj1430 and Cj1428 convert GDP-d-glycero-4-keto-α-d-lyxo-heptose (2) to GDP-d-glycero-β-l-gluco-heptose (4) as illustrated in Scheme 2.6, 8, 19 However, the proposed catalytic activities of Cj1430 and Cj1428 have only been demonstrated with the 6-deoxy surrogate substrate, since GDP-d-glycero-4-keto-α-d-lyxo-heptose (2) has not previously been available. Here GDP-d-glycero-α-d-manno-heptose (1) was incubated with Cj1427, Cj1430, Cj1428 in the presence of α-KG and an NADPH/NADP+ recycling system to enzymatically synthesize GDP-d-glycero-β-l-gluco-heptose (4) in high yield. The 1H NMR spectrum of the GDP-d-glycero-α-d-manno-heptose (1) starting material is presented in Figure 2a and the isolated GDP-d-glycero-β-l-gluco-heptose (4) product is presented in Figure 2b.
Figure 2.
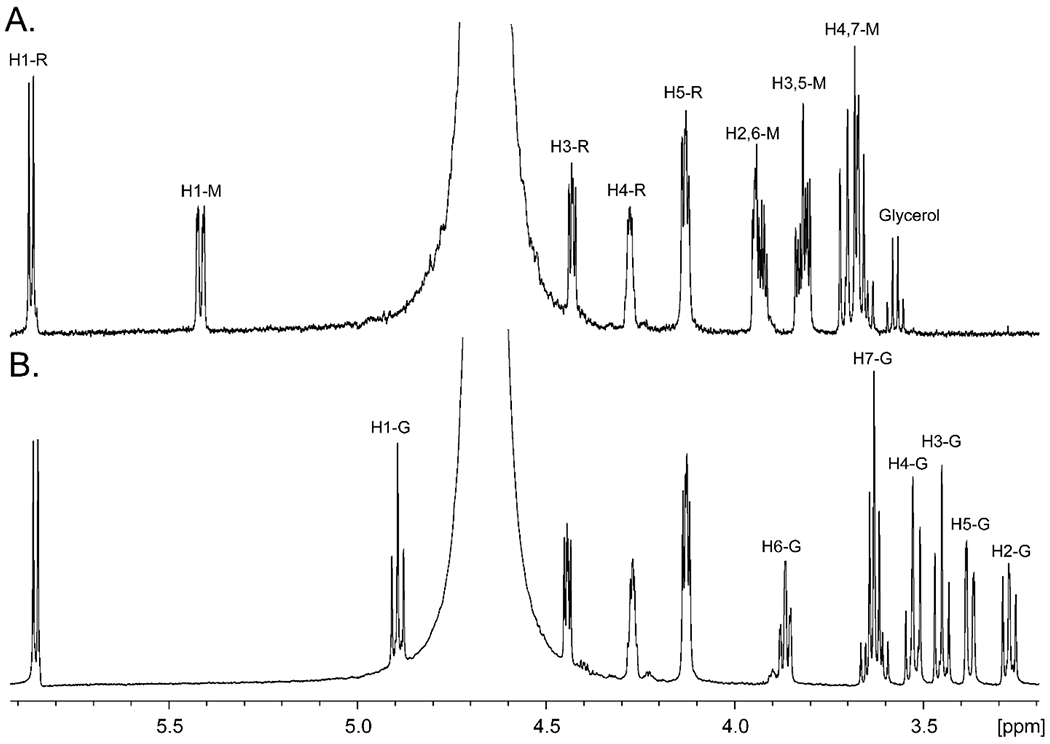
1H NMR spectra of GDP-d-glycero-α-d-manno-heptose (1) and GDP-d-glycero-β-d-gluco-heptose GDP-heptose (4). (A) GDP-d-glycero-α-d-manno-heptose (1). (B) GDP-d-glycero-β-l-gluco-heptose (4) synthesized enzymatically from GDP-d-glycero-α-d-manno-heptose (1) using the catalytic activities of Cj1427, Cj1428, and Cj1430 in the presence of α-KG, and NADPH. Resonances for the hydrogens labeled with an “R” correspond to the ribose moiety of GDP, those labeled with an “M” correspond to those of the mannoheptose moiety and those labeled with a “G” correspond to those of the l-gluco-heptose moiety.
Intermediates in Reaction Catalyzed by Cj1430.
The reaction catalyzed by Cj1430 requires the isomerization of stereochemistry at C3 and C5 within the product (2) of the reaction catalyzed by Cj1427. This reaction is facilitated by the enhanced acidity of the hydrogens at C3 and C5 due to the newly formed ketone at C4. When Cj1427 is added to GDP-d-glycero-α-d-manno-heptose (1) in the presence of α-KG, the newly formed product can easily be detected by the change in chemical shift for the anomeric hydrogen at C1 that goes from a doublet of doublets at ~5.42 ppm (with coupling constants of 7.8 Hz to the adjacent phosphorus and 1.9 Hz with the hydrogen at C2) to a broad doublet at ~5.54 ppm for the 4-keto product 2 (Figure 3a). After addition of Cj1430 to this reaction mixture three new resonances are observed (Figure 3b). There is a triplet at ~4.97 ppm, another triplet at ~5.25 ppm, and a broader unresolved multiplet at ~5.45 ppm. All three of these new resonances disappear after the addition of Cj1428 and NADPH. Formation of the final GDP-d-glycero-β-l-gluco-heptose product (4) is shown by the appearance of a new triplet at ~4.91 ppm with coupling constants of 7.8 Hz to the adjacent phosphorus and the hydrogen at C2 (Figure 3c).
Figure 3.
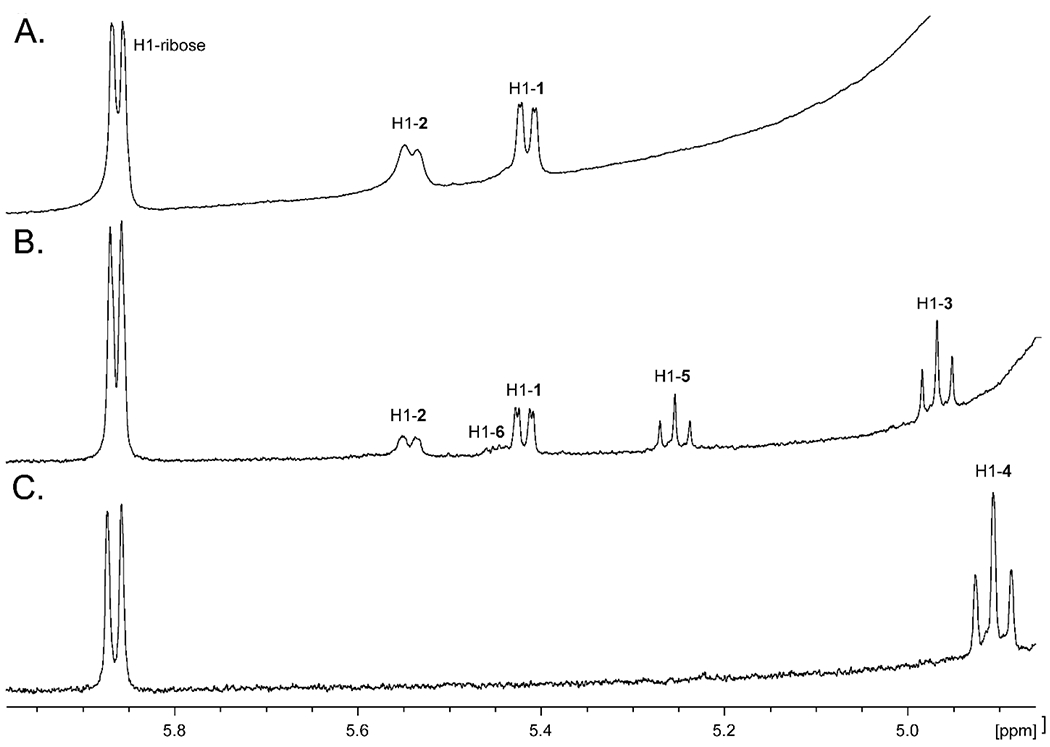
1H NMR spectra of products formed from GDP-d-glycero-α-d-manno-heptose (1) by the addition of Cj1427, Cj1430 and Cj1428. (A) Reaction product (2) formed by the catalytic activity of Cj1427 with GDP-d-glycero-α-d-manno-heptose (1) and α-KG. (B) Reaction products formed by the catalytic activity of Cj1427 and Cj1430. (C) Reaction product (4) formed by the catalytic activity of with Cj1427, Cj1430, and Cj1428. Additional details are found in the text.
The probable intermediates formed during the reaction catalyzed by Cj1430 are presented in Scheme 3. The epimerization at C3 and C5 can occur in either a random or ordered fashion. The epimerization at C5 would initially form GDP-d-glycero-4-keto-β-l-ribo-heptose (5), whereas the epimerization at C3 would form GDP-d-glycero-4-keto-α-d-arabino-heptose (6). When the enzymes (Cj1427 and Cj1430) in the reaction mixture shown in Figure 3b are removed by filtration, followed by the addition of Cj1428 and NADPH, the resonance at ~4.97 ppm disappears and a new resonance for the GDP-d-glycero-β-l-gluco-heptose GDP-heptose (4) appears at ~4.91 ppm. This experiment demonstrates that the resonance at 4.97 ppm is for the doubly epimerized product 3 and that the singly epimerized intermediates (5 and 6) are not substrates for Cj1428. To distinguish between intermediates 5 and 6 from one another, we added the C3 epimerase Cjj1430 from C. jejuni 81-176 to the reaction mixture shown in Figure 3a. This epimerase has been previously shown to catalyzed the epimerization only at C3.19 Under these conditions the only new resonance is the broad multiplet at ~5.45 ppm and thus this represents intermediate 6 (Figure 4). This leaves the resonance that appears as a triplet at ~5.25 ppm for intermediate 5.
Scheme 3:
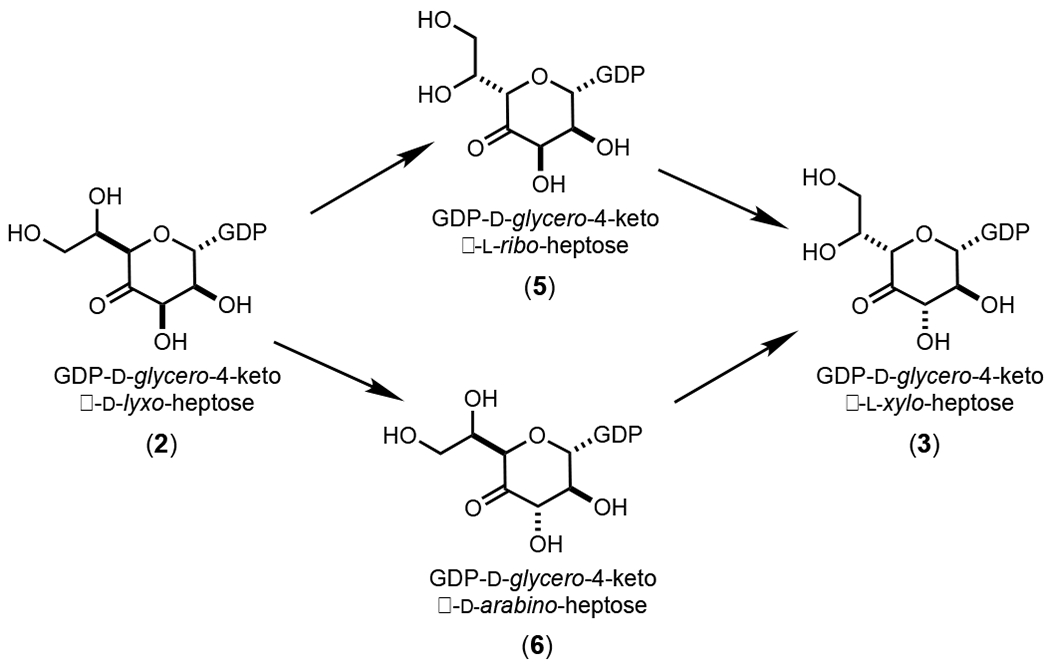
Proposed intermediates in the reaction catalyzed by Cj1430.
Figure 4.
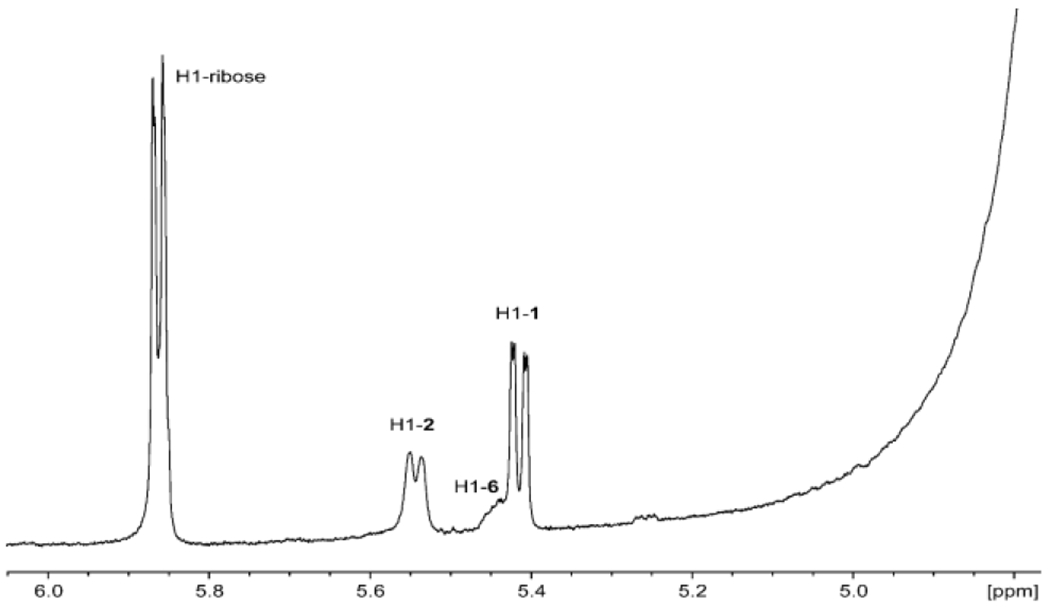
Portion of the 1H NMR spectrum after the addition of Cj1427 and Cjj1430 to GDP-d-glycero-α-d-manno-heptose (1). Additional details are provided in the text.
Kinetic Constants for Reactions Catalyzed by Cj1430 and Cj1428.
The kinetic constants for the reaction catalyzed by Cj1430 were determined using Cj1428 to reduce the product (3) in the presence of added NADPH. The substrate for the reaction catalyzed by Cj1430 was made enzymatically using Cj1427 and GDP-d-glycero-α-d-manno-heptose (1) in the presence of α-KG. The concentration of the substrate (2) was determined by integration of the NMR spectrum after removal of Cj1427. The kinetic constants were determined to be: kcat = 3.5 ± 0.2 s−1; Km = 136 ± 16 μM; and kcat/Km = 25700 ± 3400 M−1 s−1. For Cj1428 the kinetic constants were determined by following the oxidation of NADPH. The apparent kinetic constants for the reduction of 3 at a fixed concentration of 300 μM NADPH were determined to be: kcat = 6.1 ± 0.2 s−1; Km = 121 ± 10 μM; and kcat/Km = 50400 ± 4500 M−1 s−1.
Structure of Cj1430.
The initial structure of Cj1430 was solved with crystals of the wild-type enzyme/GDP complex. Unfortunately, it was never possible to refine the protein model to an R-factor of below 26% most likely due to the many disordered surface loops. The packing of the dimers in the unit cell was therefore examined to determine if there were any surface mutations that could be engineered to disrupt the crystalline packing and lead to new crystal forms with better diffraction properties. Two contact points were identified that both contained a pair of charged amino acids: K128 and E129; and E139 and E141. The K128A/E129A and the E139A/E141A site-directed mutant proteins were prepared and the K128A/E129A mutant led to crystals that diffracted to 2.1 Å resolution. It was subsequently possible to refine the protein model to an overall R-factor of 20.5% to 2.1 Å resolution.
The K128A/E129A mutant variant/GDP complex crystallized with an asymmetric unit containing three dimers. Only subunit F had a disordered surface loop between Cys 90 to Tyr 97 that connects β-strand eight to β-strand nine. Other than this region, the electron densities for all six subunits were continuous from Met 1 to Thr 177. Note that the K128 and E129A mutations are approximately 19 Å and 16 Å, respectively from the active site. Their positions in relationship to the active site are shown in Figure 5. Note that the α-carbons for the six subunits in the asymmetric unit superimpose with root-mean-square deviations of between 0.28 Å – 0.46 Å. For the sake of simplicity, the following discussion refers only to the first dimer in the X-ray coordinate file (subunits A and B).
Figure 5.
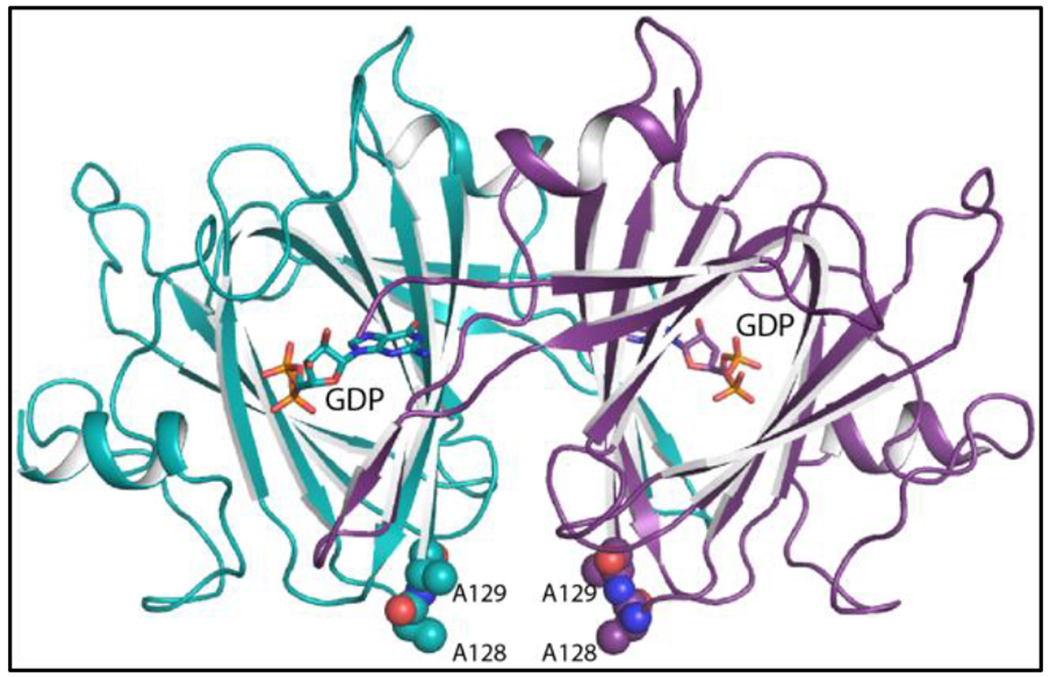
Ribbon representation of the Cj1430 dimer. Subunits A and B are highlighted in purple and teal, respectively. The GDP ligands are displayed in stick representations. The positions the two site-directed mutations utilized to produce crystals with improved diffraction properties are depicted in sphere representations.
Each subunit of the Cj1430 dimer folds into 12 β-strands and two α-helices. The N-terminal residues Met 1 to Glu 4, and β-strands 3 (Phe 24 – Asp 26) and 4 (Gly 29 – Ile 31) from one subunit reach over to form a pocket in the active site of the other subunit in a classical domain swapping interaction. The architecture of the Cj1430 subunits places it into the “cupin” superfamily. Members in this superfamily are remarkably diverse with some having enzymatic and others non-enzymatic functions 20.
To probe the active site of Cj1430, the next structure solved in this investigation was that of the K128A/E129A protein/GDP-d-glycero-β-l-gluco-heptose complex. The model was refined to 1.85 Å resolution with an overall R-factor of 19.2%. Again, the asymmetric unit contained six subunits with their α-carbons superimposing with root-mean-square deviations of between 0.19 Å – 0.56 Å. The electron densities for the six polypeptide chains were continuous from Met 1 to Thr 177. In addition, a histidine residue at position 0 from the purification tag was visible in subunits A, B, C, E, and F. Not surprisingly, the α-carbons for the Cj1430/GDP versus the Cj1430/GDP-linked sugar models superimpose with a root-mean-square deviation of 0.4 Å, and the structures are nearly identical with the exception of several water molecules that are expelled from the active site upon nucleotide-linked sugar binding.
All six subunits contained well-ordered electron density for the GDP-sugar ligand. Shown in Figure 6a is the electron density corresponding to the GDP-d-glycero-β-l-gluco-heptose bound to subunit B. The ribose of the ligand adopts the C-2′ endo pucker whereas the pyranosyl moiety is in the 4C1 conformation. A close-up view of the active site is provided in Figure 6b. The guanine ring is anchored into the active site by hydrogen bonds with Asn 22 and Thr 33 from subunit A, Lys 54 from subunit B, and two ordered water molecules. In addition to a water molecule, the side chain of His 0 from subunit A, which is part of the purification tag, lies within 3.2 Å of the ribosyl C-3 hydroxyl. The aromatic ring of Phe 24 from subunit A provides a platform for binding the ribose. This type of interaction is common. The pyrophosphoryl group is surrounded by four waters and the guanidinium groups of Arg 28 from subunit A, and Arg 64 and Arg 172 from subunit B. The sugar moiety forms extensive interactions with five water molecules and the following side chains: His 67, Lys 74, Asn 121, His 123, and Gln 148. Strikingly Ile 66, which abuts one side of the active site, adopts a cis-peptide conformation.
Figure 6.
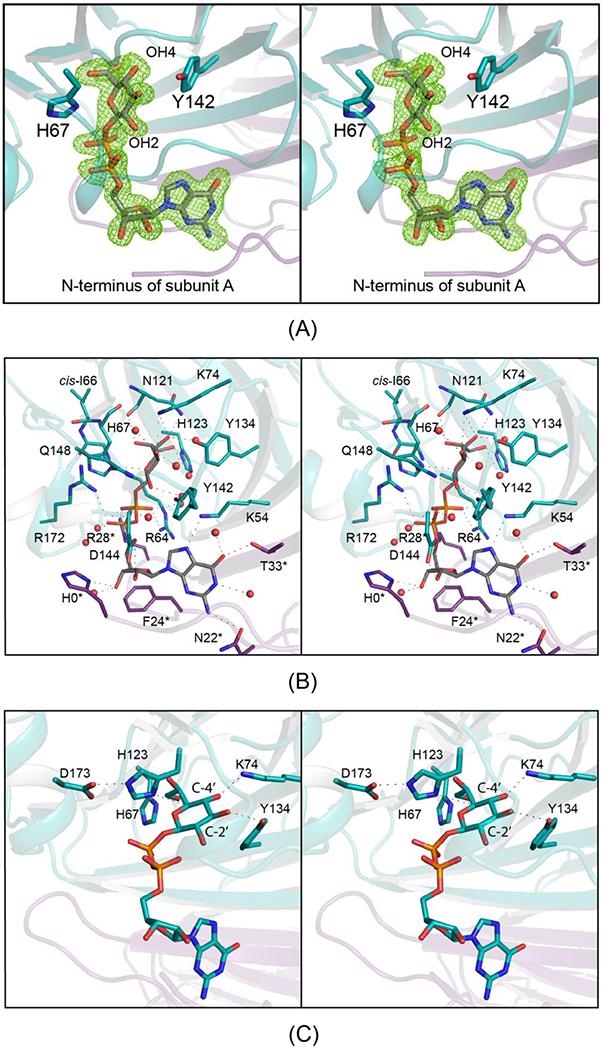
The active site of Cj1430 with bound GDP-d-glycero-β-l-gluco-heptose All panels are in stereo. Shown in (A) is the observed electron density in subunit B for the GDP-sugar ligand. The electron density map was calculated with (Fo-Fc) coefficients and contoured at 3σ. The ligands were not included in the X-ray coordinate file used to calculate the omit map, and thus there is no model bias. A close-up view of the active site is presented in (B). Side chains highlighted in violet and teal are contributed by subunits A and B, respectively. Note that Ile 66 adopts a cis-peptide conformation. Possible hydrogen bonding interactions, within 3.2 Å, are indicated by the dashed lines. Water molecules are represented as red spheres. The position of those residues in Cj1430 that are conserved amongst the 3,5 sugar epimerases and that are thought to play key catalytic roles are shown in (C). All panels were prepared by PyMol.23
The first example of a sugar epimerase adopting a cupin-like fold was reported in 2000 from the Naismith laboratory, namely the Salmonella enterica 3,5-epimerase from the dTDP-l-rhamnose pathway.21 Subsequent studies have shown that there are four conserved catalytic residues in the 3,5-epimerases, a histidine, a lysine, an aspartate and a tyrosine.22 In Cj1430, these correspond to His 67, Lys 74, Asp 173, and Tyr 134. The position of these residues with respect to the bound GDP-sugar ligand are shown in Figure 6c. Their roles in the catalytic mechanism will be discussed further below.
Structure of Cj1428.
The crystals employed for the structure analysis of Cj1428 were grown in the presence of NADPH and GDP and contained a dimer in the asymmetric unit. The model was refined to an overall R-factor of 16.4% at 1.5 Å resolution. The α-carbons for the two subunits of the dimer superimpose with a rather high root-mean-square deviation of 1.6 Å. This is most likely due to the conformational flexibity exhibited by the C-terminal domain with respect to the N-terminal (or core) domain as will be described below. If residues Thr 169 to Trp 231 are omitted from the calculation, the root-mean-square deviation for the α-carbons is lowered to 0.9 Å.
A ribbon representation of the dimer is presented in Figure 7a. Each subunit is distinctly bilobal and folded around 11 β-strands and 12 α-helices. The N-terminal or core domain is dominated by a six-stranded parallel β-sheet flanked on either side by three α-helices. The dinucleotide cofactor extends across the C-terminal end of this β-sheet . The subunit:subunit interface is formed by α-helices three and four. Proline 118, which is located at the surface in a loop connecting β-strand four to α-helix four, adopts a cis-peptide conformation. The overall architecture of the Cj1428 subunit places it into the well-characterized short chain dehydrogenase/reductase or SDR superfamily.24
Figure 7.
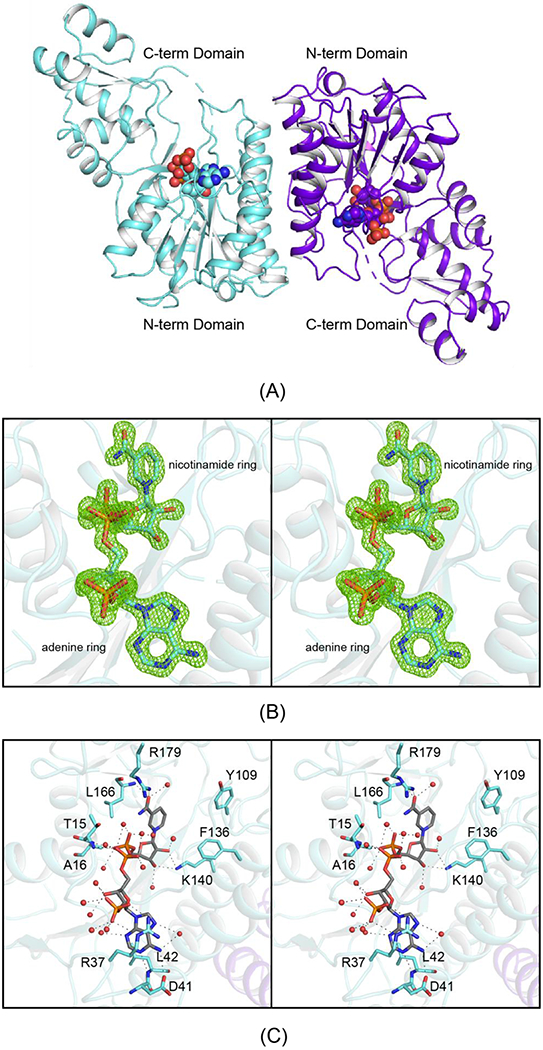
The molecular architecture of Cj1428. Shown in (A) is a ribbon representation of the Cj1428 dimer. Each subunit folds into two distinct domains with the active site wedged between them. The observed electron density in subunit A for the NADPH cofactor is displayed in stereo in (B). The electron density map was calculated with (Fo-Fc) coefficients and contoured at 3σ. A close-up view of the active site for subunit B is presented in (C). Side chains highlighted in teal. Possible hydrogen bonding interactions, within 3.2 Å, are indicated by the dashed lines. Water molecules are represented as red spheres. Proteins that belong to the SDR superfamily typically have the characteristic signature sequence of YXXXK. In Cj1428, the tyrosine has been replaced with Phe 136. All panels were prepared by PyMol 23.
Although the crystals were grown in the presence of 5 mM GDP there was no evidence in the electron density map for the ligand. The electron density for the bound dinucleotide was unambiguous, however, as can be seen in Figure 7b. Both riboses of the dinucleotide adopt the C-2′ endo pucker. A close-up view of the binding interactions between the protein and the cofactor is provided in Figure 7c. As can be seen, most of the interactions between the protein and the cofactor are mediated by water molecules. Indeed, 16 water molecules surround the ligand. The guanidinium group of Arg 37 forms a classical π-cation interaction with the adenine ring. It also provides electrostatic interactions with the ribosyl phosphate group. In addition to Arg 37, the side chain of Asp 41 and the backbone amide group of Leu 42 also provide interactions to anchor the adenine ring into the active site. Other than the backbone amides of Thr 15 and Ala 16 that form hydrogen bonding interactions with two of the phosphoryl oxygens, the others are simply surrounded by water molecules. The nicotinamide ring, which adopts the syn conformation, is positioned into the active site by interactions with the side chain of Arg 179, the backbone amide of Leu 166 and a water molecule. Most members of the SDR superfamily have a characteristic signature sequence of YXXXK with the conserved tyrosine playing an important catalytic role as either an acid or base depending upon its protonation state. Surprisingly, in Cj1428, the sequence is FGAAK. The positions of Phe 136 and Lys 140 are shown in Figure 7c.
DISCUSSION
Reaction Mechanism of Cj1430.
Through this combined functional and structural analysis, the kinetic properties of both Cj1430 and Cj1428 have been determined and their three-dimensional architectures defined. The observed kinetic properties for Cj1430 are in keeping with previously determined values for both the mono- and di-epimerases that belong to the cupin superfamily.25–27 The first biochemically characterized 3,5-epimerase belonging to this group was that from Salmonella enterica Serovar Typhimurium LT2 in 1999 25. On the basis of numerous crystal structure determinations, a detailed reaction mechanism for this 3,5-epimerase was subsequently put forward which takes into account various conformations of the pyranosyl ring.22 There are four key residues involved in the reaction mechanism that are apparently conserved amongst the 3,5-epimerases. Their roles are outlined in Scheme 4 in an abbreviated version for the sake of simplicity. Assuming that the first epimerization occurs at C-5′, it has been proposed that the His 65/Asp 171 dyad functions to remove the C-5′ proton, that Lys 74 provides an oxyanion hole to stabilize the enolate, and that Tyr 134 serves as the active site acid to deliver a proton on the opposite face of the pyranosyl ring. The position of these residues in Cj1430 can be seen in Figure 6c. After this first epimerization or perhaps during the C-5′ proton transfer, there is a ring flip that allows the epimerization at C-3′ to occur using the same amino acid residues. The manner in which the protonation states of the histidine and tyrosine are reset is not clear. Additionally, it was initially proposed that the orientation of the conserved tyrosine dictated whether the enzyme functions as a mono- or di-epimerase but this was subsequently disproven.26, 28
Scheme 4:
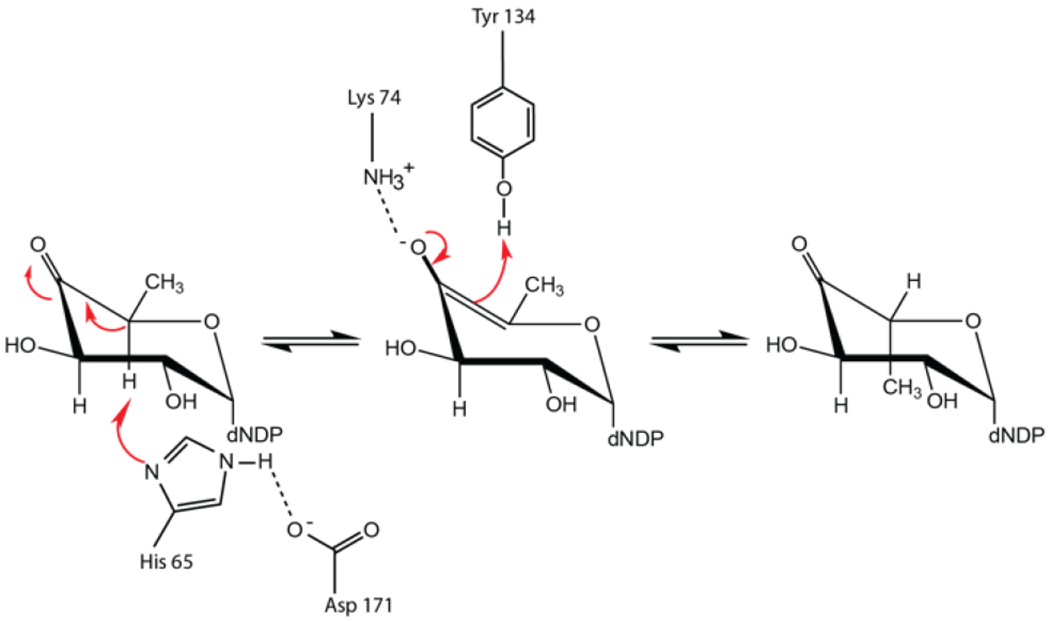
Proposed catalytic mechanism for the first epimerization step catalyzed by RmlC.
A superposition of the models for Cj1430 and the Pseudomonas aeruginosa RmlC (PDB entry 2IXK ) near the active site region is shown in Figure 9. The α-carbons for these two proteins superimposed with a root-mean-square deviation of 1.6 Å. As can be seen, the key residues involved in catalysis are structurally conserved. Interestingly, the cis peptide observed for Ile 66 in Cj1430 is conserved as a cis peptide in RmlC (Leu 64). The key difference between these two enzymes is the substitution of Phe 123 in RlmC for His 123 in Cj1430, which participates in a hydrogen bonding interaction with the C-7′ hydroxy group of the GDP-sugar.
Figure 9.
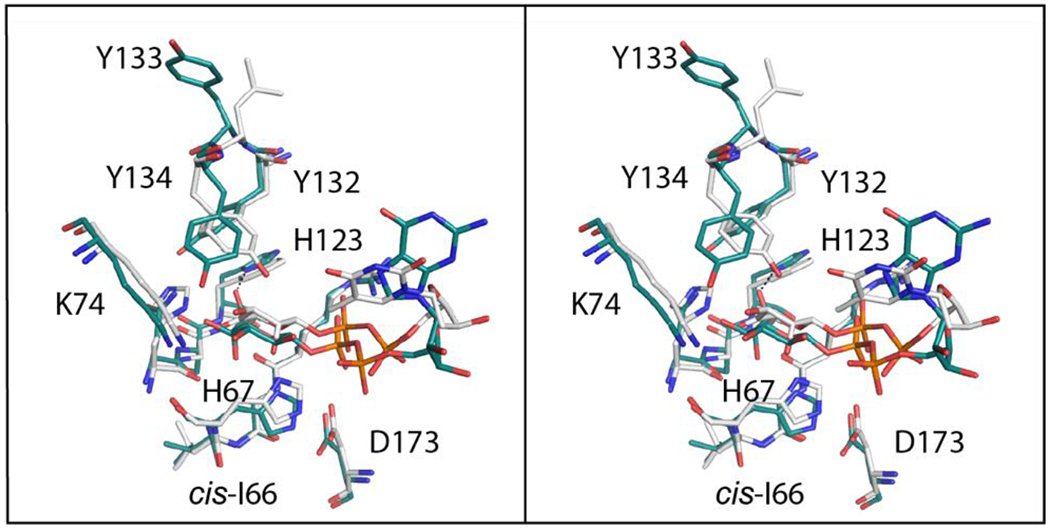
Superposition of the Cj1430 and RmlC active site regions in stereo. Those residues belonging to Cj1430 and RmlC are highlighted in teal and white, respectively. The labels correspond to residues in Cj1430.
Reaction Mechanism of Cj1428.
We were able to obtain the three-dimensional structure of Cj1428 in the presence of NADPH, but unfortunately not in the presence of the carbohydrate-based oxidized substrate (3) or reduced product (4). In most members of the SDR superfamily with the YXXXK signature sequence the conserved tyrosine residue functions as a general acid during the reduction of the keto-substrate to the corresponding alcohol.24 In Cj1428 this residue is a phenylalanine rather than tyrosine and thus another residue within Cj1428 must serve this function. Inspection of the active site identifies two possibilities and these include Tyr 109 and/or Ser 107. Unfortunately, it was not possible through computational docking to provide a clear understanding of how Cj1428 recognizes the specific stereochemical arrangements of the carbohydrate-based substrate at C3 and C5. The homologues of Cj1428 found in the different strains of C. jejuni catalyze the C4 reduction of various heptose substrates with alterations in stereochemistry at C3 through C6 and this recognition must be embedded within the active site. This distinction will ultimately be determined from the structural and functional characterization of additional homologues of Cj1428 and Cj1430.
Recently, Naismith and Creuzenet reported the three-dimentional structures of Cj1428 (aka MlghC) in the presence of NADP (PDB id: 7ANC) and Cj1430 (aka MlghB) in the presence of GDP-mannose (PDB id: 7AN4).29 These two proteins were functionally characterized using GDP-mannose and GDP-6-deoxy-manno-heptose based substrates in contrast to the more physiological GDP-d-glycero-d-manno-heptose based substrates employed in this investigation. Nevertheless, that investigation clearly demonstrated that MlghB catalyzes the epimerization at C3 and C5 of the 4-keto-mannose moiety of the substrate and that MlghC catalyzes the reduction of the corresponding reaction product generating GDP-6-deoxy-l-gluco-heptose.
Unfortunately, the coordinates deposited under PDB id: 7AN4 show only bound GDP, not GDP-mannose. Additionally, the model was determined to a lower resolution of 2.6 Å versus that determined in this investigation at 1.85 Å. Consequently, the structure presented here for Cj1430 represents a more complete, and detailed molecular description of the enzyme. With respect to the models of Cj1428 determined in the presence of NADPH and MlghC solved in the presence of NADP+, they are basically identical (α-carbons superimpose with a root-mean-square deviation of 0.2 Å). Whereas NADPH was specifically included in the crystallization trials for Cj1430, the presented data in Barnawi et al., do not specifically address the oxidation state of the cofactor in the crystals.29
CONCLUSION
We have shown that the combined activities of Cj1427, Cj1430, and Cj1428 from C. jejuni NCTC 11168 catalyze the synthesis of GDP-d-glycero-β-l-gluco-heptose (4) starting from GDP-d-glycero-α-d-manno-heptose (1). In the presence of α-KG, Cj1427 catalyzes the oxidation at C4 of GDP-glycero-α-d-manno-heptose (1) to form GDP-d-glycero-4-keto-α-d-lyxo-heptose (2) while Cj1430 catalyzes the subsequent epimerization of this product at C3 and C5 to generate GDP-d-glycero-4-keto-β-l-xylo-heptose (3). Cj1428, in the presence of NADPH, catalyzes the reduction of this product at C4 to form GDP-d-glycero-β-l-gluco-heptose (4). The X-ray crystal structure of Cj1430 in the presence of GDP-d-glycero-β-l-gluco-heptose was solved as was the structure of Cj1428 solved in the presence of NADPH. The three-dimensional structure Cj1430 has helped to elucidate the underlying contraints that govern the ability of this enzyme to function as a 3,5-epimerase. The structure of Cj1428 has laid the groundwork for understanding how homologues of the C4 reductase in the various strains of C. jejuni are able to catalyze the formation of heptoses with a host of stereochemical modifications at C3, C4, C5, and C6. We suggest that Cj1428 be named GDP-d-glycero-l-gluco-heptose synthase and that Cj1430 be named GDP-d-glycero-4-keto-d-lyxo-heptose 3,5-epimerase.
Funding
This work was supported in part by grants from the Robert A. Welch Foundation (A-840) and the National Institutes of Health (GM 122825) and (R35 GM134643 to HMH).
Footnotes
REFERENCES
- [1].García-Sánchez L, Melero B, and Rovira J (2018) Campylobacter in the Food Chain, In Advances in Food and Nutrition Research (Rodriquez-Lazaro D, Ed.) pp 215–252, Academic Press. [DOI] [PubMed] [Google Scholar]
- [2].Burnham PM, and Hendrixson DR (2018) Campylobacter jejuni: collective components promoting a successful enteric lifestyle, Nat Rev Microbiol 16, 551–565. [DOI] [PubMed] [Google Scholar]
- [3].Gaudet RG, and Gray-Owen SD (2016) Heptose sounds the alarm: innate sensing of a bacterial sugar stimulates immunity, PLoS Pathog 12, e1005807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Monteiro MA, Baqar S, Hall ER, Chen YH, Porter CK, Bentzel DE, Applebee L, and Guerry P (2009) Capsule polysaccharide conjugate vaccine against diarrheal disease caused by Campylobacter jejuni, Infect Immun 77, 1128–1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].St Michael F, Szymanski CM, Li J, Chan KH, Khieu NH, Larocque S, Wakarchuk WW, Brisson JR, and Monteiro MA (2002) The structures of the lipooligosaccharide and capsule polysaccharide of Campylobacter jejuni genome sequenced strain NCTC 11168, Eur J Biochem 269, 5119–5136. [DOI] [PubMed] [Google Scholar]
- [6].Huddleston JP, and Raushel FM (2020) Functional characterization of Cj1427, a unique ping-pong dehydrogenase responsible for the oxidation of GDP-d-glycero-alpha-d-manno-heptose in Campylobacter jejuni, Biochemistry 59, 1328–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].McCallum M, Shaw GS, and Creuzenet C (2013) Comparison of predicted epimerases and reductases of the Campylobacter jejuni d-altro- and l-gluco-heptose synthesis pathways, J Biol Chem 288, 19569–19580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huddleston JP, Anderson TK, Spencer KD, Thoden JB, Raushel FM, and Holden HM (2020) Structural analysis of Cj1427, an essential NAD-dependent dehydrogenase for the biosynthesis of the heptose residues in the capsular polysaccharides of Campylobacter jejuni, Biochemistry 59, 1314–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Thoden JB, and Holden HM (2005) The molecular architecture of human N-acetylgalactosamine kinase, J Biol Chem 280, 32784–32791. [DOI] [PubMed] [Google Scholar]
- [10].Minor W, Cymborowski M, Otwinowski Z, and Chruszcz M (2006) HKL-3000: the integration of data reduction and structure solution-from diffraction images to an initial model in minutes, Acta Crystallogr D Biol Crystallogr 62, 859–866. [DOI] [PubMed] [Google Scholar]
- [11].Cowtan K (2006) The Buccaneer software for automated model building. 1. Tracing protein chains, Acta Crystallogr D Biol Crystallogr 62, 1002–1011. [DOI] [PubMed] [Google Scholar]
- [12].Somers WS, Stahl ML, and Sullivan FX (1998) GDP-fucose synthetase from Escherichia coli: structure of a unique member of the short-chain dehydrogenase/reductase family that catalyzes two distinct reactions at the same active site, Structure 6, 1601–1612. [DOI] [PubMed] [Google Scholar]
- [13].Shornikov A, Tran H, Macias J, Halavaty AS, Minasov G, Anderson WF, and Kuhn ML (2017) Structure of the Bacillus anthracis dTDP-l-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose 3,5-epimerase (RfbC), Acta Crystallogr F Struct Biol Commun 73, 664–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Emsley P, and Cowtan K (2004) Coot: model-building tools for molecular graphics, Acta Crystallogr D Biol Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- [15].Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010) Features and development of Coot, Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Murshudov GN, Vagin AA, and Dodson EJ (1997) Refinement of macromolecular structures by the maximum-likelihood method, Acta Crystallogr D Biol Crystallogr 53, 240–255. [DOI] [PubMed] [Google Scholar]
- [17].McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007) Phaser crystallographic software, J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Laskowski RA, Moss DS, and Thornton JM (1993) Main-chain bond lengths and bond angles in protein structures, J Mol Biol 231, 1049–1067. [DOI] [PubMed] [Google Scholar]
- [19].McCallum M, Shaw SD, Shaw GS, and Creuzenet C (2012) Complete 6-deoxy-d-altro-heptose biosynthesis pathway from Campylobacter jejuni: more complex than anticipated, J Biol Chem 287, 29776–29788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dunwell JM, Purvis A, and Khuri S (2004) Cupins: the most functionally diverse protein superfamily?, Phytochemistry 65, 7–17. [DOI] [PubMed] [Google Scholar]
- [21].Giraud MF, Leonard GA, Field RA, Berlind C, and Naismith JH (2000) RmlC, the third enzyme of dTDP-l-rhamnose pathway, is a new class of epimerase, Nat Struct Biol 7, 398–402. [DOI] [PubMed] [Google Scholar]
- [22].Dong C, Major LL, Srikannathasan V, Errey JC, Giraud MF, Lam JS, Graninger M, Messner P, McNeil MR, Field RA, Whitfield C, and Naismith JH (2007) RmlC, a C3′ and C5′ carbohydrate epimerase, appears to operate via an intermediate with an unusual twist boat conformation, J Mol Biol 365, 146–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Delano W (2002) The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA. [Google Scholar]
- [24].Persson B, Kallberg Y, Bray JE, Bruford E, Dellaporta SL, Favia AD, Duarte RG, Jornvall H, Kavanagh KL, Kedishvili N, Kisiela M, Maser E, Mindnich R, Orchard S, Penning TM, Thornton JM, Adamski J, and Oppermann U (2009) The SDR (short-chain dehydrogenase/reductase and related enzymes) nomenclature initiative, Chem Biol Interact 178, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Graninger M, Nidetzky B, Heinrichs DE, Whitfield C, and Messner P (1999) Characterization of dTDP-4-dehydrorhamnose 3,5-epimerase and dTDP-4-dehydrorhamnose reductase, required for dTDP-l-rhamnose biosynthesis in Salmonella enterica serovar Typhimurium LT2, J Biol Chem 274, 25069–25077. [DOI] [PubMed] [Google Scholar]
- [26].Kubiak RL, Phillips RK, Zmudka MW, Ahn MR, Maka EM, Pyeatt GL, Roggensack SJ, and Holden HM (2012) Structural and functional studies on a 3′-epimerase involved in the biosynthesis of dTDP-6-deoxy-d-allose, Biochemistry 51, 9375–9383. [DOI] [PubMed] [Google Scholar]
- [27].Salinger AJ, Brown HA, Thoden JB, and Holden HM (2015) Biochemical studies on WbcA, a sugar epimerase from Yersinia enterocolitica, Protein Sci 24, 1633–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Merkel AB, Major LL, Errey JC, Burkart MD, Field RA, Walsh CT, and Naismith JH (2004) The position of a key tyrosine in dTDP-4-Keto-6-deoxy-d-glucose-5-epimerase (EvaD) alters the substrate profile for this RmlC-like enzyme, J Biol Chem 279, 32684–32691. [DOI] [PubMed] [Google Scholar]
- [29].Barnawi H, Woodward L, Fava N, Roubakha M, Shaw SD, Kubinec C, Naismith JH, and Creuzenet C (2021) Structure-function studies of the C3/C5 epimerases and C4 reductases of the Campylobacter jejuni capsular heptose modification pathways, J. Biol Chem 296, 100352. [DOI] [PMC free article] [PubMed] [Google Scholar]


