Abstract
Background/Aim: Fucoxanthinol (FxOH) is a marine carotenoid metabolite with potent anti-cancer activity. However, little is known about the efficacy of FxOH in pancreatic cancer. In the present study, we investigated the inhibitory effect of FxOH on six types of cells cloned from N-nitrosobis(2-oxopropyl)amine (BOP)-induced hamster pancreatic cancer (HaPC) cells. Materials and Methods: FxOH action and its molecular mechanisms were investigated in HaPC cells using flow-cytometry, comprehensive gene array, and western blotting analyses. Results: FxOH (5.0 μM) significantly suppressed the growth of four out of six types of HaPC cells. Moreover, FxOH significantly suppressed cell cycle, chemokine, integrin, actin polymerization, microtubule organization and PI3K/AKT and TGF-β signals, and activated caspase-3 followed by apoptosis and anoikis induction in HaPC-5 cells. Conclusion: FxOH may have a high potential as a cancer chemopreventive agent in a hamster pancreatic carcinogenesis model.
Keywords: Apoptosis, carotenoid, fucoxanthinol, hamster pancreatic cancer, CXCR7
Pancreatic cancer is one of the most lethal cancers worldwide because it is treatment resistant and has aggressive potential for metastasis/invasion with resultant poor prognosis. From the GLOBOCAN 2018 estimates, 432,242 pancreatic cancer deaths occur per year (4.5% of total) (1), and the 5-year survival rate remains poor at 10% (2). Accumulating evidence suggests that aberrant network integrity of gene mutation, gene methylation, transcriptome, microRNA, non-coding RNA, proteome, tumor microenvironment, and immune cells are crucial for human pancreatic cancer development. In particular, highly carcinogenic point mutations in driver genes, such as KRAS, CDKN2A, TP53, and SMAD4, are observed in many specimens (3-7). Pancreatic intraepithelial neoplasia (PanIN) is a premalignant lesion in pancreatic carcinogenesis and has the stepwise progress graded as four types from mild to severe. KRAS, CDKN2A, TP53 and SMAD4 are somatically mutated in turn along with the malignant progression of PanIN, followed by the cancer progression (8).
N-nitrosobis(2-oxopropyl)amine (BOP)-treated Syrian golden hamsters are a chemical carcinogenesis model that represents human pancreatic cancer because it induces PanIN, and pancreatic ductal adenocarcinoma resembles human pancreatic cancer, which also includes similar genetic mutations such as in K-ras, CDKN2A, and SMAD4 (9). Therefore, the BOP-induced hamster pancreatic cancer model is a useful model to investigate the mechanism of carcinogenesis and in the identification of chemopreventive agents against pancreatic cancer. Several cancer prevention experiments using BOP-treated hamsters revealed that natural dietary materials, such as fermented brown rice, 4-methylthio-3-butenyl isothiocyanate, benzyl isothiocyanate, sulforaphane, green tea polyphenols, and β-carotene may be candidate cancer chemopreventive agents; however, the anti-cancer mechanisms involved remain elusive (10-13).
Fucoxanthin (Fx) is a highly polar carotenoid that has a distinctive allene and a 5,6-monoepoxide. Fx predominantly accumulates in marine brown algae, some of which are used in foods. Dietary Fx is converted to its deacetylated form fucoxanthinol (FxOH) mainly in the intestine of humans as well as in mice (14,15).
To date, human interventional studies aimed at preventing cancer with Fx or FxOH have been limited. On the other hand, many reports have shown that Fx has anti-cancer activity in various cancers in vitro and in vivo (16-23). Regarding FxOH, it suppressed tumorigenesis in immunodeficient NOD-SCID mice (24). It also induced apoptosis in colon cancer cells and colon cancer stem-like spheroids through attenuation of integrin, mitogen-activated protein kinase (MAPK), nuclear factor-ĸB, phosphatidylinositol-3 kinase/protein kinase B (PI3K/AKT), peroxisome proliferator-activated receptor, signal transducers and activators of transcription (STAT), chloride intracellular channel 4 (CLIC4), and caspase signaling (25-28). These molecules are also involved in migration, invasion, epithelial-mesenchymal transition, and cell-cycle arrest. However, little information in available on the anti-cancer function of FxOH in pancreatic cancer.
Herein, we showed the apoptosis-inducing effect of FxOH on a cell line cloned from pancreatic ductal adenocarcinoma in a BOP-treated hamster and elucidated its molecular mechanisms.
Materials and Methods
Chemicals. All-trans-FxOH (purity, ≥98%) was extracted and purified from algal lipids by Dr. Hayato Maeda (Hirosaki University, Japan). Anti-C-X-C chemokine receptor type 4 (CXCR4) and anti-CXCR7 antibodies were purchased from BioVision (Milpitas, CA, USA) and Novus Biologicals (Littleton, CO, USA), respectively. Anti-Akt (pan), anti-cyclin B1, anti-phosphorylated focal adhesion kinase [pFAK(Tyr397)], anti-integrin α5, anti-integrin β1, anti-integrin β4, and anti-caspase-3 antibodies were obtained from GeneTex (Irvine, CA, USA). Anti-cyclin D1, anti-pMEK1/2(Ser217/221), anti-pERK1/2(Thr202/Tyr204), anti-pAkt(Ser473), and anti-pAkt(Thr308) antibodies were from Cell Signaling Technology (Danvers, MA, USA). Anti-cyclin D2 and anti-integrin β8 antibodies were purchased from Bioss Antibodies (Beijing, PR China) and R&D Systems (Minneapolis, MN, USA), respectively. Anti-pPaxillin (Tyr31) and anti-p53 antibodies were obtained from Novex (San Diego, CA, USA) and Thermo Scientific (Waltham, MA, USA), respectively. The cells were routinely maintained in Dulbecco’s modified Eagle medium (DMEM, Wako Pure Chemicals, Osaka, Japan) containing 10% heat-inactivated fetal bovine serum (FBS), 4 mM L-glutamine, 40,000 U/l penicillin, and 40 mg/l streptomycin. All other reagents and solvents used were of analytical grade.
Establishment of cell lines. Female Syrian golden hamsters (5-week-old, Japan SLC, Shizuoka, Japan) were acclimated for a week and injected subcutaneously with BOP (Nacalai Tesque, Kyoto, Japan) 4 times (on days 1, 3, 5, and 7) at a dose of 10 mg/kg body weight. CE-2 pellets (CLEA Japan, Shizuoka, Japan) were used as a standard diet, and 1 group of hamsters was fed Quick Fat pellets (QF) (CLEA Japan, Shizuoka, Japan). Hamsters were sacrificed with deep anesthesia at 38-90 weeks of age, and then the pancreas from each hamster was taken and part of the tumor collected. Most pancreatic tissue was placed in 10% formalin/phosphate-buffered saline for 2-3 days. Histopathologic diagnosis of pancreatic tissue in the hamster was performed by a highly proficient pathologist. The animal experiments were approved by the Institutional Guidelines for Animal Care and Use in the National Cancer Center Research Institute.
The tumor sections were minced using scissor and cultured in 5% FBS/RPMI-1640 (Wako Pure Chemicals, Osaka, Japan) medium in 24-well plates. When the cultured cells reached confluence, they were seeded in another dish (6-well plates), and then in a larger dish (100-mm in diameter). The established cell lines were designated hamster pancreatic cancer (HaPC)-1, -2, -3, -4, and -5 derived from a hamster given CE-2, and HaPC-6 from a hamster given QF.
Cell viability assay. HaPC-1–6 cells were adhered at a density of 5×104 cells/ml into a 24-well plate in 10% FBS/DMEM medium for 3.5 h. Cells were then incubated in 1% FBS/DMEM medium with FxOH (final concentrations, 1.0 and 5.0 μM) or vehicle alone [dimethylsulfoxide (DMSO)] for 1 day. Cell viability was determined using a WST-1 reagent assay. The absorbance was monitored using an ELISA reader at 450 nm (TECAN Japan, Tokyo, Japan).
Cell cycle analysis. HaPC-5 cells were adhered at a density of 5×104 cells/ml into 100-mm dishes in 10% FBS/DMEM medium for 3.5 h. Cells were then incubated in 1% FBS/DMEM medium with FxOH (final concentration, 5.0 μM) or vehicle alone (DMSO) for 2 days. The cells were trypsinized, fixed with 70% ethanol, and then treated with ribonuclease A (Nacalai Tesque, Kyoto, Japan). Nuclei in the cells were stained with propidium iodide (Sigma-Aldrich, St Louis, MO, USA), and the cells were suspended with 0.1% bovine serum albumin (BSA)/phosphate-buffered saline. The ratios of Sub-G1 (apoptosis-like cells), G1, S, and G2/M phases were determined using a FACSAria-III flow cytometer (BD Biosciences).
Total RNA preparation. HaPC-5 cells were adhered at a density of 5×104 cells/ml into 100-mm dishes in 10% FBS/DMEM medium for 3.5 h. Cells were then incubated in 1% FBS/DMEM medium with FxOH (final concentration, 5.0 μM) or vehicle alone (DMSO) for 1 day. Total RNA from HaPC-5 cells with or without 5.0 μM FxOH treatment was isolated using an RNeasy Mini Kit with RNase-Free DNase Set and QIAshredder (QIAGEN, Valencia, CA, USA) in accordance with the manufacturer’s instructions. The concentration of total RNA was measured using Nanodrop ND-1000 (NanoDrop, Wilmington, DE, USA). Subsequently, quantitation of total RNA was determined using an Agilent 2100 bioanalyzer (Agilent, Santa Clara, USA). Total RNA from FxOH-treated HaPC-5 cells or control cells was prepared as a sample with equivalently mixed mRNAs with triplicate experiments and then subjected to next-generation sequencing.
Transcriptome analysis. Whole transcriptome analysis was performed in accordance with the TruSeq Standard mRNA Reference Guide Document #1000000040498 v00 and the next generation sequencer NovaSeq 6000 System User Guide Document #1000000019358 v02. In brief, total RNA was purified into mRNA, fragmented, and then prepared as double-stranded cDNA. Subsequently, libraries from the cDNA template were prepared using a TruSeq standard mRNA LT Sample Prep kit (Illumina, San Diego, CA, USA) and sequenced using a NovaSeq 6000 S4 Reagent kit (Illumina, San Diego, CA, USA). The sequencing was carried out using a NovaSeq 6000 system (Illumina, San Diego, CA, USA) equipped with sequencing control software (version 1.4.0). Differentially expressed genes between FxOH-treated HaPC5 cells and the control cells were detected using ≥2.0- and ≤–2.0-fold cutoff and p-values <0.05 (exact test using edgeR). Gene profiles were displayed using volcano plots and hierarchy clustering maps. Gene annotation was performed using the NCBI reference sequence database Mesocricetus auratus Annotation Release 102 (ncbi.nlm.nih.gov/genome/annotation_euk/Mesocricetus_auratus/102/). Functional interpretation analysis was performed using the g:Profiler tool (https://biit.cs.ut.ee/gprofiler), based on the gene ontology (GO) database (http://www.geneontology.org/).
Quantitative-polymerase chain reaction (qPCR). The cDNA was synthesized from total RNA using a High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific, Waltham, MA). Gene primer and probe sequences were as follows: Ackr3-forward (5’-AGG TAG GTA TCA GGC AGA G-3’), Ackr3-reverse (5’-CAG CAC CTC CAG CTA TAA GAA G-3’), glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-forward (5’-GTT GGA ACC CAG TGC ATA GA-3’), GAPDH-reverse (5’-GGG TGT GAA CCA TGA CAA GTA-3’), Ackr3 probe (5’-/56-FAM/TGT TGT CTG/ZEN/CAT CTT GGT GTG GCT/3lABkFQ/-3’) and GAPDH probe (5’-/56-FAM/CTG CAC CAC/ZEN/CAA CTG GCT GAA ATG/3lABkFQ/-3’) (Integrated DNA Technologies, Coralville, IA).
The cDNA template (10 ng), primers (final 500 pM)/probe (final 250 pM) sets (Integrated DNA Technologies, Coralville, IA, USA), PrimeTime Gene Expression Master Mix (Integrated DNA Technologies), and distilled water were mixed (total volume, 20 μl). qPCR was performed as follows: initial denaturation for 5 min at 95˚C, followed by 40 cycles of 15 s at 95˚C and 45 s at 60˚C. qPCR was performed using a LightCycler®Nano real-time PCR system (Roche Diagnostics, Mannheim, Germany).
Western blotting. HaPC-5 cells were adhered at a density of 5×104 cells/ml in 100-mm dishes in 10% FBS/DMEM medium for 3.5 h. Cells were then incubated in 1% FBS/DMEM medium with FxOH (final concentration, 5.0 μM) or vehicle alone (DMSO) for 1 day. HaPC-5 cells with or without 5.0 μM FxOH treatment were harvested and lysed in lysis buffer. The protein concentration in whole cell lysates was determined using the Bradford assay (Bio-Rad, Hercules, CA, USA). Ten μg of protein was separated using a sodium dodecyl sulfate-10% polyacrylamide gel. Gels were electroblotted onto a PVDF membrane (Amersham Bioscience, Chalfont St. Giles, UK). The membrane was incubated in Tris-buffered saline containing 0.1% polyoxyethylene (20) sorbitan monolaurate with 1% BSA (1% BSA/Tris-buffered saline containing 0.1% Tween 20 [TBS-T]) at room temperature for 1 h, and probed with each of the primary antibodies (1:1,000 dilution) in 1% BSA/TBS-T at 4˚C overnight. The membranes were then probed with horseradish peroxidase-conjugated anti-mouse or anti-rabbit secondary antibody (1:2,000 dilution) in 1% BSA/TBS-T at room temperature for 1 h. Protein bands were visualized using a chemiluminescence reagent (Millipore, Billerica, MA, USA).
Statistics analysis. All the values are expressed as the mean±standard error (SE). All differences were examined using the Student’s t-test or exact test with edgeR between two groups, and one-way ANOVA with Tukey–Kramer post-hoc tests for multiple comparisons. Significant differences were presented at *p<0.05, **p<0.01 or exact p-values.
Results
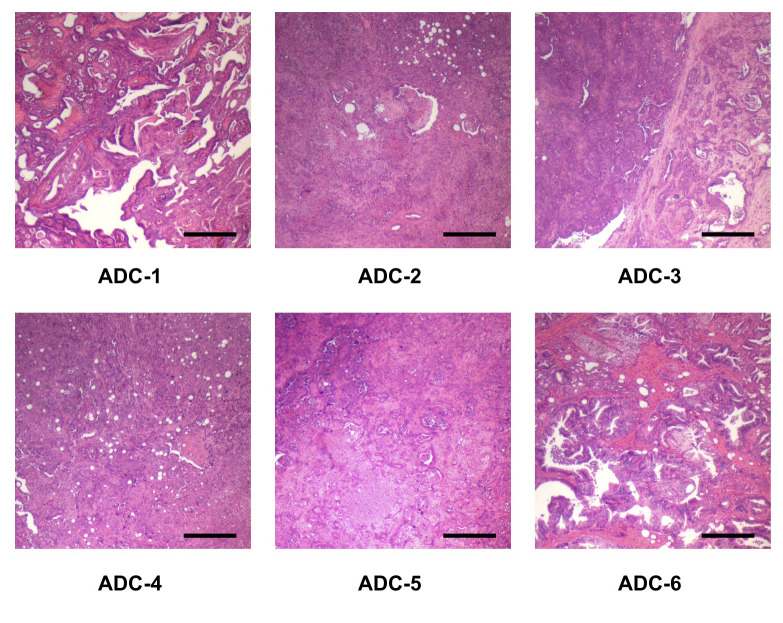
Characterization of six types of pancreatic ductal adenocarcinoma cancers in BOP-treated hamster. Pathological findings revealed that several types of pancreatic cancers exist in BOP-treated hamsters. These were pathologically diagnosed as papillary adenocarcinoma, well differentiated tubular adenocarcinoma, moderately differentiated tubular adenocarcinoma, and poorly differentiated adenocarcinoma. These cells cloned from the six types of pancreatic adenocarcinomas were designated as HaPC-1–6 (Figure 1 and Table I).
Figure 1. Representative histopathology of six pancreatic ductal adenocarcinomas developed in BOP-treated hamsters. ADC, adenocarcinoma. Bar, 400 μm.
Table I. Pathological findings for six cell lines cloned from pancreatic ductal adenocarcinoma of BOP-initiated Hamster.
BOP, N-Nitrosobis(2-oxopropyl)amine; HaPC, Hamster pancreatic cancer. aName of cell lines cloned from each pancreatic tumor. bPap ADC, papillary adenocarcinoma. cTub2, moderately differentiated tubular adenocarcinoma. dPor, poorly differentiated adenocarcinoma. eTub1, well differentiated tubular adenocarcinoma.
Effect of FxOH on cell growth in HaPC-1–6 cells. The growth of HaPC-1, -4, -5, and -6 cells was significantly decreased in a dose-dependent manner by FxOH treatment. Little significant difference in the cell growth of HaPC-2 and -3 cells was observed with FxOH treatment. The percentages of cell growth (control 100%) were as follows: 1.0 μM FxOH, 89.8±2.8%; 5.0 μM FxOH, 88.3±2.0% in HaPC-1 cells; 1.0 μM FxOH, 97.4±6.0%; 5.0 μM FxOH, 85.6±1.7% in HaPC-4 cells; 1.0 μM FxOH, 82.5±2.8%; 5.0 μM FxOH, 76.6±2.2% in HaPC-5 cells; and 1.0 μM FxOH, 98.0±1.1%; 5.0 μM FxOH, 93.1±2.0% in HaPC-6 cells (Figure 2).
Figure 2. Effects of fucoxanthinol (FxOH) on cell growth in pancreatic cancer HaPC-1–6 cells. HaPC-1-6 cells were treated with 1.0 and 5.0 μM FxOH for 1 day. Cell viability was measured using WST-1 reagent assay. The cell viability of control cells was set as 100%. Means±SE (n=6). *p<0.05 vs. control cells (vehicle only).
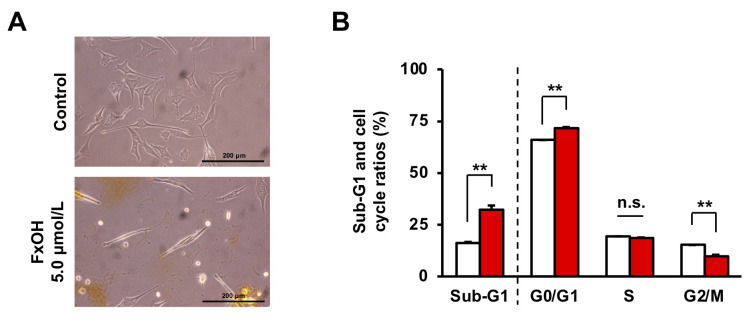
Effect of FxOH on apoptosis induction and cell-cycle arrest in HaPC-5 cells. Treatment of HaPC-5 cells with 5.0 μM FxOH showed drastic morphological changes from an elongated cell form to a thin spindle form (Figure 3A). Treatment with 5.0 μM FxOH significantly augmented the ratio of Sub-G1 (control cells, 16.4±0.3% and FxOH-treated cells, 32.3±2.0%) in HaPC-5 cells. The ratio of HaPC-5 cells in each cell cycle phase was significantly changed by 5.0 μM FxOH treatment: G0/G1 phase, control cells, 65.8±0.1% and FxOH-treated cells, 71.7±0.4%; G2/M phase, control cells, 15.0±0.2% and FxOH-treated cells, 10.0±0.4%. The proportion of HaPC-5 cells in S phase did not significantly differ between control and FxOH-treated cells (Figure 3B).
Figure 3. Effects of fucoxanthinol (FxOH) on apoptosis induction in pancreatic cancer HaPC-5 cells. HaPC-5 cells were treated with 5.0 μM FxOH for 2 days. (A) Phase contrast microscopy images. Bar, 200 μm. (B) Proportion of sub-G1 phase (apoptotic-like cells) and cells in each cell-cycle phase (G1, S and G2/M) in FxOH-treated and control HaPC-5 cells, which were evaluated using a FACSAria-III flow cytometer are shown. Means±SE (n=3). **p<0.01.
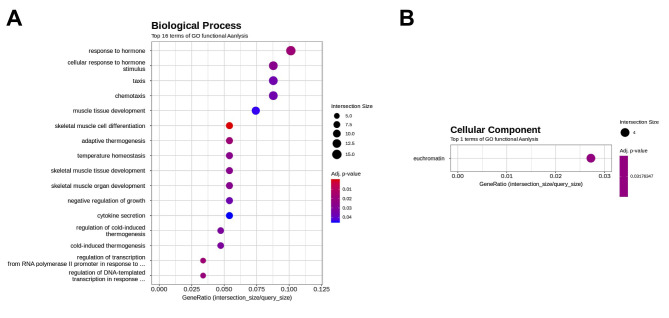
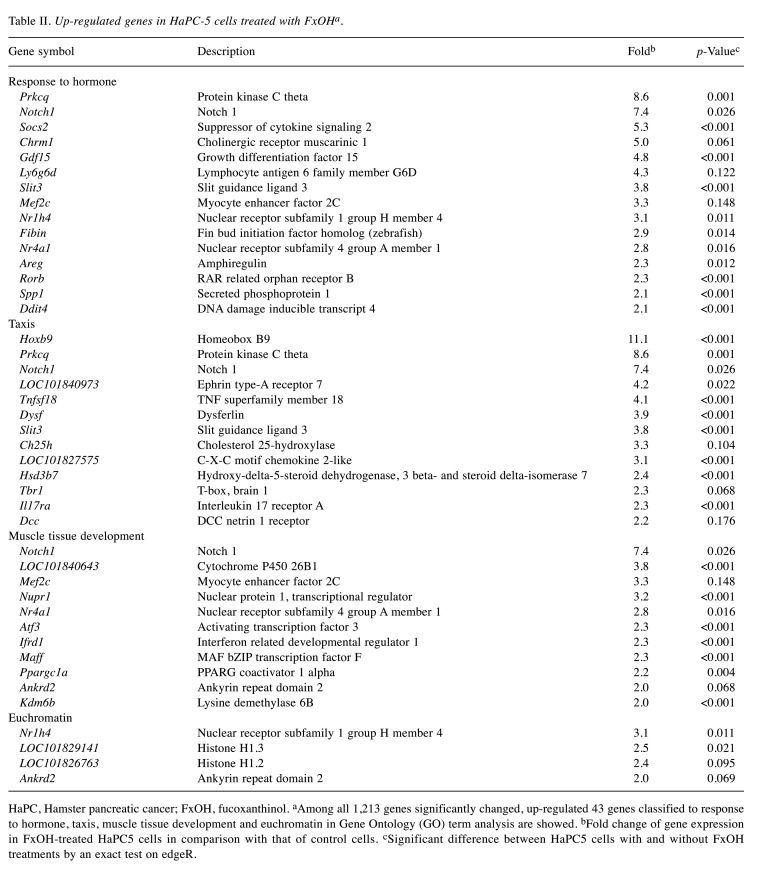
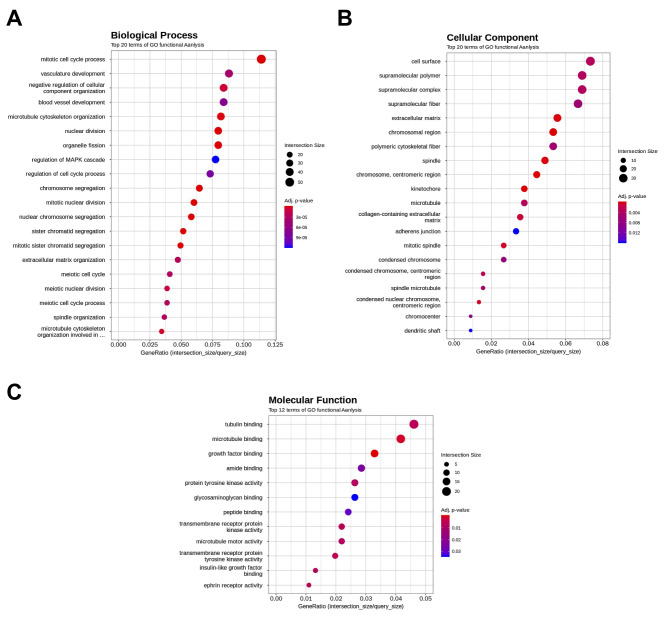
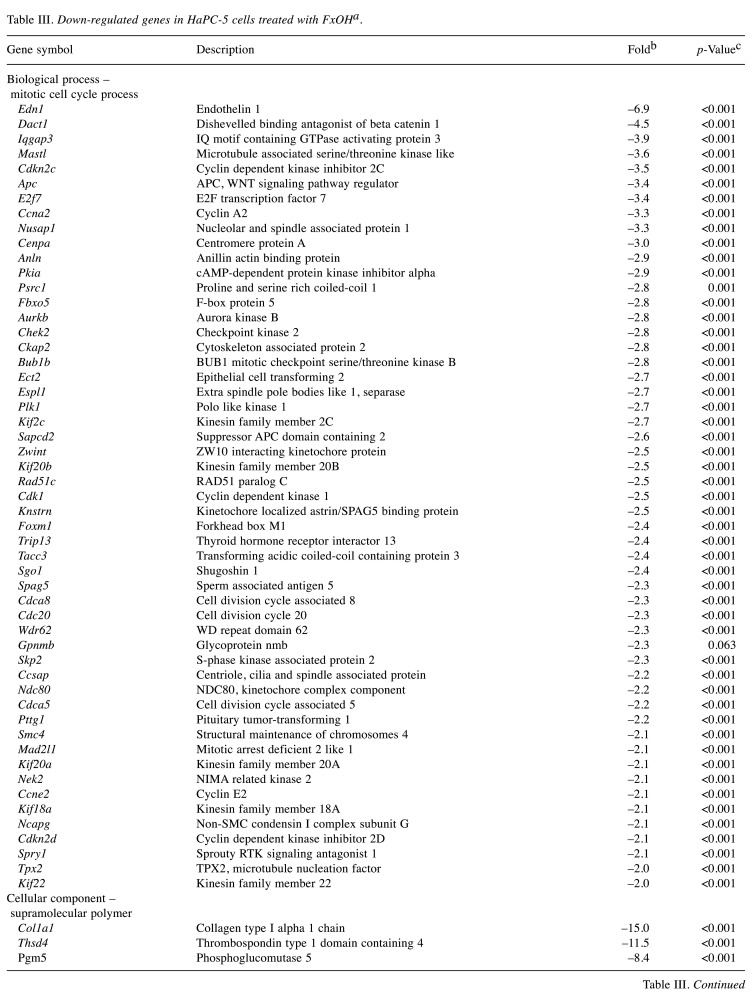
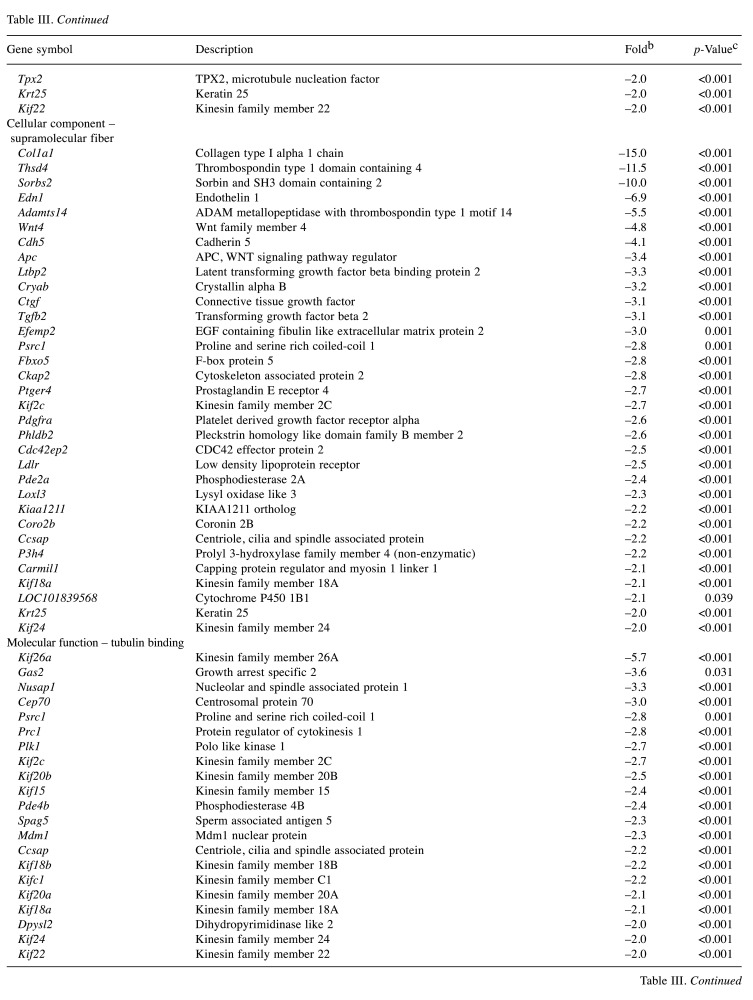
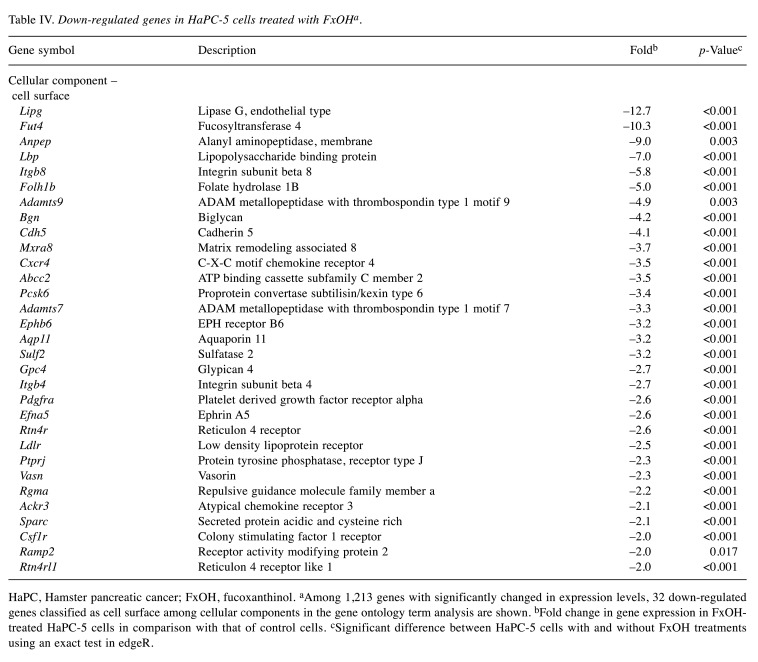
Effect of FxOH on the transcriptome in HaPC-5 cells. Transcriptome alterations in HaPC-5 cells after 5.0 μM FxOH treatment for 1 day were investigated. As a result, volcano plots showed that the number of down-regulated genes was greater than that of up-regulated genes in both fold-change and p-value (Figure 4A). Heat maps were used to display one-way hierarchical clustering of the 1,213 genes that showed differences between the two groups (Figure 4B). Overall, 344 up-regulated and 869 down-regulated genes (total 1,213 genes) were altered in FxOH-treated HaPC-5 cells compared to control cells (Figure 4C). The top 16 GO terms for biological processes and 1 GO term for cellular components were significantly enriched in the 344 up-regulated genes. The genes in the GO terms on response to hormone (15 genes), taxis (13 genes), muscle tissue development (11 genes), and euchromatin (4 genes) were mainly associated with growth and inflammation, although few up-regulated genes involved in apoptosis induction were observed (Figure 5 and Table II). The genes for cellular response to hormone stimulus and chemotaxis are not shown in Table II, because the genes contained in these were similar for responses to hormone and chemotaxis, respectively. The top 20 GO terms for biological processes and cellular components and the top 12 GO terms for molecular function were significantly enriched in the 869 down-regulated genes. The GO terms for mitotic cell-cycle process (53 genes), cell surface (33 genes), supramolecular polymer (30 genes), supramolecular complex (30 genes), supramolecular fiber organization (33 genes), tubulin binding (21 genes), and microtubule binding (19 genes) were mainly correlated with many signals as follows: cell cycle, cell division, chemokine, cadherin, extracellular matrix, integrin, actin polymerization, microtubule organization, Ras, transforming growth factor beta (TGF-β) and wingless/integrated (Wnt). Moreover, a GO term for regulation of the MAPK cascade for biological processes was decreased by FxOH treatment (Figure 6, Table III and Table IV).
Figure 4. Effects of fucoxanthinol (FxOH) on the transcriptome profile in HaPC-5 cells. HaPC-5 cells were treated with 5.0 μM FxOH for 1 day. Gene alterations between FxOH-treated HaPC-5 cells and control cells were analyzed using a next-generation sequencer NovaSeq 6000 system and sequencing control software (version 1.4.0). Levels of gene expression with ≥2.0 and ≤–2.0 -fold with cutoff p-value <0.05 in FxOH-treated HaPC-5 cells and control cells are presented as a sample with equivalently mixed mRNAs with triplicate experiments. (A) Volcano plots between the two groups. (B) Hierarchical clustering analysis for 1,213 genes with significant expression level differences between the two groups. (C) Number of up- (≥2.0-fold), and down-regulated (≤-2.0-fold) genes between the two groups. Yellow, up-regulated genes. Blue, down-regulated genes.
Figure 5. Gene ontology (GO) enrichment profiles of genes up-regulated by fucoxanthinol (FxOH) treatment in HaPC-5 cells. The functional interpretation of genes up-regulated by ≥2.0-fold and cutoff p-value <0.05 were performed using g:Profiler. The top 16 GO terms in more than four gene sizes are shown. (A) Sixteen GO terms in a biological process category. (B) One GO term in a cellular component category.
Table II. Up-regulated genes in HaPC-5 cells treated with FxOHa.
HaPC, Hamster pancreatic cancer; FxOH, fucoxanthinol. aAmong all 1,213 genes significantly changed, up-regulated 43 genes classified to response to hormone, taxis, muscle tissue development and euchromatin in Gene Ontology (GO) term analysis are showed. bFold change of gene expression in FxOH-treated HaPC5 cells in comparison with that of control cells. cSignificant difference between HaPC5 cells with and without FxOH treatments by an exact test on edgeR.
Figure 6. Gene ontology (GO) enrichment profiles of genes down-regulated by fucoxanthinol (FxOH) treatment in HaPC-5 cells. The functional interpretation of genes down-regulated by ≤–2.0-fold and cutoff p-value <0.05 were performed using g:Profiler. The top 20 GO terms in more than five gene sizes are shown. (A) Twenty GO terms in a biological process category. (B) Twenty GO terms in a cellular component category. (C) Twelve GO terms in a molecular function.
Table III. Down-regulated genes in HaPC-5 cells treated with FxOHa.
HaPC, Hamster pancreatic cancer; FxOH, fucoxanthinol. aAmong all 1,213 genes significantly changed, down-regulated genes classified to biological process [1 gene ontology (GO) term], cellular component (3 GO terms) and molecular function (2 GO terms) in GO term analysis are showed. bFold change of gene expression in FxOH-treated HaPC5 cells in comparison with that of control cells. cSignificant difference between HaPC5 cells with and without FxOH treatments by an exact test on edgeR.
Table IV. Down-regulated genes in HaPC-5 cells treated with FxOHa.
HaPC, Hamster pancreatic cancer; FxOH, fucoxanthinol. aAmong 1,213 genes with significantly changed in expression levels, 32 down-regulated genes classified as cell surface among cellular components in the gene ontology term analysis are shown. bFold change in gene expression in FxOHtreated HaPC-5 cells in comparison with that of control cells. cSignificant difference between HaPC-5 cells with and without FxOH treatments using an exact test in edgeR.
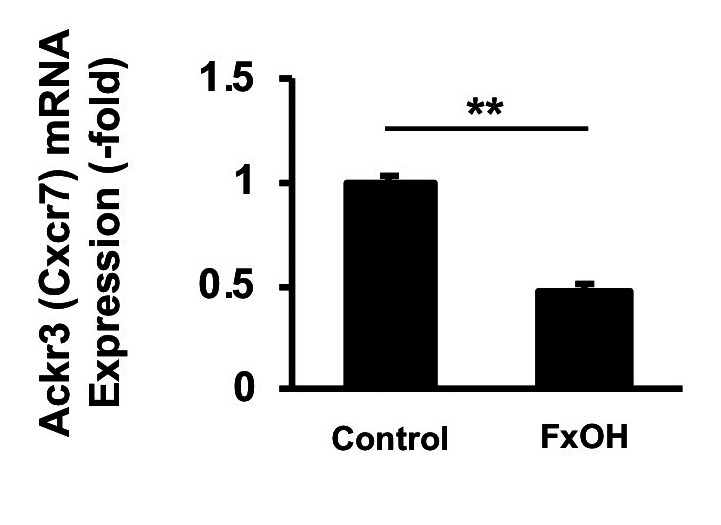
Effect of FxOH on Ackr3 (Cxcr7) gene expression in HaPC-5 cells. The gene expression of Ackr3 (Cxcr7) in HaPC-5 cells after 5.0 μM FxOH treatment for 1 day was evaluated. The mRNA expression of Ackr3 decreased significantly to 0.5-fold after 5.0 μM FxOH treatment in HaPC-5 cells compared with untreated control cells (Figure 7).
Figure 7. Effect of fucoxanthinol (FxOH) on Ackr3 gene expression in HaPC-5 cells. HaPC-5 cells were treated with 5.0 μM FxOH for 1 day. Ackr3 mRNA expression was determined using quantitative PCR. Expression was normalized to Gapdh expression levels. Ackr3 mRNA expression in the control cells was set as 1.0. Means±SE (n=3). **p<0.01.
Effect of FxOH on protein expressions in HaPC-5 cells. Based on the cell-cycle arrest and transcriptome analysis, the effect of FxOH on protein expression and activation in HaPC-5 cells was determined. FxOH treatment decreased the expression levels of cyclin D1, cyclin B1, CXCR7, integrin α5, pFAK(Tyr397), pPaxillin(Tyr31), pAKT(Ser473), and pSmad2(Ser465/467) and increased that of pERK1/2(Thr202/Tyr204) in HaPC-5 cells. Expression of cleaved caspase-3 (p17/p19), the active form of caspase-3, was increased in HaPC-5 cells after FxOH treatment. Little difference between FxOH-treated HaPC-5 cells and control cells was observed for cyclin D2, CXCR4, integrin β1, integrin β4, integrin β8, pAKT(Thy308), AKT(pan), pMEK1/2 (Ser217/221), Smad2, pro-caspase-3, and p53 (Figure 8).
Figure 8. Effect of fucoxanthinol (FxOH) on protein expression levels in HaPC-5 cells. HaPC5 cells were treated with 5.0 μM FxOH for 1 day, and protein expression levels were evaluated using western blotting. Levels of indicated protein are shown with β-actin (internal control).
Discussion
The present study demonstrated that FxOH induced apoptosis in HaPC-5 cells through suppression of many genes and signal transduction pathways. This is the first study suggesting the anti-proliferating effect of FxOH on cell lines from a hamster pancreatic cancer model.
No associations between pathological findings and growth inhibition in FxOH-treated HaPC-1–6 were observed (Table I and Figure 2). However, FxOH significantly suppressed the growth of HaPC-1, -4, -5, and -6 cells among the six types of HaPC cells (Figure 2). Thus, we decided to elucidate the molecular mechanisms of the growth inhibitory effects using FxOH-treated HaPC-5 cells, which showed the highest growth inhibition. For example, 5.0 μM FxOH treatment significantly induced apoptosis in HaPC-5 cells with G1 phase arrest (Figure 3B).
Transcriptome analysis revealed that FxOH treatment significantly inhibited gene sets for cell cycle, cell division, chemokine, cadherin, extracellular matrix, integrin, actin polymerization, microtubule organization, and the pathways of the renin–angiotensin system, MAPK, TGF-β, and Wnt in HaPC-5 cells (Figure 6, Tables III and IV). On the other hand, few up-regulated genes involved in apoptosis induction were found (Figure 5 and Table II). Based on these transcriptome profiles, we confirmed alterations in the levels of proteins related to cell cycle, chemokines, adhesion, apoptosis, and pathways for PI3K/AKT, MAPK, and TGF-β by western blotting analysis. Western blotting analysis showed that the levels of cyclin D1, cyclin B1, CXCR7, integrin α5, pFAK(Tyr397), pPaxillin(Tyr31), pAKT(Ser473), and pSmad2(Ser465/467) were decreased, and pERK1/2(Thr202/Tyr204) and cleaved caspase-3(p17/p19) levels were increased in FxOH-treated HaPC-5 cells (Figure 8).
In the present study, FxOH treatment inhibited the cell cycle in HaPC-5 cells (Figures 3B and 8). The effects of cell cycle arrest by FxOH or Fx in cancer cells were previously found in various cancer cell lines (20,28,29). As was observed in pancreatic cancer cells, cell-cycle arrest followed by apoptosis induction may be a common feature of various cancer cells.
In the present experiments, we especially focused on the expression levels of CXC motif chemokine receptor 4 (Cxcr4) and atypical chemokine receptor 3 (Ackr3, also called as Cxcr7) with the GO term for cell surface in cellular component. Regarding CXCR7, a significant decrease in Ackr3 (Cxcr7) gene expression was also identified (Figure 7). FxOH treatment also decreased CXCR7 expression but not CXCR4 in HaPC-5 cells (Figure 8). CXCR7, a member of the chemokine receptors, is known to interact with homeostatic and inflammatory chemokines, although CXCR4 interacts only with homeostatic chemokines (30). CXCR7 has been reported to enhance cell growth, migration and metastasis, induction of angiogenesis, homing of immune cells, recruitment of β-arrestin, release of interleukin, and activation of the vascular endothelial growth factor, PI3K/AKT, mammalian target or rapamycin, and MAPK pathways (31,32). CXCR7 is highly expressed in both pancreatic cancer tissue and pancreatic cancer cell lines (33). This protein expression is positively associated with poor prognosis in pancreatic cancer patients (32,34-36).
FxOH treatment also decreased the expression of integrin α5, and the activation of FAK and Paxillin, which are down-stream regulators of integrins, in HaPC-5 cells (Figure 8). Anoikis is caspase-dependent apoptosis that happens after the detachment of cancer cells from the extracellular matrix via attenuation of integrin signaling along with the suppression of PI3K/AKT, MAPK, and TGF-β signals (37-39). Our previous studies demonstrated that Fx and FxOH induce anoikis in murine colorectal tissue and in colon cancer DLD-1 cells, respectively, through suppression of integrin signals (25,26). FxOH is also known to down-regulate many cytoskeletal genes in HaPC-5 cells (Figure 6, Tables III and IV). Taking into consideration that integrins interact with the extracellular matrix and intracellular cytoskeleton, and promote migration/metastasis in cancer cells (40-42), it was suggested that FxOH may suppress firstly CXCR7 and integrin α5 on cellular membrane, and then alter the down-streams of PI3K/AKT, FAK/Paxillin, TGF-β and cell cycle signals, actin polymerization and microtubule organization in HaPC-5 cells, followed by apoptosis and anoikis inductions.
A previous study revealed the growth inhibition by Fx in a human pancreatic cancer MIA PaCa-2 cell (43). On the other hands, crocetinic acid, a carotenoid having two carboxylic acids, induced apoptosis in human pancreatic cancer MIA PaCa-2 cells and suppressed a tumorigenesis in the xenograft model mice by inhibiting EGFR and AKT pathways (44). Lycopene, a hydrophobic carotenoid, could induce apoptosis in human pancreatic cancer PANC-1 cells by inhibiting the activation of NF-ĸB signals through suppression of reactive oxygen species (45). Further studies are needed to elucidate the effects of FxOH in pancreatic cancer cells.
In conclusion, FxOH modified the expression levels of 1,213 genes and induced apoptosis in a pancreatic ductal adenocarcinoma HaPC-5 cell cloned from a pancreatic cancer hamster model. Moreover, the protein expression and activation levels of cyclin D1, cyclin B1, CXCR7, integrin α5, pFAK(Tyr397), pPaxillin(Tyr31), pAKT(Ser473), and pSmad2(Ser465/467), which play central roles in cell cycle, chemokine, adhesion, and TGF-β signals were significantly suppressed. Our findings suggested that FxOH may have high potential as a cancer chemopreventive agent in a hamster pancreatic carcinogenesis model.
Conflicts of Interest
No conflicts of interest.
Authors’ Contributions
M. Terasaki conceived and designed the study and wrote the paper. M. Terasaki, Y.N., W. M., T. T. and M. Takahashi performed the experiments. A. K., H. K., M. K., H. M., K. M., M. M. and M. Takahashi reviewed and edited the manuscript. All Authors read and approved the final manuscript.
Acknowledgements
This work was supported in part by the Japan Society for the Promotion of Science KAKENHI (Grant Number 20K05879, 19H05652 and 19K07150).
References
- 1.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144(8):1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society Cancer Facts and Figures 2020. American Cancer Society, Atlanta, GA USA. Available at: https://www.cancer.org/research/cancer-facts-statistics/all-cancerfacts-figures/cancer-facts-figures-2020.html [Last accessed on February 12, 2021] [DOI] [PMC free article] [PubMed]
- 3.Puleo F, Nicolle R, Blum Y, Cros J, Marisa L, Demetter P, Quertinmont E, Svrcek M, Elarouci N, Iovanna J, Franchimont D, Verset L, Galdon MG, Devière J, de Reyniès A, Laurent-Puig P, Van Laethem JL, Bachet JB, Maréchal R. Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology. 2018;155(6):1999–2013.e3. doi: 10.1053/j.gastro.2018.08.033. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Electronic address andrew_aguirre@dfci.harvard.edu. , Cancer Genome Atlas Research Network Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32(2):185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey P, Chang DK, Nones K, Johns AL, Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC, Nourse C, Murtaugh LC, Harliwong I, Idrisoglu S, Manning S, Nourbakhsh E, Wani S, Fink L, Holmes O, Chin V, Anderson MJ, Kazakoff S, Leonard C, Newell F, Waddell N, Wood S, Xu Q, Wilson PJ, Cloonan N, Kassahn KS, Taylor D, Quek K, Robertson A, Pantano L, Mincarelli L, Sanchez LN, Evers L, Wu J, Pinese M, Cowley MJ, Jones MD, Colvin EK, Nagrial AM, Humphrey ES, Chantrill LA, Mawson A, Humphris J, Chou A, Pajic M, Scarlett CJ, Pinho AV, Giry-Laterriere M, Rooman I, Samra JS, Kench JG, Lovell JA, Merrett ND, Toon CW, Epari K, Nguyen NQ, Barbour A, Zeps N, Moran-Jones K, Jamieson NB, Graham JS, Duthie F, Oien K, Hair J, Grützmann R, Maitra A, Iacobuzio-Donahue CA, Wolfgang CL, Morgan RA, Lawlor RT, Corbo V, Bassi C, Rusev B, Capelli P, Salvia R, Tortora G, Mukhopadhyay D, Petersen GM, Australian Pancreatic Cancer Genome Initiative , Munzy DM, Fisher WE, Karim SA, Eshleman JR, Hruban RH, Pilarsky C, Morton JP, Sansom OJ, Scarpa A, Musgrove EA, Bailey UM, Hofmann O, Sutherland RL, Wheeler DA, Gill AJ, Gibbs RA, Pearson JV, Waddell N, Biankin AV, Grimmond SM. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 6.Witkiewicz AK, McMillan EA, Balaji U, Baek G, Lin WC, Mansour J, Mollaee M, Wagner KU, Koduru P, Yopp A, Choti MA, Yeo CJ, McCue P, White MA, Knudsen ES. Whole-exome sequencing of pancreatic cancer defines genetic diversity and therapeutic targets. Nat Commun. 2015;6:6744. doi: 10.1038/ncomms7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321(5897):1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cicenas J, Kvederaviciute K, Meskinyte I, Meskinyte-Kausiliene E, Skeberdyte A, Cicenas J. KRAS, TP53, CDKN2A, SMAD4, BRCA1, and BRCA2 mutations in pancreatic cancer. Cancers (Basel) 2017;9(5):42. doi: 10.3390/cancers9050042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi M, Hori M, Mutoh M, Wakabayashi K, Nakagama H. Experimental animal models of pancreatic carcinogenesis for prevention studies and their relevance to human disease. Cancers (Basel) 2011;3(1):582–602. doi: 10.3390/cancers3010582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kuno T, Takahashi S, Tomita H, Hisamatsu K, Hara A, Hirata A, Kobayashi H, Mori H. Preventive effects of fermented brown rice and rice bran against N-nitrosobis (2-oxopropyl) amine-induced pancreatic tumorigenesis in male hamsters. Oncol Lett. 2015;10(6):3377–3384. doi: 10.3892/ol.2015.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okamura T, Umemura T, Inoue T, Tasaki M, Ishii Y, Nakamura Y, Park EY, Sato K, Matsuo T, Okamoto S, Nishikawa A, Ogawa K. Chemopreventive effects of 4-methylthio-3-butenyl Isothiocyanate (Raphasatin) but not curcumin against pancreatic carcinogenesis in hamsters. J Agric Food Chem. 2013;61(9):2103–2108. doi: 10.1021/jf3003174. [DOI] [PubMed] [Google Scholar]
- 12.Kuroiwa Y, Nishikawa A, Kitamura Y, Kanki K, Ishii Y, Umemura T, Hirose M. Protective effects of benzyl isothiocyanate and sulforaphane but not resveratrol against initiation of pancreatic carcinogenesis in hamsters. Cancer Lett. 2006;241(2):275–280. doi: 10.1016/j.canlet.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 13.Majima T, Tsutsumi M, Nishino H, Tsunoda T, Konishi Y. Inhibitory effects of beta-carotene, palm carotene, and green tea polyphenols on pancreatic carcinogenesis initiated by N-nitorsobis(2-oxopropyl)amine in Syrian golden hamsters. Pancreas. 1998;16(1):13–18. doi: 10.1097/00006676-199801000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Asai A, Yonekura L, Nagao A. Low bioavailability of dietary epoxyxanthophylls in humans. Br J Nutr. 2008;100(2):273–277. doi: 10.1017/S0007114507895468. [DOI] [PubMed] [Google Scholar]
- 15.Yonekura L, Kobayashi M, Terasaki M, Nagao A. Keto-carotenoids are the major metabolites of dietary lutein and fucoxanthin in mouse tissues. J Nutr. 2010;140(10):1824–1831. doi: 10.3945/jn.110.126466. [DOI] [PubMed] [Google Scholar]
- 16.Nishino H, Murakoshi M, Tokuda H, Satomi Y. Cancer prevention by carotenoids. Arch Biochem Biophys. 2009;483(2):165–168. doi: 10.1016/j.abb.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Chen W, Zhang H, Liu Y. Anti-inflammatory and apoptotic signaling effect of fucoxanthin on benzo(A)pyrene-induced lung cancer in mice. J Environ Pathol Toxicol Oncol. 2019;38(3):239–251. doi: 10.1615/JEnvironPatholToxicolOncol.2019030301. [DOI] [PubMed] [Google Scholar]
- 18.Garg S, Afzal S, Elwakeel A, Sharma D, Radhakrishnan N, Dhanjal JK, Sundar D, Kaul SC, Wadhwa R. Marine carotenoid fucoxanthin possesses anti-metastasis activity: Molecular evidence. Mar Drugs. 2019;17(6):338. doi: 10.3390/md17060338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jin Y, Qiu S, Shao N, Zheng J. Fucoxanthin and tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) synergistically promotes apoptosis of human cervical cancer cells by targeting PI3K/Akt/NF-ĸB signaling pathway. Med Sci Monit. 2018;24:11–18. doi: 10.12659/msm.905360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu RX, Yu RT, Liu Z. Inhibition of two gastric cancer cell lines induced by fucoxanthin involves downregulation of Mcl-1 and STAT3. Hum Cell. 2018;31(1):50–63. doi: 10.1007/s13577-017-0188-4. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Zheng J, Zhang Y, Wang Z, Yang Y, Bai M, Dai Y. Fucoxanthin activates apoptosis via inhibition of PI3K/Akt/mTOR pathway and suppresses invasion and migration by restriction of p38-MMP-2/9 pathway in human glioblastoma cells. Neurochem Res. 2016;41(10):2728–2751. doi: 10.1007/s11064-016-1989-7. [DOI] [PubMed] [Google Scholar]
- 22.Lopes FG, Oliveira KA, Lopes RG, Poluceno GG, Simioni C, Gabriel DSP, Bauer CM, Maraschin M, Derner RB, Garcez RC, Tasca CI, Nedel CB. Anti-cancer effects of fucoxanthin on human glioblastoma cell line. Anticancer Res. 2020;40(12):6799–6815. doi: 10.21873/anticanres.14703. [DOI] [PubMed] [Google Scholar]
- 23.Kotake-Nara E, Terasaki M, Nagao A. Characterization of apoptosis induced by fucoxanthin in human promyelocytic leukemia cells. Biosci Biotechnol Biochem. 2005;69(1):224–227. doi: 10.1271/bbb.69.224. [DOI] [PubMed] [Google Scholar]
- 24.Terasaki M, Maeda H, Miyashita K, Tanaka T, Miyamoto S, Mutoh M. A marine bio-functional lipid, fucoxanthinol, attenuates human colorectal cancer stem-like cell tumorigenicity and sphere formation. J Clin Biochem Nutr. 2017;61(1):25–32. doi: 10.3164/jcbn.16-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terasaki M, Maeda H, Miyashita K, Mutoh M. Induction of anoikis in human colorectal cancer cells by fucoxanthinol. Nutr Cancer. 2017;69(7):1043–1052. doi: 10.1080/01635581.2017.1339814. [DOI] [PubMed] [Google Scholar]
- 26.Terasaki M, Iida T, Kikuchi F, Tamura K, Endo T, Kuramitsu Y, Tanaka T, Maeda H, Miyashita K, Mutoh M. Fucoxanthin potentiates anoikis in colon mucosa and prevents carcinogenesis in AOM/DSS model mice. J Nutr Biochem. 2019;64:198–205. doi: 10.1016/j.jnutbio.2018.10.007. [DOI] [PubMed] [Google Scholar]
- 27.Terasaki M, Mima M, Kudoh S, Endo T, Maeda H, Hamada J, Osada K, Miyashita K, Mutoh M. Glycine and succinic acid are effective indicators of the suppression of epithelial-mesenchymal transition by fucoxanthinol in colorectal cancer stem-like cells. Oncol Rep. 2018;40(1):414–424. doi: 10.3892/or.2018.6398. [DOI] [PubMed] [Google Scholar]
- 28.Yokoyama R, Kojima H, Takai R, Ohta T, Maeda H, Miyashita K, Mutoh M, Terasaki M. Effects of CLIC4 on fucoxanthinol-induced apoptosis in human colorectal cancer cells. Nutrition and. 2020;Cancer:1–10. doi: 10.1080/01635581.2020.1779760. [DOI] [PubMed] [Google Scholar]
- 29.Mei C, Zhou S, Zhu L, Ming J, Zeng F, Xu R. Antitumor effects of laminaria extract fucoxanthin on lung cancer. Mar Drugs. 2017;15(2):39. doi: 10.3390/md15020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nomiyama H, Osada N, Yoshie O. A family tree of vertebrate chemokine receptors for a unified nomenclature. Dev Comp Immunol. 2011;35(7):705–715. doi: 10.1016/j.dci.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 31.Huynh C, Dingemanse J, Meyer Zu Schwabedissen HE, Sidharta PN. Relevance of the CXCR4/CXCR7-CXCL12 axis and its effect in pathophysiological conditions. Pharmacol Res. 2020;161:105092. doi: 10.1016/j.phrs.2020.105092. [DOI] [PubMed] [Google Scholar]
- 32.Guo JC, Li J, Zhou L, Yang JY, Zhang ZG, Liang ZY, Zhou WX, You L, Zhang TP, Zhao YP. CXCL12-CXCR7 axis contributes to the invasive phenotype of pancreatic cancer. Oncotarget. 2016;7(38):62006–62018. doi: 10.18632/oncotarget.11330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heinrich EL, Lee W, Lu J, Lowy AM, Kim J. Chemokine CXCL12 activates dual CXCR4 and CXCR7-mediated signaling pathways in pancreatic cancer cells. J Transl Med. 2012;10:68. doi: 10.1186/1479-5876-10-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan H, Wang W, Yan J, Xiao L, Yang L. Prognostic significance of CXCR7 in cancer patients: a meta-analysis. Cancer Cell Int. 2018;18:212. doi: 10.1186/s12935-018-0702-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Teng XY, Meng XP, Wang BS. Expression of stromal cell-derived factor 1 and CXCR7 ligand receptor system in pancreatic adenocarcinoma. World J Surg Oncol. 2014;12:348. doi: 10.1186/1477-7819-12-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gebauer F, Tachezy M, Effenberger K, von Loga K, Zander H, Marx A, Kaifi JT, Sauter G, Izbicki JR, Bockhorn M. Prognostic impact of CXCR4 and CXCR7 expression in pancreatic adenocarcinoma. J Surg Oncol. 2011;104(2):140–145. doi: 10.1002/jso.21957. [DOI] [PubMed] [Google Scholar]
- 37.Buchheit CL, Weigel KJ, Schafer ZT. Cancer cell survival during detachment from the ECM: multiple barriers to tumour progression. Nat Rev Cancer. 2014;14(9):632–641. doi: 10.1038/nrc3789. [DOI] [PubMed] [Google Scholar]
- 38.Paoli P, Giannoni E, Chiarugi P. Anoikis molecular pathways and its role in cancer progression. Biochim Biophys Acta. 2013;1833(12):3481–3498. doi: 10.1016/j.bbamcr.2013.06.026. [DOI] [PubMed] [Google Scholar]
- 39.Horbinski C, Mojesky C, Kyprianou N. Live free or die: tales of homeless (cells) in cancer. Am J Pathol. 2010;177(3):1044–1052. doi: 10.2353/ajpath.2010.091270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DeMali KA, Wennerberg K, Burridge K. Integrin signaling to the actin cytoskeleton. Curr Opin Cell Biol. 2003;15(5):572–582. doi: 10.1016/s0955-0674(03)00109-1. [DOI] [PubMed] [Google Scholar]
- 41.Ridley AJ, Schwartz MA, Burridge K, Firtel RA, Ginsberg MH, Borisy G, Parsons JT, Horwitz AR. Cell migration: integrating signals from front to back. Science. 2003;302(5651):1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 42.Ali MRK, Wu Y, Tang Y, Xiao H, Chen K, Han T, Fang N, Wu R, El-Sayed MA. Targeting cancer cell integrins using gold nanorods in photothermal therapy inhibits migration through affecting cytoskeletal proteins. Proc Natl Acad Sci USA. 2017;114(28):E5655–E5663. doi: 10.1073/pnas.1703151114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuboi M, Etoh H, Kato K, Nakatugawa H, Kato H, Maejima Y, Matsumoto G, Mori H, Hosokawa M, Miyashita K, Tokuda H, Suzuki N, Maoka T. Nitrocapsanthin and nitrofucoxanthin, respective products of capsanthin and fucoxanthin reaction with peroxynitrite. J Agric Food Chem. 2011;59(19):10572–10578. doi: 10.1021/jf203493k. [DOI] [PubMed] [Google Scholar]
- 44.Rangarajan P, Subramaniam D, Paul S, Kwatra D, Palaniyandi K, Islam S, Harihar S, Ramalingam S, Gutheil W, Putty S, Pradhan R, Padhye S, Welch DR, Anant S, Dhar A. Crocetinic acid inhibits hedgehog signaling to inhibit pancreatic cancer stem cells. Oncotarget. 2015;6(29):27661–27673. doi: 10.18632/oncotarget.4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeong Y, Lim JW, Kim H. Lycopene Inhibits Reactive Oxygen Species-Mediated NF-ĸB Signaling and Induces Apoptosis in Pancreatic Cancer Cells. Nutrients. 2019;11(4) doi: 10.3390/nu11040762. [DOI] [PMC free article] [PubMed] [Google Scholar]