Abstract
Background: Colorectal cancer is the most common gastrointestinal carcinoma in western countries. Prognosis of metastatic colorectal cancer has improved in the last decades, but the disease continues to carry an adverse outcome in most cases. An improved understanding of molecular pathogenesis has provided incremental benefits in survival outcomes with the introduction of targeted therapies for specific sub-types and gives hope for further improvements. Materials and Methods: Publicly available data from genomic series of colorectal cancer published by the TCGA were analyzed with the aim of characterizing the sub-set of colorectal cancers carrying amplifications of chromosome 20q11.21, compared with cancers with no amplifications in this locus. Associations of 20q11.21-amplified cancers with other molecular lesions commonly observed in colorectal cancer were explored. mRNA expression of genes from the locus in amplified cases was analyzed. An exploratory survival analysis was also performed. Results: Amplifications of genes at chromosome arm 20q are observed in 7% to 9% of colorectal cancers, representing the most commonly amplified loci in this type of cancer. The 20q11.21 presents the highest amplification rate in the 20q arm. 20q11.21 amplified cancers display concomitant mutations in the KRAS pathway and SMAD4 less often than non-amplified cancers. Mutations in DNA repair genes are also less often encountered in 20q11.21 amplified colorectal cancers than non-amplified ones. Conclusion: Amplification of genes at locus 20q11.21, representing the most frequently amplified locus in colorectal cancers, is associated with specific molecular characteristics and may have therapeutic implications.
Keywords: Colorectal cancer, amplifications, copy number alterations, mutations, chromosomal instability
Colorectal cancer is among the most prevalent cancers and a major cause of cancer-related morbidity and mortality (1). Colon and rectal cancers accounted for over 1.8 million cases worldwide in 2018 and 860,000 deaths. Improvements in survival of metastatic colorectal cancer have been accomplished in the recent decades with incremental benefits obtained by the introduction of more effective chemotherapies and targeted therapies. Targeted therapies based on an improved understanding of the molecular biology of colorectal cancer hold the promise for further improvement in outcomes.
The molecular biology of colorectal cancer has been elucidated and in-depth studies of the trajectory of neoplastic lesions, from adenoma to in situ carcinoma and invasive carcinoma with associated molecular defects accumulating during these transitions, paints the picture of colorectal cancer as a multistep process. Molecular lesions in different cancer-associated pathways have been described in detail and include mutations in genes encoding for tumor suppressors such as APC, p53 and SMAD4 as well as activating mutations of the KRAS oncogene (2). Some colorectal cancers present with increased microsatellite instability due to mismatch repair defects akin to the hereditary non-polyposis colorectal cancer syndrome. Other cancers, which are more common, have increased chromosomal instability with numerous gains and losses. Genomic studies have established the heterogeneity of colorectal cancer with several overlapping sub-types (3-8). A meta-analysis of genomic studies has provided a unified classification of genomic profiles in four consensus molecular sub-types (9,10).
Loci at chromosome arm 20q are frequently amplified in colorectal cancers. Commonly these amplifications encompass several dozen genes located in the chromosome arm, even though the amplified areas are not always continuous. This suggests that fragments of 20q are independently and concomitantly amplified in colorectal cancer. Genes at the 20q11.21 locus top the list of amplified genes in colorectal cancer in published genomic series (11,12). This study analyzed colorectal cancers with 20q11.21 amplifications and compared their characteristics with those of cancers with no amplification of this locus.
Materials and Methods
Genomic data of colorectal cancer patients from The Cancer Genome Atlas (TCGA) study cohort and the Memorial Sloan Kettering Cancer Center (MSKCC) IMPACT (MSK-IMPACT) cohort were extracted and analyzed. The cBioCancer Genomics Portal (http://www.cbioportal.org) was utilized for the analysis (13). The platform maintained by MSKCC and other academic institutions is freely available on-line and provides a user-friendly interface. Sub-sets of colorectal cancers with or without amplifications of genes at the 20q11.21 locus were analyzed for defining clinical and genomic characteristics of each sub-set. The two series used in the analysis employ different genomic protocols (11,12). TCGA employs whole exome sequencing, while the MSK-COMPACT study uses a more targeted sequencing protocol that includes 341 cancer-associated genes (11,12). TCGA presents an aneuploidy score (AS), which is the sum of the number of chromosome arms in each patient sample included in the study that display copy number alterations (gains or losses). A chromosome arm is considered copy number altered based on the length of alterations as calculated by the ABSOLUTE algorithm from Affymetrix 6.0 SNP arrays (14). The definition of a somatic copy number alteration was set at more than 80% of the length of the arm. Alterations in 20% to 80% of a given arm length were considered inadequate to call, while chromosomal arms with somatic copy number alterations in less than 20% of the arm length were considered not altered. The OncoKB knowledgebase, a database of cancer-related genes is used for classification of the genes as cancer-related and for information on specific cancer roles of the genes from the 20q11.21 locus (15).
Statistical comparisons of categorical data were carried using Fisher’s exact test or the χ2 test. The Log-Rank test was used to compare Kaplan-Meier survival curves. All statistical comparisons were considered significant if p<0.05.
Results
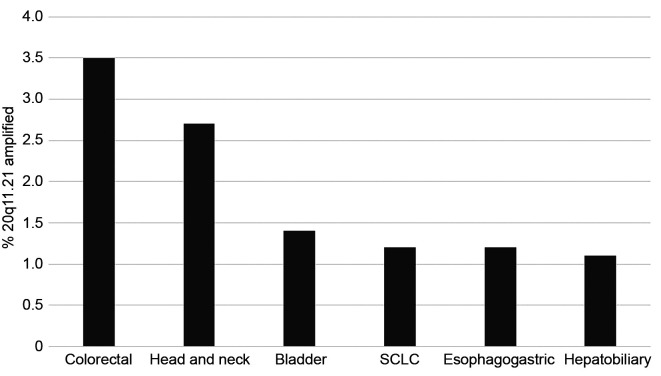
The chromosome locus 20q11.21 is the most commonly amplified genomic area in colorectal cancer. The frequency of amplification of genes located in this locus ranges from 8.1% to 9.3% in the TCGA cohort. The three representative genes from the locus that are included in the analysis of the cohort of metastatic colorectal cancers from MSKCC show a lower amplification rate (BCL2L1: 4.9%, ASXL1: 3.8%, DNMT3B: 3.4%). Various other cancers in the MSK-IMPACT study show a low rate of amplification of the 20q11.21 locus (as represented by amplification of the ASXL1 gene) of about 1% (Figure 1). The highest amplification rate was observed in colorectal cancer (3.5% in this study) and in head and neck carcinomas (2.5%).
Figure 1. Frequency of amplification of 20q11.21 (as defined by amplification of ASXL1) in various cancers. Cancers with 20q11.21 amplification in more than 1% of cases are shown. Data are derived from the MSK-IMPACT study (12).
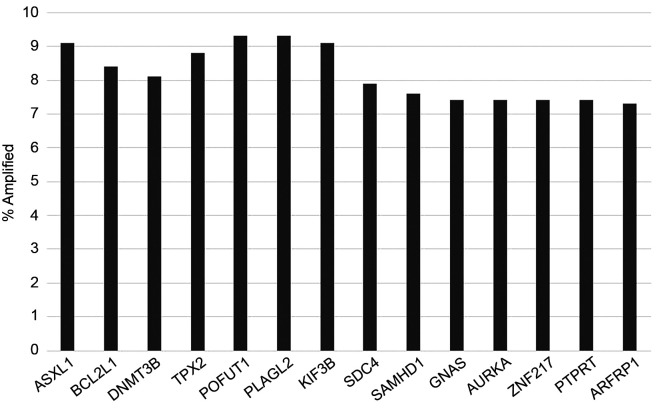
The three genes at 20q11.21 locus that are listed at OncoKB database (BCL2L1, ASXL1 and DNMT3B) are co-amplified in a high percentage of cases in the TCGA colorectal cancer group. Among cases with BCL2L1 amplifications, 94% and 86% of cases have co-amplifications of ASXL1 and DNMT3B, respectively. Other genes across the entire 20q arm are also amplified in lower frequencies overall (Figure 2). Co-amplification of these genes with genes at 20q11.21 occur less frequently. For example, AURKA and GNAS genes located at 20q13.2 and 20q13.32 are amplified in 7.4% of colorectal cancers but only 52% of BCL2L1 amplified cases contain co-amplifications in these genes.
Figure 2. Frequency of amplification of representative genes from chromosome arm 20q in colorectal cancer. Data are derived from the TCGA colorectal cancer cohort.
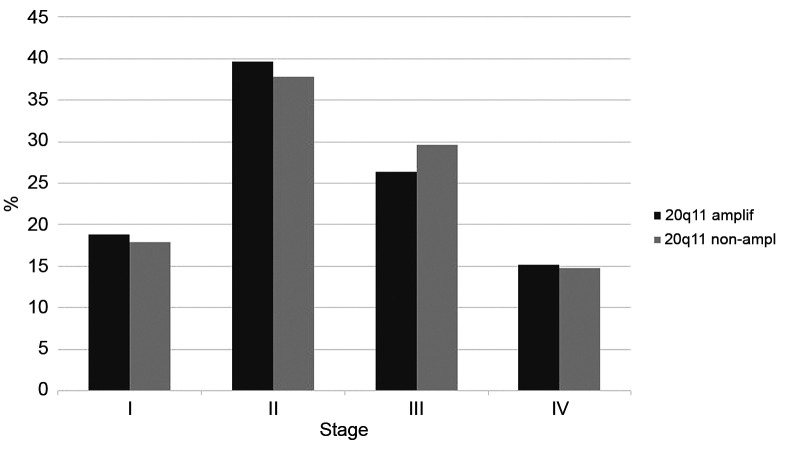
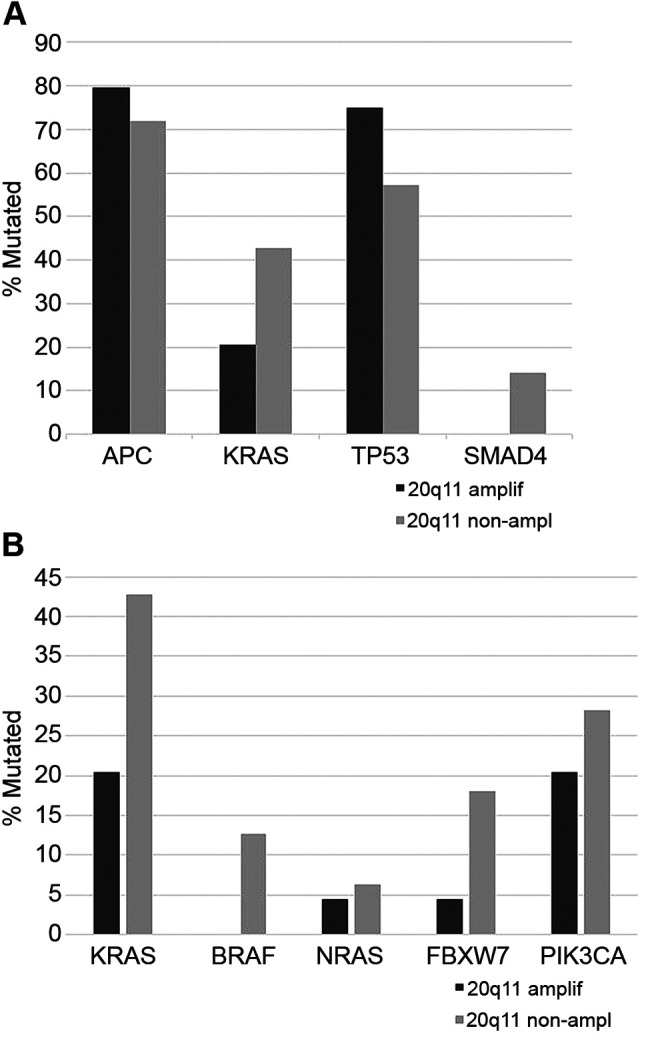
Colorectal cancers with 20q11.21 amplifications did not differ significantly from non-amplified cancers regarding stage at presentation (χ2 p=0.9, Figure 3). The prevalence of mutations in the four most commonly mutated genes in colorectal cancer, the KRAS oncogene and the APC, TP53 and SMAD4 tumor suppressors displayed a balanced distribution, with KRAS and SMAD4 mutations being more frequent in non-amplified cases (42.7% and 14.1%, respectively) than in amplified ones (20.5% and none, respectively Figure 4A). In contrast, APC and TP53 mutations were more frequent in amplified cases (79.5% and 75%, respectively) than in non-amplified ones (71.8% and 57.3%, respectively). These differences were statistically significant in the case of KRAS, SMAD4 and TP53 (Fisher’s exact test p=0.003, 0.01 and 0.02, respectively) but not for APC (Fisher’s exact test p=0.37). Besides KRAS, other genes of the RAS-RAF-MEK and RAS-PI3K-Akt pathway are also more commonly mutated in 20q11.21 non-amplified cases than in 20q11.21 amplified colorectal cancers (Fisher’s exact test p=0.007, Figure 4B). These genes are less frequently mutated than KRAS in colorectal cancers, but several are included in clinical panels as markers of anti-EGFR1 drug resistance.
Figure 3. Distribution of stage at presentation in colorectal cancers with and without 20q11.21 amplifications. Data are derived from the TCGA colorectal cancer cohort.
Figure 4. Percentage of mutations: A. in the most common colorectal cancer associated genes and B. in genes of the RAS-RAF-PI3K cascade in colorectal cancers with and without 20q11.21 amplifications. Data are derived from the TCGA colorectal cancer cohort.
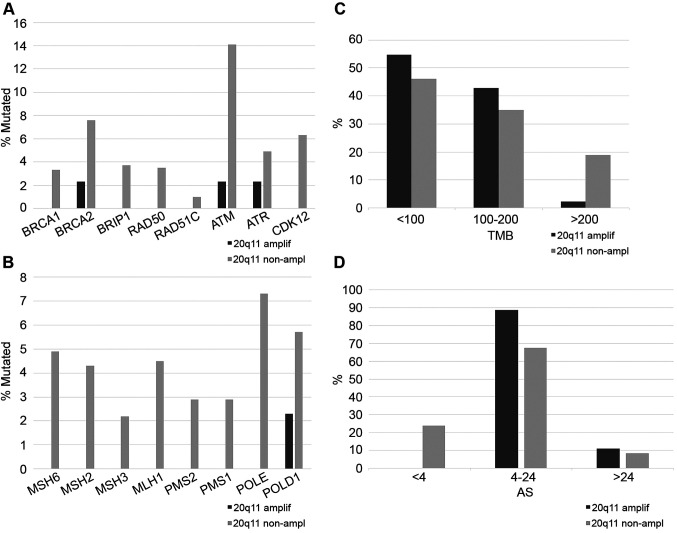
Mutations in BRCA1 and BRCA2 and other genes involved in DNA damage response (DDR) are individually comparatively rare in colorectal cancer. In the colorectal cancer TCGA cohort, BRCA1 mutations were observed in 3% of cases and BRCA2 mutations in 7.1%. However, in cancers with no 20q11.21 amplifications, mutations in one or more of the DDR genes are collectively present in 24% of cases and are more common than DDR gene mutations in 20q11.21 amplified colorectal cancers (Fisher’s exact test p=0.005, Figure 5A). Likewise, mutations in one or more of the genes associated with microsatellite instability (MSI) or mutations in the proofreading polymerases epsilon and delta1 (POLE and POLD1) are present in 18.2% of colorectal cancers without 20q11.21 amplifications but only in 2.3% of cases with 20q11.21 amplifications (Fisher’s exact test p=0.003, Figure 5B). MSI related proteins and proofreading polymerases are associated with higher tumor mutation burden (TMB) and, consistent with their higher prevalence in 20q11.21 non-amplified cancers, these cancers have a TMB above 200 in 18.9% of cases while only 2.4% of 20q11.21 amplified colorectal cancers have TMB above 200 (Fisher’s exact test p=0.003, Figure 5C). In contrast, 20q11.21 amplified colorectal cancers present a higher prevalence of chromosomal instability (CIN) with all the cases having an Aneuploidy Score (AS) of 4 or above, while cancers without 20q11.21 amplifications are CIN low with an AS score below 4 in 23.9% of cases (Figure 5D).
Figure 5. A. Percentage of mutations of genes involved in DNA damage response and repair in colorectal cancers with and without 20q11.21 amplifications. B. Percentage of mutations of genes involved in mismatch repair (MMR) or encoding for the proof-reading polymerases ε (POLE) and δ (POLD1) in colorectal cancers with and without 20q11.21 amplifications. C. Percentage of total mutation burden (TMB) levels in colorectal cancer patients with and without 20q11.21 amplifications. D. Percentage of patients with different Aneuploidy Score (AS) levels in colorectal cancer patients with and without 20q11.21 amplifications. Data are from the TCGA colorectal cancer cohort.
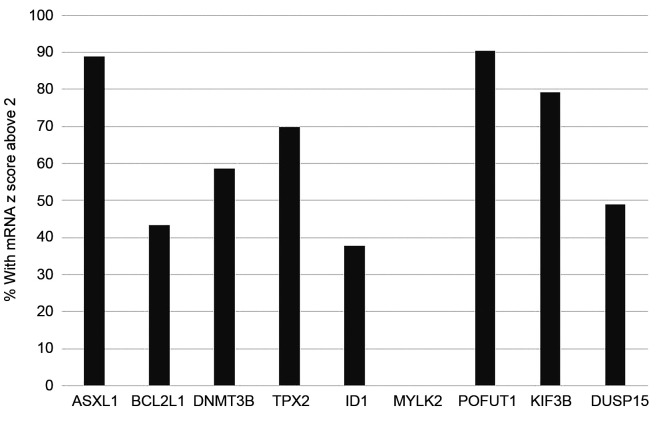
Expression of 20q11.21 genes at the mRNA level varies widely in cases with amplification of the locus. The genes with the higher expression at the mRNA level in 20q11.21 amplified colorectal cancers include the gene for fucosyl-transferase POFUT1 which shows a z score above 2 compared with diploid samples in 90.5% of 20q11.21 amplified cases and ASXL1 with a z score above 2 compared to diploid samples in 88.9% of 20q11.21 amplified cases (Figure 6). BCL2L1 and DNMT3B show mRNA overexpression only in 43.4% and 58.5% of 20q11.21 amplified colorectal cancer cases.
Figure 6. Percentage of patients with mRNA over-expression (z score above 2) of genes at the 20q11.21 locus among colorectal cancers with 20q11.21 amplification.
Genes of interest at locus 20q11.21 including BCL2L1, DNMT3B, TPX2 and POFUT1 are expressed in colorectal cancer tissues according to the human protein atlas, albeit with variable staining intensity. ASXL1 expression has not been investigated in human colorectal cancer tissues by immunohistochemistry in the atlas but is expressed in the colorectal cancer cell line CACO2.
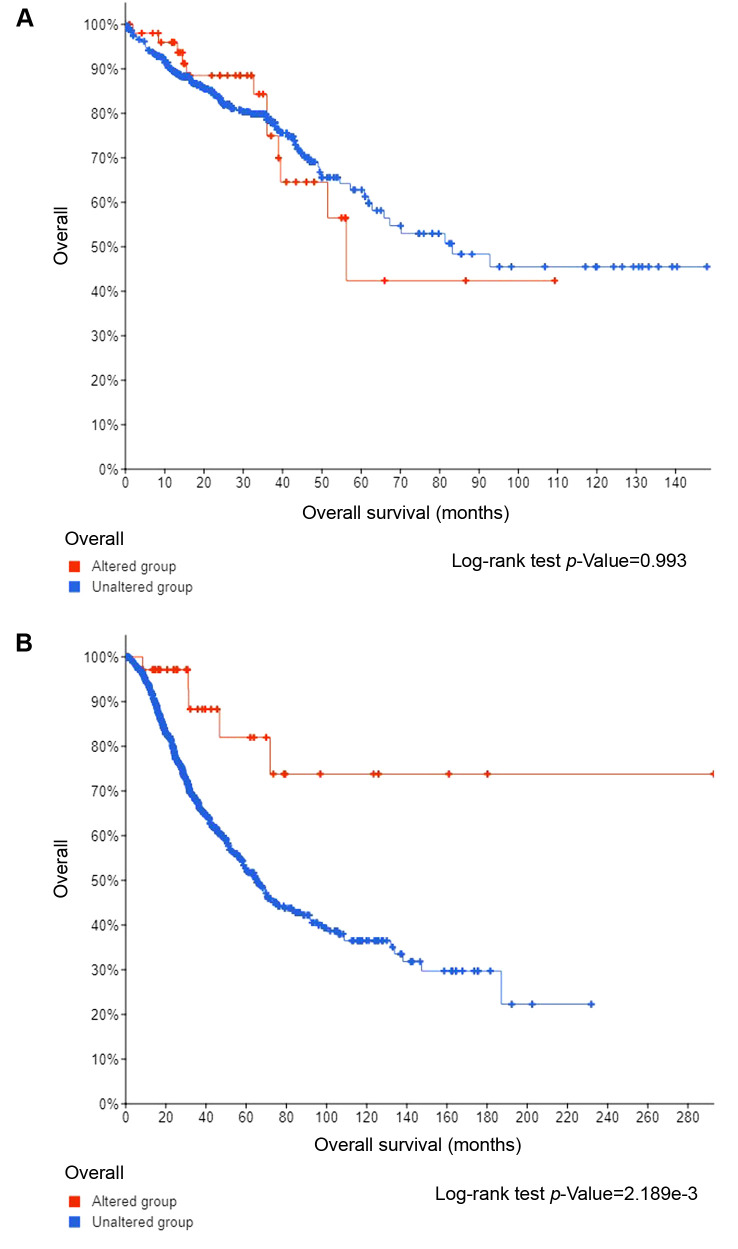
Overall Survival (OS) of patients with 20q11.21 amplified tumors was no different from the OS of patients without 20q11.21 amplifications in TCGA colorectal cancer cohort (Log-rank test p=0.99, Figure 7A). However, in the MSK-IMPACT series that included only metastatic colorectal cancer patients the group with 20q11.21 amplifications had a better OS than the non-amplified group (Figure 7B).
Figure 7. (A) Overall Survival curves of patients with 20q11.21 amplifications (altered group) and without 20q11.21 amplifications (unaltered group) in TCGA. Log-rank p=0.99. B. Overall Survival curves of patients with 20q11.21 amplifications (altered group) and without 20q11.21 amplifications (unaltered group) in the MSK-IMPACT cohort. Log-rank p=0.002.
Discussion
Chromosome locus 20q11.21 is the most frequently amplified locus in colorectal cancers, albeit with variable frequencies in different series, possibly due to the variable purity of samples included (16). Cancer related genes that map at this locus and show the highest frequency of amplification in the TCGA colorectal cancer data include BCL2L1, ASXL1 and DNMT3B. Other genes of potential interest located at 20q11.21 include TPX2, POFUT1 and KIF3B. BCL2L1 (also known as Bcl-x) is a BCL2 family member and a major apoptosis regulator (17). The gene encodes for two alternative protein isoforms, through alternative mRNA splicing. The longer isoform is anti-apoptotic and is called Bcl-xL while the shorter is a pro-apoptotic isoform, called Bcl-xS (18). Up-regulation of Bcl-xL provides a pro-survival signal for cancer cells. Induction of BCL2L1 and other proteins of the intrinsic apoptosis pathway results from DNA damage and oxidative stress. The long isoform is preferentially expressed over the shorter isoform when the BCL2L1 gene is induced in neoplastic cells (18). An increased Bcl-xL to Bcl-xS ratio contributes to mitochondrial outer membrane potential maintenance and prevents cytochrome c from spilling into the cytoplasm and from activating caspases, the executioners of apoptosis enzymes (19).
The two other genes of the locus listed in the OncoKB database as cancer associated genes, ASXL1 and DNMT3B encode epigenetic chromatin modifiers. ASXL1, one of the three human homologues of Drosophila ASX gene is a member of the Polycomb group repressor complex and is involved in methylation of histone 3 at position 27 (H3K27) which leads to transcription suppression (20,21). DNMT3B is a DNA methyl-transferase that regulates CpG island promoter methylation of various genes modulating transcription (22). Up-regulation of DNMT3B results in epigenetic modifications of expression of several proteins across the colorectal neoplasia stages from adenoma to carcinoma, including the Wnt pathway regulator SFRP2, the CDK inhibitor p16 and the mismatch repair protein MLH1 (23). Thus, altered expression of ASXL1 and DNMT3B in colorectal cancers with 20q11.21 amplifications cause a global epigenetic dysregulation.
TPX2 is not listed at the OncoKB database but has been discussed as a player in colorectal cancer, notably in connection with Aurora A kinase (AURKA), whose gene is also located in arm 20q, at the 20q13.2 locus (24). The expression of the two genes at the protein level correlates with their amplification and promotes transition from adenoma to carcinoma. The two genes positively influence anchorage independent growth and invasion of colorectal cancer cell lines (24). TPX2 is a microtubule associated protein and directly interacts with Aurora A kinase during mitosis, positioning the kinase on spindle microtubules where it promotes microtubule formation and stabilization. Dysregulation of the TPX2-AURKA functional unit leads to defects in chromosome segregation and chromosomal instability. The kinesin family member KIF3B, whose gene is also located at 20q11.21, is a microtubule associated protein and, interestingly, interacts with Wnt/ β-catenin regulator APC, which is commonly mutated in colorectal cancers (25).
The fucosyltransferase POFUT1 located at 20q11.21 regulates Notch signaling through fucosylation of the Notch receptor (26). POFUT1 fucosylates serines or threonines between the second and third cysteine in proteins with EGF-like domains and, thus, may have other targets regulating additional transduction pathways involved in colorectal cancer (27). POFUT1 has been found to be up-regulated in high-risk adenomas, suggesting a role in early pre-neoplastic lesions and progression to colorectal cancer (28). Another gene located next to POFUT1 at 20q11.21, pleomorphic adenoma gene-like 2 (PLAGL2) is a regulator of the Wnt pathway and may be co-regulated with POFUT1 by sharing the same promoter (29). Combined increased expression of the two genes affects stem cell maintenance, given that both Notch and Wnt are critical stem cell pathways (30).
Colorectal cancers are categorized to four consensus subsets according to their general molecular characteristics based on a meta-analysis of molecular subtype studies (9,10). These consensus molecular subtypes (CMS) include MSI/ immune (CMS1), the canonical subtype (CMS2) which is the most common (37% of all cases), the metabolic subtype (CMS3) and the mesenchymal subtype (CMS4). Colorectal cancers with 20q11.21 amplifications align with the CMS2 canonical subtype characterized by high CIN and activated WNT pathway, as underlined by high frequency APC mutations. Moreover, MSI-associated abnormalities and BRAF mutations that are observed in CMS1 and SMAD4 mutations leading to TGFβ pathway activation as observed in the mesenchymal CMS4 subtype are absent in 20q11.21 amplified colorectal cancers. KRAS mutations, which are more common in the CMS3 metabolic subtype, show a lower prevalence in 20q11.21 amplified colorectal cancers. CMS2 cancers have an intermediate overall prognosis, in between CMS1,(which have a favorable prognosis) and CMS4 mesenchymal cancers (which have the worst prognosis). The lack of prognostic implications of 20q11.21 amplifications in TCGA is consistent with their alignment with the intermediate prognosis CMS2 group. In contrast, in metastatic disease, CMS1 cancers behave more aggressively, and thus, CMS2 cancers and, among them cancers with 20q11.21 amplifications, have the best prognosis, as confirmed in the MSK-IMPACT cohort that includes only metastatic colorectal cancers. A study from MSK-IMPACT that included both gains and amplifications of the whole chromosome 20q showed better overall survival of the altered patient group compared to 20q diploid patients (16).
The common occurrence of amplifications in 20q11.21 in colorectal carcinomas suggests that high dosage of one or more of the genes located in the area is beneficial for cancer cells and may be a driver event. This assumption would be strengthened if mRNA up-regulation accompanies gene amplification. mRNA up-regulation of transcripts from the 20q11.21 locus in cancers with 20q11.21 amplifications is variable and for some genes the percentage of mRNA up-regulation among amplified cases is low. In contrast, other genes including ASXL1 and POFUT1 display a high prevalence of mRNA up-regulation in 20q11.21 amplified cases. This suggests that these genes may be important in the pathogenesis of colorectal cancers with the amplification. However, other cancer associated genes such as BCL2L1 and DNMT3B, which are overexpressed in a lower percentage of amplified cases, are not completely ruled out as contributors on the pro-tumorigenic effect of amplification. Moreover, additive effects of multiple genes are also a possibility, as illustrated by the TPX2-AURKA case discussed above. A feasible hypothesis is that the amplification of the 20q11.21 area is selected due to one or more resident genes that favor cancer initiation or progression and neighboring co-amplified genes may or may not contribute; they are often co-amplified due to their inclusion in an area that is contained between two breakage-prone points.
The association of 20q11.21 amplified metastatic colorectal cancers with a better survival argues for the presence of additional molecular lesions in these cancers that improve outcomes and counterbalance the higher dosage of genes at 20q11.21. Indeed, the alignment of 20q11.21 amplified cancers with the CMS2 canonical subtype suggests that they constitute a subset of a more extensive group of colorectal cancers characterized by increased CIN, absence of MMR/ POLE alterations, lower TMB, a lower frequency of KRAS pathway mutations and absence of lesions in DDR genes. This group may respond differentially to treatments. Advanced colorectal cancers with increased chromosomal alterations have been reported to respond better to capecitabine/ irinotecan-based chemotherapy compared with counterparts bearing a lower number of chromosomal alterations (31). Given the low prevalence of KRAS pathway mutations, 20q11.21 amplified colorectal cancers would be expected to respond to combination treatments that include EGFR targeting therapies, such as cetuximab and panitumumab. A study of 11 colorectal cancer patients with 20q gains or amplifications that received cetuximab or panitumumab showed only three responses (16). However, this study considered amplifications of the whole arm and included patients with gains in 20q, in addition to amplifications. Other targeted therapies against targets over-expressed from locus 20q11.21 are worth investigating in 20q11.21 amplified colorectal cancers. The c-Met inhibitor tivantinib has pro-apoptotic effects mediated by inhibition of Bcl-xL and another BCL2 family anti-apoptotic protein, Mcl1 and could be effective in cancers with over-expression of these members (32). BCL2 itself is currently a therapeutic target following the introduction of the inhibitor venetoclax in chronic lymphocytic leukemia and has been further investigated in other malignancies. The putative dependence of cancers with 20q11.21 amplifications to inhibition of the intrinsic apoptosis pathway would also be worth exploring therapeutically, especially in view of the absence of KRAS pathway activation in these cancers. When activated by oncogenic mutations, the KRAS pathway provides additional anti-apoptotic signals (33,34).
Amplifications of the microtubule associated protein TPX2 and interacting AURKA kinase could be a marker of sensitivity to AURKA inhibitors. The two interacting proteins have also been proposed to co-amplify with the c-MYC oncogene and participate in oncogenesis in c-MYC dependent colorectal cancers (35). Thus, AURKA inhibitors could offer a therapeutic target in c-MYC amplified cancers that are currently not directly targetable. In addition, Bcl-xL antagonizes c-MYC induced apoptosis; inhibition of Bcl-xL was able to restore apoptosis in cells of various types of cancers, including colorectal cancer, with low c-MYC activity (36).
Fucosyltransferase POFUT1 activates the Notch signal cascade, a pathway implicated in proliferation and inhibition of cancer cell differentiation (37). Thus, POFUT1 amplification and up-regulation could sensitize colorectal cancers to inhibitors of the Notch pathway. Notch signaling leads to aggressive colorectal cancers in mice with increased metastatic potential (38).
In conclusion, the presence of 20q11.21 amplifications in a sub-set of colorectal cancers may provide opportunities for development of targeted treatments. Candidate therapies need to be studied in conjunction with the sub-set of cancers that carry the corresponding target lesion. Designing exploratory studies that pair targeted therapies with the cancers that carry the target lesion could be the most rational way to further advance targeted cancer therapeutics.
Conflicts of Interest
None to be declared.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Voutsadakis IA. Pathogenesis of colorectal carcinoma and therapeutic implications: the roles of the ubiquitin-proteasome system and Cox-2. J Cell Mol Med. 2007;11(2):252–285. doi: 10.1111/j.1582-4934.2007.00032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schlicker A, Beran G, Chresta CM, McWalter G, Pritchard A, Weston S, Runswick S, Davenport S, Heathcote K, Castro DA, Orphanides G, French T, Wessels LF. Subtypes of primary colorectal tumors correlate with response to targeted treatment in colorectal cell lines. BMC Med Genomics. 2012;5:66. doi: 10.1186/1755-8794-5-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Budinska E, Popovici V, Tejpar S, D’Ario G, Lapique N, Sikora KO, Di Narzo AF, Yan P, Hodgson JG, Weinrich S, Bosman F, Roth A, Delorenzi M. Gene expression patterns unveil a new level of molecular heterogeneity in colorectal cancer. J Pathol. 2013;231(1):63–76. doi: 10.1002/path.4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.De Sousa E Melo F, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, de Jong JH, de Boer OJ, van Leersum R, Bijlsma MF, Rodermond H, van der Heijden M, van Noesel CJ, Tuynman JB, Dekker E, Markowetz F, Medema JP, Vermeulen L. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 6.Marisa L, de Reyniès A, Duval A, Selves J, Gaub MP, Vescovo L, Etienne-Grimaldi MC, Schiappa R, Guenot D, Ayadi M, Kirzin S, Chazal M, Fléjou JF, Benchimol D, Berger A, Lagarde A, Pencreach E, Piard F, Elias D, Parc Y, Olschwang S, Milano G, Laurent-Puig P, Boige V. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med. 2013;10(5):e1001453. doi: 10.1371/journal.pmed.1001453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LC, Lannon WA, Grotzinger C, Del Rio M, Lhermitte B, Olshen AB, Wiedenmann B, Cantley LC, Gray JW, Hanahan D. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19(5):619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roepman P, Schlicker A, Tabernero J, Majewski I, Tian S, Moreno V, Snel MH, Chresta CM, Rosenberg R, Nitsche U, Macarulla T, Capella G, Salazar R, Orphanides G, Wessels LF, Bernards R, Simon IM. Colorectal cancer intrinsic subtypes predict chemotherapy benefit, deficient mismatch repair and epithelial-to-mesenchymal transition. Int J Cancer. 2014;134(3):552–562. doi: 10.1002/ijc.28387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, Bot BM, Morris JS, Simon IM, Gerster S, Fessler E, De Sousa E Melo F, Missiaglia E, Ramay H, Barras D, Homicsko K, Maru D, Manyam GC, Broom B, Boige V, Perez-Villamil B, Laderas T, Salazar R, Gray JW, Hanahan D, Tabernero J, Bernards R, Friend SH, Laurent-Puig P, Medema JP, Sadanandam A, Wessels L, Delorenzi M, Kopetz S, Vermeulen L, Tejpar S. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang W, Kandimalla R, Huang H, Zhu L, Li Y, Gao F, Goel A, Wang X. Molecular subtyping of colorectal cancer: Recent progress, new challenges and emerging opportunities. Semin Cancer Biol. 2019;55:37–52. doi: 10.1016/j.semcancer.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yaeger R, Chatila WK, Lipsyc MD, Hechtman JF, Cercek A, Sanchez-Vega F, Jayakumaran G, Middha S, Zehir A, Donoghue MTA, You D, Viale A, Kemeny N, Segal NH, Stadler ZK, Varghese AM, Kundra R, Gao J, Syed A, Hyman DM, Vakiani E, Rosen N, Taylor BS, Ladanyi M, Berger MF, Solit DB, Shia J, Saltz L, Schultz N. Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell. 2018;33(1):125–136.e3. doi: 10.1016/j.ccell.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taylor AM, Shih J, Ha G, Gao GF, Zhang X, Berger AC, Schumacher SE, Wang C, Hu H, Liu J, Lazar AJ, Cancer Genome Atlas Research Network , Cherniack AD, Beroukhim R, Meyerson M. Genomic and functional approaches to understanding cancer aneuploidy. Cancer Cell. 2018;33(4):676–689.e3. doi: 10.1016/j.ccell.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chakravarty D, Gao J, Phillips SM, Kundra R, Zhang H, Wang J, Rudolph JE, Yaeger R, Soumerai T, Nissan MH, Chang MT, Chandarlapaty S, Traina TA, Paik PK, Ho AL, Hantash FM, Grupe A, Baxi SS, Callahan MK, Snyder A, Chi P, Danila D, Gounder M, Harding JJ, Hellmann MD, Iyer G, Janjigian Y, Kaley T, Levine DA, Lowery M, Omuro A, Postow MA, Rathkopf D, Shoushtari AN, Shukla N, Voss M, Paraiso E, Zehir A, Berger MF, Taylor BS, Saltz LB, Riely GJ, Ladanyi M, Hyman DM, Baselga J, Sabbatini P, Solit DB, Schultz N. OncoKB: A precision oncology knowledge base. JCO Precis Oncol. 2017;2017:PO.17.00011. doi: 10.1200/PO.17.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ptashkin RN, Pagan C, Yaeger R, Middha S, Shia J, O’Rourke KP, Berger MF, Wang L, Cimera R, Wang J, Klimstra DS, Saltz L, Ladanyi M, Zehir A, Hechtman JF. Chromosome 20q amplification defines a subtype of microsatellite stable, left-sided colon cancers with wild-type RAS/RAF and better overall survival. Mol Cancer Res. 2017;15(6):708–713. doi: 10.1158/1541-7786.MCR-16-0352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voutsadakis IA. Apoptosis and the pathogenesis of lymphoma. Acta Oncol. 2000;39(2):151–156. doi: 10.1080/028418600430707. [DOI] [PubMed] [Google Scholar]
- 18.Stevens M, Oltean S. Modulation of the apoptosis gene Bcl-x function through alternative splicing. Front Genet. 2019;10:804. doi: 10.3389/fgene.2019.00804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramesh P, Medema JP. BCL-2 family deregulation in colorectal cancer: potential for BH3 mimetics in therapy. Apoptosis. 2020;25(5-6):305–320. doi: 10.1007/s10495-020-01601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Micol JB, Abdel-Wahab O. The role of additional sex combs-like proteins in cancer. Cold Spring Harb Perspect Med. 2016;6(10):a026526. doi: 10.1101/cshperspect.a026526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Katoh M. Functional and cancer genomics of ASXL family members. Br J Cancer. 2013;109(2):299–306. doi: 10.1038/bjc.2013.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huidobro C, Urdinguio RG, Rodríguez RM, Mangas C, Calvanese V, Martínez-Camblor P, Ferrero C, Parra-Blanco A, Rodrigo L, Obaya AJ, Suárez-Fernández L, Astudillo A, Hernando H, Ballestar E, Fernández AF, Fraga MF. A DNA methylation signature associated with aberrant promoter DNA hypermethylation of DNMT3B in human colorectal cancer. Eur J Cancer. 2012;48(14):2270–2281. doi: 10.1016/j.ejca.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 23.Ibrahim AE, Arends MJ, Silva AL, Wyllie AH, Greger L, Ito Y, Vowler SL, Huang TH, Tavaré S, Murrell A, Brenton JD. Sequential DNA methylation changes are associated with DNMT3B overexpression in colorectal neoplastic progression. Gut. 2011;60(4):499–508. doi: 10.1136/gut.2010.223602. [DOI] [PubMed] [Google Scholar]
- 24.Sillars-Hardebol AH, Carvalho B, Tijssen M, Beliën JA, de Wit M, Delis-van Diemen PM, Pontén F, van de Wiel MA, Fijneman RJ, Meijer GA. TPX2 and AURKA promote 20q amplicon-driven colorectal adenoma to carcinoma progression. Gut. 2012;61(11):1568–1575. doi: 10.1136/gutjnl-2011-301153. [DOI] [PubMed] [Google Scholar]
- 25.Jimbo T, Kawasaki Y, Koyama R, Sato R, Takada S, Haraguchi K, Akiyama T. Identification of a link between the tumour suppressor APC and the kinesin superfamily. Nat Cell Biol. 2002;4(4):323–327. doi: 10.1038/ncb779. [DOI] [PubMed] [Google Scholar]
- 26.Germot A, Maftah A. POFUT1 and PLAGL2 gene pair linked by a bidirectional promoter: the two in one of tumour progression in colorectal cancer. EBioMedicine. 2019;46:25–26. doi: 10.1016/j.ebiom.2019.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Shao L, Shi S, Harris RJ, Spellman MW, Stanley P, Haltiwanger RS. Modification of epidermal growth factor-like repeats with O-fucose. Molecular cloning and expression of a novel GDP-fucose protein O-fucosyltransferase. J Biol Chem. 2001;276(43):40338–40345. doi: 10.1074/jbc.M107849200. [DOI] [PubMed] [Google Scholar]
- 28.Komor MA, de Wit M, van den Berg J, Martens de Kemp SR, Delis-van Diemen PM, Bolijn AS, Tijssen M, Schelfhorst T, Piersma SR, Chiasserini D, Sanders J, Rausch C, Hoogstrate Y, Stubbs AP, de Jong M, Jenster G, Carvalho B, Meijer GA, Jimenez CR, Fijneman RJA, NGS-ProToCol Consortium Molecular characterization of colorectal adenomas reveals POFUT1 as a candidate driver of tumor progression. Int J Cancer. 2020;146(7):1979–1992. doi: 10.1002/ijc.32627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li D, Lin C, Li N, Du Y, Yang C, Bai Y, Feng Z, Su C, Wu R, Song S, Yan P, Chen M, Jain A, Huang L, Zhang Y, Li X. PLAGL2 and POFUT1 are regulated by an evolutionarily conserved bidirectional promoter and are collaboratively involved in colorectal cancer by maintaining stemness. EBioMedicine. 2019;45:124–138. doi: 10.1016/j.ebiom.2019.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sonbol MB, Ahn DH, Bekaii-Saab T. Therapeutic targeting strategies of cancer stem cells in gastrointestinal malignancies. Biomedicines. 2019;7(1):17. doi: 10.3390/biomedicines7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Postma C, Koopman M, Buffart TE, Eijk PP, Carvalho B, Peters GJ, Ylstra B, van Krieken JH, Punt CJ, Meijer GA. DNA copy number profiles of primary tumors as predictors of response to chemotherapy in advanced colorectal cancer. Ann Oncol. 2009;20(6):1048–1056. doi: 10.1093/annonc/mdn738. [DOI] [PubMed] [Google Scholar]
- 32.Lu S, Török HP, Gallmeier E, Kolligs FT, Rizzani A, Arena S, Göke B, Gerbes AL, De Toni EN. Tivantinib (ARQ 197) affects the apoptotic and proliferative machinery downstream of c-MET: role of Mcl-1, Bcl-xl and Cyclin B1. Oncotarget. 2015;6(26):22167–22178. doi: 10.18632/oncotarget.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carné Trécesson S, Souazé F, Basseville A, Bernard AC, Pécot J, Lopez J, Bessou M, Sarosiek KA, Letai A, Barillé-Nion S, Valo I, Coqueret O, Guette C, Campone M, Gautier F, Juin PP. BCL-XL directly modulates RAS signalling to favour cancer cell stemness. Nat Commun. 2017;8(1):1123. doi: 10.1038/s41467-017-01079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zaanan A, Okamoto K, Kawakami H, Khazaie K, Huang S, Sinicrope FA. The mutant KRAS gene up-regulates BCL-XL protein via STAT3 to confer apoptosis resistance that is reversed by BIM protein induction and BCL-XL antagonism. J Biol Chem. 2015;290(39):23838–23849. doi: 10.1074/jbc.M115.657833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Takahashi Y, Sheridan P, Niida A, Sawada G, Uchi R, Mizuno H, Kurashige J, Sugimachi K, Sasaki S, Shimada Y, Hase K, Kusunoki M, Kudo S, Watanabe M, Yamada K, Sugihara K, Yamamoto H, Suzuki A, Doki Y, Miyano S, Mori M, Mimori K. The AURKA/TPX2 axis drives colon tumorigenesis cooperatively with MYC. Ann Oncol. 2015;26(5):935–942. doi: 10.1093/annonc/mdv034. [DOI] [PubMed] [Google Scholar]
- 36.Topham C, Tighe A, Ly P, Bennett A, Sloss O, Nelson L, Ridgway RA, Huels D, Littler S, Schandl C, Sun Y, Bechi B, Procter DJ, Sansom OJ, Cleveland DW, Taylor SS. MYC is a major determinant of mitotic cell fate. Cancer Cell. 2015;28(1):129–140. doi: 10.1016/j.ccell.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tyagi A, Sharma AK, Damodaran C. A review on notch signaling and colorectal cancer. Cells. 2020;9(6):1549. doi: 10.3390/cells9061549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jackstadt R, van Hooff SR, Leach JD, Cortes-Lavaud X, Lohuis JO, Ridgway RA, Wouters VM, Roper J, Kendall TJ, Roxburgh CS, Horgan PG, Nixon C, Nourse C, Gunzer M, Clark W, Hedley A, Yilmaz OH, Rashid M, Bailey P, Biankin AV, Campbell AD, Adams DJ, Barry ST, Steele CW, Medema JP, Sansom OJ. Epithelial NOTCH signaling rewires the tumor microenvironment of colorectal cancer to drive poor-prognosis subtypes and metastasis. Cancer Cell. 2019;36(3):319–336.e7. doi: 10.1016/j.ccell.2019.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]