Abstract
Background/Aim: Cisplatin is a potent anticancer drug for treating several types of cancer, including non-small-cell lung cancer (NSCLC). In this study, we investigated the cytotoxicity and mechanism of action of cisplatin in the human NSCLC cell line PC9. Materials and Methods: PC9 cells were treated with cisplatin for 72 h and then evaluated by a cell viability assay, DAPI staining, Giemsa staining, apoptosis assay, membrane permeability assay, cell cycle assay, ROS assay, SA-β-gal staining, TUNEL assay and Western blotting. Results: Our findings revealed that the cytotoxic activity was associated with an apoptotic signaling pathway in response to DNA damage. Cisplatin exerted a significant concentration-dependent antiproliferative effect on PC9 cells. Cells subjected to cisplatin treatment showed morphological indications of apoptosis. Cell cycle arrest was related to the restriction of E2F-1 action by the cyclin-dependent protein kinase inhibitor p21WAF1/CIP1. Cisplatin induced apoptosis of PC9 cells by upregulating Fas, FasL, Bak, and tBID expression and PARP proteolytic cleavage. Cisplatin also reduced the mitochondrial membrane potential (MMP) and initiated a caspase cascade. Furthermore, the apoptotic impact of cisplatin depended on reactive oxygen species (ROS), as confirmed by ROS generation. Conclusion: Cisplatin induced anticancer effects through cell cycle arrest, ROS generation and caspase activation, resulting in cell apoptosis. Overall, the results show the mechanism by which cisplatin works as an anticancer drug in the treatment of NSCLC.
Keywords: Cisplatin, ROS, ATR, cell cycle arrest, apoptosis, NSCLC
Lung cancer is a leading cause of death worldwide (1,2). EGFR mutations have been identified (3,4) in 10%-30% of non-small-cell lung cancers (NSCLCs). Cisplatin is a standard drug for first-line treatment after surgery (5). Indeed, cisplatin chemotherapy was found to have reduced the rate of lung cancer-related death by 6.9% after 5 years compared with an untreated control group (6). However, the precise antineoplastic mechanism of cisplatin is not fully understood (7).
In multicellular organisms, apoptosis is a vital process of modified cell death. This cellular process acts as a barrier to cancer by removing damaged or unwanted cells (8,9). Various stressors, including DNA damage, reactive oxygen species (ROS), viral disease, serum deprivation, hypoxia, and heat shock, initiate apoptosis. ROS are considered toxic to cell metabolism and regulate many physiological processes (10). In recent years, numerous studies have shown that oxidative stress can cause cell apoptosis through intrinsic (mitochondria) and extrinsic (death receptor) pathways (11,12).
The essential molecular sensors of DNA damage incorporate the kinase ataxia telangiectasia mutated (ATM) and Rad3-related (ATR) checkpoint kinase (13-15). Due to genotoxic stress or DNA damage, these protein kinases accumulate at the site of DNA damage, forming nuclear foci (13,16,17). This process is supported by the enrollment and activation of other checkpoint molecules, including checkpoint kinase 1 (Chk1) and Chk2, triggering cell cycle arrest or apoptosis (13,18,19). Importantly, these checkpoint kinases can phosphorylate p53 at various serine sites (13,18). Despite the general understanding of the DNA damage response, the molecular mechanisms underlying DNA damage and cell death caused by platinum-based cancer drugs is unclear (20,21).
The p53-encoding TP53 gene is the most frequently mutated gene in human cancer (22,23). The two main routes by which p53 hinders tumor cell development are growth arrest and apoptosis induction. p53 plays a crucial role in genotoxic stress-induced apoptosis following exposure to DNA-damaging agents (24). However, the mechanism by which p53 causes apoptosis is unclear. It has been proposed that p53 can control the expression of some proteins that participate in apoptosis and cell cycle arrest, such as Bax, Bak, and p21WAF1/CIP1 (25,26). At the same time, senescence appears to play a central role in the anticancer impact of various anticancer agents and ionizing radiation (27-29). Another prominent event during apoptosis is the proteolytic cleavage of PARP into 89- and 24-kDa fragments by caspases (30-32). PARP breakdown induced by DNA damage contributes to DNA repair and participates in cell cycle arrest, cell death, and cell survival signaling mechanisms (33,34).
In the present study, we investigated the mechanism underlying the anticancer effects of cisplatin on PC9 cells. We herein report the impact of cisplatin on growth inhibition in PC9 cells.
Materials and Methods
Cell line and cell culture. The PC9 human NSCLC cell line (EGFR exon 19 deletion), derived from a previously untreated adenocarcinoma patient (35) was kindly provided by Professor K. Hayata (Tokyo Medical College, Tokyo, Japan). PC9 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS; Invitrogen, Carlsbad, CA, USA), in 5% CO2 at 37˚C.
Drug formulation and administration. Cisplatin [PtCl2(NH3)2] was purchased from FUJIFILM Wako Pure Chemical Corporation (Osaka, Japan). The drug was dissolved in DMSO for the in vitro study.
Cell proliferation assay. The cytotoxicity of various cisplatin concentrations in PC9 cells was assessed using a water-soluble tetrazolium salt (WST-1) assay (Cell Proliferation Reagent WST-1; Roche, Tokyo, Japan). Into each well of a 96-well microtiter plate, 100 μl of a growing cell suspension (4×103 cells/well) was seeded, and 100 μl of cisplatin solution at various concentrations was added to each well (35). After incubation for 72 h at 37˚C in 5% CO2, 10 μl of WST-1 solution was added to each well, and the plates were incubated at 37˚C for an additional 4 h (35). The absorbance was measured at 450 nm with a microplate enzyme-linked immunosorbent assay reader (Multiskan FC; Thermo Scientific, Tokyo, Japan).
Giemsa staining. Cells were treated with various cisplatin concentrations or DMSO (as control) for 72 h. After collection, the cells were resuspended in phosphate buffer solution and centrifuged in a Cytospin at 800 rpm for 8 min to adhere to a microscopic slide. After air-drying, the cells were fixed with methanol, stained with Giemsa (Merck KGaA, Darmstadt, Germany) for 20 min, rinsed in water, and air-dried again. Under a light microscope, morphological changes were observed to assess apoptosis.
Fluorescent staining of nuclei. PC9 cells from exponentially growing cultures were seeded in 12-well plates. After cisplatin treatment for 72 h, cells were washed with phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde phosphate buffer solution (FUJIFILM Wako Pure Chemical Corporation) for 7 min at room temperature. Subsequently, cells were permeabilized using 0.1% Triton X-100 (Sigma-Aldrich, St. Louis, MO, USA) in PBS for 10 min at room temperature. Cells were then stained with 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI; Invitrogen) for 10 min at room temperature and examined using a BZ-X710 All-in-One Fluorescence Microscope (Keyence, Osaka, Japan).
ROS assay. To measure ROS in cells by flow cytometry, we used an ab186029 (Abcam, Tokyo, Japan) Cellular Reactive Oxygen Species Detection Assay Kit (Deep Red Fluorescence), following the manufacturer’s instructions. After 72 h of drug treatment, cells were collected to stain with ROS deep red dye working solution. Subsequently, cells were incubated at 37˚C for 60 min before being subjected to a flow cytometry analysis.
Terminal deoxynucleotidyl transferase dUTP nick-end labeling (TUNL) assay. To quantify DNA fragmentation in apoptotic cells, we used the in situ Direct DNA Fragmentation (TUNEL) Assay Kit (ab66108, Abcam), following the manufacturer’s instructions. In brief, cells were fixed with 1% paraformaldehyde in PBS and placed on ice for 15 min. Following this, the samples were treated with a staining solution and incubated at 37˚C for 60 min. After addition of rinse buffer in propidium iodide/RNase A solution, cells were resuspended and incubated at room temperature for 30 min for a flow cytometry analysis.
Western blotting. Cells were treated with several concentrations of cisplatin (or DMSO for control) for 72 h. After treatment, whole-cell lysates were prepared using a mammalian protein extraction reagent M-PER; Thermo Scientific) accompanied by a phosphatase inhibitor cocktail and protease inhibitor cocktail (Sigma-Aldrich) for 30 min at 4˚C. The BCA protein assay (Thermo Scientific) was used to evaluate protein concentrations. Total cellular protein (40 μg) was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene fluoride membranes (Bio-Rad, USA). Milk-blocked blots were incubated at 4˚C overnight with primary antibodies against the following proteins: ATR (Cell Signaling Technology, Beverly, MA, USA), p-ATR (Ser 428) (Cell Signaling Technology), ATM (Cell Signaling Technology), p-ATM (Ser 1981) (Cell Signaling Technology), Chk1 (Cell Signaling Technology), p-Chk1 (Ser 345) (Cell Signaling Technology), Chk2 (Cell Signaling Technology), p-Chk2 (Thr 68) (Cell Signaling Technology), p-p53 (Ser 15) (Cell Signaling Technology), p-p53 (Ser 46) (Cell Signaling Technology), γ-H2AX (Phospho S140) (Abcam, Tokyo, Japan), Fas (Cell Signaling Technology), FasL (Cell Signaling Technology), Bak (Cell Signaling Technology), caspase-9 (Cell Signaling Technology), caspase-8 (Cell Signaling Technology), Bax (Cell Signaling Technology), BID (Cell Signaling Technology), cleaved caspase-7 (Asp 198) (Cell Signaling Technology), cleaved caspase-3 (Asp 175) (Cell Signaling Technology), cleaved caspase-6 (Asp 162) (Cell Signaling Technology), PARP (Cell Signaling Technology), cyclin D1 (BioLegend, San Diego, CA, USA), CDK4 (Cell Signaling Technology), E2F-1 (Cell Signaling Technology), p21WAF1/CIP1 (Cell Signaling Technology), p53 (Cell Signaling Technology), Rb (Cell Signaling Technology), p-Rb (Ser 807/811) (Cell Signaling Technology) and β-actin (Cell Signaling Technology), followed by the appropriate horseradish peroxidase-conjugated secondary antibodies (Cell Signaling Technology). Proteins of interest were visualized using the SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Fisher Scientific, Rockford, IL, USA) and the Invitrogen iBright FL1000 Imaging System (Thermo Fisher Scientific).
MMP evaluation. The MMP was evaluated using the JC-10 Mitochondrial Membrane Potential Assay Kit-Flow Cytometry (ab112133; Abcam). Cells were seeded in 75-cm2 flasks (Falcon, USA), treated as indicated in the results, trypsinized, washed with PBS, and incubated with 1X JC-10 dye-loading solution at room temperature for 30 min. Cell fluorescence was measured using a BD FACSCanto™ II (BD Biosciences, San Jose, CA, USA).
Apoptosis assay. For 72 h, PC9 cells were treated with cisplatin at various concentrations (or DMSO as a control). FITC Annexin V Apoptosis Detection Kit with Pl Protocol (BioLegend) was used to evaluate the apoptotic cell percentage by flow cytometry analysis.
Flow cytometry for cell cycle analysis. For 72 h, PC9 cells were treated with cisplatin at several concentrations (or DMSO as a control). A cell cycle analysis was conducted following the Propidium Iodide Cell Cycle Staining Protocol (BioLegend). The DNA content was determined with a FACSCanto™ II. The data were analyzed using an FCS Express 7 (De Novo Software, Pasadena, CA, USA).
Senescence-associated β-galactosidase staining. Cells (1×106) were cultured in 25-cm2 flasks (Falcon) and treated with various concentrations of cisplatin (or DMSO as a control) for 72 h. For senescence-activated β-galactosidase (SA-β-gal) staining, we used the Senescence-β-gal Staining Kit (Cell Signaling Technology) following the manufacturer’s instructions.
Statistical analysis. Graphs were generated using the GraphPad PRISM software, v. 7.0 (GraphPad Software Inc., San Diego, CA, USA). Results are presented as the mean±standard deviation (SD) of three independent experiments and were analyzed by one-way analysis of variance (ANOVA) with Bonferroni’s multiple comparison test. Results were considered to be statistically significant when p<0.05.
Results
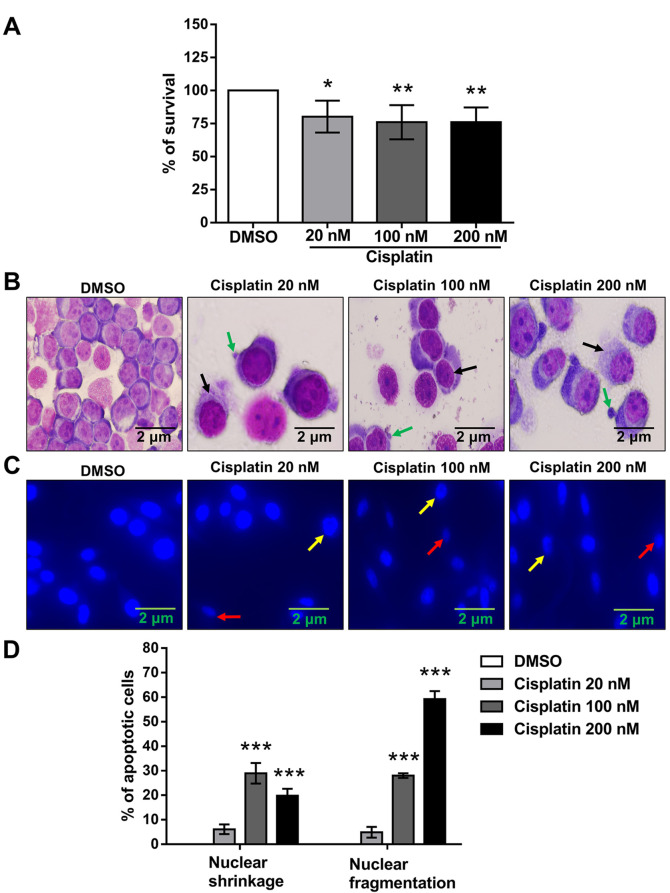
Impact of cisplatin on the viability of PC9 cells: morphological and nuclear changes. The water-soluble tetrazolium salt (WST-1) assay was performed to examine the effects of cisplatin on the viability of PC9 cells. Cisplatin treatment reduced the cell viability in a concentration-dependent manner (Figure 1A). Cells were stained with Giemsa and DAPI to investigate morphological changes induced by cisplatin. Cells going through cisplatin treatment showed morphological indications of apoptosis, such as cytoplasmic vacuolization (black arrows) and membrane blebbing (green arrows) (Figure 1B). These alterations were combined with nuclear shrinkage (red arrows) and nuclear fragmentation (yellow arrows) (Figure 1C).
Figure 1. Impact of cisplatin on the viability and morphology of PC9 cells. (A) After 72 h of treatment with various cisplatin concentrations, the cell viability was analyzed by the WST-1 cell proliferation assay. Data were acquired and presented as the mean±SD from six independent experiments. *p<0.05 and **p<0.01 compared with the DMSO-treated group using one-way ANOVA with Bonferroni’s multiple comparison test. (B) Cells were treated with different concentrations of cisplatin (or DMSO as control) for 72 h and stained with Giemsa to detect morphological changes. Cells with cytoplasmic vacuolization (black arrows) and membrane blebbing (green arrows) were found among the cisplatin-treated cells but not the DMSO-treated controls. The images are representative results of three independent experiments. (C) Cells were treated at different concentrations of cisplatin (or with DMSO alone) for 72 h. Cells with condensed (red arrows) and fragmented (yellow arrows) fluorescent nuclei are seen among the cisplatin-treated cells but not the DMSO-treated control group. (D) The percentage of cells with nuclear shrinkage and nuclear fragmentation was increased in the cisplatin-treated groups but not in the DMSO-treated control cohort. Data were analyzed by one-way ANOVA with Bonferroni’s multiple comparison test. ***p<0.001 compared with the DMSO-treated group.
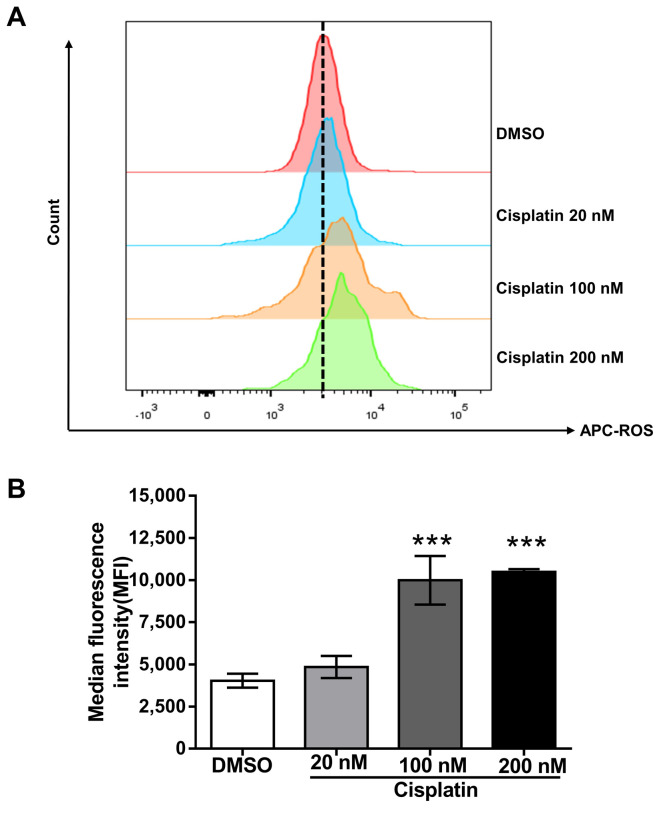
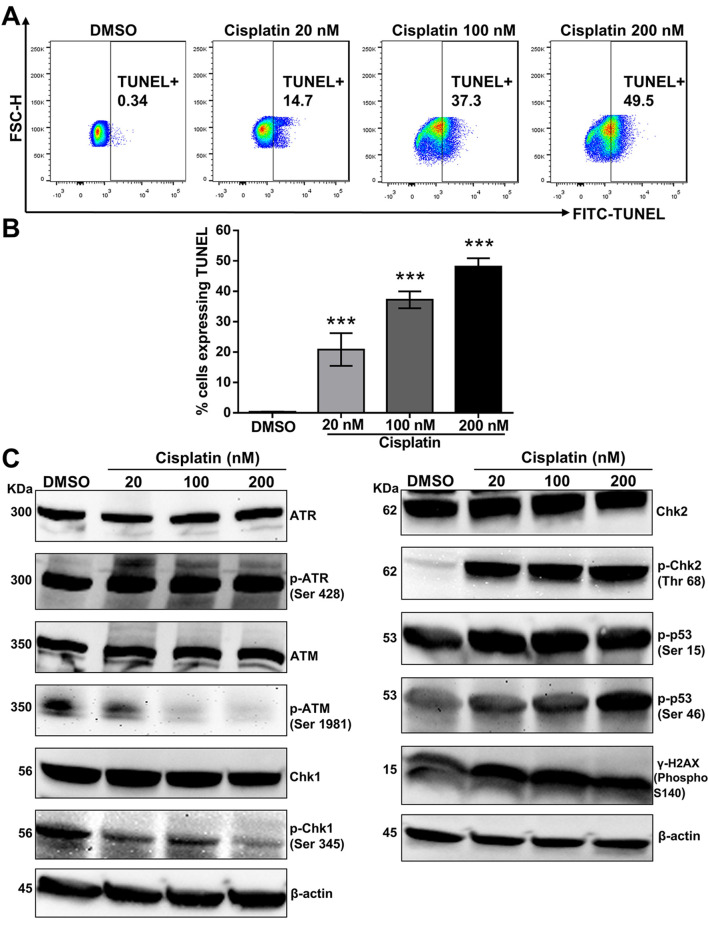
Impact of cisplatin on ROS generation and DNA fragmentation. Increased ROS production can lead to mitochondrial dysfunction through depolarization of the mitochondrial membrane potential (MMP) (36-39). We examined whether cisplatin increased ROS production, which can trigger apoptosis. Indeed, compared to cells treated with the vehicle alone, cisplatin increased ROS production in a dose-dependent manner (Figure 2). These outcomes indicate that ROS might be involved in apoptosis of cisplatin-treated PC9 cells. These changes are related to DNA fragmentation, a key indicator of apoptosis (Figure 3A, 3B).
Figure 2. The impact of cisplatin on ROS generation. (A) PC9 cells incubated with cisplatin at different concentrations (or DMSO as control) for 72 h were stained with ROS deep red dye. The fluorescent signal was measured by flow cytometry, and the median fluorescence intensity (MFI) was determined. The histogram shows that the ROS levels increased in a concentration-dependent manner. (B) The bar diagram shows that the ROS production increased in a dose-dependent manner. Data (mean±SD) are representative of three technical replicates. Data were analyzed by one-way ANOVA with Bonferroni’s multiple comparison test. ***p<0.001 compared with the DMSO-treated group.
Figure 3. The impact of cisplatin on DNA fragmentation and the DNA damage signaling pathway. (A) PC9 cells were stained with fluorescein isothiocyanate-dUTP dye after incubation with cisplatin at different concentrations for 72 h. Fluorescence was measured by flow cytometry. DNA fragmentation increased in a concentration-dependent manner, as represented by the dot plot. (B) The percentage of cells expressing TUNEL increased in a dose-dependent manner, as represented in the bar graph. Results are the mean±SD of three independent experiments. ***p<0.001 compared with the DMSO-treated group, using one-way ANOVA with Bonferroni’s multiple comparison test. (C) Total cell lysates from cisplatintreated PC9 cells for 72 h were subjected to a Western blot analysis with the following antibodies: ATR, p-ATR (Ser 428), ATM, p-ATM (Ser 1981), Chk1, p-Chk1 (Ser 345), Chk2, p-Chk2 (Thr 68), p-p53 (Ser 15), p-p53 (Ser 46), and γ-H2AX (Phospho S140). β-actin served as a loading control. Representative Western blots are shown from three independent experiments.
Impact of cisplatin on the DNA damage signaling pathway. After treatment with cisplatin, we analyzed the expression of DNA damage-related proteins. Oxidative DNA damage causes ATM phosphorylation at Ser 1981 and ATR phosphorylation at Ser 428 (40). The phosphorylation activation of these proteins promotes the phosphorylation of different downstream proteins (including H2AX, p53, Chk1, Chk2, etc.) and ultimately promotes cell cycle arrest and apoptosis (41,42). ATR can phosphorylate a broad scope of protein substrates during genotoxic stress, bringing about the activation of complex signaling cascades (14,16). A key sign of ATR activation during genotoxic stress is ATR accumulation in nuclear foci, where signaling proteins aggregate and connect due to DNA damage (17,43). Genotoxic stress incited by camptothecin brings about the decay of Chk1 through the proteasomal pathway (44). As presented in Figure 3C, cisplatin treatment increased the protein expression of p-ATR (Ser 428), p-Chk2 (Thr 68), γ-H2AX (Phospho S140), p-p53 (Ser 15) and p-p53 (Ser 46) but decreased the protein expression of p-ATM (Ser 1981) and p-Chk1 (Ser 345). These results suggest that ATR-Chk2 signaling plays a crucial role in p53 activation and the DNA damage response during cisplatin treatment in PC9 cells.
Impact of cisplatin on MMP. Once generated, ROS can cause mitochondrial membrane permeabilization (45). Using the fluorescent cation dye JC-10, we evaluated the depolarization of the MMP. The intact MMP of non-apoptotic cells allows JC-10 to accumulate in the mitochondria, forming red fluorescent "J aggregates." In contrast, the MMP is broken down in apoptotic cells, resulting in JC-10 staying in the cytoplasm with a green fluorescent monomer structure (37). In cisplatin-treated PC9 cells, at low MMP (lower-right quadrant, Q3), JC-10 is predominantly a monomer that produces green fluorescence, indicating that cisplatin impairs mitochondrial integrity (as assessed by MMP loss) (Figure 4).
Figure 4. The impact of cisplatin on the MMP. (A) PC9 cells were incubated with various cisplatin concentrations for 72 h to examine the MMP, stained with JC-10 dye, and analyzed by flow cytometry. Representative results are shown from three independent experiments. (B) Quantitative data are presented by green fluorescence (depolarized MMP) of JC-10 monomer. Results are presented as the mean±SD of three independent measurements. *p<0.05 and ***p<0.001 compared with the DMSO-treated group, using one-way ANOVA with Bonferroni’s multiple comparison test.
The impact of cisplatin on the induction of apoptosis in PC9 cells. To confirm that cisplatin-induced cell death was due to apoptosis, we used annexin V and PI staining to conduct a flow cytometry analysis. As shown (Figure 5A), after 72 h of exposure to cisplatin, the proportion of late-apoptotic cells (upper right quadrant 2, Annexin V/PI positive) increased from 3.15% to 22.6%. Flow cytometry results showed that cisplatin-induced apoptosis was dose-dependent (Figure 5B). Western blotting experiments targeting apoptosis marker proteins were performed to examine the molecular mechanism underlying cell death due to apoptosis following cisplatin treatment. As shown in Figure 5C, the expression of Fas and FasL increased with cisplatin treatment in a dose-dependent manner. Compared to the control group, the expression of the proapoptotic Bak protein was also increased.
Figure 5. The impact of cisplatin on the induction of apoptosis in PC9 cells. (A) Apoptosis was assessed by Annexin V/PI dual staining. Flow cytometry was used to evaluate the number of Annexin V- and PI-positive cells. Quadrant 1 shows necrotic cells, quadrant 2 late-apoptotic cells, quadrant 3 early-apoptotic cells and quadrant 4 viable cells. (B) A bar diagram illustrates the percentage of apoptotic cells. The values were calculated as the mean±SD from three independent experiments. Significance was determined by one-way ANOVA with Bonferroni’s multiple comparison test: *p<0.05, **p<0.01 and ***p<0.001 compared with the DMSO-treated group. (C) To assess the expression of Fas, FasL, Bak, caspase-9, caspase-8, Bax, BID, cleaved caspase-7 (Asp 198), cleaved caspase-3 (Asp 175), cleaved caspase-6 (Asp 162), and PARP proteins, Western blotting was performed. β-actin served as a loading control. Representative immunoblots from three independent experiments are shown.
Compared with the control group, cisplatin treatment increased the expression of truncated BID (tBID). tBID is located in the mitochondrial membrane, where it promotes the release of cytochrome c and enhances the process of apoptosis (11,12). Compared to the untreated control, cisplatin also increased the levels of cleaved caspase-3 (Asp 175), caspase-9, caspase-8, caspase-6 (Asp 162) and caspase-7 (Asp 198), in a dose-dependent manner. Exposure of PC9 cells to cisplatin also resulted in cleavage of PARP (Figure 5C), a standard substrate of caspase-3; PARP cleavage is considered an indicator of cell apoptosis. These results suggested that cisplatin induced PC9 cell apoptosis.
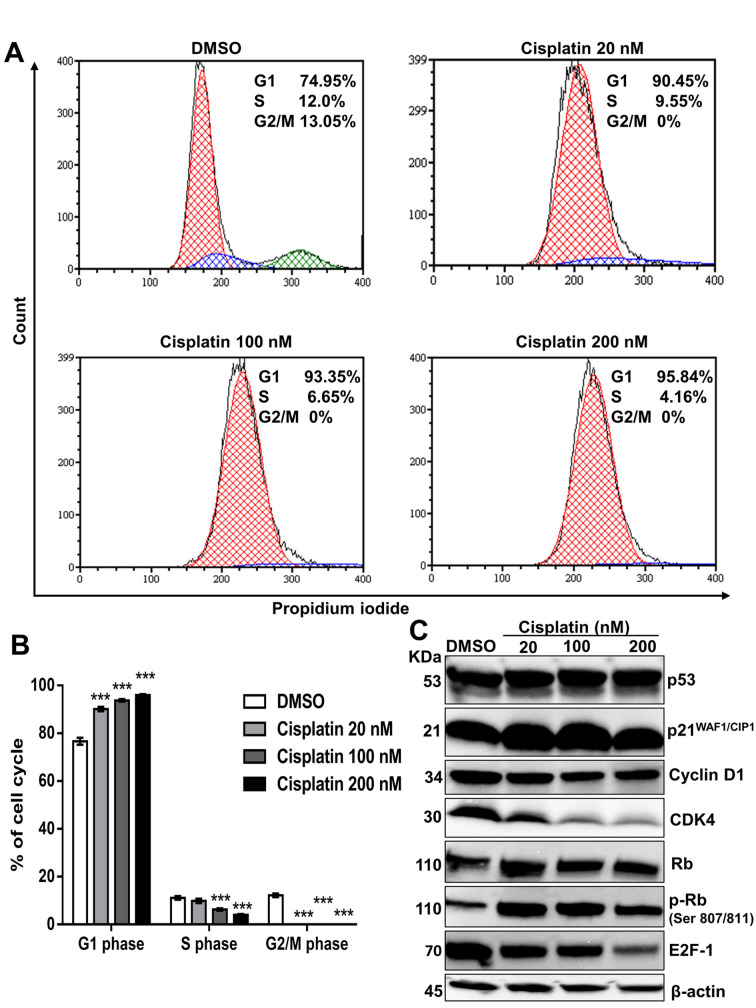
Impact of cisplatin on cell cycle regulation and senescence induction in PC9 cells. Cell cycle arrest is an important cause of cell death. To study the cell cycle arrest induced by cisplatin, we used flow cytometry to analyze the cell cycle distribution in PC9 cells in order to evaluate the DNA content in each cell cycle phase. As shown (Figure 6A, 6B), compared to DMSO-treated control, cisplatin treatment resulted in a dose-dependent increase in the proportion of G1 phase cells, while the proportions of S and G2/M phase cells decreased. The data show that cisplatin induced G1 phase arrest.
Figure 6. The impact of cisplatin on cell cycle regulation in PC9 cells. (A) PC9 cells were treated with DMSO or cisplatin (20 nM, 100 nM and 200 nM) for 72 h and stained with propidium iodide (PI) before a flow cytometric analysis. Representative results from three independent experiments are shown. (B) The results are presented as the mean±SD of three independent experiments. ***p<0.001 compared with the DMSO-treated group, using one-way ANOVA with Bonferroni’s multiple comparison test. (C) For the Western blot analysis, PC9 cells were treated with DMSO or cisplatin (20 nM, 100 nM, 200 nM) for 72 h. β-actin served as a loading control. Representative immunoblots from three independent trials are shown.
Having shown that cisplatin induced cell cycle arrest, we endeavored to characterize the mechanisms by which this outcome was achieved. Activated cyclin D1, CDK4, and CDK6 enable cells to enter S phase from G1, during cell cycle progression (46,47). The cyclin D1-CDK4/6 complex is responsible for DNA synthesis, while CKIs, such as p21WAF1/CIP1 and p27KIP1, negatively regulate this process (48,49). Western blotting (Figure 6C) revealed that cisplatin treatment significantly reduced cyclin D1, CDK4, and E2F-1 protein expression and led to the upregulation of the p21WAF1/CIP1 expression, accompanied by p53 activation.
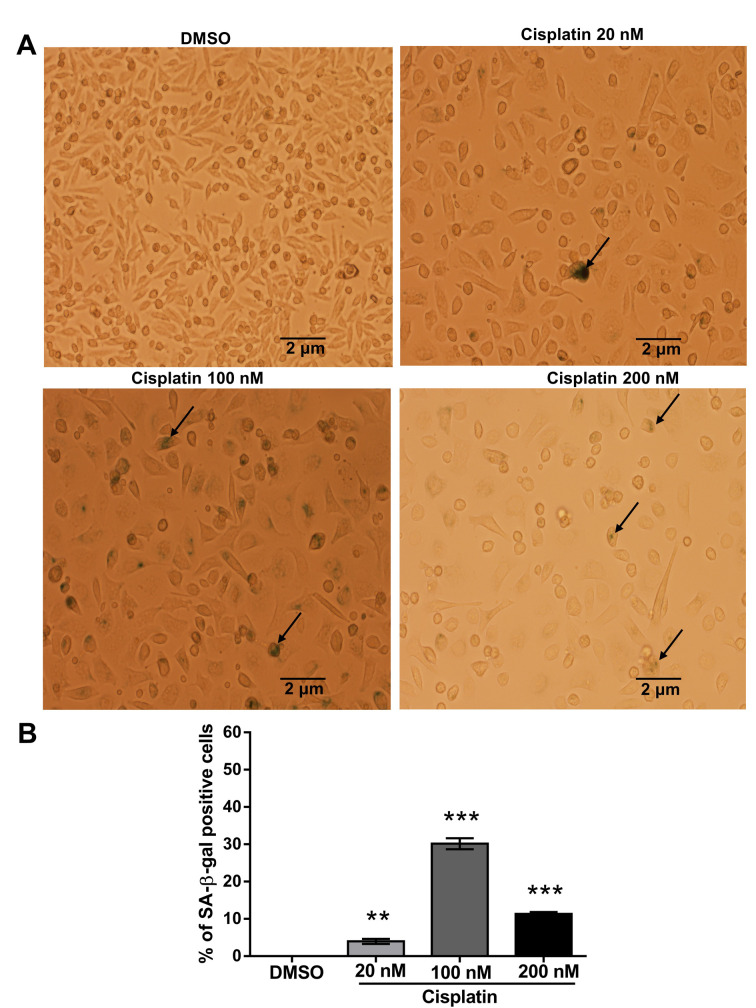
Since senescence is considered irreversible cell cycle arrest, especially in the G1 phase (50,51), we observed the induction of senescence-associated beta-galactosidase (SA-β-gal) activity by cisplatin treatment. We found that, at a relatively high dose (200 nM) of cisplatin, the proportion of SA-β-gal-positive cells (black arrows indicate senescent cells) was lower and the staining weaker than in cells treated with 100 nM cisplatin (Figure 7). At this high dose, cellular damage may be too high for the cells to respond actively, resulting in significant growth inhibition and decreased cell viability.
Figure 7. Impact of cisplatin on senescence induction in PC9 cells. (A) Before staining, PC9 cells were treated with different concentrations of cisplatin (DMSO-treated control) for 72 h. The cells showing SA-β-gal activity were stained green (black arrows). (B) The bar diagram represents the percentage of senescent cells. Values are the mean±SD of three independent experiments. Significance was determined by one-way ANOVA with Bonferroni’s multiple comparison test: **p<0.01 and ***p<0.001 compared with the DMSO-treated group.
Discussion
In the present study, we evaluated the cytotoxic impacts of cisplatin in PC9 cells. The results demonstrated that cisplatin caused intense growth inhibitory effects against PC9 cells. Cisplatin induced G1 phase arrest via an increase in p21WAF1/CIP1 and decrease in cyclin D1, CDK4, and E2F-1 activities in PC9 cells. Further analysis showed that an upregulated Bak, tBID, Fas, and FasL expression, increased ROS production, and activated caspase cascade are associated with cisplatin-induced apoptosis.
ROS are constantly produced and eliminated. In both homeostasis and infection, ROS play a vital role. In mitochondria, increased ROS generation plays an essential role in apoptosis (52,53). The oxidative pressure created by ROS can damage parts of the cell, including DNA and proteins (54). Many studies have confirmed that ROS can disrupt cell cycle progression and induce apoptosis by activating the ATM and ATR pathways (55). The present study showed that exposure to cisplatin causes ROS production, enhancing the cytotoxic effects associated with DNA damage.
The DNA damage response (DDR) is a complex signaling pathway (17). It includes sensor proteins that detect DNA damage and activate signal transducer proteins including ATM and ATR, resulting in the activation of effector proteins (Chk1 and Chk2) that control DNA repair, cell cycle arrest, and apoptosis (13,14,17). In this study, we show that ATR but not ATM was activated during cisplatin treatment (Figure 3C).
Activated ATR can phosphorylate H2AX in response to single-stranded DNA breaks. The increase in ATR activity during cisplatin therapy is an essential factor in evaluating therapeutic efficacy (16). The ATR-mediated Chk1 phosphorylation at serine 345 is necessary to achieve maximum kinase activity (14). Chk2 is not homologous to Chk1 but participates in DNA structure modification (56). In response to genotoxic stress, Chk2 is phosphorylated at threonine 68 (56). After DNA damage, the monomeric Chk2 undergoes dimerization through phosphorylation, promoting kinase activity (56). DNA double-strand breaks induced ATM-Chk2 pathway activation (56,57). However, ATR-dependent Chk2 activation has also been reported (58). Our present results showed ATR-dependent Chk2 phosphorylation at threonine 68 during cisplatin treatment (Figure 3C).
Mitochondria react to numerous death stimuli, including those related to the Bcl-2 group of proapoptotic proteins, such as Bax and Bak, which cause permeabilization of the mitochondrial membrane and release of apoptotic molecules (59-62). Furthermore, Bak can induce cell apoptosis in a Bax-dependent or independent manner (60,63,64). Conversely, Fas receptor and tumor necrosis factor receptor 1 (TNFR1) can activate caspase-8 and release the active sections of caspase-8 (p18 and p10) (65-67). Activated caspase-8 cleaves and activates downstream effectors like BID, caspase-3, 6, 7 and 9, eventually prompting the morphological signs of apoptosis. In the present study, cisplatin increased the Bak, Fas, and FasL expression as well as the proteolysis of BID protein in PC9 cells (Figure 5C). Previous studies have shown that mitochondrial phosphorylation of p53 to Ser 15 can mediate Bak activation (68). The phosphorylation of p53 to Ser 46 controls the ability of p53 to cause apoptosis (69). Activated caspase-9 prompts other caspases to activate, including caspase-3, 6 and 7, and start a caspase cascade, resulting in apoptosis (70-73). Caspase-3 is essential because it contributes to the proteolytic cleavage of many key proteins, similar to the nuclear PARP enzyme (74). Our study provides evidence in support of this conclusion.
After cisplatin treatment, the viability of PC9 cells decreased in a concentration-dependent manner, demonstrating the strong cytotoxic activity of cisplatin, which was related to an increase in the proportion of cells in G1. This indicated a cell cycle block. Cell cycle phases are controlled through various cyclins and their complex development with CDKs (75). The activity of cyclin D-CDK4/6 drives cells through the early G1 phase of the cell cycle, whereas cyclin E-Cdk2 complexes control the transition from the G1 phase into the S phase (76). In the present study, a flow cytometry analysis showed that cisplatin induced PC9 cell arrest in G1 phase by inhibiting the activity of CDK4, cyclin D1, and E2F-1. Members of the CIP/KIP family (p21WAF1/CIP1, p27KIP1 and p57KIP2) and INK4 family (p15INK4b, p16INK4a, p19INK4d and p18INK4c) can inhibit the activities of cyclin-CDK complexes (77). p21WAF1/CIP1 negatively regulates cyclin-CDK complexes for G1 phase arrest. Previous studies have shown that anticancer drugs can cause cell cycle arrest through a p53-dependent or independent manner (78,79). In the present study, cisplatin upregulated the p21WAF1/CIP1 expression in PC9 cells, accompanied by an increase in the p53 protein level. p16INK4a (80) and p21WAF1/CIP1 (81) cause cellular senescence (82-84). The retinoblastoma (Rb) tumor suppressor protein plays an important role at decision points in the G1 phase of the cell cycle (85). E2F-1 is an essential transcription factor that controls cell cycle progression and cell proliferation. E2F-1, 2, and 3 bind to pRb, and these connections are under the control of the cell cycle (86,87). The present findings suggest a possible mechanism by which cisplatin upregulates the p21WAF1/CIP1 expression, leading to G1 arrest by inhibiting the E2F-1 activity.
Conclusion
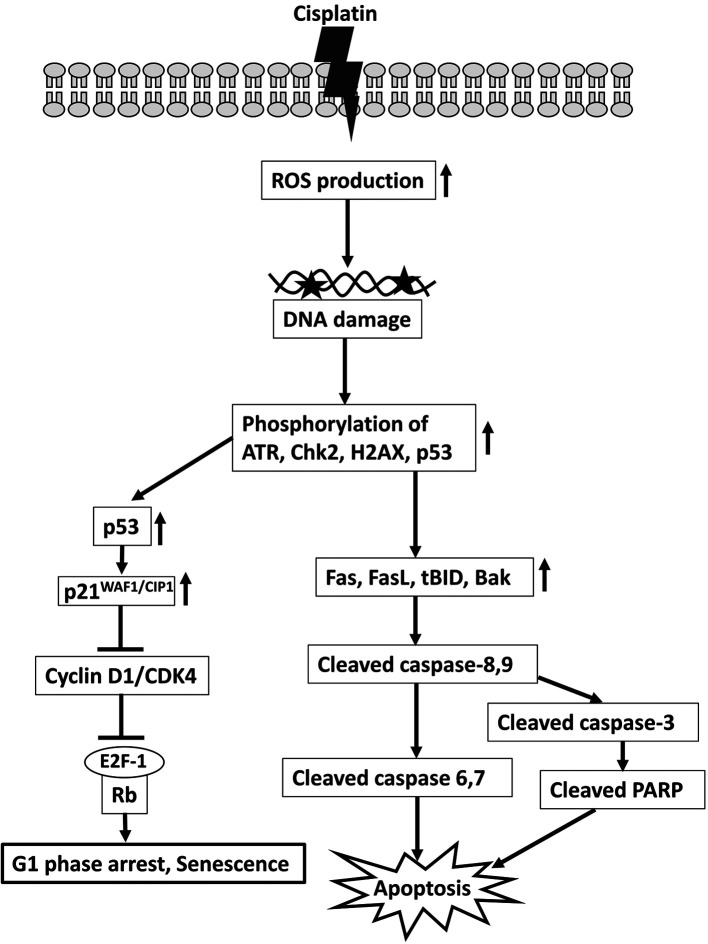
Cisplatin can inhibit PC9 cell proliferation through oxidative damage caused by ROS generation, cell cycle arrest at the G1 phase, and activation of a caspase cascade to induce apoptosis. Overall, our outcomes clarify how cisplatin can be used as a therapeutic agent against PC9 cells, a representative NSCLC cell line. We have proposed a model of the major mechanism of the induction of apoptosis and cell cycle arrest by cisplatin in PC9 cells (Figure 8).
Figure 8. A schematic illustration demonstrates the effects and mechanism of cisplatin on apoptosis and cell cycle distribution in PC9 cells. Cisplatin enhances ROS production, triggers DNA damage response signaling, and activates a caspase cascade to induce apoptosis. It also demonstrates that cisplatin up-regulates p21WAF1/CIP1 and inhibits the activities of cyclin D1, CDK4, and E2F-1 to induce G1 phase arrest in PC9 cells.
Conflicts of Interest
The Authors declare no competing financial interests.
Authors’ Contributions
Md Mohiuddin and Kazuo Kasahara conceived this study; Md Mohiuddin carried out the experiments; Md Mohiuddin and Kazuo Kasahara discussed and interpreted the results; Md Mohiuddin wrote the manuscript; Kazuo Kasahara supervised the experiments and project.
Acknowledgements
The Authors would like to thank Ms. Miki Kashiwano (Department of Respiratory Medicine, Graduate School of Medical Sciences, Kanazawa University) for her technical assistance. This work was supported by a Grant-in-Aid for Scientific Research (C) (JSPS KAKENHI Grant Number 17K09606) to K.K. The funders had no role in the study design, data collection or interpretation, or decision to submit the work for publication.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jänne PA, Johnson BE. Effect of epidermal growth factor receptor tyrosine kinase domain mutations on the outcome of patients with non-small cell lung cancer treated with epidermal growth factor receptor tyrosine kinase inhibitors. Clin Cancer Res. 2006;12(14 Pt 2):4416s–4420s. doi: 10.1158/1078-0432.CCR-06-0555. [DOI] [PubMed] [Google Scholar]
- 5.Dasari S, Tchounwou PB. Cisplatin in cancer therapy: molecular mechanisms of action. Eur J Pharmacol. 2014;740:364–378. doi: 10.1016/j.ejphar.2014.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pignon JP, Tribodet H, Scagliotti GV, Douillard JY, Shepherd FA, Stephens RJ, Dunant A, Torri V, Rosell R, Seymour L, Spiro SG, Rolland E, Fossati R, Aubert D, Ding K, Waller D, Le Chevalier T, LACE Collaborative Group Lung adjuvant cisplatin evaluation: a pooled analysis by the LACE Collaborative Group. J Clin Oncol. 2008;26(21):3552–3559. doi: 10.1200/JCO.2007.13.9030. [DOI] [PubMed] [Google Scholar]
- 7.Galluzzi L, Vitale I, Michels J, Brenner C, Szabadkai G, Harel-Bellan A, Castedo M, Kroemer G. Systems biology of cisplatin resistance: past, present and future. Cell Death Dis. 2014;5:e1257. doi: 10.1038/cddis.2013.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fulda S, Gorman AM, Hori O, Samali A. Cellular stress responses: cell survival and cell death. Int J Cell Biol. 2010;2010:214074. doi: 10.1155/2010/214074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Strasser A, Vaux DL. Cell death in the origin and treatment of cancer. Mol Cell. 2020;78(6):1045–1054. doi: 10.1016/j.molcel.2020.05.014. [DOI] [PubMed] [Google Scholar]
- 10.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48(2):158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang NL, Kang YJ, Sung B, Hwang SY, Jang JY, Oh HJ, Ahn YR, Kim DH, Kim SJ, Ullah S, Hossain MA, Moon HR, Chung HY, Kim ND. MHY451 induces cell cycle arrest and apoptosis by ROS generation in HCT116 human colorectal cancer cells. Oncol Rep. 2017;38(3):1783–1789. doi: 10.3892/or.2017.5836. [DOI] [PubMed] [Google Scholar]
- 12.Sinha K, Das J, Pal PB, Sil PC. Oxidative stress: the mitochondria-dependent and mitochondria-independent pathways of apoptosis. Arch Toxicol. 2013;87(7):1157–1180. doi: 10.1007/s00204-013-1034-4. [DOI] [PubMed] [Google Scholar]
- 13.Roos WP, Kaina B. DNA damage-induced cell death by apoptosis. Trends Mol Med. 2006;12(9):440–450. doi: 10.1016/j.molmed.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Yu Y, Hamrick HE, Duerksen-Hughes PJ. ATM, ATR and DNA-PK: initiators of the cellular genotoxic stress responses. Carcinogenesis. 2003;24(10):1571–1580. doi: 10.1093/carcin/bgg137. [DOI] [PubMed] [Google Scholar]
- 15.Champeris Tsaniras S, Villiou M, Giannou AD, Nikou S, Petropoulos M, Pateras IS, Tserou P, Karousi F, Lalioti ME, Gorgoulis VG, Patmanidi AL, Stathopoulos GT, Bravou V, Lygerou Z, Taraviras S. Geminin ablation in vivo enhances tumorigenesis through increased genomic instability. J Pathol. 2018;246(2):134–140. doi: 10.1002/path.5128. [DOI] [PubMed] [Google Scholar]
- 16.Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes Dev. 2001;15(17):2177–2196. doi: 10.1101/gad.914401. [DOI] [PubMed] [Google Scholar]
- 17.Sancar A, Lindsey-Boltz LA, Unsal-Kaçmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev Biochem. 2004;73:39–85. doi: 10.1146/annurev.biochem.73.011303.073723. [DOI] [PubMed] [Google Scholar]
- 18.Norbury CJ, Zhivotovsky B. DNA damage-induced apoptosis. Oncogene. 2004;23(16):2797–2808. doi: 10.1038/sj.onc.1207532. [DOI] [PubMed] [Google Scholar]
- 19.Sen T, Rodriguez BL, Chen L, Corte CMD, Morikawa N, Fujimoto J, Cristea S, Nguyen T, Diao L, Li L, Fan Y, Yang Y, Wang J, Glisson BS, Wistuba II, Sage J, Heymach JV, Gibbons DL, Byers LA. Targeting DNA damage response promotes antitumor immunity through STING-Mediated T-cell activation in small cell lung cancer. Cancer Discov. 2019;9(5):646–661. doi: 10.1158/2159-8290.CD-18-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22(47):7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 21.Wang D, Lippard SJ. Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov. 2005;4(4):307–320. doi: 10.1038/nrd1691. [DOI] [PubMed] [Google Scholar]
- 22.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54(18):4855–4878. [PubMed] [Google Scholar]
- 23.Pavlakis E, Stiewe T. p53’s extended reach: The mutant p53 secretome. Biomolecules. 2020;10(2) doi: 10.3390/biom10020307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bates S, Vousden KH. p53 in signaling checkpoint arrest or apoptosis. Curr Opin Genet Dev. 1996;6(1):12–18. doi: 10.1016/s0959-437x(96)90004-0. [DOI] [PubMed] [Google Scholar]
- 25.el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75(4):817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 26.Miyashita T, Reed JC. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell. 1995;80(2):293–299. doi: 10.1016/0092-8674(95)90412-3. [DOI] [PubMed] [Google Scholar]
- 27.Wainwright LJ, Lasorella A, Iavarone A. Distinct mechanisms of cell cycle arrest control the decision between differentiation and senescence in human neuroblastoma cells. Proc Natl Acad Sci USA. 2001;98(16):9396–9400. doi: 10.1073/pnas.161288698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mansilla S, Piña B, Portugal J. Daunorubicin-induced variations in gene transcription: commitment to proliferation arrest, senescence and apoptosis. Biochem J. 2003;372(Pt 3):703–711. doi: 10.1042/BJ20021950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han Z, Wei W, Dunaway S, Darnowski JW, Calabresi P, Sedivy J, Hendrickson EA, Balan KV, Pantazis P, Wyche JH. Role of p21 in apoptosis and senescence of human colon cancer cells treated with camptothecin. J Biol Chem. 2002;277(19):17154–17160. doi: 10.1074/jbc.M112401200. [DOI] [PubMed] [Google Scholar]
- 30.Lazebnik YA, Kaufmann SH, Desnoyers S, Poirier GG, Earnshaw WC. Cleavage of poly(ADP-ribose) polymerase by a proteinase with properties like ICE. Nature. 1994;371(6495):346–347. doi: 10.1038/371346a0. [DOI] [PubMed] [Google Scholar]
- 31.Nicholson DW, Ali A, Thornberry NA, Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle M, Lazebnik YA. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376(6535):37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 32.Gobeil S, Boucher CC, Nadeau D, Poirier GG. Characterization of the necrotic cleavage of poly(ADP-ribose) polymerase (PARP-1): implication of lysosomal proteases. Cell Death Differ. 2001;8(6):588–594. doi: 10.1038/sj.cdd.4400851. [DOI] [PubMed] [Google Scholar]
- 33.Oliver FJ, de la Rubia G, Rolli V, Ruiz-Ruiz MC, de Murcia G, Murcia JM. Importance of poly(ADP-ribose) polymerase and its cleavage in apoptosis. Lesson from an uncleavable mutant. J Biol Chem. 1998;273(50):33533–33539. doi: 10.1074/jbc.273.50.33533. [DOI] [PubMed] [Google Scholar]
- 34.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83(3):354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 35.Sakai A, Kasahara K, Ohmori T, Kimura H, Sone T, Fujimura M, Nakao S. MET increases the sensitivity of gefitinib-resistant cells to SN-38, an active metabolite of irinotecan, by up-regulating the topoisomerase I activity. J Thorac Oncol. 2012;7(9):1337–1344. doi: 10.1097/JTO.0b013e31825cca4c. [DOI] [PubMed] [Google Scholar]
- 36.Khan M, Ding C, Rasul A, Yi F, Li T, Gao H, Gao R, Zhong L, Zhang K, Fang X, Ma T. Isoalantolactone induces reactive oxygen species mediated apoptosis in pancreatic carcinoma PANC-1 cells. Int J Biol Sci. 2012;8(4):533–547. doi: 10.7150/ijbs.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S, Dong P, Wang J, Zhang J, Gu J, Wu X, Wu W, Fei X, Zhang Z, Wang Y, Quan Z, Liu Y. Icariin, a natural flavonol glycoside, induces apoptosis in human hepatoma SMMC-7721 cells via a ROS/JNK-dependent mitochondrial pathway. Cancer Lett. 2010;298(2):222–230. doi: 10.1016/j.canlet.2010.07.009. [DOI] [PubMed] [Google Scholar]
- 38.Sang H, Zhang L, Li J. Anti-benzopyrene-7,8-diol-9,10-epoxide induces apoptosis via mitochondrial pathway in human bronchiolar epithelium cells independent of the mitochondria permeability transition pore. Food Chem Toxicol. 2012;50(7):2417–2423. doi: 10.1016/j.fct.2012.04.041. [DOI] [PubMed] [Google Scholar]
- 39.Zhou Y, Tian L, Long L, Quan M, Liu F, Cao J. Casticin potentiates TRAIL-induced apoptosis of gastric cancer cells through endoplasmic reticulum stress. PLoS One. 2013;8(3):e58855. doi: 10.1371/journal.pone.0058855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith J, Tho LM, Xu N, Gillespie DA. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv Cancer Res. 2010;108:73–112. doi: 10.1016/B978-0-12-380888-2.00003-0. [DOI] [PubMed] [Google Scholar]
- 41.Boohaker RJ, Xu B. The versatile functions of ATM kinase. Biomed J. 2014;37(1):3–9. doi: 10.4103/2319-4170.125655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Turner T, Caspari T. When heat casts a spell on the DNA damage checkpoints. Open Biol. 2014;4:140008. doi: 10.1098/rsob.140008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst) 2007;6(7):953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 44.Zhang YW, Otterness DM, Chiang GG, Xie W, Liu YC, Mercurio F, Abraham RT. Genotoxic stress targets human Chk1 for degradation by the ubiquitin-proteasome pathway. Mol Cell. 2005;19(5):607–618. doi: 10.1016/j.molcel.2005.07.019. [DOI] [PubMed] [Google Scholar]
- 45.Daugas E, Susin SA, Zamzami N, Ferri KF, Irinopoulou T, Larochette N, Prévost MC, Leber B, Andrews D, Penninger J, Kroemer G. Mitochondrio-nuclear translocation of AIF in apoptosis and necrosis. FASEB J. 2000;14(5):729–739. [PubMed] [Google Scholar]
- 46.Bockstaele L, Bisteau X, Paternot S, Roger PP. Differential regulation of cyclin-dependent kinase 4 (CDK4) and CDK6, evidence that CDK4 might not be activated by CDK7, and design of a CDK6 activating mutation. Mol Cell Biol. 2009;29(15):4188–4200. doi: 10.1128/MCB.01823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aggarwal P, Vaites LP, Kim JK, Mellert H, Gurung B, Nakagawa H, Herlyn M, Hua X, Rustgi AK, McMahon SB, Diehl JA. Nuclear cyclin D1/CDK4 kinase regulates CUL4 expression and triggers neoplastic growth via activation of the PRMT5 methyltransferase. Cancer Cell. 2010;18(4):329–340. doi: 10.1016/j.ccr.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy S, Gu M, Ramasamy K, Singh RP, Agarwal C, Siriwardana S, Sclafani RA, Agarwal R. p21/Cip1 and p27/Kip1 Are essential molecular targets of inositol hexaphosphate for its antitumor efficacy against prostate cancer. Cancer Res. 2009;69(3):1166–1173. doi: 10.1158/0008-5472.CAN-08-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Perisanidis C, Perisanidis B, Wrba F, Brandstetter A, El Gazzar S, Papadogeorgakis N, Seemann R, Ewers R, Kyzas PA, Filipits M. Evaluation of immunohistochemical expression of p53, p21, p27, cyclin D1, and Ki67 in oral and oropharyngeal squamous cell carcinoma. J Oral Pathol Med. 2012;41(1):40–46. doi: 10.1111/j.1600-0714.2011.01071.x. [DOI] [PubMed] [Google Scholar]
- 50.Smith JR, Pereira-Smith OM. Replicative senescence: implications for in vivo aging and tumor suppression. Science. 1996;273(5271):63–67. doi: 10.1126/science.273.5271.63. [DOI] [PubMed] [Google Scholar]
- 51.Stein GH, Dulić V. Origins of G1 arrest in senescent human fibroblasts. Bioessays. 1995;17(6):537–543. doi: 10.1002/bies.950170610. [DOI] [PubMed] [Google Scholar]
- 52.Redza-Dutordoir M, Averill-Bates DA. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim Biophys Acta. 2016;1863(12):2977–2992. doi: 10.1016/j.bbamcr.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 53.Di Meo S, Reed TT, Venditti P, Victor VM. Role of ROS and RNS sources in physiological and pathological conditions. Oxid Med Cell Longev. 2016;2016:1245049. doi: 10.1155/2016/1245049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8(3):193–204. doi: 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 55.Sahu RP, Batra S, Srivastava SK. Activation of ATM/Chk1 by curcumin causes cell cycle arrest and apoptosis in human pancreatic cancer cells. Br J Cancer. 2009;100(9):1425–1433. doi: 10.1038/sj.bjc.6605039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ahn J, Urist M, Prives C. The Chk2 protein kinase. DNA Repair (Amst) 2004;3(8-9):1039–1047. doi: 10.1016/j.dnarep.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 57.Hirao A, Cheung A, Duncan G, Girard PM, Elia AJ, Wakeham A, Okada H, Sarkissian T, Wong JA, Sakai T, De Stanchina E, Bristow RG, Suda T, Lowe SW, Jeggo PA, Elledge SJ, Mak TW. Chk2 is a tumor suppressor that regulates apoptosis in both an ataxia telangiectasia mutated (ATM)-dependent and an ATM-independent manner. Mol Cell Biol. 2002;22(18):6521–6532. doi: 10.1128/mcb.22.18.6521-6532.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joe Y, Jeong JH, Yang S, Kang H, Motoyama N, Pandolfi PP, Chung JH, Kim MK. ATR, PML, and CHK2 play a role in arsenic trioxide-induced apoptosis. J Biol Chem. 2006;281(39):28764–28771. doi: 10.1074/jbc.M604392200. [DOI] [PubMed] [Google Scholar]
- 59.Hu W, Wang F, Tang J, Liu X, Yuan Z, Nie C, Wei Y. Proapoptotic protein Smac mediates apoptosis in cisplatin-resistant ovarian cancer cells when treated with the anti-tumor agent AT101. J Biol Chem. 2012;287(1):68–80. doi: 10.1074/jbc.M111.271205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hemmati PG, Güner D, Gillissen B, Wendt J, von Haefen C, Chinnadurai G, Dörken B, Daniel PT. Bak functionally complements for loss of Bax during p14ARF-induced mitochondrial apoptosis in human cancer cells. Oncogene. 2006;25(50):6582–6594. doi: 10.1038/sj.onc.1209668. [DOI] [PubMed] [Google Scholar]
- 61.Nie C, Tian C, Zhao L, Petit PX, Mehrpour M, Chen Q. Cysteine 62 of Bax is critical for its conformational activation and its proapoptotic activity in response to H2O2-induced apoptosis. J Biol Chem. 2008;283(22):15359–15369. doi: 10.1074/jbc.M800847200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000;14(16):2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 63.Chandra D, Choy G, Daniel PT, Tang DG. Bax-dependent regulation of Bak by voltage-dependent anion channel 2. J Biol Chem. 2005;280(19):19051–19061. doi: 10.1074/jbc.M501391200. [DOI] [PubMed] [Google Scholar]
- 64.Kandasamy K, Srinivasula SM, Alnemri ES, Thompson CB, Korsmeyer SJ, Bryant JL, Srivastava RK. Involvement of proapoptotic molecules Bax and Bak in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced mitochondrial disruption and apoptosis: differential regulation of cytochrome c and Smac/DIABLO release. Cancer Res. 2003;63(7):1712–1721. [PubMed] [Google Scholar]
- 65.Boldin MP, Goncharov TM, Goltsev YV, Wallach D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell. 1996;85(6):803–815. doi: 10.1016/s0092-8674(00)81265-9. [DOI] [PubMed] [Google Scholar]
- 66.Fernandes-Alnemri T, Armstrong RC, Krebs J, Srinivasula SM, Wang L, Bullrich F, Fritz LC, Trapani JA, Tomaselli KJ, Litwack G, Alnemri ES. In vitro activation of CPP32 and Mch3 by Mch4, a novel human apoptotic cysteine protease containing two FADD-like domains. Proc Natl Acad Sci USA. 1996;93(15):7464–7469. doi: 10.1073/pnas.93.15.7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, Mann M, Krammer PH, Peter ME, Dixit VM. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death—inducing signaling complex. Cell. 1996;85(6):817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 68.Nieminen AI, Eskelinen VM, Haikala HM, Tervonen TA, Yan Y, Partanen JI, Klefström J. Myc-induced AMPK-phospho p53 pathway activates Bak to sensitize mitochondrial apoptosis. Proc Natl Acad Sci USA. 2013;110(20):E1839–E1848. doi: 10.1073/pnas.1208530110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Oda K, Arakawa H, Tanaka T, Matsuda K, Tanikawa C, Mori T, Nishimori H, Tamai K, Tokino T, Nakamura Y, Taya Y. p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell. 2000;102(6):849–862. doi: 10.1016/s0092-8674(00)00073-8. [DOI] [PubMed] [Google Scholar]
- 70.Deveraux QL, Roy N, Stennicke HR, Van Arsdale T, Zhou Q, Srinivasula SM, Alnemri ES, Salvesen GS, Reed JC. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998;17(8):2215–2223. doi: 10.1093/emboj/17.8.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Slee EA, Harte MT, Kluck RM, Wolf BB, Casiano CA, Newmeyer DD, Wang HG, Reed JC, Nicholson DW, Alnemri ES, Green DR, Martin SJ. Ordering the cytochrome c-initiated caspase cascade: hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J Cell Biol. 1999;144(2):281–292. doi: 10.1083/jcb.144.2.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun XM, MacFarlane M, Zhuang J, Wolf BB, Green DR, Cohen GM. Distinct caspase cascades are initiated in receptor-mediated and chemical-induced apoptosis. J Biol Chem. 1999;274(8):5053–5060. doi: 10.1074/jbc.274.8.5053. [DOI] [PubMed] [Google Scholar]
- 73.MacFarlane M, Cain K, Sun XM, Alnemri ES, Cohen GM. Processing/activation of at least four interleukin-1beta converting enzyme-like proteases occurs during the execution phase of apoptosis in human monocytic tumor cells. J Cell Biol. 1997;137(2):469–479. doi: 10.1083/jcb.137.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fernandes-Alnemri T, Litwack G, Alnemri ES. CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme. J Biol Chem. 1994;269(49):30761–30764. [PubMed] [Google Scholar]
- 75.Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25(38):5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- 76.Topacio BR, Zatulovskiy E, Cristea S, Xie S, Tambo CS, Rubin SM, Sage J, Kõivomägi M, Skotheim JM. Cyclin D-Cdk4,6 drives cell-cycle progression via the retinoblastoma protein’s C-Terminal helix. Mol Cell. 2019;74(4):758–770.e4. doi: 10.1016/j.molcel.2019.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hwang HJ, Kang YJ, Hossain MA, Kim DH, Jang JY, Lee SH, Yoon JH, Moon HR, Kim HS, Chung HY, Kim ND. Novel dihydrobenzofuro[4,5-b][1,8]naphthyridin-6-one derivative, MHY-449, induces apoptosis and cell cycle arrest in HCT116 human colon cancer cells. Int J Oncol. 2012;41(6):2057–2064. doi: 10.3892/ijo.2012.1659. [DOI] [PubMed] [Google Scholar]
- 78.Chatterjee SJ, Datar R, Youssefzadeh D, George B, Goebell PJ, Stein JP, Young L, Shi SR, Gee C, Groshen S, Skinner DG, Cote RJ. Combined effects of p53, p21, and pRb expression in the progression of bladder transitional cell carcinoma. J Clin Oncol. 2004;22(6):1007–1013. doi: 10.1200/JCO.2004.05.174. [DOI] [PubMed] [Google Scholar]
- 79.Le NT, Richardson DR. The role of iron in cell cycle progression and the proliferation of neoplastic cells. Biochim Biophys Acta. 2002;1603(1):31–46. doi: 10.1016/s0304-419x(02)00068-9. [DOI] [PubMed] [Google Scholar]
- 80.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366(6456):704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 81.Hunter T. Braking the cycle. Cell. 1993;75(5):839–841. doi: 10.1016/0092-8674(93)90528-x. [DOI] [PubMed] [Google Scholar]
- 82.Hara E, Smith R, Parry D, Tahara H, Stone S, Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16(3):859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Noda A, Ning Y, Venable SF, Pereira-Smith OM, Smith JR. Cloning of senescent cell-derived inhibitors of DNA synthesis using an expression screen. Exp Cell Res. 1994;211(1):90–98. doi: 10.1006/excr.1994.1063. [DOI] [PubMed] [Google Scholar]
- 84.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88(5):593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 85.Sherr CJ. Cancer cell cycles. Science. 1996;274(5293):1672–1677. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 86.Takahashi Y, Rayman JB, Dynlacht BD. Analysis of promoter binding by the E2F and pRB families in vivo: distinct E2F proteins mediate activation and repression. Genes Dev. 2000;14(7):804–816. [PMC free article] [PubMed] [Google Scholar]
- 87.Wu L, Timmers C, Maiti B, Saavedra HI, Sang L, Chong GT, Nuckolls F, Giangrande P, Wright FA, Field SJ, Greenberg ME, Orkin S, Nevins JR, Robinson ML, Leone G. The E2F1-3 transcription factors are essential for cellular proliferation. Nature. 2001;414(6862):457–462. doi: 10.1038/35106593. [DOI] [PubMed] [Google Scholar]