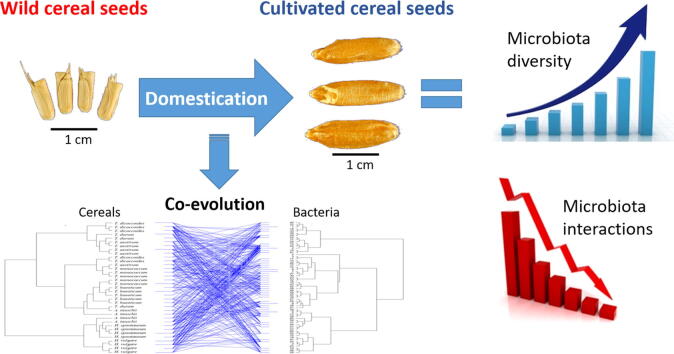
Graphical abstract
Keywords: Seed microbiome, Cereals, Random Amplified Polymorphic DNA - RAPD, 16S metabarcoding, Co-evolution, Domestication
Abstract
Introduction
The seed-associated microbiome has a strong influence on plant ecology, fitness, and productivity. Plant microbiota could be exploited for a more responsible crop management in sustainable agriculture. However, the relationships between seed microbiota and hosts related to the changes from ancestor species to breeded crops still remain poor understood.
Objectives
Our aims were i) to understand the effect of cereal domestication on seed endophytes in terms of diversity, structure and co-occurrence, by comparing four cereal crops and the respective ancestor species; ii) to test the phylogenetic coherence between cereals and their seed microbiota (clue of co-evolution).
Methods
We investigated the seed microbiota of four cereal crops (Triticum aestivum, Triticum monococcum, Triticum durum, and Hordeum vulgare), along with their respective ancestors (Aegilops tauschii, Triticum baeoticum, Triticum dicoccoides, and Hordeum spontaneum, respectively) using 16S rRNA gene metabarcoding, Randomly Amplified Polymorphic DNA (RAPD) profiling of host plants and co-evolution analysis.
Results
The diversity of seed microbiota was generally higher in cultivated cereals than in wild ancestors, suggesting that domestication lead to a bacterial diversification. On the other hand, more microbe-microbe interactions were detected in wild species, indicating a better-structured, mature community. Typical human-associated taxa, such as Cutibacterium, dominated in cultivated cereals, suggesting an interkingdom transfers of microbes from human to plants during domestication. Co-evolution analysis revealed a significant phylogenetic congruence between seed endophytes and host plants, indicating clues of co-evolution between hosts and seed-associated microbes during domestication.
Conclusion
This study demonstrates a diversification of the seed microbiome as a consequence of domestication, and provides clues of co-evolution between cereals and their seed microbiota. This knowledge is useful to develop effective strategies of microbiome exploitation for sustainable agriculture.
Introduction
Endophytes are harmless microorganisms living inside plant tissues. Microbial endophytes colonize all plant organs [1], including seeds. The presence of endophytes in the seeds of several plant species has been previously reported, including cereals [2], [3], [4], [5]. Microorganisms were hypothesized to have co-evolved with their host plants and animals, and developed symbiotic relationships with the hosts over the years [6]. It has been demonstrated that seed-borne microorganisms facilitate germination by protecting seeds from predation and attack by pathogens [7], [8] and by reducing abiotic stresses [9], [10]. Moreover, these microbes have a role in plant growth promotion and biocontrol of phytopathogens [4], [11], [12]. Seed endophytes can later be critical in shaping the root microbiota by ‘priority effect’ [13] as they are able to colonize very efficiently the rhizosphere [14]. This vertical transmission of seed endophytes was reported for some cereal species, such as maize [2], wheat [15], and barley [3]. However, the underlying evolutionary principles of these interactions remain to be elucidated. This knowledge is necessary to implement effective strategies of microbiome integration into the responsible management of soil and resources, to achieve a more sustainable modern agriculture [16], [17].
Wheat and barley are considered as the earliest domesticated crop plants and are, respectively, the first and the fourth most cultivated cereals in the world (FAO - Statistical pocketbook 2018: www.fao.org/3/CA1796EN/ca1796en.pdf). Different varieties of barley and wheat were domesticated from their wild ancestors about 10,000 years ago [18]. Throughout the domestication period, wild plants were transformed into food crops as a result of conscious and unconscious genetic selection of important traits, such as grain size and shape, and seed hull elimination [19]. As the plants evolved, their associated microbiomes are supposed to have undergone substantial changes, too, for instance, because of the loss of the fruit shell [2]. Several studies investigated the influence of plant genotype, crop rotation, fertilizer inputs, fungicide and herbicide application, and cultural practices on the composition of seed endophytes [2], [20], [21]. Modern plant cultivars may have missed some of the characteristics required to attract beneficial microbes compared to their wild relatives, which are more adapted to pre-agricultural soils [22], [23], [24].
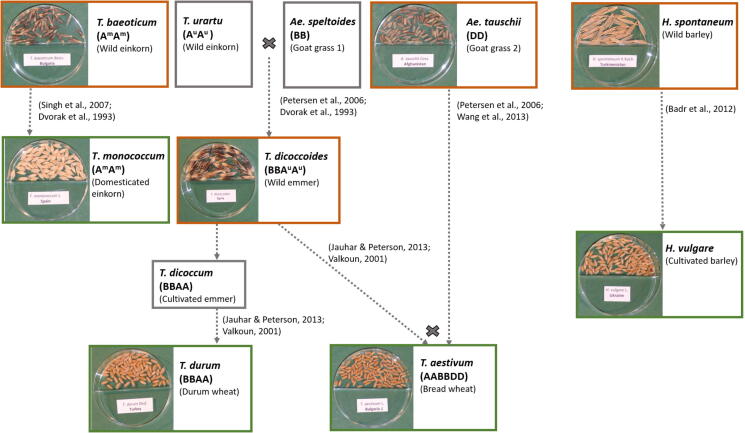
Regardless of these evolutionary changes, grains of currently cultivated crops appeared to carry similar microbiota as their wild relatives [2], [25]. However, to what extent domestication affected the diversity of seed microbiota and whether these bacterial communities preserve the same traits than in wild forms remains unclear. In this work, to explain the effect of genetic selection and domestication on cereal seed microbiota, wheat and barley were selected because of their historical, economic, and agricultural value. The species analyzed here include three cultivated wheats, Triticum aestivum L. ssp. aestivum (hereafter “Triticum aestivum”), Triticum monoccocum L. ssp. monoccocum (hereafter “Triticum monococcum”), Triticum durum Desf. ssp. durum (hereafter “Triticum durum”), and the three corresponding wild ancestors, Aegilops tauschii Coss. ssp. tauschii (hereafter “Triticum tauschii”), Triticum baeoticum Boiss. ssp. baeoticum (hereafter “Triticum baeoticum”), and Triticum dicoccoides Schweinf. ssp. dicoccoides (hereafter “Triticum dicoccoides”), as well as the cultivated barley Hordeum vulgare L. ssp. vulgare (hereafter “Hordeum vulgare”) and its ancestor Hordeum vulgare K.Koch. ssp. spontaneum (Coss.) Thell. (hereafter “Hordeum spontaneum”) (Fig. 1).
Fig. 1.
The evolutionary history of wheat and barley crops (green boxes) and their wild relatives, both used (orange boxes) and not used (gray boxes) in this work. Dotted arrows show the parental lines of domesticated forms. Crosses indicate cross-breeding events.
In order to confirm genetic bounds with corresponding ancestors of wheat and barley, genetic distances need to be measured. Several molecular methods can be used for assessing plant genetic distances, including Amplified Fragment Length Polymorphism (AFLP), Restriction Fragment Length Polymorphism (RFLP), Random Amplified Polymorphic DNA (RAPD), Inter-Simple Sequence Repeat (ISSR), Simple Sequence Repeat (SSR), among others. RAPD analysis was selected for this study because it is an effective and established method to measure genetic polymorphisms in cereals [26], [27], [28].
The objectives of this study were: i) to investigate the effect of cereal domestication on seed endophytes in terms of diversity, structure and co-occurrence, by comparing four crops and the four respective ancestor species; ii) to test the phylogenetic coherence between cereals and their seed microbiota, by comparing the genetic relatedness between cereals with that between the seed-associated bacteria (clue of co-evolution). We hypothesized that: i) a more diverse bacterial microbiota is associated with the seeds of current cultivars of wheat and barley compared to their ancestors, due to an ongoing process of microbiome diversification; ii) the dominant species will be different in the cultivated crops due to the effect of domestication; iii) more correlations (representing potential microbial interactions) will be found in the wild species, evolutionary older and therefore associated to a better-structured microbiota; and iv) cereal evolution has been coupled with a coherent evolution of their associated seed microbiota during the domestication period.
Materials and methods
Seed samples used
Cultivars of three wheat species, Triticum aestivum, Triticum monococcum, Triticum durum, as well as barley, Hordeum vulgare, and their corresponding ancestors (Aegilops tauschii, Triticum baeoticum, Triticum turgidum, and Hordeum spontaneum) were used. Aegilops tauschii (2n = 2x = 14, DD genomes) is one of the three wild diploid progenitors of the hexaploid bread wheat (Triticum aestivum, 2n = 6x = 42, AABBDD genome), which has three sets of homologous chromosomes, AABBDD, where D chromosomes derive from Aegilops tauschii and AABB from Triticum dicoccoides [29], [30]. Aegilops tauschii is distributed in eastern Turkey, Azerbaijan, Iran, Syria, and around the Caspian Sea [31].
Triticum baeoticum (2n = 2x = 14) is the wild ancestor of the einkorn wheat Triticum monococcum (2n = 2x = 14). It occurs in Southeast Europe and Turkey's mountainous regions [32].
Tetraploid Triticum dicoccoides (2n = 4x = 28, AABB genomes), is the ancestor of the durum wheat Triticum durum [33]. The A and B chromosomes of the tetraploid Triticum dicoccoides derive from an earlier hybridization between Triticum urartu [34] and Aegilops speltoides [29] (Fig. 1). Durum wheat is predominantly cultivated in the Middle East [35]. Hordeum spontaneum is the progenitor of currently cultivated Hordeum vulgare, first domesticated in the Israel-Jordan region [36] and predominantly cultivated in temperate areas.
Viable seeds were obtained from the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK), Germany. Five different accessions for each plant species were used (Tab. S1; Fig. S1). All seeds were produced, collected, and stored under the same conditions at the IPK. Therefore, the only factor expected to generate differences in the microbiota was the plant genotype. Once arrived at our laboratory, the seeds were stored at 4 °C until analysis.
Seed surface sterilization and DNA extraction from seeds
The analysis was performed on two seed subsamples per cereal accession, for two reasons: i) to increase the robustness of the sequencing results and ii) to account for the low number of bacterial sequences usually obtained by metabarcoding of seed microbiomes [5], [37]. Prior to the genomic DNA extraction, two aliquots of 10–15 seeds per each wheat or barley accession (total number of samples: 80) were surface-sterilized under room temperature by immersion into 70% ethanol for 2 min and then 2.5% sterilization solution (30 g NaCl, 1.5 g NaOH and 1 g Na2CO3) for 15 min [38]. Thereafter, seeds were washed with sterile distilled water four times for increasing intervals (5, 15, 25, and 45 min) and under shaking at 100 rpm. Surface sterilized seeds (2 samples per plant accession, 80 samples in total) were grounded using sterile pestle and mortar in liquid nitrogen. The DNA was isolated from 300–500 mg of grounded samples. Initially, each sample was added into a 2 ml screw-cap tube containing 200 µl of sterile glass beads. Then, each sample received 800 µl of extraction buffer (2.5 g l−1 SDS, 0.2 M sodium phosphate buffer, 50 mM EDTA and 0.1 M NaCl, pH 8). Cells were disrupted for 2 min at 30 Hz using a cell disrupter MM400 (Retsch GmbH, Haan, Germany) and then centrifuged (Heraeus Fresco, Thermo Fisher Scientific Inc., Waltham, USA) at 12,000 × g for 5 min at 4 °C. The supernatant was transferred into a new 2 ml microcentrifuge tube (Laborhaus Scheller GmbH & Co KG, Euerbach, Germany). The cell disruption step was repeated by adding another 700 µl of extraction buffer to the pellet of the same sample. Before moving to the next step, RNA was digested by adding 5 µl RNAse per 1 ml supernatant, and incubated at 37 °C for 30 min. The RNA digestion was followed by 500 µl of phenol/chloroform/isoamyl alcohol (25:24:1) addition. The tube was then centrifuged again at 16,000 × g for 5 min at 4 °C and then the upper, aqueous phase was transferred into the new 2 ml tube. Then 500 µl chloroform/isoamyl alcohol (24:1) was added, mixed well by inverting, and centrifuged at 16,000 × g for 5 min at 4 °C. Again, the upper, aqueous phase was collected into a new tube. One ml of precipitation buffer [20% polyethylene glycol, 2.5 M NaCl] was added and incubated at room temperature for 30 min and finally centrifuged at 16,000 × g for 30 min at 4 °C. The precipitated DNA was washed with 800 µl ice-cold 75% ethanol, dried beside the Bunsen burner flame, and dissolved in 30 µl nuclease-free water. The DNA was quantified by NanoDrop™ 2000 Spectrophotometer (Peqlab, Erlangen, Germany) and then stored at −20 °C until further analysis. This DNA was used for both, the RAPD analysis and the 16S rRNA gene library construction for IonTorrent sequencing.
Ion Torrent sequencing of prokaryotic 16S rRNA gene libraries
High-throughput sequencing is a state-of-the-art method to analyze the structure and diversity of microbiomes [39]. Here, we used the IonTorrent metabarcoding of 16S rRNA gene libraries, using a peptide nucleic acid (PNA) probe to reduce the amplification of plant mitochondrial and plastid DNA [40]. The V4 and V5 regions of the 16S rRNA genes were PCR amplified from the 80 seed samples using the primer 520F and 907 R [41], [42]. Fifteen µl of PCR reaction included 10 ng of seed DNA, 1 X KAPAHiFi (KAPA Biosystems, Wodurn, MA) buffer, KAPA dNTP mix 200 µM each, primer 5 pM each, 15 µM of chloroplast-PNA [40] and mitochondrial-PNAII (AAACCAATTCACTTGAGT, designed in this work to replace the mt-PNA of Lundberg et al., [40], which was not suitable due to the different position of the forward primer), and KAPAHiFi polymerase 0.3 units. The PCR was performed using a MycyclerTM (Bio-Rad, USA) for 20 cycles with the initial denaturation for 3 min at 95 °C, cyclic denaturation for 20 sec at 98 °C, PNA annealing for 30 sec at 65 °C, primer annealing for 30 sec at 55 °C, an extension for 30 sec at 72 °C and a final extension at 72 °C for 5 min. The second PCR was prepared with primer 520F and 907 R comp, adapter, and barcodes. The final volume of 50 µl contained 2 µl of the first PCR product, 10 µl of 5X KAPAHiFi buffer, KAPA dNTP mix 600 µM, primer 5 pM and KAPAHiFi polymerase 1 unit. The PCR was performed using MycyclerTM (Bio-Rad, USA) for 8 cycles with the initial denaturation for 3 min at 95 °C, cyclic denaturation for 20 sec at 98 °C, annealing for 30 sec at 55 °C, an extension for 30 sec at 72 °C and a final extension at 72 °C for 7 min.
Final PCR products were eluted and purified from agarose gel using NucleoSpin PCR purification kit (MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany), followed by primer-dimers removal using NucleoMag® beads (NGS clean-up kit, MACHEREY-NAGEL GmbH & Co. KG, Düren, Germany). The concentration of the purified PCR products was quantified using Qubit dsDNA HS assay kit by Qubit® 3.0 fluorometer (Life Technologies, Carlsbad, USA) and then adjusted to 1 µM. Two independent DNA extractions and PCRs were done for each seed accession. The PCR products were then pooled and the final concentration was again adjusted to 26 pM. The pooled product was used for emulsion PCR with Ion One Touch 2 (Ion PGM Hi-Q View OT2 kit, Life Technologies, Carlsbad, USA). The quality of the final product was assessed using Ion Sphere Quality Control Kit (Life Technologies, Carlsbad, USA) and loaded on a 314 or 318 chip for sequencing with an Ion PGM sequencer (Life Technologies, Carlsbad, USA).
Analysis of Ion Torrent sequencing data
Ion Torrent sequencing data were analyzed using QIIME 1.9 [43]. The reads of each two replicate samples per accession were pooled together (number of samples analyzed = 40). The sequences were length (200–500 nucleotides) and quality (threshold: 20) filtered, then chimeric sequences were removed using VSEARCH [44]. OTUs were generated at a sequence similarity level of 97% using the SUMACLUST method [45] and the “SUMACLUST exact” option (a sequence is assigned to the best matching OTU rather than the first OTU passing the similarity threshold). Taxonomy was assigned using the reference sequences of the SILVA 132 database, release: April 2018 [46]. OTUs identified as plastids or mitochondria, as well as singleton OTUs, were removed from the dataset.
Statistical analyses were performed in R using the OTU table generated from QIIME. Taxa summary plots were created using RStudio 1.1.463 [47], package ggplot2 [48]. To compare the alpha diversity indices (Shannon, Simpson, Dominance, and Equitability) and the relative abundances of taxa between wild ancestors and cultivated crops, the Student’s t-test on a normalized data set (sequencing depth: 1000 reads per sample) was used, after false discovery rate – FDR – adjustment of the p-values (Benjamini-Hochberg method). Beta diversity was calculated based on non-metric multidimensional scaling (NMDS) of weighted Bray-Curtis dissimilarities, calculated on a normalized data set (sequencing depth: 1000 reads per sample); the statistical significance of the factors “cultivation form” and “species” was assessed using the ADONIS test included in the R package ‘vegan’ [49].
The sequences were submitted to EMBL (www.ebi.ac.uk/ena) under the project number PRJEB36663.
Co-occurrence network analysis
Co-occurrence analysis using high-throughput sequencing data is used to detect potential microbe-microbe interactions as well as to identify hub species [50], [51], [52]. Studies on cereal seed endophytes have been carried out focusing on identification, microbial composition, and community structure [15], [53] and function [4], [5]. However, a complex network of interactions within the seed microbiota and the influence of evolutionary patterns on microbe-microbe interactions were not yet investigated. To investigate the effect of domestication on the microbial interaction network, the co-occurrence analysis was performed with the Co-occurrence Network inference software (CoNet) [54], using not-normalized data as recommended to reduce the compositional effect [55]. Only OTUs occurring in at least 10 samples were considered. Pairwise scores were calculated for four measures: the Bray-Curtis and Kullback–Leibler similarities, and the Pearson and Spearman correlations. For each measure and edge, 100 permutations (with row shuffling re-sampling and re-normalization for correlation measures), as well as the bootstrap scores, were generated. Unstable edges (outside the 2.5–97.5 percentiles of the bootstrap distribution) were deleted. The individual p-values generated by the four measures were merged using Brown's method. Only edges with false discovery rate (FDR)-corrected P-values below 0.05 and supported by at least three measures, were retained. The network layout was generated automatically with the “edge-forced spring embedded” algorithm, which leads to unbiased networks showing interconnected nodes closer to each other and less-linked ones placed in the outside position. Network legends were created with the Cytoscape Add-on “Legend creator” (http://apps.cytoscape.org/apps/legendcreator).
Random Amplified Polymorphic DNA (RAPD) analysis
Twenty RAPD primers (10-mer) (Integrated DNA Technologies, Inc. Coralville, USA) were tested for screening wheat and barley genotypes based on the quantity of the polymorphism they produced. Finally, five of them (OPA-17, OPH-19 [56], OPJ-18 [57], OPO-06 [58] and OPH-13 [59]) were selected for the analysis (Tab. S2).
RAPD assay protocol was adapted from Mantzavinou et al. [58], by further optimizing annealing temperature, using gradient temperature-PCR protocol and MgCl2 concentration for each primer. DNA sample concentration was adjusted at 100 ng µl−1. PCR was performed using MycyclerTM (Bio-Rad, Hercules, USA) in a reaction volume of 25 µl containing 1 µl DNA template, 1X KAPAHiFi Buffer (KAPA Biosystems, Wodurn, USA), 0.4 µM each 10-mer primer, 2.5 mM KAPA MgCl2, 200 µM KAPA dNTPs mix and 0.625 units Taq DNA polymerase. RAPD was amplified using following thermal profile: 5 min at 94 °C, 40 cycles of 30 sec at 94 °C, 1 min at 30–40 °C (depending on primer, Tab. S2), 1 min at 72 °C, and 10 min at 72 °C for final elongation. The amplification products were separated on 1.5% (w/v) Agarose gel containing 5 µl 100 ml−1 DNA dye HDGreen™ (Intas, Göttingen, Germany) in 0.5 X TBE buffer. Both 1 kb and 100 bp DNA ladders (Quick-Load® Purple, New England BioLabs Inc., Ipswich, USA) were used for size comparison. RAPD fragments were illuminated under UV light and images were captured using Gel Doc 2000 (Bio-Rad, Hercules, USA).
Gel images were analyzed using the software GelCompar II version 5.10 (Applied Maths, NV). The five fingerprints for each seed accession were linked to form a composite data set (Fig. S2). The dendrogram was constructed using similarity coefficients based on the number of different bands (optimization: 1%, position tolerance: 1%) with the unweighted paired group method of cluster analysis with arithmetic averages (UPGMA).
Co-evolution analysis
To test the co-evolution between cereals and associated seed microbiota, we measured their phylogenetic congruence. A cophylogeny analysis was performed between cereal plants and the corresponding bacterial OTUs, using the host distance matrix obtained from RAPD analysis and the bacterial distance matrix calculated from the high-throughput sequencing. Cophylogeny analysis identifies the effect of evolution on diversification patterns of two or more ecologically associated species [60], [61]. To date, cophylogeny studies have been mainly used to study host-parasite relationships or vertically transmitted symbionts [62]. In this study, we established the use of cophylogeny assessment to study the co-evolution of seed microbiota from wild progenitors to modern cultivars of wheat and barley. The various techniques available for cophylogenetic assessment are divided into two categories: event-based and topology-based (global-fit) methods [60]. In this study, we used a global-fit method because it can afford large-scale cophylogenetic analyses and because the quantity of phylogenetic congruence generated by the cophylogenetic assessment can be associated with the significance of co-evolution in the studied scheme [60]. The test was a global goodness-of-fit test performed with 1000 permutations, using the functions cophyloplot and ParaFit in the ‘paco’ [60] and ‘ape’ [63] R packages. A tanglegram was created for the visual representation of the shared branching events. ParaFit requires the phylogeny of the host, the phylogeny of bacterial OTUs, and a matrix of connections as input. It compares the observed host and the bacterial distance matrices, and then tests for random associations between the two taxa groups, by randomizing the matrix of association. So, it generates P-values to calculate the contribution of each host-bacteria association to the global statistic testing (ParaFitGlobal) for each random association test between hosts and bacterial OTUs [64]. A global sum of squared residuals, called m2XY, is calculated, which represents the sum of all connection distances in the tanglegram. The observed m2XY value is statistically compared to the 1000 values generated by random permutations [65], to assess the significance of the phylogenetic congruence: the lower this observed m2XY value, the higher the statistical significance of the phylogenetic congruence.
Results
High-throughput sequencing analysis and taxonomic composition of the bacterial microbiota
A PCR product was obtained from 78 out of 80 samples; both replicates of one seed accession (T. durum TRI_13547, Tab. S1) did not produce a PCR product. A total of 6,595,794 sequence reads were produced and, after filtration of 15,696 sequences, removal of all plant-originated sequences (5,870,289), and singletons (1,744), 708,065 high-quality prokaryotic 16S rRNA gene sequences (1,004 to 93,702 reads per sample) remained. These sequences were grouped into 423 OTUs at 97% similarity level.
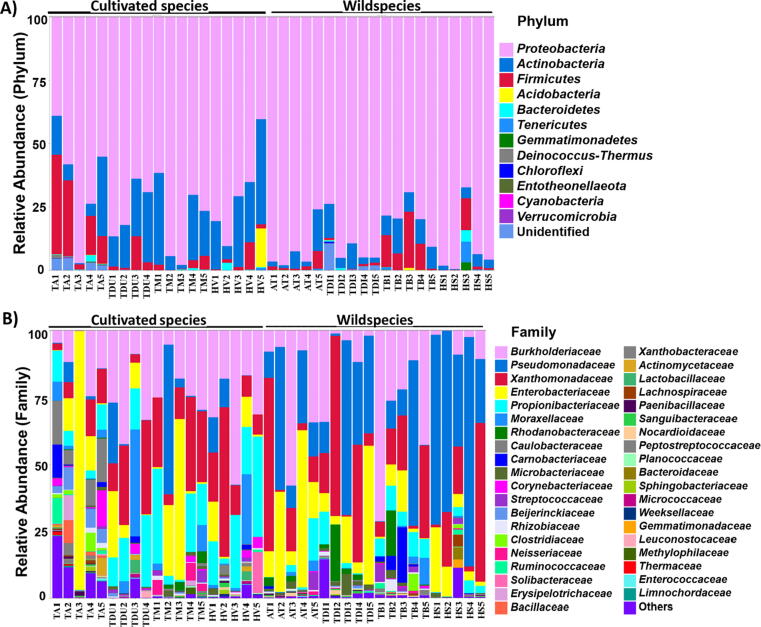
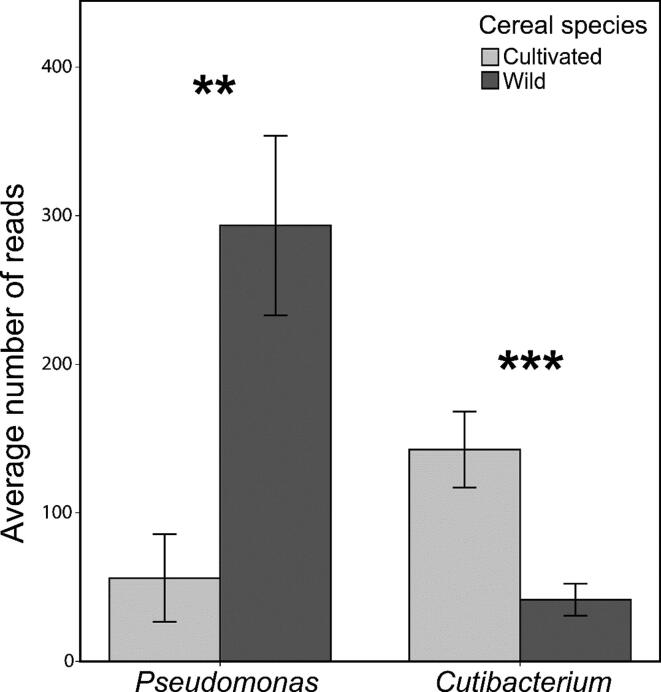
Proteobacteria, Actinobacteria, and Firmicutes were the predominant phyla (Fig. 2A); Actinobacteria and Firmicutes were found comparatively higher in cultivated species. Burkholderiaceae, Pseudomonadaceae, and Xanthomonadaceae were the major families. Twenty families were significantly different between wild and cultivated cereals (FDR-adjusted p value < 0.05); the most abundant were Pseudomonadaceae, more abundant in wild species, and Propionibacteriaceae, more abundant in cultivated crops (Fig. S3). In particular, at the genus level, we found a statistically significant higher abundance of Pseudomonas in wild species, while Cutibacterium was more abundant in cultivated crops (Fig. 3). When considering individually each couple of wild ancestor and cultivated derivate, Caulobacteraceae was found abundant in Triticum aestivum compared to Aegilops tauschii. Pseudomonadaceae, Enterobacteriaceae, and Xanthomonadaceae were found abundant in Triticum dicoccoides, while Propionibacteriaceae, Burkholderiaceae, and Xanthomonadaceae were more abundant in Triticum durum. Pseudomonadaceae was found a major abundant family in Hordeum spontaneum, while Xanthomonadaceae, Burkholderiaceae, and Propionibacteriaceae were the major families in Hordeum vulgare. Finally, Pseudomonadaceae was more abundant in Triticum baeoticum compared to Triticum monococcum (Fig. 2B).
Fig. 2.
Bacterial taxonomic composition of seed endophytes of wild and cultivated species, at Phylum (A) and Family (B) levels. Triticum aestivum (TA), Triticum durum (TDU), Triticum monococcum (TM), Aegilops tauschii (AT), Triticum diccocoides (TDI), Triticum baeoticum (TB), Hordeum vulgare (HV) and Hordeum spontaneum (HS). Relative abundance of major taxa only (>1% of total reads) according to 16S rRNA gene metabarcoding.
Fig. 3.
Relative abundance of the genera Pseudomonas and Cutibacterium in the wild and cultivated cereals, as resulted from metabarcoding analysis, calculated on the rarefied dataset (sequence depth = 1000 reads per sample). ** p = 0.0013; *** p < 0.001.
Alpha- and beta-diversity, shared taxa and co-occurrence analysis
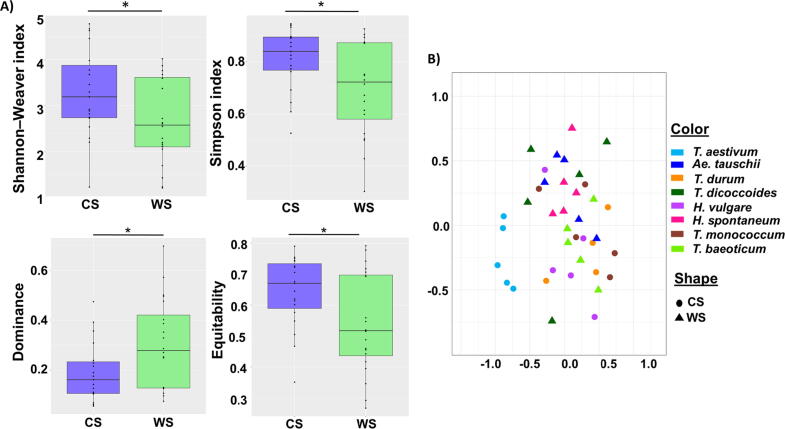
All the four calculated alpha diversity indices were significantly different between wild species and cultivar species (t-test, P < 0.05). Shannon-Weaver, Simpson, and Equitability indices were higher and Dominance was lower in cultivated species compared to wild species (Fig. 4A). We calculated the relative increment % of each cultivated species to the corresponding wild ancestor (Tab. S3). This value was positive for all of them, except for the couple T. baeoticum/T. monococcum (but at a lower absolute extent than any other couple). Interestingly, Tb-Tm (having a genetic similarity higher than the other couples, see Fig. 1) appears as the most closely related couple also concerning the structure (beta-diversity): in fact, T.monococcum and T.baeoticum samples are the only ones that largely overlap in the beta-diversity plot (Fig. 4B).
Fig. 4.
Alpha and beta diversity metrics of seed endophyte microbiota. (A) Shannon–Weaver, Simpson, Dominance and Equitability indices of bacterial microbiota (OTU 97%), grouped by cultivation form (t-test, P < 0.05), according to 16S rRNA gene metabarcoding. CS = Cultivated species; WS = Wild species. (B) Non-metric multidimensional scaling plot for bacterial microbiota structure based on weighted Bray-Curtis distances. Samples are colored by plant species and shaped by cultivation form. ADONIS significance test: R2 = 0.078, P = 0.003 for the factor “cultivation form”; R2 = 0.32, P < 0.001 for the factor “species”; stress value: 0.1495.
Non-metric multidimensional scaling plot based on weighted Bray-Curtis distances were significantly influenced by factors, cultivation form (ADONIS, R2 = 0.078, P = 0.003), plant species (ADONIS, R2 = 0.32, P < 0.001) (Fig. 4B), plant varieties (ADONIS, R2 = 0.36, P < 0.001), and sets of homologous chromosomes (ADONIS, R2 = 0.094, P < 0.001), but not by the factor “country of origin” (ADONIS, R2 = 0.58, P = 0.106).
The number of exclusive OTUs was higher in cultivated species (43%) than in wild species (24%), which is coherent with the higher alpha-diversity. 33% of the OTUs were shared (Fig. S4A). The ten most abundant OTUs shared between wild and cultivated cereals were Pseudomonas, Stenotrophomonas, Cutibacterium, Kosakonia cowanii, Burkholderiaceae, Stenotrophomonas, Ralstonia, Pantoea, Delftia, and Acinetobacter radioresistens. OTUs identified as Pseudomonas, Stenotrophomonas, Kosakonia cowanii, and Delftia were found higher in wild species, while OTUs identified as Cutibacterium, Burkholderiaceae, Stenotrophomonas, Ralstonia, Pantoea, and Acinetobacter radioresistens were higher in cultivated species (Fig. S4B). OTUs exclusively found in cultivated cereals belonged to several genera, including Cutibacterium and Methylobacterium (Fig. S5).
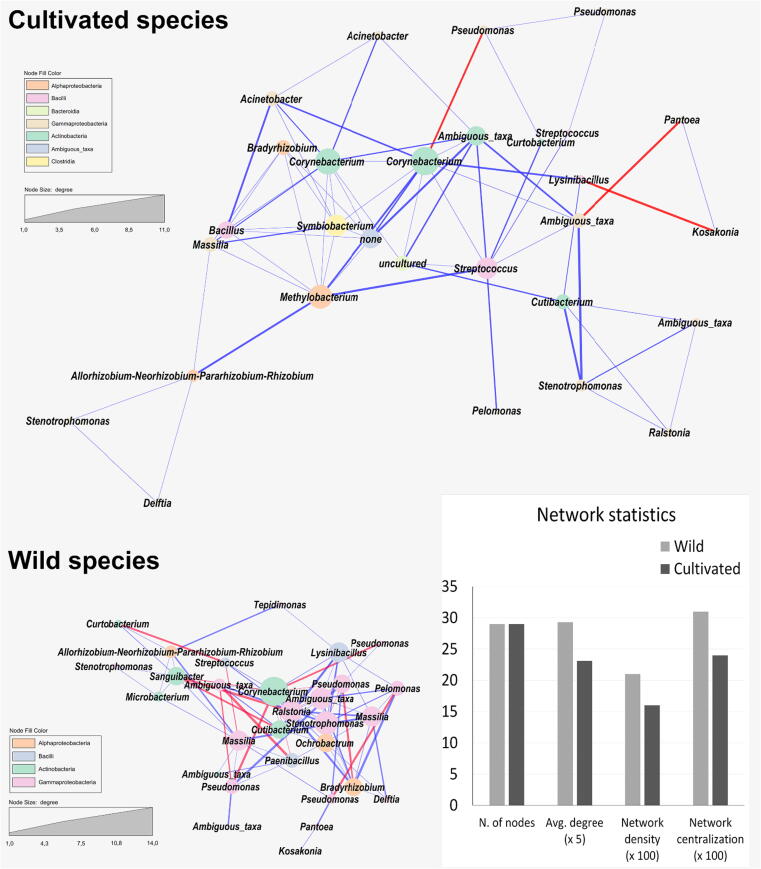
Co-occurrence analysis showed that the microbiota of wild species had higher connectivity than cultivated species (Fig. 5). In particular, despite the number of connected nodes was the same, the microbiota of wild cereals had a higher average number of neighbors (“degree”) and higher network density and centralization, with respect to the cultivated species (Fig. 5).
Fig. 5.
Co-occurrence network of OTUs, calculated for wild and cultivated cereal species separately. Nodes are colored by taxonomy and sized by degree (=n. of connections). Edges are colored by correlation type (blue: positive; red: negative) and the thickness represents merged FDR-corrected P-values (the thicker, the more significant).
RAPD analysis of genetic distances between cereal species
Of the 40 initial seed samples, four ones (one of each T. durum, Ae. tauschii, H. vulgare, and H. spontaneum) did not give bands after RAPD PCR, therefore sample number reduced to 36 (Fig. 6, Fig. S2). The UPGMA dendrogram was divided into two main clusters, separating barley and wheat species (Fig. 6, Fig. S2). In the barley cluster, cultivated and wild barley appeared as sister clades. In the wheat cluster, there was a further separation of Triticum and Aegilops genera. Within the Triticum species, Triticum baeoticum and Triticum monococcum formed a monophyletic group and appeared as sister clades (Fig. 6, Fig. S2). Triticum durum and Triticum dicoccoides did not cluster together but were mixed with Triticum aestivum. However, the accessions of Triticum aestivum were placed in the expected position with respect to the ancestor Aegilops tauschii (Fig. 6, Fig. S2).
Fig. 6.
UPGMA dendrogram showing the genetic similarity between cultivars of wheat and barley and their corresponding ancestors, calculated on RAPD data. The dendrogram was constructed using similarity coefficients based on the proportion of different bands.
Co-evolution analysis
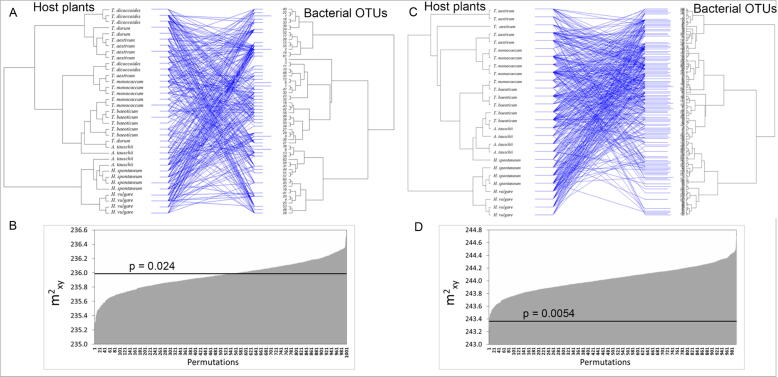
A co-evolution analysis was performed using the host distance matrix obtained by the RAPD analysis and the bacterial OTU distance matrix obtained by metabarcoding analysis. Co-evolution analysis was performed on 35 seed samples since one sample did not give a PCR product for metabarcoding and four ones did not give RAPD profiles. The evolutionary relationships between host species and bacterial OTUs were analyzed by the goodness-of-fit test. The global fit of the regression of bacterial OTUs phylogeny to the host phylogeny was evaluated using m2XY as a sample statistics, which is determined by a randomization procedure and shows the strength of associations between organisms from different phylogenetic groups. The goodness-of-fit test of phylogenic association between the bacteria and the host species phylogenies revealed a significant topological congruence (m2XY = 235. 98; P = 0.024; 1000 permutations) (Fig. 7A). 52.5% of the 1000 randomizations had a lower m2XY than the observed one (Fig. 7B). Here, 62 OTUs (14.6% of all OTUs) significantly contributed to the coherence of the tree topologies (Fig. 7A).
Fig. 7.
Co-evolution analysis. (A) Tanglegram between the cereal phylogeny, based on RAPD analysis, and the bacterial phylogeny (at OTU level), based on the metabarcoding analysis. Blue lines indicate plant–bacterial associations that were more significant than expected by chance, according to the ParaFitGlobal statistic. (B) Visualization of the global goodness-of-fit statistics over 1000 permutation (P = 0.024); the black line indicates the observed global sum of squared residuals (m2XY) value, while the grey columns indicate the m2XY value of each of the 1000 randomizations, sorted by ascending values. (C) and (D) Same than (A) and (B), calculated after removal of T. durum and T. dicoccoides samples.
To test whether the unresolved RAPD clustering of the species T. durum and T. dicoccoides might have affected the co-evolution assessment, we deleted these two species and repeated the analysis: indeed, a more significant topological congruence was obtained (m2XY = 243.36; P = 0.0054; 1000 Permutations) (Fig. 7C). Only two (0.2%) of the 1000 randomizations resulted in a lower m2XY than the observed one (Fig. 7D). Here, 160 OTUs (37.8% of all OTUs) significantly contributed to the coherence of the tree topologies (Fig. 7C).
Discussion
Domestication and breeding of plants have resulted in productive cultivars, but also in significant changes in plant microbiota with compositional shifts, as already reported for different crops [23], [50], [66], [67], [68], [69]. The idea behind our current study was to analyze the changes from species A to B, from species C to D, E to F, and G to H (four ancestors and their four descendant cultivated cereal species, respectively), and to test whether there were common traits in these changes, which would then suggest a common effect of domestication. The four individual wild species and the four individual cultivated species were treated as “replicates” for the factor “cultivation form” in our experimental design. We therefore intended to go behind the pairwise comparisons between individual species (largely tested in literature) and to assess further potential drivers of the microbiome that could be important at the (co–)evolutionary level, such as domestication.
We found a more diverse microbiota associated to the seeds of modern cereals compared to the wild ancestors. This suggests that cereal breeding lead to a compositional shift in the plant-associated microbiome. This finding is in line with previous studies that showed higher microbial diversity in the rhizosphere of modern crops than wild ancestors [50], [66], [70]. Suggested drivers of these changes were agricultural soil conditions, crop management methods, and changes in root exudates in wheat [71], since breeding of modern crops resulted in increased root exudation of organic compounds [71], [72]. Other factors, such as host genotype [2] and environmental circumstances were indicated as further possible drivers [20], [21], [22].
So far, the influence of domestication on bacterial diversity was studied mainly in the root system [22], [50], [66], [70]; a few studies specifically focused on the effect of domestication on seed endophytes and reported minor effects of domestication on community richness [2], [69]. Compared to these studies, we observed the effect of domestication in a larger set of species originally derived from areas of different continents (Tab. S1), which can explain the higher microbiota diversification found in our study. The relative increment % of each cultivated species to the corresponding wild ancestor is positive for all of them, except for the couple T. baeoticum/T. monococcum (but at a lower absolute extent than any other couple). Coherently, Tb-Tm appears as the most closely related couple in Fig. 6, which suggests that perhaps the microbial diversity is not high between them because the genetic similarity between T. boeticum and T. monococcum is higher than the other couples (Fig. 1). Moreover, this genetic similarity between the plants appears to be reflected not only in the microbial diversity but also in the structure: in fact, T. monococcum and T. baeoticum samples are the only ones that largely overlap in the beta-diversity plot (Fig. 4B). However, we argue that this observation even supports our conclusion that cereal domestication might lead to a general increase in diversity, which is associated to the genetic distance.
Our beta-diversity analysis supports this idea and shows that domesticated and wild species differ in their microbiota by plant genotype (Fig. 4B). In fact, both factors (“species” and “variety”) relate to the host genotype, which is known to be one of the main factors affecting the plant-associated microbiome, and therefore it is not surprising that the effect of the factors “plant species” and “plant varieties” was stronger compared to “cultivation form”. Indeed, our aim was not to demonstrate that “cultivation form” (in the sense of “cultivated” or “wild/ancestor”) is the most important factor affecting the seed microbiota. Instead, we tested and demonstrated that cereal domestication implied a certain level of a compositional shift in the seed-associated microbiome. This fact is not trivial, since it has important potential implications on crop ecology and plant–microbe interactions in an (co–)evolutionary framework. Moreover, we argue that the common shift from ancestors to cultivated forms can be somehow masked by the strong genotype effect, which drives the microbiome changes in an independent way.
Overall, domestication-related traits, as a factor for plant long-term adaptation, appear to determine a compositional shift in microbiomes of modern crops. Although the seeds of both wild and cultivated plants were dominated by similar bacterial phyla (Proteobacteria, Firmicutes, and Actinobacteria), there was a clear difference between cultivated crops and wild progenitors, which may link to the domestication effect. Among the enriched bacterial taxa in cultivated cereals, we found the genus Cutibacterium (family Propionibacteriaceae), a dominant member of the human skin microbiota. This could result from the human manipulation of seeds and plants during cereal domestication. It is also possible to assume that the presence of genus Cutibacterium (family Propionibacteriaceae) in our samples could be a contamination due to sample mishandling. However, recent studies endorse more the idea of an interkingdom exchange of microbes during plant domestication than the possibility of contamination. For example, Kuźniar and colleagues [73] studied eight wheat seeds microbiota in different compartments of seed (the embryo, endosperm, and the seed coat) and they found Cutibacterium in all the parts of the studied cultivars. Many other recent studies also found Propionibacterium as a member of the core microbiota of cereal seeds such as wheat, barley, maize and rice [20], [73], [74], [75]. Interestingly, Campisano and colleagues [76] found a subspecies of Propionibacterium acnes in grapevine (Vitis vinifera L.), which evolved from the human-associated strain since a time comparable with the beginning of grapevine domestication. The authors concluded that there was an event of interkingdom exchange between humans and plants during grapevine domestication. Likewise, Yousaf and colleagues [77] investigated the relationships between human and animal pathogens (HAP) with plants. They identified Propionibacterium and other HAPs in the grapevine endosphere in both stems and leaves, and concluded that human and animal pathogens can be integrated within plant tissues, adapt to the plants, and finally become plant symbionts, for at least one stage of their life cycle. In our work, we found a similar situation; therefore we suggest that such exchange of microbes from humans to plants (and, perhaps, vice-versa) might be an effect of plant domestication more common than currently supposed.
We also found a higher abundance of Pseudomonas in the seeds of wild species compared to cultivars. Although some species of this genus are pathogenic to plants, several studies showed that plant-originated Pseudomonas ssp. have the ability to promote plant health and productivity by different mechanisms [11], [78], [79]. Some Pseudomonas spp. are also regarded as biocontrol agents against several fungal pathogens [11]. Rahman and colleagues [5] demonstrated that a Pseudomonas sp., isolated from barley seeds, has beneficial effects for the host, especially under harsh environmental conditions. This ability to cope with biotic and abiotic stresses of Pseudomonas and Stenotrophomonas species, which were isolated from wild beetroots, was also documented by Zachow and colleagues [67]. Another dominant OTU found in wild cereals belonged to Acinetobacter, which was previously found in rice seeds [80] and was shown to possess nitrogen fixation, siderophore production, and mineral solubilization abilities [81]. This evidences suggest that wild plants, often living under stressed conditions, can be supported by microbes to cope with abiotic and biotic stresses [5], [67].
We identified a shared microbiome among seeds of wild and modern cereals from various accessions coming from a range of geographic locations. The presence of a shared microbiome preserved across plant species and geographical locations suggests that the seed-associated microbiome, intimately associated with the host, is in some cases preserved during plant domestication. These observations are consistent with other studies, showing that maize seed-associated endophytic bacteria were preserved from the progenitor species teosinte, growing in different geographical places [2]. The majority of bacterial OTUs of the shared cereal microbiome were related to Pantoea, Pseudomonas, Acinetobacter, Burkholderiaceae and Stenotrophomonas, which were reported as core microbiota of different plant seeds [80], [82]. This suggests that such preserved endophytes are well adapted to the internal seed habitat (high osmotic pressure, low moisture and nutrient deficiency, in mature seeds), and likely resulted from long-term selection and adaptation to the seed microhabitat. However, before being analyzed in our study, all the cereal species were propagated and maintained for several years on the same site. It is therefore possible that some of the shared OTUs are derived from the common soil/site. Nevertheless, in our study, the difference between cereal species, as well as between wild and cultivated cereals, have been identified as significant factors for the variation of the microbiome. Therefore, the existing differences can only be considered as dependent either on the host genotype or the cultivation form.
Interestingly, although the wild cereals harbor a lower bacterial diversity in their seeds, a higher level of connectivity was found by co-occurrence analysis. This means that certain microbial species may have better adapted to the seed habitat and had longer time to develop mutual interactions. This indicates a higher level of “maturity” of the microbiome associated with wild cereals, which suggests co-evolution with the host and vertical transmission across plant generations [5]. In contrast, the microbiota associated with the cultivated cereals did not have enough time yet to establish a solid network of microbial interactions, compared to the wild species.
All the above-discussed evidences strongly suggest a co-evolution of the seed microbiota with the host plants, across the period of cereal domestication. Seed inhabiting microbes are among the most intimate partners of the plant, and they are transmitted to the next plant generations [24]. Therefore, seed endophytes can be regarded as one of the most adapted and specific part of the plant microbiota, if compared to other plant habitats (rhizosphere, phyllosphere, etc.), which are more influenced by external factors and are usually colonized by microbial species recruited from the surrounding environment. Coherently with our conclusion, Wassermann and colleagues [83] found that eight wild plants growing under the same environmental conditions for centuries showed a unique microbiota, and shared just a very small core microbiome, in their seeds. This is surprising, considering that they grew intermixed for decades, and suggests a strong co-evolution. Therefore, we aimed to demonstrate the co-evolution between seed endophytes and cereals by a co-evolution analysis, using RAPD genetic distances of cereals and phylogenetic distances of the associated bacterial OTUs.
The dendrogram based on RAPD profiles showed a clear division of wheat and barley genotype. However, Triticum aestivum, Triticum durum and Triticum dicoccoides were not well discriminated. This phenomenon can be explained by the behavior of the different chromosome sets and polymorphisms of repeated nucleotide sequences, the analysis of which showed close relationships between Triticum durum and Triticum aestivum. This is likely due to the fact that Triticum urartu is the donor of the A genome of these polyploid wheat sorts [48]; indeed, many studies revealed genetic similarity between Triticum aestivum and Triticum durum [36], [84]. Three sets of homologous chromosomes of Triticum aestivum derive from the alloploidization of wild DD diploid Aegilops tauschii and wild AABB tetraploid Triticum dicoccoides, whereas the A and B chromosomes of Triticum dicoccoides derive from the wild AA diploid Triticum urartu [34] and BB diploid genome donor Aegilops speltoides [29]. Triticum diccocoides is therefore equally genetically related to both polyploid species of wheat.
The cophylogenetic analysis revealed a significant coherence of phylogenies between seed microbiota and corresponding cereal hosts, from the wild ancestors to the recently cultivated crops, which is a clear clue of co-evolution. This phylogenetic concordance suggests a plant–microbe co-adaptation related to the plant genotype since a stronger effect of the plant genotype on the endophytic bacterial community than on the root-associated bacterial communities was found previously [2], [22]. The topology of the cereal tree in the tanglegram (Fig. 7) is not totally coherent with that obtained by the RAPD analysis (Fig. 6); the differences arise from the clustering method applied by the specific R-script for the co-phylogeny analysis: this is a principal component analysis, which is not the best method for clustering RAPD profiles. The correct method is UPGMA, like that applied for Fig. 6, where the topology follows well the expected clustering (with the exception of T. dicoccoides and T. durum that were not discriminated, while for the other species the topology is coherent with the known phylogeny, at both genus and species level). However, the aim of this kind of cophylogeny analysis is not to get a perfect clustering, but instead to test whether there is significant coherence between the two components (hosts and microbes). We found such significant coherence, which suggests that the two associated components have a certain level of co-evolution. The significance increased drastically when the not resolved species (T. dicoccoides and T. durum) were removed from the analysis, highlighting the importance to have a well-discriminated analysis of the hosts' genetic distances for performing the co-evolution test.
The approach of identifying interactions and comparing between seed-associated microbial communities and host plants provides the opportunity to move beyond the linear assessments of plant-microbial associations towards a more thorough knowledge of how endophytes are related to host inherited traits. To fully comprehend the processes responsible for these associations, future studies on functional properties and investigations of the impacts of host characteristics on the development of associated microbiomes will be needed.
In this study, we used cereal cultivars and their wild relatives as a model to analyse the effect of plant domestication on bacterial seed endophytes, and to test whether seed endophytes might have co-evolved with their hosts. We are aware that our dataset of four pairs of cereal plants is actually relatively limited; therefore our findings cannot be considered as definitely conclusive, but rather provides: i) indications for a certain level of a compositional shift in the seed-associated microbiome due to domestication, and ii) clues of co-evolution. These intriguing findings, which are in part supported by a limited number of previous studies, need to be tested on further plant species to verify whether they can be generalized or not.
Our understanding of the development of endophytic microbial associations at an evolutionary time scale is presently very restricted. Our work provides new insights into complex microbial interactions and highlights the importance of integrating bacterial seed endophytes into both microbial ecology and applied agricultural microbiology research. From an applied point of view, this knowledge is of paramount importance to develop effective strategies of biofertilization and biocontrol, which are urgently needed to increase sustainability and responsible use of soil resources in modern agriculture.
Compliance with Ethics Requirements
This article does not contain any study with human or animal subjects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank all the laboratory members, particularly Bellinda Schneider (Giessen, Germany) for her technical support in molecular works. We also thank IPK staff for providing us with a generous amount of seed. We thank Andreas Schwiertz, Institut für Mikroökologie (Herborn, Germany) for allowing us to use the IonTorrent sequencing facility. This work was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) — project number 390838017.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.12.008.
Contributor Information
Yulduzkhon Abdullaeva, Email: abdullaeva.yulduzkhon@umwelt.uni-giessen.de.
Binoy Ambika Manirajan, Email: binoyam@gmail.com.
Bernd Honermeier, Email: bernd.honermeier@agrar.uni-giessen.de.
Sylvia Schnell, Email: sylvia.schnell@umwelt.uni-giessen.de.
Massimiliano Cardinale, Email: massimiliano.cardinale@unisalento.it.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Hardoim P.R., van Overbeek L.S., van Elsas J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008;16:463–471. doi: 10.1016/j.tim.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 2.Johnston-Monje D., Raizada M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0020396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardoim P.R., Hardoim C.C.P., van Overbeek L.S., van Elsas J.D. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0030438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Díaz Herrera S., Grossi C., Zawoznik M., Groppa M.D. Wheat seeds harbour bacterial endophytes with potential as plant growth promoters and biocontrol agents of Fusarium graminearum. Microbiol Res. 2016;186–187:37–48. doi: 10.1016/j.micres.2016.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Rahman M.M., Flory E., Koyro H.W., Abideen Z., Schikora A., Suarez C. Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.) Syst Appl Microbiol. 2018;41:386–398. doi: 10.1016/j.syapm.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Zilber-Rosenberg I., Rosenberg E. Role of microorganisms in the evolution of animals and plants: The hologenome theory of evolution. FEMS Microbiol Rev. 2008;32:723–735. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 7.Dalling J.W., Davis A.S., Schutte B.J., Elizabeth A.A. Seed survival in soil: Interacting effects of predation, dormancy and the soil microbial community. J Ecol. 2011;99:89–95. [Google Scholar]
- 8.Nelson E.B. The seed microbiome: Origins, interactions, and impacts. Plant Soil. 2018;422:7–34. [Google Scholar]
- 9.Yandigeri M.S., Meena K.K., Singh D., Malviya N., Singh D.P., Solanki M.K. Drought-tolerant endophytic actinobacteria promote growth of wheat (Triticum aestivum) under water stress conditions. Plant Grow Reg. 2012;68:411–420. [Google Scholar]
- 10.Murphy B.R., Jadwiszczak M.J., Soldi E., Hodkinson T.R. Endophytes from the crop wild relative Hordeum secalinum L. improve agronomic traits in unstressed and salt-stressed barley. Cogent Food Agric. 2018;4:1549195. [Google Scholar]
- 11.Mercado-Blanco J., Alós E., Rey M.D., Prieto P. Pseudomonas fluorescens PICF7 displays an endophytic lifestyle in cultivated cereals and enhances yield in barley. FEMS Microbiol Ecol. 2016;92:1–13. doi: 10.1093/femsec/fiw092. [DOI] [PubMed] [Google Scholar]
- 12.Robinson R.J., Fraaije B.A., Clark I.M., Jackson R.W., Hirsch P.R., Mauchline T.H. Wheat seed embryo excision enables the creation of axenic seedlings and Koch’s postulates testing of putative bacterial endophytes. Sci Rep. 2016;6:1–9. doi: 10.1038/srep25581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ridout M.E., Schroeder K.L., Hunter S.S. Priority effects of wheat seed endophytes on a rhizosphere symbiosis. Symbiosis. 2019;78:19–31. [Google Scholar]
- 14.Kaga H., Mano H., Tanaka F., Watanabe A., Kaneko S., Morisaki H. Rice seeds as sources of endophytic bacteria. Microb Environ. 2009;24:154–162. doi: 10.1264/jsme2.me09113. [DOI] [PubMed] [Google Scholar]
- 15.Huang Y., Kuang Z., Wang W., Cao L. Exploring potential bacterial and fungal biocontrol agents transmitted from seeds to sprouts of wheat. Biol Control. 2016;98:27–33. [Google Scholar]
- 16.Hartmann A., Fischer D., Kinzel L., Chowdhury S.P., Hofmann A., Baldani J.I. Assessment of the structural and functional diversities of plant microbiota: Achievements and challenges–A review. J Adv Res. 2019;19:3–13. doi: 10.1016/j.jare.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hegazi N., Hartmann A., Ruppel S. The plant microbiome: Exploration of plant-microbe interactions for improving agricultural productivity. J Adv Res. 2019;19:1–2. doi: 10.1016/j.jare.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zohary D., Hopf M. Oxford University Press; Oxford, UK: 1993. Domestication of plants in the Old World: the origin and spread of cultivated plants in West Asia, Europe and the Nile Valley. [Google Scholar]
- 19.Eckardt N.A. Evolution of domesticated bread wheat. Plant Cell. 2010;22:993. doi: 10.1105/tpc.110.220410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang L., Danzberger J., Schöler A., Schröder P., Schloter M., Radl V. Dominant groups of potentially active bacteria shared by barley seeds become less abundant in root associated microbiome. Front Plant Sci. 2017;8:1–12. doi: 10.3389/fpls.2017.01005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klaedtke S., Jacques M.A., Raggi L., Préveaux A., Bonneau S., Negri V. Terroir is a key driver of seed-associated microbial assemblages. Environ Microbiol. 2016;18:1792–1804. doi: 10.1111/1462-2920.12977. [DOI] [PubMed] [Google Scholar]
- 22.Bulgarelli D., Garrido-Oter R., Münch P.C., Weiman A., Dröge J., Pan Y. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17:392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pérez-Jaramillo J.E., Mendes R., Raaijmakers J.M. Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Molecul Biol. 2016;90:635–644. doi: 10.1007/s11103-015-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berg G., Raaijmakers J.M. Saving seed microbiomes. ISME J. 2018;12:1167–1170. doi: 10.1038/s41396-017-0028-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Truyens S., Weyens N., Cuypers A., Vangronsveld J. Bacterial seed endophytes: Genera, vertical transmission and interaction with plants. Environ Microbiol Rep. 2014;7:40–50. [Google Scholar]
- 26.Reif J.C., Zhang P., Dreisigacker S., Warburton M.L., Van Ginkel M., Hoisington D. Wheat genetic diversity trends during domestication and breeding. Theor Appl Gen. 2005;110:859–864. doi: 10.1007/s00122-004-1881-8. [DOI] [PubMed] [Google Scholar]
- 27.Joshi C.P., Nguyen H.T. Application of the random amplified polymorphic DNA technique for the detection of polymorphism among wild and cultivated tetraploid wheats. Genome. 1993;36:602–609. doi: 10.1139/g93-081. [DOI] [PubMed] [Google Scholar]
- 28.Fernández M.E., Figueiras A.M., Benito C. The use of ISSR and RAPD markers for detecting DNA polymorphism, genotype identification and genetic diversity among barley cultivars with known origin. Theor Appl Gene. 2002;104:845–851. doi: 10.1007/s00122-001-0848-2. [DOI] [PubMed] [Google Scholar]
- 29.Petersen G., Seberg O., Yde M., Berthelsen K. Phylogenetic relationships of Triticum and Aegilops and evidence for the origin of the A, B, and D genomes of common wheat (Triticum aestivum) Mol Phylo Evol. 2006;39:70–82. doi: 10.1016/j.ympev.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Wang J., Luo M.C., Chen Z., You F.M., Wei Y., Zheng Y. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013;198:925–937. doi: 10.1111/nph.12164. [DOI] [PubMed] [Google Scholar]
- 31.Bektas H., Hohn CE., Waines J.G. Characteristics of the root system in the diploid genome donors of hexaploid wheat (Triticum aestivum L.) Gen Res Crop Evol. 2017;64:1641–1650. [Google Scholar]
- 32.Singh K., Ghai M., Garg M., Chhuneja P., Kaur P., Schnurbusch T. An integrated molecular linkage map of diploid wheat based on a Triticum boeoticum x T. monococcum RIL population. Theor Appl Gen. 2007;115:301–312. doi: 10.1007/s00122-007-0543-z. [DOI] [PubMed] [Google Scholar]
- 33.Jauhar P.P., Peterson T.S. Synthesis and characterization of advanced durum wheat hybrids and addition lines with Thinopyrum chromosomes. J Hered. 2013;104:428–436. doi: 10.1093/jhered/ess143. [DOI] [PubMed] [Google Scholar]
- 34.Dvorak J., DiTerlizzi P., Zhang H.B., Resta P. The evolution of polyploid wheats: Identification of the A genome donor species. Genome. 1993;36:21–31. doi: 10.1139/g93-004. [DOI] [PubMed] [Google Scholar]
- 35.Valkoun J.J. Wheat pre-breeding using wild progenitors. Euphytica. 2001;119:17–23. [Google Scholar]
- 36.Badr A., Salamini F.M.K., Pozzi C., Effgen S., Rohde W. On the origin and domestication history of barley (Hordeum vulgare) Mol Biol Evol. 2012;17:499–510. doi: 10.1093/oxfordjournals.molbev.a026330. [DOI] [PubMed] [Google Scholar]
- 37.Alibrandi P., Cardinale M., Rahman M.M., Strati F., Ciná P., de Viana M.L. The seed endosphere of Anadenanthera colubrina is inhabited by a complex microbiota, including Methylobacterium spp. and Staphylococcus spp. with potential plant-growth promoting activities. Plant Soil. 2018;422:81–99. [Google Scholar]
- 38.Hurek T., Reinhold-Hurek B., Van Montagu M., Kellenberger E. Root colonization and systemic spreading of Azoarcus sp. strain BH72 in grasses. J Bacteriol. 1994;176:1913–1923. doi: 10.1128/jb.176.7.1913-1923.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mardis E.R. The impact of next-generation sequencing technology on genetics. Trends Genet. 2008;24:133–141. doi: 10.1016/j.tig.2007.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Lundberg D.S., Yourstone S., Mieczkowski P., Jones C.D., Dangl J.L. Practical innovations for high-throughput amplicon sequencing. Nature Meth. 2013;10:999–1002. doi: 10.1038/nmeth.2634. [DOI] [PubMed] [Google Scholar]
- 41.Claesson M.J., O’Sullivan O., Wang Q., Nikkilä J., Marchesi J.R., Smidt H. Comparative analysis of pyrosequencing and a phylogenetic microarray for exploring microbial community structures in the human distal intestine. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0006669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Engelbrektson A., Kunin V., Wrighton K.C., Zvenigorodsky N., Chen F., Ochman H. Experimental factors affecting PCR-based estimates of microbial species richness and evenness. ISME J. 2010;4:642–647. doi: 10.1038/ismej.2009.153. [DOI] [PubMed] [Google Scholar]
- 43.Caporaso J.G., Fierer N., Peña A.G., Goodrich J.K., Gordon J.I., Huttley G.A. QIIME allows analysis of high-throughput community sequencing data. Nature Meth. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rognes T., Flouri T., Nichols B., Quince C., Mahé F. VSEARCH: a versatile open source tool for metagenomics. Peer J. 2016;4 doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mercier C., Boyer F., Bonin A., Coissac E. In Programs and Abstracts of the SeqBio 2013 workshop. Abstract. 2013. November). SUMATRA and SUMACLUST: fast and exact comparison and clustering of sequences; pp. 27–29. [Google Scholar]
- 46.Quast C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.RStudio Team. (2016) RStudio: integrated development for R. [WWW document] URL http://www.rstudio.com/.
- 48.Wickham H. ggplot2 - Elegant Graphics for Data Analysis. J Stat Softw. 2017;77:2–5. [Google Scholar]
- 49.Oksanen, J., Kindt, R., and Legendre, P.O.B. (2017) vegan: Community Ecology Package. In: R package version 2.4–4 [WWW document] URL http://cran.r-project.org/ package=vegan.
- 50.Cardinale M., Grube M., Erlacher A., Quehenberger J., Berg G. Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ Microbiol. 2015;17:239–252. doi: 10.1111/1462-2920.12686. [DOI] [PubMed] [Google Scholar]
- 51.Agler M.T., Ruhe J., Kroll S., Morhenn C., Kim S.T., Weigel D. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016;14:1–31. doi: 10.1371/journal.pbio.1002352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toju H., Yamamoto S., Tanabe A.S., Hayakawa T., Ishii H.S. Network modules and hubs in plant-root fungal biomes. Interface. 2016;13:20151097. doi: 10.1098/rsif.2015.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Coombs J.T., Franco C.M.M. Isolation and identification of Actinobacteria from surface-sterilized wheat roots. Appl Environ Microbiol. 2003;69:5603–5608. doi: 10.1128/AEM.69.9.5603-5608.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Faust K., Raes J. CoNet app: Inference of biological association networks using Cytoscape. F1000Res. 2016;5:15–19. doi: 10.12688/f1000research.9050.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berry D., Widder S. Deciphering microbial interactions and detecting keystone species with co-occurrence networks. Front Microbiol. 2014;5:1–14. doi: 10.3389/fmicb.2014.00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sipahi H., Akar T., Yildiz M.A., Sayim I. Determination of genetic variation and relationship in Turkish barley cultivars by hordein and RAPD markers. Turkish J Field Crops. 2010;15:108–113. [Google Scholar]
- 57.Genetic diversity of Tunisian accessions of Aegilops geniculata Roth and durum wheats (Triticum durum Desf.) using RAPD markers. Mahjoub A., Abdellaoui R., Ben Naceur M., Ben, Brahim N., editors. Acta Bot Gall. 2010;157:3–12. [Google Scholar]
- 58.Mantzavinou A., Bebeli P.J., Kaltsikes P.J. Estimating genetic diversity in Greek durum wheat landraces with RAPD markers. Aust J Agric Res. 2005;56:1355–1364. [Google Scholar]
- 59.Abdellaoui R., Kadri K., Ben Naceur M., Ben Kaab L.B. Genetic diversity in some Tunisian barley landraces based on rapd markers. Pakistan J Bot. 2010;42:3775–3782. [Google Scholar]
- 60.Balbuena J.A., Míguez-Lozano R., Blasco-Costa I. PACo: A novel procrustes application to cophylogenetic analysis. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0061048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Althoff D.M., Segraves K.A., Johnson M.T.J. Testing for coevolutionary diversification: Linking pattern with process. Trends Ecol Evol. 2014;29:82–89. doi: 10.1016/j.tree.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 62.Doña J., Sweet A.D., Johnson K.P., Serrano D., Mironov S., Jovani R. Cophylogenetic analyses reveal extensive host-shift speciation in a highly specialized and host-specific symbiont system. Mol Phylogen Evol. 2017;115:190–196. doi: 10.1016/j.ympev.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 63.Paradis E., Schliep K. Ape 5.0: An environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics. 2019;35:526–528. doi: 10.1093/bioinformatics/bty633. [DOI] [PubMed] [Google Scholar]
- 64.Sweet A.D., Boyd B.M., Johnson K.P. Cophylogenetic patterns are uncorrelated between two lineages of parasites on the same hosts. Biol J Linnean Society. 2016;118:813–828. [Google Scholar]
- 65.Hutchinson M.C., Cagua E.F., Balbuena J.A., Stouffer D.B., Poisot T. paco: implementing Procrustean Approach to Cophylogeny in R. Meth Ecol and Evol. 2017;8:932–940. [Google Scholar]
- 66.Germida J.J., Siciliano S.D. Taxonomic diversity of bacteria associated with the roots of modern, recent and ancient wheat cultivars. Biol Fertil Soils. 2001;33:410–415. [Google Scholar]
- 67.Zachow C., Müller H., Tilcher R., Berg G. Differences between the rhizosphere microbiome of Beta vulgaris ssp. maritima-ancestor of all beet crops-and modern sugar beets. Front Microbiol. 2014;5:1–13. doi: 10.3389/fmicb.2014.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pérez-Jaramillo J.E., Carrión V.J., Bosse M., Ferrão L.F.V., De Hollander M., Garcia A.A.F. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017;11:2244–2257. doi: 10.1038/ismej.2017.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Leff J., Lynch R., Kane N., Fierer N. Plant domestication and the assembly of bacterial and fungal communities associated with strains of the common sunflower, Helianthus annuus. New Phytol. 2016;214:412–423. doi: 10.1111/nph.14323. [DOI] [PubMed] [Google Scholar]
- 70.Pérez-jaramillo J.E., Hollander M. De., Ramírez C.A., Mendes R., Raaijmakers J.M. Deciphering rhizosphere microbiome assembly of wild and modern common bean (Phaseolus vulgaris) in native and agricultural soils from Colombia. Microbiome. 2019;7:114. doi: 10.1186/s40168-019-0727-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Iannucci A., Fragasso M., Beleggia R., Nigro F., Papa R. Evolution of the crop rhizosphere: Impact of domestication on root exudates in tetraploid wheat (Triticum turgidum L.). Front Plant Sci. 2017;8:2124. doi: 10.3389/fpls.2017.02124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rengel Z. Genetic control of root exudation. Plant Soil. 2002;245:59–70. [Google Scholar]
- 73.Kuźniar A., Włodarczyk K., Grządziel J., Woźniak M., Furtak K., Gałązka A. New insight into the composition of wheat seed microbiota. Int J Mol Sci. 2020;21:1–18. doi: 10.3390/ijms21134634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu Y., Yan H., Zhang X., Zhang R., Li M., Xu T. Investigating the endophytic bacterial diversity and community structures in seeds of genetically related maize (Zea mays L.) genotypes. Biotech. 2020;10:1–10. doi: 10.1007/s13205-019-2034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raj G., Shadab M., Deka S., Das M., Baruah J., Bharali R. Seed interior microbiome of rice genotypes indigenous to three agroecosystems of Indo-Burma biodiversity hotspot. BMC Genomics. 2019;20:1–16. doi: 10.1186/s12864-019-6334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Campisano A., Ometto L., Compant S., Pancher M., Antonielli L., Yousaf S. Interkingdom transfer of the acne-causing agent, Propionibacterium acnes, from human to grapevine. Molecul Biol Evol. 2014;31:1059–1065. doi: 10.1093/molbev/msu075. [DOI] [PubMed] [Google Scholar]
- 77.Yousaf S., Bulgari D., Bergna A., Pancher M., Quaglino F., Casati P. Pyrosequencing detects human and animal pathogenic taxa in the grapevine endosphere. Front Microbiol. 2014;5:1–9. doi: 10.3389/fmicb.2014.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lugtenberg B., Kamilova F. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol. 2009;63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 79.Lidbury I.D.E.A., Murphy A.R.J., Scanlan D.J., Bending G.D., Jones A.M.E., Moore J.D. Comparative genomic, proteomic and exoproteomic analyses of three Pseudomonas strains reveals novel insights into the phosphorus scavenging capabilities of soil bacteria. Environ Microbiol. 2016;18:3535–3549. doi: 10.1111/1462-2920.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J., Zhang C., Yang J., Zhang R., Gao J., Zhao X. Insights into endophytic bacterial community structures of seeds among various Oryza sativa L. rice genotypes. Plant Grow Regul. 2019;38:93–102. [Google Scholar]
- 81.Sachdev D., Nema P., Dhakephalkar P., Zinjarde S., Chopade B. Assessment of 16S rRNA gene-based phylogenetic diversity and promising plant growth-promoting traits of Acinetobacter community from the rhizosphere of wheat. Microbiol Res. 2010;165:627–638. doi: 10.1016/j.micres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 82.Links M.G., Demeke T., Gräfenhan T., Hill J.E., Hemmingsen S.M., Dumonceaux T.J. Simultaneous profiling of seed-associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on Triticum and Brassica seeds. New Phytol. 2014;202:542–553. doi: 10.1111/nph.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wassermann B., Cernava T., Müller H., Berg C., Berg G. Seeds of native alpine plants host unique microbial communities embedded in cross-kingdom networks. Microbiome. 2019;7:1–12. doi: 10.1186/s40168-019-0723-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Castagna R., Gnocchi S., Perenzin M., Heun M. Genetic variability of the wild diploid wheat Triticum urartu revealed by RFLP and RAPD markers. Theor Appl Gen. 1997;94:424–430. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.