Abstract
Social science genetics is concerned with understanding if, how, and why genetic differences between human beings are linked to differences in behaviours and socio-economic outcomes. Our review discusses the goals, methods, challenges and implications of this research endeavor. We survey how the recent developments in genetics are beginning to provide social scientists with a powerful new toolbox they can use to better understand environmental effects and illustrate this with several substantive examples. Furthermore, we examine how medical research can benefit from genetic insights into social-scientific outcomes and vice versa. Finally, we discuss the ethical challenges of this work and clarify several common misunderstandings and misinterpretations of genetic research on individual differences.
According to the dictionary definition, social science focuses on understanding “the institutions and functioning of human society and … the interpersonal relationships of individuals as members of society.”1 The phrase “social science genetics” might, therefore, appear oxymoronic, as genetics traditionally focuses on phenomena (such as the translation of DNA sequence differences into proteins) that are squarely outside the typical purview of the social sciences. As an economist and a psychologist who both study human behaviour and social phenomena, such as people’s risky behaviours, we would consider the actions and interactions among sub-atomic particles as obviously irrelevant to our work – what do quarks have to do with understanding, for example, who starts a new business? What, then, makes DNA any different?
The difference is that decades of twin research – and more recent research using genome wide measures of DNA – have shown that nearly every aspect of human individual differences is partly heritable.2,3 That is, differences between people in personality, educational attainment, income, risk tolerance, well-being, occupational choice, financial decision making, political ideology, sexual behaviour, as well as physical and psychiatric health, longevity, and number of children are all affected in some way by differences in their inherited DNA sequence variation.3–6 Genetic differences between people, therefore, have incontrovertible relevance for all branches of the social sciences that are concerned with or affected by individual differences in behaviour and outcomes.
Research in human molecular genetics has made tremendous progress in the past decade.5 Much of this work has focused on health outcomes, but these developments have begun to influence the social sciences as well.7 Attempts to link genetics to social and behavioural outcomes are often met with greater skepticism and concerns about potential consequences than medical applications of genetic research (Box 1).8 These concerns have to be taken seriously, partly because they are based on a long, troublesome history of abusing genetic research to justify discrimination and atrocities, including forced sterilization and even genocide. Continued vigilance about the misappropriation of genetics is necessary (Box 2).
Box 1. Three Common Concerns about Social Science Genetics.
(1). Do social scientists have the training to work with genetic data?
The word “genetics” might conjure up scientific activities that most social scientists consider alien to their training. But the uses of genetic data that lend themselves most readily to social science research actually resemble the typical tools of social science, with a heavy reliance on statistical methods and natural experiments. Unfortunately, social scientists still often miss training on how they can utilize genetic data and methods. Curricula that fill these knowledge gaps are needed, and graduate programs at an increasing number of top research universities are moving in this direction.
(2). Does genetic research imply bio-determinism?
The goal of the natural sciences is often to identify universal “laws of nature” that are invariant across time and place. In contrast, nearly all of the causal regularities discovered by social scientists are (a) probabilistic rather than deterministic, and (b) exception-ridden rather than universal. The observation that human behaviour is influenced by genetic differences between people should not be misinterpreted as bio-determinism.12 Although DNA variants do not change after conception, their potential influence on social and behavioural outcomes can vary across different environments.
(3). Is genetic research a threat to social justice?
A major obstacle to admitting genetics into the social sciences is fear that their integration “runs along a knife edge, with cliffs of eugenic risk on either side.”8 This fear is well-founded, as genetic research has been used to justify numerous crimes against humanity.13 But, as the political philosopher John Rawls summarized,14 “The natural distribution is neither just nor unjust…. These are simply natural facts. What is just and unjust is the way that institutions deal with these facts.” We would expand this idea: Which genetic variants are present in which people, and what they do within a person’s cellular machinery, are natural facts. How these genetic variants are associated, in a particular time and a particular place, with outcomes such as education or wealth are social facts. How these natural and social facts should be used – by governments, schools, businesses, hospitals, etc. – is the appropriate locus of social justice concerns.
Box 2. Genetics and Scientific Racism.
Scientific racism invokes genetic differences to explain racial disparities in health, wealth, power, and life opportunities as inevitable and insurmountable.34,35 Peeling apart genetic inquiry from scientific racism requires attention to four important points:
(1). Race and genetic ancestry are not the same thing.
Racial and ethnic categories are correlated with genetic ancestry, but race is not reducible to genetics.36 The U.S. Census categories for race/ethnicity do not have a neat 1:1 correspondence with genetic diversity. More importantly, what is considered a “race” is socially-defined and culturally-specific.37
(2). Genetic research has a profound Euro-centric bias.
The vast majority of social science research is conducted with “the WEIRDest people in the world” (Western, Educated, Industrialized, Rich, and Democratic),38 and social science genetics is no exception. Over 75% of participants in GWAS are from European populations, and the exclusion of non-European participants from genomic research has the potential to exacerbate health disparities.39
(3). Polygenic scores are not comparable or perfectly portable across ancestry groups.
If the environments of two groups are not identical, the same genetic endowment may give rise to drastically different outcomes. And, differences in ancestry can lead to differences in LD patterns and minor allele frequencies, such that GWAS results from one ancestry group are, at best, only partially portable to another. For example, a PGS that captures ≈11% of the variation of educational attainment in white Americans captures only ≈2% of the variation among African Americans.40 Neither can average PGSs be meaningfully compared across populations.41
(4). Geneticists have a special responsibility to communicate their results responsibly.
Extremist groups pay close attention to developments in genetic research and can sometimes show a surprising level of technical sophistication.42 Given the potential for rampant and pernicious misinformation, researchers have an ethical responsibility to communicate those results clearly to the general public.43
We believe that modern social science genetics can and should play a central role in overcoming these misconceptions, by illuminating how genetic variation contributes to human diversity and by showcasing the myriad ways in which genetic and environmental factors are entangled with each other and interact. Furthermore, integrating molecular genetics into the social sciences can deliver richer, more precise answers to old questions in psychology, sociology, economics, and related fields. Ultimately, the greatest impact from integrating genetics into the social sciences will probably not come from simply applying new tools to old questions, but from the potential to change how people think about the world around them, allowing them to ask new questions and to pursue new answers that would not have been feasible before. For example, the realization that success in life is partly the result of a genetic lottery raises new questions not only about underlying mechanisms, but also about fairness and what a desirable distribution of wealth in a society should look like. In this sense, genetics is akin to other new tools that have made inroads into the social sciences in the past few decades (e.g., experiments, game theory, or neuroscience), all of which have contributed to a more realistic understanding of human behaviour and the functioning of societies.
In this review, we will describe the main tools of statistical genetics, discuss some of the underlying assumptions and implications of these tools, and illustrate their current use in the social sciences with concrete examples. We restrict our discourse here to genetics but note that there is also a growing literature that uses other types of biological data (e.g., the metabolome, the epigenome, hormones, neurotransmitters) to address social-scientific research questions.
The toolbox of statistical genetics
The ambition to bring the insights and tools of genetics into the social sciences is not new. For decades, behavioural genetics (a psychology-focused subfield of the larger field of social science genetics) has been estimating the heritability of psychological traits, such as personality, intelligence, and psychopathology.3 The conclusion of this line of research was simple: Everything is heritable2, even ostensibly “environmental” variables such as life stress, divorce, or harsh parenting. The ubiquitous heritability of individual differences can pose a serious threat to inferences about the impact of specific environments, as these environments, particularly when they are provided by genetic relatives of the focal person, cannot be considered exogenous to the genotype of the person.9 This critique was – and continues to be – valid. Now, the combination of genetic data and family data (e.g., genotyped samples of trios) offers exciting new possibilities to tackle this and many other challenges.10,11
Genome-wide association studies (GWAS)
Thanks to rapid technological progress, people can now be cheaply genotyped on arrays that measure specific DNA sequences that commonly vary between people. This advance has led to the emergence of large-scale biobanks15 and consumer genetics companies,16 explosively increasing the sample sizes available for genetic research.5 As sample sizes increased, human molecular genetics and social science genetics went through a painful – but, ultimately, highly productive – paradigm change. During the mid-2000s, many researchers embraced a “candidate gene” approach, focusing on a small handful of genetic variants which were selected on the basis of a priori reasoning about their possible functional significance. However, as genotyping became cheaper and more people were genotyped, it became clear that most studies reporting associations between single genetic variants and behavioural phenotypes did not replicate.7,17 Clearly, something was wrong.
But thanks to growing data availability, an alternative approach became feasible – genome-wide association studies (GWAS). A GWAS systematically scans the entire genome for possible associations with an outcome, examining millions of single nucleotide polymorphisms (SNPs), i.e. variations in individual DNA “letters,” or base pairs. GWASs are typically conducted in samples with similar ancestries, most commonly people of European descent (see Box 2). Researchers run a separate regression of the outcome of interest on each SNP separately to deal with the fact that there are typically much more SNPs than individuals in a particular dataset, thereby ignoring any correlations between SNPs (i.e., linkage disequilibrium, or LD18). This approach yields some association signal from all observed SNPs that are in LD with potentially causal genetic variants, which may or may not be observed directly in the available genetic data.19
GWASs typically control for technical parameters, sex, age, and – importantly – multiple principal components (PCs) of the genetic data, which are supposed to act as proxies for historical migration patterns and long-term ancestry (i.e., genetic population structure).20 This is intended to control for spurious genetic associations with outcomes that vary for non-genetic reasons in sub-populations that also vary in gene frequencies. Large-scale GWAS initiatives are often based on pre-registered analysis plans; they rigorously control for multiple testing by imposing extremely stringent p-value thresholds, and they typically report replication results for novel findings based on evidence from different, independently collected samples in the same paper. All of the above dramatically decrease the risk of finding false positives in GWAS compared to earlier candidate gene studies.
The first GWASs of social scientific outcomes delivered humbling results, either coming up empty-handed or only finding a few genes, each of which accounted for mere fractions of a percent of variance.21 This ostensible failure, however, was key to understanding why the candidate gene approach was flawed: Social and behavioural outcomes, like fertility, education, personality, and risk taking, are massively polygenic.22 That is, they are influenced by thousands upon thousands of genetic loci scattered throughout the genome, each with a tiny effect. In contrast, candidate gene studies were operating under the wrong assumption that a few genes with large effects are responsible for the heritability of most traits, and were therefore conducted with sample sizes that were hopelessly underpowered.22
Over the past 5 years, GWAS sample sizes have rapidly grown from tens of thousands to millions. As a result of this growing statistical power to detect tiny effects on highly polygenic traits, the GWAS approach has now yielded hundreds of replicable associations of specific genetic markers with social scientific outcomes.5,6 The flood of discoveries have motivated big online repositories23 and interactive atlases comparing the genetic architecture of thousands of traits.24,25 Scientists are (finally) beginning to open the black box of heritability.26
However, the threat of finding spurious genetic associations due to unobserved variable bias remains a serious challenge.27–29 There is no guarantee that using samples with similar ancestry and adding genetic PCs as control variables will eliminate all forms of spurious genetic signal. For example, genetic PCs don’t capture rare variants that are only weakly correlated with common SNPs, they seem to perform less well in small samples,27 and they can both under- and overcontrol for potential confounds that are associated with genetic variation among people.30
New statistical methods get constantly developed to tackle the challenge of spurious associations more rigorously, such as linear mixed models (which were previously used in the animal breeding literature).31,32 Yet, even the most advanced methods rely on assumptions such as additivity that can be violated in practice. The gold standard to correct for confounds in genetic association studies are research designs that use family data and exploit the random assortment of alleles during meiosis for the identification of causal genetic effects (e.g., using dizygotic twins, siblings with the same biological parents, or trios of mother-father-child).10,33 The availability of such data keeps growing and will enable within-family GWAS and follow-up analyses for many heritable traits in the future. At the moment, however, the vast majority of GWAS are based on population samples or case-control cohorts that are not entirely immune to unobserved variable bias.
Genetic correlations
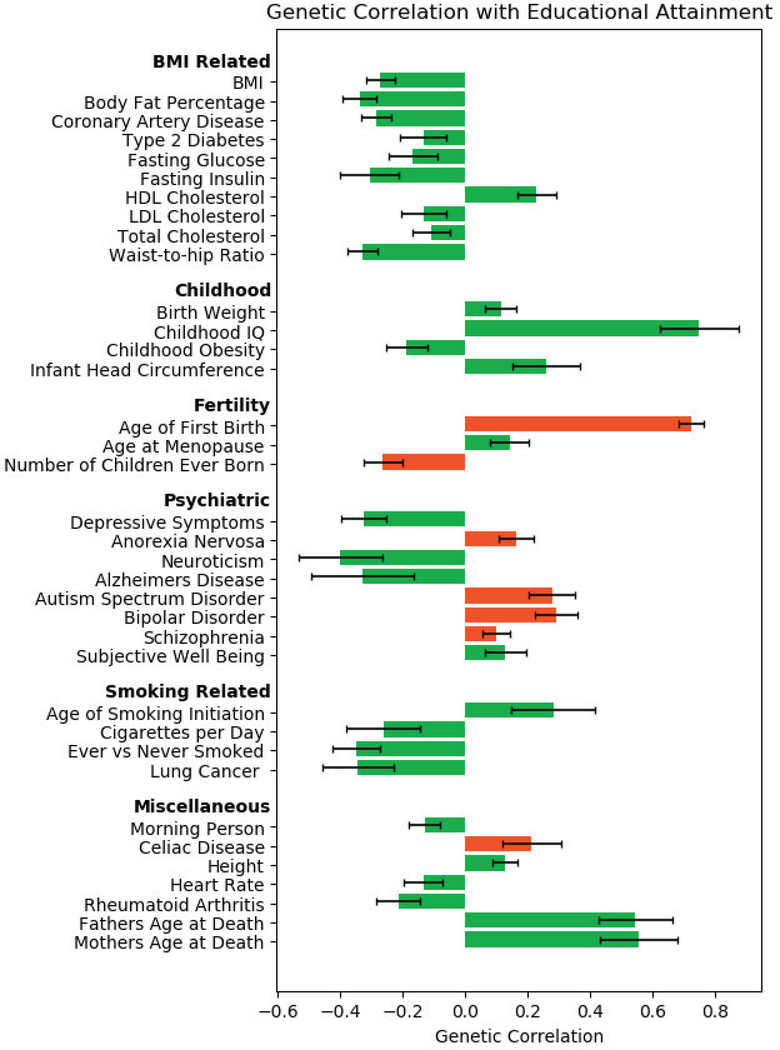
GWAS data makes it possible to calculate genetic correlations, including – surprisingly – between pairs of traits that have never been measured in the same sample. Specifically, the technique of bivariate LD-score regression uses the GWAS summary statistics from two traits to estimate co-heritability in a remarkably robust way.44,45 As an illustration, Figure 1 summarizes genetic correlations between educational attainment and a variety of other traits from LD Hub, an online repository of genetic correlations.46 EA-associated genes40 are associated with traits across the lifespan, from birthweight (+), infant head circumference (+) and childhood IQ (+), to BMI-related traits (−), depressive symptoms (−), neuroticism (−), smoking (−) in adulthood, all the way to risk for lung cancer (−), Alzheimer’s disease (−), and longevity (+) (as implied by parent’s age at death). However, the effects of EA-increasing genes are not universally positive: They are also linked to reduced reproductive success (fewer children born, most likely due to reproductive decisions rather than decreased fecundity) and an increased risk for several psychiatric disorders.
Figure 1. Genetic correlations of educational attainment with traits across the entire lifespan.
Note: Genetic correlations of educational attainment (EA)40 with 196 traits were computed using bivariate LD-score regression and GWAS summary statistics with varying sample sizes obtained from LDHub (http://ldsc.broadinstitute.org/ldhub/). The Figure only shows a subset of all results for EA that are significant after Bonferroni-correction (p < 0.05/196 = 2.5×10−4, two-sided tests). Error bars represent 95% confidence intervals. Green and red colors represent positive and negative genetic relationships, respectively, between EA and health.
These findings underscore how tightly social-scientific outcomes, such as education, are linked with health. At the same time, it is important to remember that genetic correlations are not, by themselves, informative about causal mechanisms, nor do they necessarily imply direct, “inside the skin” pleiotropic effects of genes on two traits.47 They may also reflect indirect, possibly environmentally-mediated pathways (e.g., high childhood IQ → higher EA → less smoking → reduced risk for lung cancer).
Moving beyond atlases of pairwise genetic correlations, multivariate approaches further capitalize on genetic similarities among traits by jointly analyzing GWAS summary statistics from several traits simultaneously. Multi-trait analysis of GWAS (MTAG) and genomic structural equation modeling (Genomic SEM) are two recently developed methods that are quickly gaining popularity.48,49 A multivariate analyses can boost the power to detect genes associated with a trait by “borrowing” relevant information from other, genetically-related traits (e.g., neuroticism as a proxy for depressive symptoms and vice versa).50 Other work is using multivariate GWAS to identify specific genes and biological pathways that confer general vulnerability to psychiatric disorders versus genes that operate uniquely on a specific symptom or syndrome.49 Overall, the shift toward multivariate GWAS parallels an earlier development in twin studies, which went from estimating the heritability of single traits in isolation, to estimating the extent to which the same (unobserved) genes influenced a variety of human phenotypes.51
Polygenic scores
Answering biological questions that might, on their face, appear irrelevant to social scientists – how many polymorphisms in the genome affect human phenotypes? – can turn out to be crucial for developing tools that will, in fact, be broadly useful for social science.52 Specifically, as it has become clear that the effects of individual variants are tiny, methods to aggregate the effects of many variants into a single composite, a polygenic score (PGS), have proliferated. In polygenic scoring, researchers take results from a GWAS of a specific trait and apply them in a new sample, weighting each person’s genetic variants by the effect size from the GWAS and summing across the variants. The resulting PGS is therefore an index that summarizes current estimates of additive genetic influences towards a particular phenotype.53,54
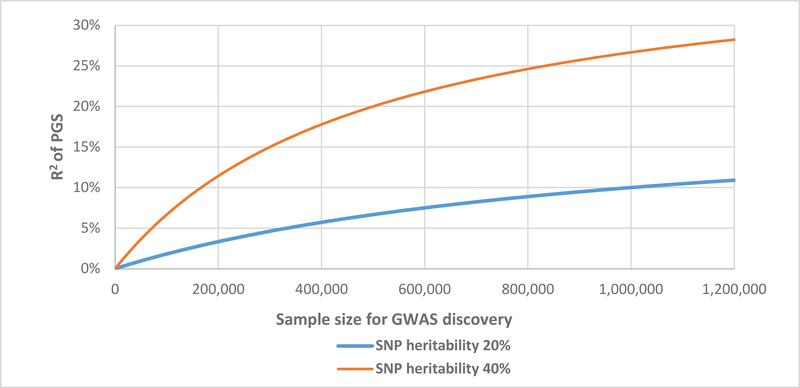
Because PGSs aggregate over many genetic markers, they capture a much larger share of the variance of the trait of interest than any one variant on its own. The accuracy of PGS primarily depends on the heritability of the trait (+), the GWAS sample size (+), the polygenicity of the trait (−), and whether the genetic architecture of the trait varies across different environments (−).53 So far, theoretical projections of the accuracy of PGS have been borne out by the data pretty well. Figure 2 illustrates this by comparing the theoretically-expected55 predictive accuracy of a PGS in a hold-out sample (assuming 200,000 independent causal SNPs) with results from studies of increasing sample size for educational attainment (EA156, EA257, EA340) and body height (Height158, Height259). As GWAS sample sizes increased, the number of genome-wide significant loci increased from three in EA156 to 1,271 in EA3.40
Figure 2: The influence of GWAS sample size on the accuracy of polygenic scores for two genetically complex traits with assumed SNP heritability of 20% and 40%.
Note: The two solid lines show theoretical expectations assuming 200,000 causal genetic markers for both traits. De Vlaming et al.55 estimate the SNP heritability of height and years of schooling with 43.3% (SE = 1.8%) and 16.4% (SE = 1.7%), respectively. In practice, the accuracy of polygenic scores depends on many technical parameters such as the number of SNPs in the score and which prediction sample is used. The empirical results for years of schooling in this figure are for PGS based on all HapMap 3 SNPs, using the Swedish Twin Registry as a prediction sample for EA1 and the Health and Retirement Study (HRS) as a prediction sample for EA2 and EA3. The empirical results for height are based on the HRS prediction sample with SNP p-value thresholds of <5×10−5 for Height1 and <0.001 for Height2.59 All statistical tests referred to here were 2-sided.
The R2 of the EA340 PGS mirrors the effect size seen for traditional social science variables, such as the relationship between family income and educational attainment. As a result, such PGSs are beginning to be useful for follow-up studies with much smaller sample sizes, including experimental studies, policy and intervention studies, and longitudinal datasets with deep phenotypes.60–63 A researcher who has a PGS that captures 5% of the variance of the phenotype she is interested in needs only ≈260 individuals if she wants to have 90% statistical power at α = 0.05 (2-sided test) to find an association of the phenotype with that PGS.
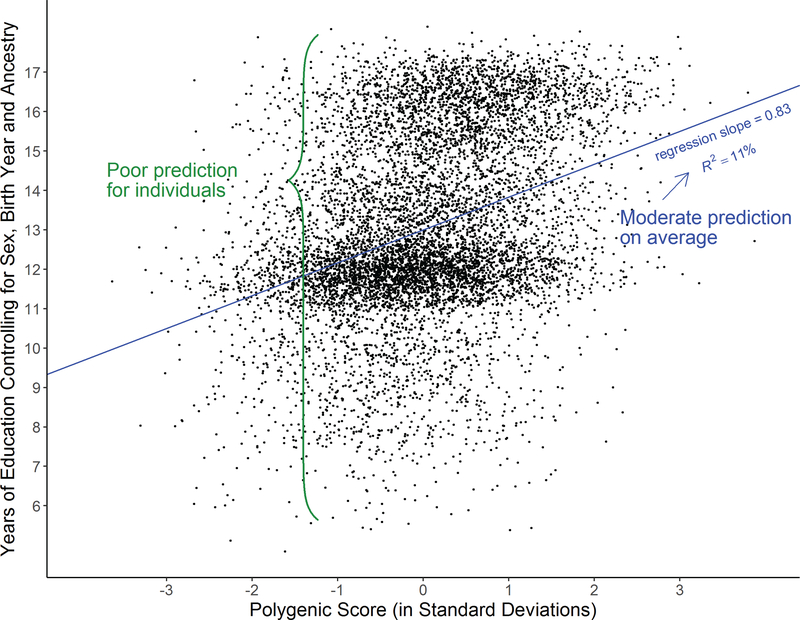
At the same time, it is important to remember that even the best currently available PGS for behavioural outcomes cannot make accurate predictions for the outcome of any specific individual. Figure 3 illustrates this by plotting the EA3 PGS against actual years of schooling in a U.S. sample of individuals with European ancestries. The relationship between the two variables is positive and statistically highly significant (p = 2.2×10−16, 2-sided test). The PGS accounts for ≈11% of the variance in the years of schooling after residualizing this variable for sex, birth year, and the first 10 genetic PCs. Yet, we also see that, for almost any single value of the PGS, almost every level of actual education is observed. Even for PGS values that are 2 standard deviations above or below the sample mean, we observe everything from high school drop-outs to people with PhDs.
Figure 3: The relationship between a polygenic score for educational attainment and actual years of schooling in the Health and Retirement Study.
Note: The polygenic score for educational attainment (EA) was constructed using GWAS estimates from a sample of N = 1,123,243 individuals.40 The prediction sample was restricted to individuals of European ancestries (N = 8,638, Health and Retirement Study). EA was measured in typical years of schooling required to obtain the highest academic degree of an individual. EA was residualized for birth year, sex, and the first 10 principal components from the genetic data and then regressed on the polygenic score.
It is also important to remember that PGS are not a “clean” way to separate biological from non-biological factors that contribute to differences in phenotypes. GWAS results are not entirely immune to unobserved (e.g., environmental) confounds, such as parenting or neighborhood characteristics, and genetic influences are often conditional on and/or mediated by environmental channels.64 Thus, PGSs may exhibit different predictive accuracy even among members of the same ancestry group that vary from each other in sex or socio-economic status.65
Nevertheless, PGS have a variety of useful and exciting applications in the social sciences, including the possibilities to add them as control variables to boost statistical power in experiments,56 reduce unobserved heterogeneity,66 investigate gene-environment interactions,61 study environmental factors mediating the effect of genes on the outcome of interest,67 and tease apart environmental and genetic channels of intergenerational transmission.10,11 One particularly useful aspect is that once genetic data has been collected in a sample, it is in principle possible to construct PGS in that sample for all traits for which GWAS have ever been conducted,6 opening up the possibility to control for and to investigate relationships that would have been practically impossible otherwise (e.g., is a genetic predisposition for Alzheimer’s disease associated with brain anatomy in infancy or with the tendency to purchase more complete health insurance?).
Although PGS are often frustrating for biologists who are interested in specific genomic mechanisms, our perhaps controversial position is that PGS can hold the most utility for social scientists, precisely because they aggregate across lower-level mechanisms and re-focus inquiry back onto the behaviour of the whole human organism in her or his environment.60 The feasibility of integrating PGSs is aided by the fact that GWAS summary statistics are often freely available in the public domain.24 And, publicly available datasets that include genetic information have begun to release PGS for several traits to researchers who do not have access to the raw genetic data or lack the expertise to construct the scores themselves.68
Integrating genetic data and family-based study designs
Although molecular genetic data is often perceived as supplanting twin/family designs as the workhorses of social science genetics, the combination of measured genotypes and samples with family structure provides the most compelling study designs.64 Two types of family relationships are particularly noteworthy. First, dizygotic (DZ) twins or siblings can be used to estimate the within-family association between genetic variants (or PGS) and an outcome. This analysis takes advantage of the fact that a parent has two copies of each genetic locus (called alleles), and these alleles are randomly assigned to offspring during the process of making eggs or sperm. Which version of the parental genotype someone inherits, versus which version a sibling inherited, is the outcome of a true natural lottery. Because genotypes are assigned randomly with respect to all other variables, an association between sibling differences in PGS and sibling differences in phenotype is powerful evidence that the PGS is tapping genetic variants with a causal influence on the phenotype. This is a good starting point for investigating relevant causal mechanisms that will might involve malleable environmental pathways.
Second, if genetic data for trios of mother, father, and offspring are available, one can decompose the parental genotypes into two parts -- the alleles transmitted to the offspring vs. the non-transmitted alleles. The non-transmitted alleles of the parent, then, are analogous to the genotype of an adoptive parent: If they are correlated with the phenotype of the offspring (an “indirect genetic effect” or “genetic nurture”), this association cannot be due to genetic transmission from parent-to-child and must be mediated via environmental channels, such as parenting style or socioeconomic advantage.10 This “virtual parent” design is conceptually similar to the logic of adoption studies or children-of-twin studies, in that it peels apart genetic and environmental pathways for intergenerational transmission.10,11 At the same time, conditional on the genotypes of the parents, the directly-transmitted alleles of the child are the outcome of a natural lottery that can be used for causal analyses, similar to the logic of within-sibling comparisons. Recent methodological work is considering the extent to which genetic nurture biases SNP-based heritability estimates and estimates from sibling fixed-effects models.69
Third, the combination of family-based study designs and genetic data offers new, powerful approaches for causal inference using Mendelian Randomization (MR)70,71 and related approaches.66,72–74 For example, results from large-scale GWAS on within-family differences would be immune to potential confounds from subtle differences in ancestry or unobserved environmental factors that correlate with family genotypes, thereby ruling out an important source of bias in MR-like analyses.75
Substantive applications
We now turn to some examples of what we have already learnt from applying the social science genetics toolbox in five substantive areas: (1) intergenerational transmission of human capital, (2) social mobility over the lifespan, (3) genetic associations with demographic variables (fertility, mortality, and migration), (4) gene × environment interactions, and (5) the interconnections between social processes and disease processes. These research areas are of broad interest to scientists working in many different fields and focus on aspects of human functioning that resist bio-reductionism and yet are illuminated with biological data.
Intergenerational transmission of human capital
Human capital is transmitted from generation to generation,76 and understanding how “nature” and “nurture” shape children’s resemblance to their parents can inform efforts to reduce inequalities that tend to persist over time. For example, studies of trios from several countries found that a PGS of the EA-associated alleles that parents did not transmit to their offspring are nevertheless associated with offspring EA, thereby providing new evidence for the importance of environmental mechanisms and parenting.10,11,77,78 Such study designs can be extended to identify specific environmental variables and processes that mediate the effects of untransmitted parental genotypes on their children,67 thereby providing scientists with a promising new strategy for understanding the mechanisms that lead to inequalities in wealth and health.
Intragenerational social mobility
The combination of measured genotypes and family structure has also been used to examine intragenerational social mobility. In these studies, a PGS of education-associated genetic variants is used as a type of tracer dye: Just as tracking the progress of ingested barium allows a radiologist to gain a higher-resolution picture of the digestive tract’s twists and turns, tracking individuals with a high or low PGS allows social scientists to gain a higher-resolution picture of the lifecourse twists and turns that ultimately produce high or low social attainments in adulthood. The developmental pathways toward greater social mobility are already evident early in childhood – the EA PGS is also associated with earlier achievement of developmental milestones and faster reading development.63 Looking across the lifespan, people born with a higher number of education-associated variants show greater upward mobility, relative to their family-of-origin, in occupational status, income, and educational attainment – even when they are being compared to their siblings.79 The EA PGS is also associated with wealth at retirement, with a one SD increase in polygenic score estimated to be worth $137,000 in 2010 dollars. This wealth association persists even controlling for education and labor income, and operates in part through better financial decision-making.80
Evidence that children’s genotypes are associated with their educations, occupations, and financial success in adulthood should not be interpreted to mean that children are genetically determined to be rich or poor (Box 1). And, following Holland’s distinction between the “effects of causes” versus the “causes of effects”,81 reverse inferences that people are poor because of their genetics are entirely unwarranted. In fact, evidence from five longitudinal studies across three continents showed that children from rich families with low PGS scores still have more socioeconomic success as adults than children from poor families with high PGS scores.79
Genetics and demography: Fertility, mortality, migration
Over generations, fertility, mortality, and migration shape the size and genetic distribution of a population. At the same time, genetic differences within a population are associated with all three of these key demographic variables. Twin studies have long established that time to reproductive maturity, reproductive behaviour, and completed fertility are all heritable,82 and more recently, large-scale GWASs have found multiple specific loci associated with fertility-relevant traits and behaviours.83–85 Genetic research on fertility has been used to illuminate the mechanisms underlying the association between parental age and offspring risk for mental disease, as genetic risk for schizophrenia has been associated with both early and late age at first birth.86,87 Earlier work using children-of-twins similarly suggested that part of the elevated burden of mental health problems in children of adolescent mothers was due to transmission of genetic liabilities affecting both fertility behaviours and psychopathology.88
One active and politically-sensitive area of research is the relationship between education and fertility. Genetic variants associated with education are also associated with a lower number of children born, resulting in declines in the average education-associated PGS in the 20th century.89,90 A variety of twin/family studies have probed whether education operates causally to delay childbearing and sexual behaviour, above and beyond shared genetic influences, with mixed results.91,92 Interestingly, two variables that are heritable and have robust epidemiological associations with fertility behaviour – marital history93 and religiousness94 – have received almost no attention in molecular genetic research, and represent an untapped opportunity for integrating genetics into classic questions in the social sciences.
At the other end of the lifespan, some genetic loci have also been discovered for mortality risk, as imputed by the lifespan of one’s parents.95 Finally, migration, either to another country or within a country, has turned out to be linked to genetic differences in intriguing, and sometimes troubling, ways. People with a higher PGS for EA were found to be more likely to immigrate to other countries from New Zealand or to leave former coal-mining areas in the U.K., perhaps to seek better educational and occupational opportunities elsewhere.63,96 In contrast, people who stayed behind in economically-depressed coal mining regions in the U.K. tend to have lower PGS scores of EA and also carry more genetic risk factors for obesity, smoking, and coronary artery disease.96 In fact, genetic differences across geographical regions in the U.K. are so systematic that one can conduct a GWAS on neighborhood characteristics (e.g., the Townsend index -- a measure derived from registry data that reflects regional variation in over-crowding, unemployment, and lack of home/car ownership) using standard adjustments against population stratification such as genetic PCs or linear mixed models, and still find many genome-wide significant loci.97
Again, this does not mean that social outcomes like neighborhood poverty are inevitable or determined by biology (Box 1). Rather, any social process that unequally concentrates educational and economic opportunity in some places, but not others, will induce geographical variation in genotypes, because people choose (or lack the capability to choose) their place of residence on the basis of their own genetically-influenced preferences, abilities, and characteristics. This coupling between genetics and geography complicates efforts to draw causal inferences about how the characteristics of places shape inequalities in health and other life outcomes.98
Gene × environment and gene × intervention interactions
Earlier iterations of gene × environment (G × E) interaction work, either using candidate genes or latent components of genetic variance estimated in a twin model, have been criticized for being vulnerable to bias and false positives.17,99,100 Recent work to examine gene × intervention (G × I) interactions is a promising reinvigoration of the more general G × E topic, as such work examines robust measures of genotype (e.g., PGSs estimated from large-scale GWAS rather than candidate genes) and interventions that can be reasonably assumed to be exogenous (e.g., policy shocks).
A G × I analysis can test whether intervention effects are larger or smaller for people who are genetically more likely to develop the outcome that is targeted by the intervention, thus addressing three key questions about the intervention: (a) does it serve a high-need population segment? (b) will it shrink existing inequalities or amplify them? and (c) can its delivery be personalized? For example, an intervention could be particularly impactful for people at high genetic risk, therefore shrinking inequalities by serving a high-need segment of the population. Alternatively, the intervention could operate equivalently across levels of genetic risk, suggesting few potential gains from targeting the intervention to specific people on the basis of genetic characteristics (although including genetic information might improve the estimate of the intervention main effect, by accounting for residual heterogeneity56). Or, the intervention could deliver the most benefit to those who are already at lower risk, a “Matthew Effect” that improves the average level of functioning in a population but also exacerbates inequalities.101 This result would point to the need to develop new interventions for high-risk segments of the population.102
One of the recent studies of G × E showed that a policy reform in the UK, which increased compulsory schooling by one year, improved obesity-related health outcomes and lung function in mid-adulthood. The reform had particularly positive effects for individuals who carried more genetic risk factors for a high BMI, effectively reducing health disparities by counteracting genetic risks.61 Beyond understanding the impact of that specific policy reform, this study is also valuable as a novel empirical illustration of an old idea: Genetic differences between people do not necessarily restrict the possibilities for intervention.103
Part of the reason that genetic associations might be dependent on environmental factors, e.g., political regime, policy interventions, economic conditions, or school environment, is that genetic differences might be expressed via modifiable, context-dependent environmental channels.12,104 For example, genetic predispositions toward height and psychomotor speed could lead to self-selection into a sports club; the training and reinforcement received by coaches, parents, and peers, then, could amplify these initial differences to produce associations between genetic variants and basketball “talent.”105 In a different macro-environmental context, where these training and reinforcement experiences are not available (i.e., in a different “cafeteria of experience”106), genetic associations would be disrupted.
Tracing these sorts of gene-environment interplay, in which people with different genotypes are systematically more likely to elicit different environmental responses, to select different peer groups, to engage in different training experiences, and more generally, to carve different environmental niches for themselves, has long been a topic of interest in the twin/family literature,107–109 and we anticipate that research with PGSs will offer new opportunities for testing hypotheses in this area.
However, we also note some methodological challenges in estimating G × E and G × I interactions. First, PGS are typically constructed from large-scale GWAS results that estimate the average effects of SNPs across samples and environments, missing or attenuating potential environment-specific genetic effects. Second, PGS tend to capture only a part of the relevant genetic influences on a trait.66 For both of these reasons, G × E and G × I will be biased towards zero and provide only a lower bound estimate of the true effects. Third, because GWAS results are typically not immune to population structure, genetic nurture, or other unobserved environmental confounds (see above), PGS may capture some of the environmental causes of an outcome. And fourth, since genes and environments are often correlated, G × E and G × I may be endogenous terms in a regression equation. All of the above makes the interpretation of G × E and G × I interactions more difficult unless GWAS results from within-family analyses are used and combined with reasonably exogenous variations in E or I.
Implications of "disease" genetics for social processes (and vice versa)
GWAS is perhaps most commonly conceptualized as a tool for understanding the biology of psychiatric and physical diseases, and advocates for investing in GWAS often emphasize the relevance of results for drug development.5,110 But genetic discoveries for “diseases” turn out to be relevant for a much broader array of social phenomena in “normal” or non-clinical populations. For example, genetic risks for physical health conditions, like coronary artery disease, are associated with long-term wealth, education, and self-rated health in hold-out samples.111 Similarly, the schizophrenia PGS is associated with the escalation of illicit drug use in typically-developing university students.112
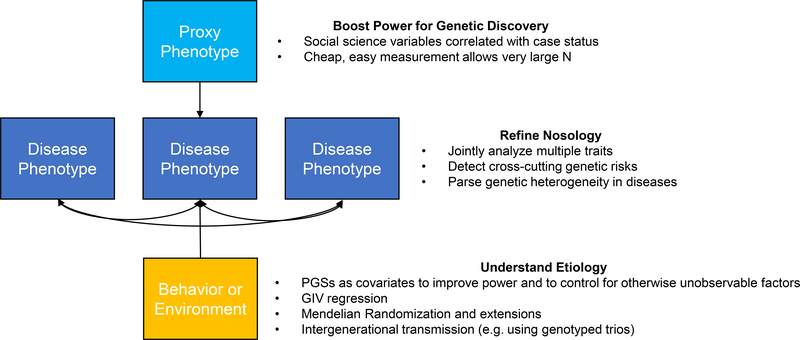
Conversely, genetic discoveries for “normal” phenotypes can be useful for understanding medical and psychiatric diseases. GWAS results for EA were used to parse the genetic heterogeneity of schizophrenia into disease subtypes with different genetic architectures and biological underpinnings, which might benefit from different treatment.62 As another example, the first robust genetic associations with major depressive disorder (MDD) among individuals of European descent were found using self-reports of subjective well-being and neuroticism as proxy-phenotypes.113,114 Recent multivariate work has investigated the joint genetic architecture of a “well-being spectrum” encompassing depression, neuroticism, life satisfaction, and positive affect.50 The genetics of subjective well-being have also been leveraged to understand its relationship to cardiometabolic health and body size.115 These results demonstrate that there is no clear boundary between social science genetics and medical genetics -- investments in either area of research will enrich the other (Figure 4).
Figure 4.
How medical science can benefit from social science genetics.
Conclusion
The rapidly increasing availability of genetic data now allows scientists to unravel the genetic underpinnings of individual differences in social, behavioural, and health outcomes. Although not without caveats, the current workhorses of statistical genetics (e.g., GWAS, LD score regression, polygenic scores) are useful for social scientists whose primary interests lie in understanding effects of environments, such as parenting, policies, or interventions, that might lead to or entrench inequalities. Genetic effects influence most dimensions of individual differences that social scientists care about, and genetic differences between people are tightly interwoven with environmental differences that social scientists study. This is both a challenge as well as an opportunity for the social science: Ignoring the relevance of genes would mean ignoring an important part of reality, which could lead to erroneous and misleading conclusions about environmental or behavioural effects. Thus, the social sciences are incomplete without genetics, and they can benefit from genetically-informed study designs in their quest for accurate and comprehensive answers to the questions they are asking. The tools and the data to do so keep emerging at a rapid pace. Now is the time to begin training social scientists to understand and use these new tools.
Clearly, the genetic revolution raises a host of new ethical, social, and legal challenges that are important and urgent to address. Genetic data are possibly the most personal piece of information about an individual that exist, and the scope of potential uses and abuses is rapidly increasing, ranging from “genetic entertainment” by learning about one’s ancestry or creating “customized” music playlists, to potential applications in insurance and labor markets, marketing campaigns, dating apps, criminal justice proceedings, the testing and selection of IVF embryos, to biohacking and engineered differences in human DNA. The genetic revolution will change our lives and our societies, whether or not we want it to. As social scientists, we have expertise in considering the ethical, cultural, political, economic, environmental, and historical forces that shape human lives and societies. As a consequence, we can make valuable contributions to the unfolding conversation about the uses and abuses of human genomics. But to do so, we need to take genetics seriously.
Acknowledgements
We thank Casper Burik for preparing Figure 1 and the Social Science Genetic Association Consortium (https://www.thessgac.org/) for Figure 3. Koellinger was financially supported by an ERC consolidator grant (647648 EdGe). Harden was supported by the Jacobs Foundation, the Templeton Foundation, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD) grants R01-HD083613 and 5-R24-HD042849 (to the Population Research Center at the University of Texas at Austin). The funders had no role in the conceptualization, preparation or decision to publish this work.
Footnotes
Competing interests
The authors declare no competing interests.
Data availability
The genetic correlations reported in Figure 1 are based on publicly available GWAS summary statistics on LDHub (http://ldsc.broadinstitute.org/ldhub/). The Health and Retirement Study data in Figure 3 can be accessed via dbGaP and the University of Michigan.
References
- 1.Definition of SOCIAL SCIENCE. Available at: https://www.merriam-webster.com/dictionary/social%20science. (Accessed: 1st November 2018)
- 2.Turkheimer E Three laws of behavior genetics and what they mean. Curr. Dir. Psychol. Sci. 9, 160–164 (2000). [Google Scholar]
- 3.Polderman TJC et al. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet. 47, 702–709 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Benjamin DJ et al. The promises and pitfalls of genoeconomics. Annu. Rev. Econom. 4, 627–662 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visscher PM et al. 10 years of GWAS discovery: Biology, function, and translation. Am. J. Hum. Genet. 101, 5–22 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buniello A et al. The NHGRI-EBI GWAS Catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. 47, D1005–D1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Freese J The arrival of social science genomics. Contemporary Sociology: A Journal of Reviews 47, 524–536 (2018). [Google Scholar]
- 8.Comfort N Nature still battles nurture in the haunting world of social genomics. Nature 553, 278 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Turkheimer E & Paige Harden K Behavior Genetic Research Methods. in Handbook of Research Methods in Social and Personality Psychology 159–187 (Cambridge University Press, 2014). [Google Scholar]
- 10.Kong A et al. The nature of nurture: Effects of parental genotypes. Science 359, 424–428 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Koellinger PD & Harden KP Using nature to understand nurture: Genetic associations show how parenting matters for children’s education. Science 359, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jencks C Heredity, environment, and public policy reconsidered. Am. Sociol. Rev. 45, 723–736 (1980). [PubMed] [Google Scholar]
- 13.Kevles DJ In the Name of Eugenics: Genetics and the Uses of Human Heredity. (Harvard University Press, 1995). [Google Scholar]
- 14.Rawls J A Theory of Justice. (Belknap Press: An Imprint of Harvard University Press, 1999). [Google Scholar]
- 15.Bycroft C et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 562, 203–209 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Servick K Can 23andMe have it all? Science 349, 1472–1477 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Duncan LE, Pollastri AR & Smoller JW Mind the gap: Why many geneticists and psychological scientists have discrepant views about gene–environment interaction (G×E) research. Am. Psychol. 69, 249–268 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reich DE et al. Linkage disequilibrium in the human genome. Nature 411, 199–204 (2001). [DOI] [PubMed] [Google Scholar]
- 19.The 1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Price AL et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Visscher PM, Brown MA, McCarthy MI & Yang J Five years of GWAS discovery. Am. J. Hum. Genet. 90, 7–24 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chabris CF, Lee JJ, Cesarini D, Benjamin DJ & Laibson DI The fourth law of behavior genetics. Curr. Dir. Psychol. Sci. 24, 304–312 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rapid GWAS of thousands of phenotypes for 337,000 samples in the UK Biobank. Neale lab Available at: http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank. (Accessed: 6th November 2018)
- 24.Kyoko Watanabe DP Atlas of GWAS Summary Statistics. GWAS Atlas (2017). Available at: http://atlas.ctglab.nl/. (Accessed: 6th November 2019)
- 25.Global Biobank Engine. Available at: https://biobankengine.stanford.edu/. (Accessed: 6th November 2019)
- 26.Manolio TA et al. Finding the missing heritability of complex diseases. Nature 461, 747–753 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sohail M et al. Polygenic adaptation on height is overestimated due to uncorrected stratification in genome-wide association studies. Elife 8, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berg JJ et al. Reduced signal for polygenic adaptation of height in UK Biobank. eLIFE 8, e39725 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haworth S et al. Apparent latent structure within the UK Biobank sample has implications for epidemiological analysis. Nat. Commun. 10, 333 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson DJ et al. Is population structure in the genetic biobank era irrelevant, a challenge, or an opportunity? Hum. Genet. (2019). doi: 10.1007/s00439-019-02014-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loh P-R et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 47, 284–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Zaitlen NA, Goddard ME, Visscher PM & Price AL Advantages and pitfalls in the application of mixed-model association methods. Nat. Genet. 46, 100–106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies NM et al. Within family Mendelian randomization studies. Hum. Mol. Genet. (2019). doi: 10.1093/hmg/ddz204 [DOI] [PubMed] [Google Scholar]
- 34.Herrnstein RJ & Murray C The Bell Curve: Intelligence and Class Structure in American Life (A Free Press s Book). (Free Press, 1996). [Google Scholar]
- 35.Jensen A How much can we boost IQ and scholastic achievement. Harv. Educ. Rev. 39, 1–123 (1969). [Google Scholar]
- 36.Yudell M, Roberts D, DeSalle R & Tishkoff S Taking race out of human genetics. Science 351, 564–565 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Smedley A & Smedley BD Race as biology is fiction, racism as a social problem is real: Anthropological and historical perspectives on the social construction of race. Am. Psychol. 60, 16–26 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Henrich J, Heine SJ & Norenzayan A The weirdest people in the world? Behav. Brain Sci. 33, 61–83; discussion 83–135 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Martin AR et al. Clinical use of current polygenic risk scores may exacerbate health disparities. Nat. Genet. 51, 584–591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee JJ et al. Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nat. Genet. 50, 1112–1121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martin AR et al. Human demographic history impacts genetic risk prediction across diverse populations. Am. J. Hum. Genet. 100, 635–649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Panofsky A & Donovan J Genetic ancestry testing among white nationalists: From identity repair to citizen science. Soc. Stud. Sci. 306312719861434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Novembre J & Barton NH Tread lightly interpreting polygenic tests of selection. Genetics 208, 1351–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bulik-Sullivan B et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 47, 1236–1241 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee JJ, McGue M, Iacono WG & Chow CC The accuracy of LD Score regression as an estimator of confounding and genetic correlations in genome-wide association studies. Genet. Epidemiol. 42, 783–795 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng J et al. LD Hub: a centralized database and web interface to perform LD score regression that maximizes the potential of summary level GWAS data for SNP heritability and genetic correlation analysis. Bioinformatics 33, 272–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gage SH, Davey Smith G, Ware JJ, Flint J & Munafò MR G = E: What GWAS can tell us about the environment. PLoS Genet. 12, e1005765 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Turley P et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 50, 229–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grotzinger AD et al. Genomic structural equation modelling provides insights into the multivariate genetic architecture of complex traits. Nature Human Behaviour 3, 513–525 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baselmans BML et al. Multivariate genome-wide analyses of the well-being spectrum. Nat. Genet. (2019). doi: 10.1038/s41588-018-0320-8 [DOI] [PubMed] [Google Scholar]
- 51.Loehlin JC The Cholesky approach: A cautionary note. Behav. Genet. 26, 65–69 (1996). [Google Scholar]
- 52.Torkamani A, Wineinger NE & Topol EJ The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Dudbridge F Power and predictive accuracy of polygenic risk scores. PLoS Genet. 9, e1003348 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wray NR et al. Research Review: Polygenic methods and their application to psychiatric traits. J. Child Psychol. Psychiatry 55, 1068–1087 (2014). [DOI] [PubMed] [Google Scholar]
- 55.de Vlaming R et al. Meta-GWAS accuracy and power (MetaGAP) calculator shows that hiding heritability is partially due to imperfect genetic correlations across studies. PLoS Genet. 13, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rietveld CA et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science 340, 1467–1471 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Okbay A et al. Genome-wide association study identifies 74 loci associated with educational attainment. Nature 533, 539–542 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wood AR et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat. Genet. 46, 1173–1186 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yengo L et al. Meta-analysis of genome-wide association studies for height and body mass index in ~700,000 individuals of European ancestry. Human Molecular Genetics 27, 3641–3649 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Belsky DW & Harden KP Phenotypic annotation: Using polygenic scores to translate discoveries from genome-wide association studies from the top down. Curr. Dir. Psychol. Sci. (2019). doi: 10.1177/0963721418807729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Barcellos SH, Carvalho LS & Turley P Education can reduce health differences related to genetic risk of obesity. Proc. Natl. Acad. Sci. U. S. A. 115, E9765–E9772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bansal V et al. Genome-wide association study results for educational attainment aid in identifying genetic heterogeneity of schizophrenia. Nat. Commun. 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Belsky DW et al. The genetics of success. Psychol. Sci. 27, 957–972 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Young AI, Benonisdottir S, Przeworski M & Kong A Deconstructing the sources of genotype-phenotype associations in humans. Science 365, 1396–1400 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mostafavi H, Harpak A, Conley D, Pritchard JK & Przeworski M Variable prediction accuracy of polygenic scores within an ancestry group. bioRxiv 629949 (2019). doi: 10.1101/629949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Diprete TA, Burik CAP, Koellinger PD & Wachter KW Genetic instrumental variable regression: Explaining socioeconomic and health outcomes in nonexperimental data. Proceedings of the National Academy of Sciences (2018). doi: 10.1073/pnas.1707388115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wertz J et al. Genetics of nurture: A test of the hypothesis that parents’ genetics predict their observed caregiving. Dev. Psychol. 55, 1461–1472 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.SSGAC Polygenic Score Data | Health and Retirement Study. Available at: https://hrs.isr.umich.edu/news/ssgac-polygenic-score-data. (Accessed: 11th November 2019)
- 69.Young AI et al. Relatedness disequilibrium regression estimates heritability without environmental bias. Nat. Genet. 50, 1304–1310 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Davey Smith G & Hemani G Mendelian randomization: Genetic anchors for causal inference in epidemiological studies. Hum. Mol. Genet. 23, R89–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brumpton B et al. Within-family studies for Mendelian randomization: avoiding dynastic, assortative mating, and population stratification biases. bioRxiv (2019). doi: 10.1101/602516 [DOI] [PMC free article] [PubMed]
- 72.O’Connor LJ & Price AL Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat. Genet. 50, 1728–1734 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhu Z et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Verbanck M, Chen C-Y, Neale B & Do R Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koellinger PD & de Vlaming R Mendelian randomization: the challenge of unobserved environmental confounds. Int. J. Epidemiol. 48, 665–671 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Black SE, Devereux PJ & Salvanes KG Why the apple doesn’t fall far: Understanding intergenerational transmission of human capital. Am. Econ. Rev. 95, 437–449 (2005). [Google Scholar]
- 77.Bates TC et al. The nature of nurture: Using a virtual-parent design to test parenting effects on children’s educational attainment in genotyped families. Twin Res. Hum. Genet. 21, 73–83 (2018). [DOI] [PubMed] [Google Scholar]
- 78.Liu H Social and genetic pathways in multigenerational transmission of educational attainment. Am. Sociol. Rev. 83, 278–304 (2018). [Google Scholar]
- 79.Belsky DW et al. Genetic analysis of social-class mobility in five longitudinal studies. Proc. Natl. Acad. Sci. U. S. A. 115, E7275–E7284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Barth D, Papageorge NW & Thom K Genetic endowments and wealth inequality. J. Polit. Econ. (2020). doi: 10.3386/w24642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Holland PW Statistics and causal inference. J. Am. Stat. Assoc. 81, 945–960 (1986). [Google Scholar]
- 82.Rodgers J & Kohler H-P The Biodemography of Human Reproduction and Fertility. (Springer Science & Business Media, 2002). [Google Scholar]
- 83.Barban N et al. Genome-wide analysis identifies 12 loci influencing human reproductive behavior. Nat. Genet. 48, 1462–1472 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Day FR et al. Genomic analyses identify hundreds of variants associated with age at menarche and support a role for puberty timing in cancer risk. Nat. Genet. 49, 834–841 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Day FR et al. Physical and neurobehavioral determinants of reproductive onset and success. Nat. Genet. 48, 617–623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mehta D et al. Evidence for genetic overlap between schizophrenia and age at first birth in women. JAMA Psychiatry 73, 497–505 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ni G, Gratten J, Wray NR, Lee SH & Schizophrenia Working Group of the Psychiatric Genomics Consortium. Age at first birth in women is genetically associated with increased risk of schizophrenia. Sci. Rep. 8, 10168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Harden KP et al. A behavior genetic investigation of adolescent motherhood and offspring mental health problems. J. Abnorm. Psychol. 116, 667–683 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beauchamp JP Genetic evidence for natural selection in humans in the contemporary United States. Proc. Natl. Acad. Sci. U. S. A. 113, 7774–7779 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kong A et al. Selection against variants in the genome associated with educational attainment. Proc. Natl. Acad. Sci. U. S. A. 114, E727–E732 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rodgers JL et al. Education and cognitive ability as direct, mediating, or spurious influences on female age at first birth: behavior genetic models fit to Danish twin data. AJS 114 Suppl, S202–32 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tropf FC & Mandemakers JJ Is the association between education and fertility postponement causal? The role of family background factors. Demography 54, 71–91 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Jocklin V, McGue M & Lykken DT Personality and divorce: a genetic analysis. J. Pers. Soc. Psychol. 71, 288–299 (1996). [DOI] [PubMed] [Google Scholar]
- 94.D’Onofrio BM, Eaves LJ, Murrelle L, Maes HH & Spilka B Understanding biological and social influences on religious affiliation, attitudes, and behaviors: a behavior genetic perspective. J. Pers. 67, 953–984 (1999). [DOI] [PubMed] [Google Scholar]
- 95.Pilling LC et al. Human longevity: 25 genetic loci associated in 389,166 UK biobank participants. Aging 9, 2504–2520 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Abdellaoui A et al. Genetic correlates of social stratification in Great Britain. Nat Hum Behav (2019). doi: 10.1038/s41562-019-0757-5 [DOI] [PubMed] [Google Scholar]
- 97.Hill WD et al. Molecular genetic contributions to social deprivation and household income in UK Biobank. Curr. Biol. 26, 3083–3089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Belsky DW et al. Genetics and the geography of health, behaviour and attainment. Nat Hum Behav 3, 576–586 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.van der Sluis S, Posthuma D & Dolan CV A note on false positives and power in G × E modelling of twin data. Behav. Genet. 42, 170–186 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Duncan LE & Keller MC A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am. J. Psychiatry 168, 1041–1049 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ceci SJ & Papierno PB The rhetoric and reality of gap closing: when the‘ have-nots’ gain but the‘ haves’ gain even more. Am. Psychol. 60, 149 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Fletcher JM Why have tobacco control policies stalled? Using genetic moderation to examine policy impacts. PLoS One 7, e50576 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goldberger AS Heritability. Economica 46, 327–347 (1979). [Google Scholar]
- 104.Bronfenbrenner U & Ceci SJ Nature-nuture reconceptualized in developmental perspective: A bioecological model. Psychol. Rev. 101, 568–586 (1994). [DOI] [PubMed] [Google Scholar]
- 105.Dickens WT & Flynn JR Heritability estimates versus large environmental effects: The IQ paradox resolved. Psychological Review 108, 346–369 (2001). [DOI] [PubMed] [Google Scholar]
- 106.Lykken DT, Bouchard TJ Jr, McGue M & Tellegen A Heritability of interests: a twin study. J. Appl. Psychol. 78, 649–661 (1993). [DOI] [PubMed] [Google Scholar]
- 107.Tucker-Drob EM & Harden KP Early childhood cognitive development and parental cognitive stimulation: evidence for reciprocal gene-environment transactions. Dev. Sci. 15, 250–259 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Klahr AM, Thomas KM, Hopwood CJ, Klump KL & Burt SA Evocative gene-environment correlation in the mother-child relationship: a twin study of interpersonal processes. Dev. Psychopathol. 25, 105–118 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Scarr S & McCartney K How people make their own environments: a theory of genotype greater than environment effects. Child Dev. 54, 424–435 (1983). [DOI] [PubMed] [Google Scholar]
- 110.Wray NR et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 50, 668–681 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wehby GL, Domingue BW & Wolinsky FD Genetic risks for chronic conditions: Implications for long-term wellbeing. J. Gerontol. A Biol. Sci. Med. Sci. 73, 477–483 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mallard TT, Harden KP & Fromme K Genetic risk for schizophrenia is associated with substance use in emerging adulthood: an event-level polygenic prediction model. Psychol. Med. 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Okbay A et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 48, 624–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Turley P et al. Multi-trait analysis of genome-wide association summary statistics using MTAG. Nat. Genet. 50, 229–237 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wootton RE et al. Evaluation of the causal effects between subjective wellbeing and cardiometabolic health: mendelian randomisation study. BMJ 362, k3788 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]