Abstract
With the rapid development of sequencing technologies, the characteristics and functions of circular RNAs (circRNAs) in different tissues, and their underlying pathophysiological mechanisms, have been identified. circRNAs are significantly enriched in the brain and are continually expressed from the embryonic stage to the adult stage in rats. Previous studies have reported that certain circRNAs are differentially expressed in glioma and regulate a number of biological processes, such as cell proliferation, metastasis and oncogenesis of glioma. Furthermore, certain circRNAs have been associated with tumor size, World Health Organization tumor grade and poor prognosis in patients with glioma. It has been hypothesized that circRNAs may be involved in the onset and progression of glioma through transcriptional regulation, protein translation and binding to microRNAs. These properties and functions suggest the potential of circRNAs as prognostic biomarkers and therapeutic targets for glioma. For the present review, published studies were examined from PubMed, Embase, Cochrane Central and the reference lists of the retrieved articles. The aim of the present review was to summarize the progress of circRNA research in glioma, discuss the potential diagnostic and prognostic values, and the roles of circRNAs in glioma, and provide a novel theoretical basis and research concepts for the prediction, diagnosis and treatment of glioma.
Keywords: circRNAs, glioma, miRNA sponges, biomarker, targeted therapy
1. Introduction
Glioma is the most prevalent type of primary tumor of the central nervous system and accounts for >75% of malignant brain tumors (1,2). Histologically, glial-derived tumors, or gliomas, share characteristics of normal glial cells, glial or neural precursors and stem cells, and are generally named based on these similarities (3–5). Recent advances in molecular understanding have provided new insights and novel molecular subclassification of gliomas. These molecular subtypes include subependymal giant cell astrocytoma, pilocytic astrocytoma, epithelioid glioblastoma, oligodendroglioma and mesenchymal glioblastoma (2,3). Early detection of gliomas may be difficult due to their deep location in the brain and lack of obvious clinical symptoms at the early stages (4,6). Despite the modest survival benefit of standard therapies, including radiation, chemotherapy and surgery, for patients with glioma, locoregional failure in the form of persistent or recurrent disease still occurs in the majority of patients (7–11). Therefore, innovative biomarkers and more specific targeted treatments for eradicating glioma are required.
Non-coding RNAs (ncRNAs) are a class of functional RNAs that do not encode proteins, including microRNAs (miRNAs/miRs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) (12–14). Previous studies have reported that the majority of miRNAs and lncRNAs have regulatory functions, including regulation of the evolution of stem cells, cardiomyocytes and epithelial cells, and regulation of the translation, stabilization and degradation of mRNA (15–19). With the development of several state-of-the-art technologies, such as the next generation of high-throughput sequencing techniques, gene silencing and gene editing, emerging evidence has demonstrated that, not only miRNAs and lncRNAs, but also circRNAs, have regulatory functions, and are associated with a variety of diseases (20–24). circRNAs can interact with protein complexes, RNA molecules and DNA molecules to exert their effects, and regulate a wide variety of physiological and pathological processes (25–27).
circRNAs are a type of ncRNA characterized by a circular configuration that consists of either prototypical spliceosome-mediated or lariat-type splicing, which provides a strong stability and resistance to RNases (28,29). Numerous studies have shown that several circRNAs are abnormally expressed in human glioma and may be associated with the carcinogenesis of glioma. For example, circ-PRKCI (30), circ-CPA4 (31), circ-CDC45 (32), circ-SCAF11 (33), circ-POSTN (34) and circ-PCMTD1 (35) have been found to act as tumor suppressors or promoters in glioma progression by regulating the proliferation, invasion, metastasis, angiogenesis and apoptosis of glioma cells. The aim of the present review was to describe the potential functional implications of circRNAs and to summarize the current knowledge on circRNA research in human glioma, including the expression, regulatory pathways and treatment potential of circRNAs, as well as the current challenges of circRNA studies in human glioma.
2. Brief overview of circRNAs
General characteristics of circRNAs
circRNAs were first identified in plants, where it was revealed that they encode subviral agents by electron microscopy in 1976 (36). At the beginning of the 21st century, researchers reported the existence of circRNAs in a wide range of eukaryotic cells and tissue samples from humans and other species (37). With the development of circRNA identification and selection techniques, it was reported that circRNAs consist of diverse combinations of sequences and domains, including intronic, exonic, intergenic and antisense chain regions (38). Subsequently, the detected types and quantity of circRNAs have increased. According to the components of the parental genes, circRNAs can be grouped into three main subtypes: Circular intronic RNAs (ciRNAs), exonic circRNAs (ecircRNAs) and exon-intron circRNAs (EIciRNAs) (39–42).
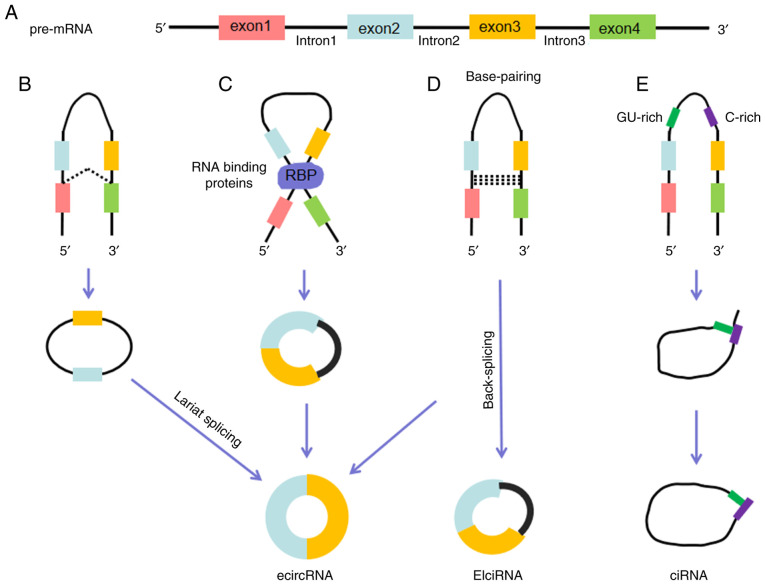
circRNAs are resistant to degradation by ribonucleases due to the lack of a free 5′ or 3′ end, which ensures their stability compared with their linear counterparts (43). circRNAs are extensively present in body fluids, and are covalently closed single-stranded RNA molecules that are generated via multiple cyclization modes, including lariat-driven cyclization, RNA-binding protein (RBP)-driven cyclization, base-pairing-driven cyclization and ciRNA biosynthesis (44,45). The cyclization modes and specific types of circRNAs are shown in Fig. 1. Selective circularization combined with alternative splicing can generate numerous circRNAs from one gene (20).
Figure 1.
Biogenesis of circRNAs. (A) Pre-mRNA contains introns and exons, and can generate a variety of circRNAs through different splicing mechanisms. (B) Lariat-driven cyclization. When a pre-mRNA is spliced, the 3′ hydroxyl of the upstream exon interacts with the 5′ phosphate of the downstream exon to form a covalent linkage, producing a lariat that contains exons and introns. The 2′ hydroxyl of the 5′ intron reacts with the 5′ phosphate of the 3′ intron, followed by an interaction between the 3′ hydroxyl of the 3′ exon and the 5′ phosphate of the 5′ exon, through which an ecircRNA is formed. (C) RBP-driven cyclization. RBPs can promote the interaction of the downstream intron and upstream intron, causing the formation of an ecircRNA. (D) Base-pairing-driven cyclization. The downstream introns and upstream introns are paired based on inverse-repeating or complementary sequences. The introns are removed or retained to form ecircRNA or EIciRNA. (E) Biosynthesis of ciRNA. Formation of ciRNAs mainly depends on a 7-nt GU-rich element and an 11-nt C-rich element to escape debranching and exonucleolytic degradation. circRNA, circular RNA; RBP, RNA-binding protein; ecircRNA, exonic circRNA; EIciRNA, exon-intron circRNA; ciRNA, circular intronic RNA.
Research on circRNAs has confirmed numerous features of circRNAs, including diversity, specificity and conservation (46). In comparison with linear RNAs, the majority of circRNAs are hyper-conserved at the nucleotide-level among different species (47). In 2021, Varela-Martinez et al (48) identified a large number of sheep circRNAs, of which 1,606 were homologous to human circRNAs. Based on RNA sequencing from differentiated cell lines and brain tissues, it was demonstrated that numerous circRNAs are tissue- and stage-specifically expressed (49,50). Due to their unusual stability, highly conserved sequence and expression specificity, circRNAs may serve as candidates for clinical biomarker research.
Biological functions of circRNAs
A growing body of studies have revealed that circRNAs can be produced in any region of the genome and can be cycled by multiple cyclization modes (51–53). Those features create multiformity in the sequence and length of circRNAs, eventually resulting in the multiplicity of circRNA functions. The biological functions of circRNAs in a wide variety of human neoplasms include acting as miRNA sponges, interacting with proteins, regulating gene splicing or transcription, translating proteins, or peptide and epigenetic regulation. The most typical role of circRNAs is to serve as miRNA sponges (54–57). Mechanistically, miRNAs can block the translation of target mRNA via complementary pairing with the target mRNA 3′-untranslated region (UTR), which impacts the stability of the target mRNA and modulates the expression in the cell nucleus via combining with gene promoters (52,53). However, circRNAs can suppress the expression of miRNAs via adsorption and, simultaneously, influence the molecular level of downstream target genes (54). Li et al (55) reported that circ_PSD3 may serve as a miR-637 sponge to modulate the expression of downstream target genes in papillary thyroid carcinoma. In addition, other circRNAs, such as circRNA_0005529 and circ-TGFBR2, have been reported to interact with miR-527 and miR-107 to regulate human gastric cancer cell proliferation and metastasis, and nasopharyngeal carcinoma cell viability, respectively (58,59). Notably, the same circRNA may even have the opposite function in different tumor cells, through targeting different miRNAs. For example, circ-FOXO3 has been reported to target miR-23a to inhibit esophageal squamous cell cancer proliferation, but may also target miR-29a-3p to promote prostate cancer progression (60,61). Notably, numerous circRNAs that do not possess miRNA-binding sites still have a wide range of biological functions, indicating that circRNAs have their own additional regulatory functions. Previous studies have revealed that circRNAs may interact with RBPs or other proteins to form circRNA-protein complexes, which may impact the function and mechanisms of action of binding proteins (62,63). For example, circ-FOKX2 may integrate with Y-box binding protein 1 and heterogeneous nuclear ribonucleoprotein K to facilitate the upregulation of the oncogenes NUF2 and pyridoxal kinase, ultimately accelerating pancreatic ductal adenocarcinoma progression (64). This form of action of circRNAs has also been reflected in the significant downregulation of circ-DCUN1D4 in lung adenocarcinoma (65). Furthermore, hsa_circ_0008367 has been shown to interact with the RBP ALKBH5, thus regulating ferroptosis in hepatocellular carcinoma cells (66).
Beyond the aforementioned two functions, circRNAs can also modulate gene transcription and alternative splicing (67,68). Conn et al (69) demonstrated that a circRNA derived from exon 6 of the SEPALLATA3 (SEP3) gene increased the abundance of the homology exon-skipped alternative splicing variant and combined with its homology DNA locus, thereby affecting the splicing efficiency of SEP3 mRNA. Furthermore, previous studies identified that EIciRNAs, such as circ-PAIP2 and circ-EIF3J, may constitute complexes with U1 small nuclear ribonucleoprotein (snRNP) via specific RNA-RNA interactions to shape EIciRNA-U1snRNP complexes, which can interact with the polymerase II transcription complex to promote transcription of the parental gene (70–72). According to recent studies, numerous circRNAs have been identified in the cytoplasm, indicating that they may be translated into proteins (73,74). Further studies revealed that cap-independent translation of circRNAs can occur via internal ribosome entry site 119 or following the incorporation of the m6A RNA modification in the 5′-UTR. Wu et al (75) demonstrated that circ-SMO encoded a fresh protein, SMO-193aa, which may drive the tumorigenicity of glioma cells. In addition, Zheng et al (76) revealed that circ-PPP1R12A can carry an open reading frame, which coded a protein known as circ-PPP1R12A-73aa. Mechanistically, circ-PPP1R12A-73aa facilitated the progression of colon cancer via stimulating the Hippo-YAP signal pathway.
3. circRNAs and glioma
Numerous studies have demonstrated that several oncogenic and anti-oncogenic circRNAs are abnormally expressed in human glioma compared with their expression levels in normal tissues or tumor-adjacent tissues. For example, the expression levels of circ-FBXW7 and circ-AKT3 were lower in glioma cell lines and cancer tissues from patients with glioma (77,78). By contrast, circ-CPA4 (31), circ-CDC45 (32) and circ-POSTN (34) have been revealed to be highly expressed in glioma cell lines and cancer tissues from patients with glioma. Mechanistically, these abnormally expressed circRNAs may modulate the migration, apoptosis, proliferation, invasion and angiogenesis of glioma, mainly through acting as a sponge targeting miRNAs. The main examples have been summarized in Table I. A meaningful role may be played by circRNAs in the carcinogenesis of glioma. The following subsections address the effect of these differentially expressed circRNAs on human glioma.
Table I.
Upregulated or downregulated circRNAs, and their functions and mechanisms in glioma.
| Authors, year | circRNA | Sample | Expression change | Function | Possible mechanism | (Refs.) |
|---|---|---|---|---|---|---|
| Yang et al, 2018 | circ-FBXW7 | Glioma tissues and glioma cells | Down | Tumorigenicity (−) | circ-FBXW7/FBXW7-185aa | (77) |
| Xia et al, 2019 | circ-AKT3 | Glioma tissues and glioma cells | Down | Tumorigenicity (−) | circ-AKT3/AKT3-174aa | (78) |
| Wu et al, 2021 | circ-SMO | Glioma tissues and glioma cells | Up | Tumorigenicity (+) | circ-SMO/SMO-193aa | (75) |
| Shi et al, 2019 | circ-0014359 | Glioma tissues and glioma cells | Up | Proliferation (+) | circ-0014359/miR-153/p-AKTser473 | (85) |
| Chen et al, 2020 | circ_PTN | Glioma cells | Up | Proliferation (+) | circ_PTN/miR-122/SOX6 | (86) |
| Peng et al, 2019 | circCPA4 | Glioma tissues and glioma cells | Up | Proliferation (+) Prognosis | circCPA4/let-7/CPA4 | (31) |
| He et al, 2020 | circ-MAPK4 | Glioma tissues and glioma cells | Up | Apoptosis (−) | circ-MAPK4/miR-125a-3p/p-p38/MAPK | (92) |
| Cao et al, 2019 | hsa_circ_0037251 | Glioma cells | Up | Apoptosis (−) | hsa_circ_0037251/miR-1229-3p/ mTOR | (93) |
| Chi et al, 2019 | circRNA-104075 | Glioma cells | Up | Apoptosis (−) | circRNA-104075/Bcl-9 | (94) |
| He et al, 2019 | circ_002136 | Glioma cells | Up | Angiogenesis (+) | circ_002136/miR-138-5p/SOX13/SPON2 | (99) |
| He et al, 2019 | circ-DICER1 | Glioma cells | Up | Angiogenesis (+) | circ-DICER1/miR-103a-3p (miR-382-5p)/ZIC4 | (100) |
| Liu et a, 2020 | circ-ATXN1 | Glioma cells | Up | Angiogenesis (+) | circ-ATXN1/miR-526b-3p/MMP2(VEGFA) | (101) |
| Zuo et al, 2019 | circ-SMAD7 | Glioma tissues and glioma cells | Up | Migration (+) Invasion (+) | circ-SMAD7/PCNA | (105) |
| Yang et al, 2019 | circ-POSTN | Glioma tissues and glioma cells | Up | Migration (+) Invasion (+) | Circ-POSTN/miR-1205 | (34) |
| Yi et al, 2019 | hsa_circ_0034642 | Glioma tissues and glioma cells | Up | Migration (+) Invasion (+) | hsa_circ_0034642/miR-1205/BATF3 | (106) |
| Su et al, 2019 | circDENND2A | Glioma tissues and glioma cells | Up | Migration (+) Invasion (+) | circDENND2A/miR-625-5p | (107) |
| Qu et al, 2019 | circ_0079593 | Glioma tissues and glioma cells | Up | Prognosis | circ_0079593 / miR-182 (miR-433) | (111) |
| Liu et al, 2020 | circCDC45 | Glioma tissues and glioma cells | Up | Prognosis | circCDC45/miR-516b (miR-527) | (32) |
| Hua et al, 2019 | circCEP128 | Glioma cells | Up | Drug resistance | circCEP128/miR-145-5p/ABCG2 | (115) |
| Zhao et al, 2019 | circATP8B4 | Glioma cells | Up | Radiosensitivity | circATP8B4/miR-766 | (116) |
| Ding et al, 2019 | circNFIX | Glioma tissues and glioma cells | Up | Targeted therapy | circNFIX/miR-378e/RPN2 | (117) |
circRNA, circular RNA.
circRNAs regulate the tumorigenicity of glioma cells
Numerous genetic studies on glioma have determined that gene mutations can promote the tumorigenicity of glioma cells (2,78,79). In general, glioma-related genes can be separated into two classes: Oncogenes and tumor suppressor genes. Activating tumor-promoting genes or deactivating tumor suppressor genes may lead to the onset and development of glioma (80,81). Furthermore, previous research on human glioma specimens has revealed that circRNAs can alter the expression of certain genes or proteins by translating proteins or sponging miRNAs to facilitate the tumorigenicity of glioma cells (82). For example, it was demonstrated that circ-FBXW7, a circRNA derived from the FBXW7 gene, encoded the protein FBXW7-185aa, which may suppress the tumorigenicity of glioma cells. Further research revealed that FBXW7-185aa reduced the half-life of c-Myc by antagonizing ubiquitin specific peptidase 28-induced c-Myc stability. Additionally, circ-FBXW7 and FBXW7-185aa levels were reduced in glioblastoma clinical samples, and the expression of circ-FBXW7 was positively associated with the overall survival of patients with glioblastoma (77). Furthermore, Xia et al (78) demonstrated that circ-AKT3 was expressed at low levels in glioblastoma tissues in contrast to the expression in normal brain tissues. Furthermore, this previous study reported that circ-AKT3 encodes a 174 amino acid (aa) non-classical protein named AKT3-174aa. Overexpression of AKT3-174aa suppressed the tumorigenicity of glioblastoma cells. Notably, Wu et al (75) revealed that circ-SMO, a circRNA derived from exons 3–6 of the SMO gene, encoded protein SMO-193aa, which is known to be essential for Hedgehog signaling activation in glioblastoma. Furthermore, in vivo and in vitro experiments verified that SMO-193aa may directly interact with SMO and enhance SMO cholesterol modification, thus promoting the tumorigenicity of glioma.
From a genetic point of view, circRNAs are vital modulators of tumor formation. During the development of glioma, circRNAs are pivotal players in the glioma-associated gene changes that cause malignant initiation and progression, as mentioned in the present review. Therefore, the role of circRNAs in glioma is considered a promising field for further investigation.
circRNAs activate and sustain proliferative signaling
Normal brain tissues regulate the generation and release of proliferative signals that instruct entrance into and development via the cell growth-and-division cycle, thereby assuring the maintenance of normal brain tissue architecture and characteristics (83). Glioma cells have been suggested to obtain the ability to induce and sustain positively acting growth-stimulatory signals in various ways (84). For example, Shi et al (85) reported that circ-0014359 exhibited high expression levels in glioma clinical tissue samples and cells. In addition, it was found that overexpression of circ-0014359 was associated with downregulation of miR-153; circ-0014359 acted as a sponge for miR-153 to facilitate glioma cell proliferation and was suggested to be involved in the PI3K axis. Further investigation revealed that high expression of miR-153 decreased the levels of phosphorylated (p)-AKTser473, whereas inhibiting miR-153 could enhance the expression of p-AKTser473. Chen et al (86) demonstrated that circ_ PTN exhibited high expression levels in glioma, and functioned as an initiator of cell proliferation via its sponging effects on the miR-122 family. Additionally, SRY-box transcription factor (SOX)6 was predicted as a target of miR-122, and high expression levels of miR-122 reduced the expression levels of SOX6. Downregulation of miR-122 reversed the tumor suppressor role of circ_PTN knockdown, whereas increased expression of SOX6 affected the apoptosis and inhibition of cell proliferation induced by miR-122 overexpression. In contrast to normal brain tissues, Peng et al (31) characterized circ-CPA4 from 28,265 differently expressed circRNAs in glioma tissues. The authors investigated the functional roles of circ-CPA4 and its downstream gene. By prediction and validation, the authors verified that circ-CPA4 interacted with let-7 and served as a sponge for let-7; circ-CPA4 may absorb let-7 to modulate the levels of CPA4, thus promoting the proliferation and progression of glioma cells (31).
Dysregulation of circRNAs may contribute to the aberrant functioning of proliferation signaling in glioma. As miRNA sponges, circRNAs may suppress the expression levels of miRNAs via adsorption and concomitantly affect the expression of downstream genes. Thus, circRNAs may provide novel insights into controlling the malignant proliferation of glioma cells.
circRNAs function in cell apoptosis
Apoptosis is finely modulated at the gene level, thus leading to the highly efficient elimination of injured cells (87). The balance between anti- and pro-apoptotic factors is essential to decide whether a cell enters apoptosis. Apoptosis is mainly modulated via the caspase family of proteases and the B-cell lymphoma 2 (Bcl-2) family of intracellular proteins. Previous studies have demonstrated that circRNAs are pivotal modulators of a number of cell processes in glioma, including apoptosis (88,89). circRNAs can alter the extrinsic and intrinsic apoptotic pathways in several ways, resulting in a decrease in apoptosis or development of apoptosis resistance; these involve the injured death receptor pathway, alteration of the balance between anti-apoptotic and pro-apoptotic proteins of the Bcl-2 family, reduced caspase function and impaired p53 function (90,91). He et al (92) demonstrated that circ-MAPK4, a circRNA that has been reported to act as a cancer-promoting gene in glioma, was upregulated and associated with the clinicopathological stage of glioma (P<0.05). In addition, this previous study revealed that knockdown of circ-MAPK4 resulted in increased levels of the cleaved form of apoptotic factors [caspase-9, caspase-3 and poly(ADP-ribose) polymerase], indicating the anti-apoptotic effect of circ-MAPK4 in glioma development. In addition, experimental validation suggested that circ-MAPK4 targeted miR-125a-3p, which activated the p38/MAPK signaling pathway to modulate cell apoptosis. Another study by Cao et al (93) revealed that hsa_circ_0037251 modulated the levels of mechanistic target of rapamycin through sponging miR-1229-3p and thus regulated cellular apoptosis. Notably, hsa_circ_0037251 was revealed to be overexpressed in glioma cells, and silencing of hsa_circ_0037251 facilitated cellular apoptosis and induced cell cycle arrest in the G1 phase. It is becoming clear that destabilizing the overall pro- or anti-apoptotic balance of the Bcl-2 family can induce dysregulated apoptosis in the affected cells. For example, Chi et al (94) reported that matrine was able to induce apoptosis via downregulating circRNA-104075 in glioma cells. circRNA-104075 was highly expressed in glioma cells and functioned as a suppressor of cell apoptosis by disrupting the equilibrium of anti-apoptotic/pro-apoptotic factors. Furthermore, this previous study demonstrated that increased expression of circRNA-104075 increased p62 and Bcl-2 expression, but reduced beclin-1, cleaved caspase-3, cleaved caspase-9 and Bax levels, as well as the light chain (LC)3-II/LC3-I ratio in matrine-treated glioma cells.
circRNAs can alter the balance of anti- and pro-apoptotic factors via multiple mechanisms, thus leading to apoptotic resistance of glioma cells (89–94). In addition, these findings may provide more evidence on the association between cell apoptosis and circRNAs to better develop novel therapeutic strategies for glioma.
circRNAs induce angiogenesis
Similar to normal tissues, tumor growth requires oxygen and nutrients, along with an ability to excrete carbon dioxide and metabolic waste products (95). Cancer cells excrete numerous pro-angiogenic regulators, which result in the creation of abnormal tumor-associated neovasculature. Tumor-associated neovasculature, which is characterized by disordered, immature and penetrable blood vessels, helps to maintain and expand tumor growth (96). Evidence has indicated that angiogenesis is induced early during the multi-step progression of glioma in humans and experimental animal models (97). Glioma cells excrete vascular endothelial growth factor (VEGF) and other pro-angiogenic regulatory factors to facilitate the development of vascular endothelial cells. Several of these angiogenic members are signaling proteins that couple with stimulating or suppressive cell surface receptors exhibited by vascular endothelial cells. In addition, glial vascular endothelial cells may excrete a considerable quantity of factors that accelerate cancer development (98). Notably, research has shown that circRNAs can regulate the interactions of these secreted factors to induce angiogenesis. He et al (99) indicated that circ_002136, spondin 2 (SPON2) and SOX13 were upregulated, whereas miR-138-5p was downregulated in glioma-exposed endothelial cells. circ_002136 served as a molecular sponge for miR-138-5p, which negatively regulated SOX13 and modulated glioma angiogenesis. By contrast, SOX13 improved the level of SPON2 and enhanced the angiogenic ability of glioma-exposed endothelial cells. Furthermore, silencing circ_002136 markedly suppressed the activity, metastasis and tubule formation of glioma-exposed endothelial cells. In another study, it was demonstrated that the level of circ-DICER1 was increased in glioma-exposed endothelial cells, and knockdown of circ-DICER1 inhibited the cell activity, metastasis and tubule-forming ability of glioma-exposed endothelial cells. Furthermore, circ-DICER1 served as a sponge for miR-382-5p or miR-103a-3p, and impaired their negative modulatory effects on zic family member 4 (ZIC4) in glioma-exposed endothelial cells. Further research verified that ZIC4 upregulated the level of its downstream gene heat shock protein (Hsp)90β, and Hsp90 subsequently promoted the cell activity, metastasis and tubule-forming ability of glioma-exposed endothelial cells via stimulating the PI3K/AKT signaling pathway (100). Additionally, Liu et al (101) demonstrated that the levels of circ-ATXN1were increased, whereas miR-526b-3p expression was decreased in glioma-associated endothelial cells. circ-ATXN1 promoted angiogenesis of glioma-associated endothelial cells by interacting with miR-526b-3p. A dual luciferase experiment elucidated that MMP2 and VEGFA were targets of miR-526b-3p. Furthermore, silencing circ-ATXN1 inhibited the viability and angiogenesis of glioma-associated endothelial cells.
Taken together, these findings indicated that circRNAs are important regulators of angiogenesis in glioma. circRNAs may regulate the interactions of pro-angiogenic factors and glioma growth factors to induce angiogenesis. Angiogenesis is the basis of hematogenous metastasis in glioma and new blood vessels have been characterized as essential channels for glioma to transport nutrients (97,98). These circRNA studies may be useful in designing approaches to improving the survival of patients with glioma.
circRNAs regulate invasion and metastasis
Metastasis is generally a late event in cancer and has limited treatment options. Invasion and migration are associated with numerous phenotypic and genotypic changes of cells that are indirectly and directly modulated via diverse extrinsic and intrinsic factors (102). The multiple-step procedures of invasion and migration have been described as a series of discontinuous steps, usually referred to as the invasion-metastasis cascade. High-grade malignant tumor tissues are hyperactive and may progress through the steps of the invasion-metastasis cascade within a short period of time (103). Glioma is an invasive disease that tends to propagate locally and migrate to other parts of the brain (104). It is increasingly apparent that dysregulation of circRNAs may contribute to the aggressive growth and migration of glioma. For example, circ-SMAD7 promoted the invasion and migration of glioma via upregulating proliferating cell nuclear antigen (PCNA), whereas knockdown of circ-SMAD7 suppressed the metastasis and invasion of glioma cells. Consistently, the mRNA and protein expression levels of PCNA were suppressed after circ-SMAD7 was downregulated in glioma cells. In addition, the levels of circ-SMAD7 were increased in glioma tissues compared with those in normal brain tissues. Furthermore, the expression of circ-SMAD7 was associated with World Health Organization tumor stage and Karnofsky Performance Scale score (105). Another study by Yang et al (34) revealed that the levels of circ-POSTN were elevated in glioma and associated with World Health Organization tumor grades, tumor dimension and poor prognosis in patients with glioma. circ-POSTN has been shown to act as a sponge of miR-1205 to accelerate glioma cell invasion and metastasis in vivo. Additionally, knockdown of circ-POSTN in glioma cells reduced cell metastasis and invasion potential (34). Moreover, Yi et al (106) demonstrated that hsa_circ_0034642 affected the migration and invasion of glioma via binding miR-1205 to regulate the basic leucine zipper transcription factor ATF-like 3 level. Notably, hsa_circ_0034642 exhibited higher levels of expression in glioma tissue and cells in comparison with in normal brain tissue. Furthermore, high hsa_circ_0034642 expression was associated with clinical severity and a worse prognosis. Su et al (107) characterized a fresh hypoxia-related circRNA, DENN domain containing 2A (DENND2A), which enhanced the invasion and metastasis of glioma cells by directly binding to miR-625-5p. In addition, glioma tissue with highly expressed hypoxia-inducible factor 1α exhibited a higher level of circ-DENND2A and a lower level of miR-625-5p.
Based on the aforementioned findings, circRNAs may affect glioma invasion and metastasis through dynamic and complex regulatory mechanisms. Invasion and metastasis of glioma cells are leading factors resulting in patient death. Therefore, further studies on the effect of circRNAs in glioma cell invasion and metastasis are expected to help with the development of treatment strategies for glioma.
circRNAs as diagnostic and prognostic biomarkers in glioma
In recent years, despite substantial advances in treatment strategy, the rate of early diagnosis of glioma remains low and it is difficult to treat patients with late-stage glioma, thus leading to the relative low survival of this disease (108). Additionally, compared with other types of cancer, the overall prognosis of patients with glioma is worse and the survival period is shorter (109). Thus, the identification of valuable biomarkers for early-stage diagnosis and prognosis is crucial for improving the survival of patients with glioma. Recent studies have demonstrated that circRNAs are strongly associated with the clinicopathological characteristics of glioma, and they may serve as diagnostic and prognostic biomarkers (32,92,110). Previous studies indicated that the closed-ring structure stabilizes circRNAs in plasma and tissues, and protects them from the enzymatic activity of exonucleases (28,50); therefore, the unique structure of circRNAs may be a potential advantage for their use as biomarkers for the diagnosis and prognosis of glioma. Qu et al (111) revealed that circ_0079593 could be considered an independent prognostic factor for patients with glioma based on multivariate analysis. Furthermore, overexpression of circ_0079593 in glioma was found to be associated with tumor size, World Health Organization tumor grade and patient survival rate. Mechanistically, circ_0079593 was revealed to facilitate glioma development via binding to miR-433 and miR-182. Another study by Liu et al (32) indicated that circ-CDC45 expression was increased in glioma compared with that in paired non-cancerous specimens. Overexpression of circ-CDC45 was closely associated with larger tumor size, higher World Health Organization tumor stage and poor prognosis. Functionally, circ-CDC45 interacted with and modulated the levels of miR-527 and miR-516b to facilitate cell proliferation and invasion. Furthermore, Peng et al (31) demonstrated that the levels of circ-CPA4 were high in glioma compared with those in paired non-cancerous specimens. Simultaneously, a high level of circ-CPA4 was positively correlated with a worse outcome of glioma.
In conclusion, due to their diverse range of functions, high stability and presence in easily obtainable body fluids, circRNAs are considered valuable biomarkers in glioma. Furthermore, the study of emerging circRNAs will improve our understanding of the molecular pathways involved, including glioma-associated gene expression, thereby improving the management of patients with glioma.
Therapeutic potential of circRNAs in glioma
Over the last decade, treatment for glioma has focused on maximum safe surgical resection alongside radiotherapy and chemotherapy (3). Radiotherapy and chemotherapy are typical and aggressive treatment approaches for numerous patients with late-stage and unresectable glioma; however, the inherent and acquired resistance of glioma cells is a major limitation to the efficacy of radiotherapy and chemotherapy (112,113). Recently, given the enhanced understanding of the etiopathogenesis of glioma, numerous signaling pathways and molecules are considered potentially suitable for targeted treatment (114). Recent studies revealed that certain circRNAs can not only modulate the resistance of glioma cells, but also block vital glioma-associated signaling pathways and molecules, thereby modulating the expression of the corresponding gene. For example, Hua et al (115) demonstrated that the levels of the circ-CEP128 were higher, whereas miR-145-5p levels were lower in temozolomide-resistant glioma cells compared with those in parental cells. Silencing circ-CEP128 suppressed cellular proliferation, reduced the level of ATP-binding cassette super-family G member 2 (ABCG2) and improved the cytotoxic function of temozolomide in glioma cells. Furthermore, a mechanistic study demonstrated that circ-CEP128 may serve as a miR-145-5p sponge to regulate the levels of the downstream target ABCG2. Additionally, Zhao et al (116) indicated that the levels of circ-ATP8B4 were significantly higher in radiation-resistant glioma cells, and indicated that circ-ATP8B4 may serve as a molecular sponge to adsorb miR-766 and to facilitate cell radioresistance. Furthermore, silencing or overexpression of circRNAs not only elucidates the mechanism underlying the initiation and development of glioma, but also provides useful information for the design of targeted treatments to modulate crucial glioma-associated signaling pathways, molecules and genes. For example, Ding et al (117) indicated that the levels of circ-NFIX were increased in glioma cells and tissues compared with those in matched control samples, and high levels of circ-NFIX markedly accelerated cell cycle progression, migration and invasion, and suppressed apoptosis. By contrast, silencing circ-NFIX led to cell cycle arrest, inhibition of cell migration and invasion, and enhanced apoptosis in glioma cells. Mechanistically, circ-NFIX positively regulated ribophorin II via binding to miR-378e in glioma cells.
These results indicated that circRNAs may provide novel insights into individualized therapeutic methods for the treatment of glioma. An appropriate combination of novel targeted and immunotherapeutic approaches that are circRNA-driven is considered an attractive strategy for the treatment of human glioma. However, compared with coding RNAs, the present knowledge on circRNAs remains inadequate, and circRNAs cannot yet be used in the clinical setting. Thus, further studies on the treatment of glioma are needed.
Summary of the regulatory pathway of circRNAs in glioma biological progression
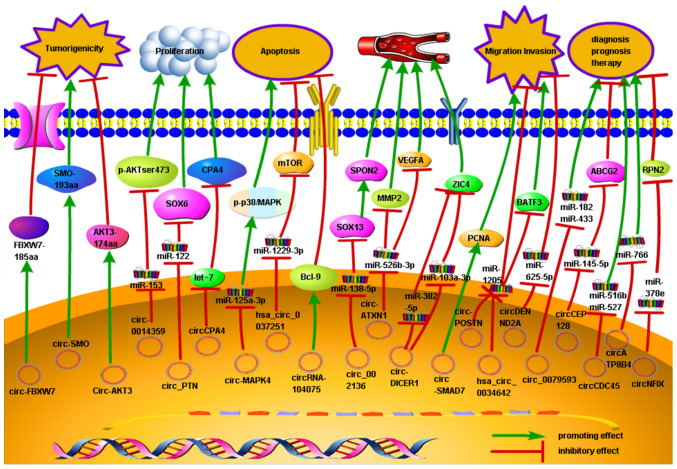
In glioma, a considerable number of circRNAs are dysregulated in neoplastic tissue, which may suppress or promote tumor progression. Notably, the understanding of circRNAs is rapidly improving, as described in the present review. In addition, numerous studies (Fig. 2) have indicated the role of circRNAs in several biological functions in human glioma, including cellular proliferation, migration, invasion, apoptosis and angiogenesis. For example, circ-0014359, circ_PTN, circ-MAPK4 and hsa_circ_0037251 bind to miR-153, miR-122, miR-125a-3p and miR-1229-3p, respectively, to modulate the apoptosis and proliferation of glioma cells (85,86,92,93). Furthermore, other circRNAs, such as circ_002136, hsa_circ_0034642 and circ-DENND2A, regulate the angiogenesis, invasion and migration of glioma cells by acting as sponges targeting miR-138-5p, miR-1205 and miR-625-5p, respectively (99,106,107).
Figure 2.
Role and regulatory pathways of glioma-related circRNAs. The schematic diagram depicts the known role of circRNAs in glioma progression and the way that circRNAs are involved in microRNA-associated gene regulatory pathways. circRNA, circular RNA.
4. Online resources
Recently, to facilitate the study of the different aspects of circRNAs, a number of circRNA research tools have been developed and improved such as circBase, CircInteractome, circAtlas, MiOncoCirc, CircR2Disease, TCSD and CSCD (118–124). These databases are currently used for circRNA identification, localization and characterization analysis, and as tools for investigating the interaction of circRNAs with targets. Online resources are essential tools for investigators to obtain data, and the number of them has rapidly increased. Some of the online databases used for the study of circRNAs are shown in Table II.
Table II.
Databases used for circRNA research.
| Authors, year | Database | URL | Annotation | (Refs.) |
|---|---|---|---|---|
| Glazar et al, 2014 | circBase | http://www.circbase.org/ | A circRNA database that contains circRNA information from human, mouse, C. elegans and Latimeria organisms | (118) |
| Panda et al, 2018 | CircInteractome | http://circinteractome.nia.nih.gov/ | Predicts binding sites of proteins to circRNAs. Predicts potential binding sites for miRNA-circRNA interactions | (119) |
| Ji et al, 2019 | circAtlas | http://circatlas.biols.ac.cn | Includes circRNA sequence information for humans and other animals | (120) |
| Vo et al, 2019 | MiOncoCirc | https://nguyenjoshvo.github.io/ | Description of targets that can be used for cancer diagnosis or treatment | (121) |
| Fan et al, 2018 | CircR2Disease | http://bioinfo.snnu.edu.cn/CircR2Disease/ | This tool delivers a broad database for circRNA dysregulation in different diseases | (122) |
| Xia et al, 2017 | TSCD | http://gb.whu.edu.cn/TSCD | TSCD distinguishes the characteristics of human and mouse tissue-specific circRNAs | (123) |
| Xia et al, 2018 | CSCD | http://gb.whu.edu.cn/CSCD | This database recognized 272,152 cancer-specific circRNAs from 228 total RNA samples Predicts cellular localization of circRNAs, RNA-binding proteins and variable splicing of related genes | (124) |
circRNA, circular RNA; miRNA, microRNA.
5. Conclusion and perspectives
Glioma is a malignancy of the human brain, which poses a major threat to the health and survival of those affected. The initiation and development of glioma include various phases and regulators; however, its pathogenesis requires further investigation. Currently, identifying novel molecules is crucial for the ongoing progress in the research of targeted treatments for glioma. Over the past few years, a large body of evidence has indicated the effect of circRNAs on the development of glioma. Databases have played a key role in bioinformatics analyses on circRNAs and have contributed to considerable advances in this field. The function of circRNAs includes a wide range of physiopathological processes. As described in the present review, circRNAs participate in glioma cell proliferation, invasion, metastasis and cell cycle progression, and are crucial for the diagnosis, prognosis and treatment of glioma. In addition, there are several recommendations for future circRNA research. Firstly, the present study on circRNAs in glioma mainly summarized studies that used glioma tissues and cells, whereas other specimens (such as cerebrospinal fluid and blood) are more easily obtained. Secondly, the basic mechanisms through which circRNAs participate in the advance of glioma await further investigation. Finally, since the final objective of circRNA research is to safely utilize circRNAs for clinical therapy of glioma, extensive clinical research is required.
In conclusion, due to the advances in technology for the assessment and screening of novel circRNAs, and the improvements in databases, circRNAs may be applied in the future in the diagnostic, therapeutic and prognostic management of glioma, thereby leading to great advances in glioma therapy.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- ABCG2
ATP-binding cassette super-family G member 2
- Bcl-2
B-cell lymphoma 2
- circRNAs
circular RNAs
- ciRNAs
circular intronic RNAs
- ecircRNAs
exonic circular RNAs
- EIciRNAs
exon-intron circular RNAs
- lncRNAs
long non-coding RNAs
- miRNAs
microRNAs
- ncRNAs
non-coding RNAs
Funding Statement
The present study was supported by The Hebei Province Key research and development projects of China (grant no. 19277737D).
Funding
The present study was supported by The Hebei Province Key research and development projects of China (grant no. 19277737D).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WS performed the literature search and drafted the manuscript. HZ conceived the idea for the article, and assisted with editing and drafting the text. XH and LH assisted with editing and drafting the text. XX participated in the critical review and substantially revised it. Data authentication is not applicable. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ng S, Zouaoui S, Bessaoud F, Rigau V, Roux A, Darlix A, Bauchet F, Mathieu-Daudé H, Trétarre B, Figarella-Branger D, et al. An epidemiology report for primary central nervous system tumors in adolescents and young adults: A nationwide population-based study in France, 2008–2013. Neuro Oncol. 2020;22:851–863. doi: 10.1093/neuonc/noz227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sminia P, van den Berg J, van Kootwijk A, Hageman E, Slotman BJ, Verbakel W. Experimental and clinical studies on radiation and curcumin in human glioma. J Cancer Res Clin Oncol. 2021;147:403–409. doi: 10.1007/s00432-020-03432-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maccallini C, Gallorini M, Cataldi A, Amoroso R. Targeting iNOS As a valuable strategy for the therapy of glioma. ChemMedChem. 2020;15:339–344. doi: 10.1002/cmdc.201900580. [DOI] [PubMed] [Google Scholar]
- 4.Daisy Precilla S, Kuduvalli SS, Thirugnanasambandhar Sivasubramanian A. Disentangling the therapeutic tactics in GBM: From bench to bedside and beyond. Cell Biol Int. 2021;45:18–53. doi: 10.1002/cbin.11484. [DOI] [PubMed] [Google Scholar]
- 5.Catalano M, D'Alessandro G, Trettel F, Limatola C. Role of infiltrating microglia/macrophages in glioma. Adv Exp Med Biol. 2020;1202:281–298. doi: 10.1007/978-3-030-30651-9_14. [DOI] [PubMed] [Google Scholar]
- 6.Zhang L, Liu F, Weygant N, Zhang J, Hu P, Qin Z, Yang J, Cheng Q, Fan F, Zeng Y, et al. A novel integrated system using patient-derived glioma cerebral organoids and xenografts for disease modeling and drug screening. Cancer Lett. 2021;500:87–97. doi: 10.1016/j.canlet.2020.12.013. [DOI] [PubMed] [Google Scholar]
- 7.Ballo MT, Urman N, Lavy-Shahaf G, Grewal J, Bomzon Z, Toms S. Correlation of tumor treating fields dosimetry to survival outcomes in newly diagnosed glioblastoma: A large-scale numerical simulation-based analysis of data from the phase 3 EF-14 randomized trial. Int J Radiat Oncol Biol Phys. 2019;104:1106–1113. doi: 10.1016/j.ijrobp.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Cloughesy TF, Mochizuki AY, Orpilla JR, Hugo W, Lee AH, Davidson TB, Wang AC, Ellingson BM, Rytlewski JA, Sanders CM, et al. Neoadjuvant anti-PD-1 immunotherapy promotes a survival benefit with intratumoral and systemic immune responses in recurrent glioblastoma. Nat Med. 2019;25:477–486. doi: 10.1038/s41591-018-0337-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roura AJ, Gielniewski B, Pilanc P, Szadkowska P, Maleszewska M, Krol SK, Czepko R, Kaspera W, Wojtas B, Kaminska B. Identification of the immune gene expression signature associated with recurrence of high-grade gliomas. J Mol Med (Berl) 2021;99:241–255. doi: 10.1007/s00109-020-02005-7. [DOI] [PubMed] [Google Scholar]
- 10.Eslahi M, Dana PM, Asemi Z, Hallajzadeh J, Mansournia MA, Yousefi B. The effects of chitosan-based materials on glioma: Recent advances in its applications for diagnosis and treatment. Int J Biol Macromol. 2021;168:124–129. doi: 10.1016/j.ijbiomac.2020.11.180. [DOI] [PubMed] [Google Scholar]
- 11.Pedretti S, Masini L, Turco E, Triggiani L, Krengli M, Meduri B, Pirtoli L, Borghetti P, Pegurri L, Riva N, et al. Hypofractionated radiation therapy versus chemotherapy with temozolomide in patients affected by RPA class V and VI glioblastoma: A randomized phase II trial. J Neurooncol. 2019;143:447–455. doi: 10.1007/s11060-019-03175-2. [DOI] [PubMed] [Google Scholar]
- 12.Saw PE, Xu X, Chen J, Song EW. Non-coding RNAs: The new central dogma of cancer biology. Sci China Life Sci. 2021;64:22–50. doi: 10.1007/s11427-020-1700-9. [DOI] [PubMed] [Google Scholar]
- 13.Ryu J, Ahn Y, Kook H, Kim YK. The roles of non-coding RNAs in vascular calcification and opportunities as therapeutic targets. Pharmacol Ther. 2021;218:107675. doi: 10.1016/j.pharmthera.2020.107675. [DOI] [PubMed] [Google Scholar]
- 14.Lu K, Yu M, Chen Y. Non-coding RNAs regulating androgen receptor signaling pathways in prostate cancer. Clin Chim Acta. 2021;513:57–63. doi: 10.1016/j.cca.2020.11.027. [DOI] [PubMed] [Google Scholar]
- 15.Uddin MN, Wang X. The landscape of long non-coding RNAs in tumor stroma. Life Sci. 2021;264:118725. doi: 10.1016/j.lfs.2020.118725. [DOI] [PubMed] [Google Scholar]
- 16.Fort V, Khelifi G, Hussein SMI. Long non-coding RNAs and transposable elements: A functional relationship. Biochim Biophys Acta Mol Cell Res. 2021;1868:118837. doi: 10.1016/j.bbamcr.2020.118837. [DOI] [PubMed] [Google Scholar]
- 17.Kalani M, Hodjati H, Ghoddusi Johari H, Doroudchi M. Memory T cells of patients with abdominal aortic aneurysm differentially expressed micro RNAs 21, 92a, 146a, 155, 326 and 663 in response to helicobacter pylori and lactobacillus acidophilus. Mol Immunol. 2021;130:77–84. doi: 10.1016/j.molimm.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Bahreini F, Rayzan E, Rezaei N. microRNA-related single-nucleotide polymorphisms and breast cancer. J Cell Physiol. 2021;236:1593–1605. doi: 10.1002/jcp.29966. [DOI] [PubMed] [Google Scholar]
- 19.Jiang S. Perspectives on MicroRNA study in oncogenesis: Where are we? Neoplasia. 2021;23:99–101. doi: 10.1016/j.neo.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao YZ, Sun JY, Chen YX, Wen CC, Wei L. The roles of circRNAs in cancers: Perspectives from molecular functions. Gene. 2021;767:145182. doi: 10.1016/j.gene.2020.145182. [DOI] [PubMed] [Google Scholar]
- 21.Yu Z, Huang Q, Zhang Q, Wu H, Zhong Z. circRNAs open a new era in the study of cardiovascular disease (review) Int J Mol Med. 2021;47:49–64. doi: 10.3892/ijmm.2020.4792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zaiou M. circRNAs signature as potential diagnostic and prognostic biomarker for diabetes mellitus and related cardiovascular complications. Cells. 2020;9:659. doi: 10.3390/cells9030659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li L, Sun D, Li X, Yang B, Zhang W. Identification of key circRNAs in non-small cell lung cancer. Am J Med Sci. 2021;361:98–105. doi: 10.1016/j.amjms.2020.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Zhang Y, Nguyen TM, Zhang XO, Wang L, Phan T, Clohessy JG, Pandolfi PP. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 2021;22:41. doi: 10.1186/s13059-021-02263-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch L. CRISPR-Cas13 targets circRNAs. Nat Rev Genet. 2021;22:68. doi: 10.1038/s41576-020-00318-4. [DOI] [PubMed] [Google Scholar]
- 26.Li Y, Feng W, Kong M, Liu R, Wu A, Shen L, Tang Z, Wang F. Exosomal circRNAs: A new star in cancer. Life Sci. 2021;269:119039. doi: 10.1016/j.lfs.2021.119039. [DOI] [PubMed] [Google Scholar]
- 27.Luo Q, Li X, Fu B, Zhang L, Fang L, Qing C, Guo Y, Huang Z, Li J. Expression profile and diagnostic value of circRNAs in peripheral blood from patients with systemic lupus erythematosus. Mol Med Rep. 2021;23:1. doi: 10.3892/mmr.2020.11639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Ye T, Liu H, Lv P, Duan C, Wu X, Jiang K, Lu H, Xia D, Peng E, et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol Cancer. 2021;20:4. doi: 10.1186/s12943-020-01300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu T, Wang M, Jiang L, Ma L, Wan L, Chen Q, Wei C, Wang Z. circRNAs in anticancer drug resistance: Recent advances and future potential. Mol Cancer. 2020;19:127. doi: 10.1186/s12943-020-01240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang X, Yang H, Zhao L, Li G, Duan Y. Circular RNA PRKCI promotes glioma cell progression by inhibiting microRNA-545. Cell Death Dis. 2019;10:616. doi: 10.1038/s41419-019-1863-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peng H, Qin C, Zhang C, Su J, Xiao Q, Xiao Y, Xiao K, Liu Q. circCPA4 acts as a prognostic factor and regulates the proliferation and metastasis of glioma. J Cell Mol Med. 2019;23:6658–6665. doi: 10.1111/jcmm.14541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Hou K, Ji H, Mi S, Yu G, Hu S, Wang J. Overexpression of circular RNA circ-CDC45 facilitates glioma cell progression by sponging miR-516b and miR-527 and predicts an adverse prognosis. J Cell Biochem. 2020;121:690–697. doi: 10.1002/jcb.29315. [DOI] [PubMed] [Google Scholar]
- 33.Meng Q, Li S, Liu Y, Zhang S, Jin J, Zhang Y, Guo C, Liu B, Sun Y. Circular RNA circSCAF11 accelerates the glioma tumorigenesis through the miR-421/SP1/VEGFA axis. Mol Ther Nucleic Acids. 2019;17:669–677. doi: 10.1016/j.omtn.2019.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y, Zhang Y, Chen B, Ding L, Mu Z, Li Y. Elevation of circular RNA circ-POSTN facilitates cell growth and invasion by sponging miR-1205 in glioma. J Cell Biochem. 2019;120:16567–16574. doi: 10.1002/jcb.28916. [DOI] [PubMed] [Google Scholar]
- 35.Zheng SQ, Qi Y, Wu J, Zhou FL, Yu H, Li L, Yu B, Chen XF, Zhang W. CircPCMTD1 acts as the sponge of miR-224-5p to promote glioma progression. Front Oncol. 2019;9:398. doi: 10.3389/fonc.2019.00398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA. 1976;73:3852–3856. doi: 10.1073/pnas.73.11.3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yin S, Tian X, Zhang J, Sun P, Li G. PCirc: Random forest-based plant circRNA identification software. BMC Bioinformatics. 2021;22:10. doi: 10.1186/s12859-020-03944-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goodall GJ, Wickramasinghe VO. RNA in cancer. Nat Rev Cancer. 2021;21:22–36. doi: 10.1038/s41568-020-00306-0. [DOI] [PubMed] [Google Scholar]
- 39.Zhang D, Ni N, Wang Y, Tang Z, Gao H, Ju Y, Sun N, He X, Gu P, Fan X. circRNA-vgll3 promotes osteogenic differentiation of adipose-derived mesenchymal stem cells via modulating miRNA-dependent integrin α5 expression. Cell Death Differ. 2021;28:283–302. doi: 10.1038/s41418-020-0600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang J, Liu Y, Shi G. The circRNA-miRNA-mRNA regulatory network in systemic lupus erythematosus. Clin Rheumatol. 2021;40:331–339. doi: 10.1007/s10067-020-05212-2. [DOI] [PubMed] [Google Scholar]
- 41.Chen J, Yang J, Fei X, Wang X, Wang K. circRNA ciRS-7: A novel oncogene in multiple cancers. Int J Biol Sci. 2021;17:379–389. doi: 10.7150/ijbs.54292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao Z, Zhang Y, Wei S, Zhang X, Guo Y, Han B. Comprehensive circRNA expression profile and function network in osteoblast-like cells under simulated microgravity. Gene. 2021;764:145106. doi: 10.1016/j.gene.2020.145106. [DOI] [PubMed] [Google Scholar]
- 43.Wu T, Li Y, Liang X, Liu X, Tang M. Identification of potential circRNA-miRNA-mRNA regulatory networks in response to graphene quantum dots in microglia by microarray analysis. Ecotoxicol Environ Saf. 2021;208:111672. doi: 10.1016/j.ecoenv.2020.111672. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z, Lei X. Identifying the sequence specificities of circRNA-binding proteins based on a capsule network architecture. BMC Bioinformatics. 2021;22:19. doi: 10.1186/s12859-020-03942-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou B, Yang H, Yang C, Bao YL, Yang SM, Liu J, Xiao YF. Translation of noncoding RNAs and cancer. Cancer Lett. 2021;497:89–99. doi: 10.1016/j.canlet.2020.10.002. [DOI] [PubMed] [Google Scholar]
- 46.Fu LY, Wang SW, Hu MY, Jiang ZL, Shen LL, Zhou YP, Guo JM, Hu YR. Circular RNAs in liver diseases: Mechanisms and therapeutic targets. Life Sci. 2021;264:118707. doi: 10.1016/j.lfs.2020.118707. [DOI] [PubMed] [Google Scholar]
- 47.Wei G, Zhu J, Hu HB, Liu JQ. Circular RNAs: Promising biomarkers for cancer diagnosis and prognosis. Gene. 2021;771:145365. doi: 10.1016/j.gene.2020.145365. [DOI] [PubMed] [Google Scholar]
- 48.Varela-Martinez E, Corsi GI, Anthon C, Gorodkin J, Jugo BM. Novel circRNA discovery in sheep shows evidence of high backsplice junction conservation. Sci Rep. 2021;11:427. doi: 10.1038/s41598-020-79781-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Z, Li X, Xu D, Chen X, Li S, Zhang L, Chan MTV, Wu WKK. An update on the roles of circular RNAs in osteosarcoma. Cell Prolif. 2021;54:e12936. doi: 10.1111/cpr.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Beilerli A, Gareev I, Beylerli O, Yang G, Pavlov V, Aliev G, Ahmad A. Circular RNAs as biomarkers and therapeutic targets in cancer. Semin Cancer Biol. 2021 Jan 9; doi: 10.1016/j.semcancer.2020.12.026. (Epub ahead of print). doi: 10.1016/j.semcancer.2020.12.026. [DOI] [PubMed] [Google Scholar]
- 51.Wang X, Cheng Z, Xu J, Feng M, Zhang H, Zhang L, Qian L. Circular RNA Arhgap12 modulates doxorubicin-induced cardiotoxicity by sponging miR-135a-5p. Life Sci. 2021;265:118788. doi: 10.1016/j.lfs.2020.118788. [DOI] [PubMed] [Google Scholar]
- 52.Khanipouyani F, Akrami H, Fattahi MR. Circular RNAs as important players in human gastric cancer. Clin Transl Oncol. 2021;23:10–21. doi: 10.1007/s12094-020-02419-2. [DOI] [PubMed] [Google Scholar]
- 53.Luo Y, Huang C. circSFMBT2 facilitates vascular smooth muscle cell proliferation by targeting miR-331-3p/HDAC5. Life Sci. 2021;264:118691. doi: 10.1016/j.lfs.2020.118691. [DOI] [PubMed] [Google Scholar]
- 54.Razavi ZS, Tajiknia V, Majidi S, Ghandali M, Mirzaei HR, Rahimian N, Hamblin MR, Mirzaei H. Gynecologic cancers and non-coding RNAs: Epigenetic regulators with emerging roles. Crit Rev Oncol Hematol. 2021;157:103192. doi: 10.1016/j.critrevonc.2020.103192. [DOI] [PubMed] [Google Scholar]
- 55.Li Z, Huang X, Liu A, Xu J, Lai J, Guan H, Ma J. circ_PSD3 promotes the progression of papillary thyroid carcinoma via the miR-637/HEMGN axis. Life Sci. 2021;264:118622. doi: 10.1016/j.lfs.2020.118622. [DOI] [PubMed] [Google Scholar]
- 56.Zhang XY, Mao L. Circular RNA circ_0000442 acts as a sponge of miR-148b-3p to suppress breast cancer via PTEN/PI3K/Akt signaling pathway. Gene. 2021;766:145113. doi: 10.1016/j.gene.2020.145113. [DOI] [PubMed] [Google Scholar]
- 57.Sun Q, Qi X, Zhang W, Li X. Knockdown of circRNA_0007534 suppresses the tumorigenesis of cervical cancer via miR-206/GREM1 axis. Cancer Cell Int. 2021;21:54. doi: 10.1186/s12935-021-01749-7. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Zhang X, Yang H, Jia Y, Xu Z, Zhang L, Sun M, Fu J. circRNA_0005529 facilitates growth and metastasis of gastric cancer via regulating miR-527/Sp1 axis. BMC Mol Cell Biol. 2021;22:6. doi: 10.1186/s12860-020-00340-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li W, Lu H, Wang H, Ning X, Liu Q, Zhang H, Liu Z, Wang J, Zhao W, Gu Y, et al. Circular RNA TGFBR2 acts as a ceRNA to suppress nasopharyngeal carcinoma progression by sponging miR-107. Cancer Lett. 2021;499:301–313. doi: 10.1016/j.canlet.2020.11.001. [DOI] [PubMed] [Google Scholar]
- 60.Xing Y, Zha WJ, Li XM, Li H, Gao F, Ye T, Du WQ, Liu YC. Circular RNA circ-Foxo3 inhibits esophageal squamous cell cancer progression via the miR-23a/PTEN axis. J Cell Biochem. 2020;121:2595–2605. doi: 10.1002/jcb.29481. [DOI] [PubMed] [Google Scholar]
- 61.Kong Z, Wan X, Lu Y, Zhang Y, Huang Y, Xu Y, Liu Y, Zhao P, Xiang X, Li L, Li Y. Circular RNA circFOXO3 promotes prostate cancer progression through sponging miR-29a-3p. J Cell Mol Med. 2020;24:799–813. doi: 10.1111/jcmm.14791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2020;98:87–97. doi: 10.1002/jnr.24356. [DOI] [PubMed] [Google Scholar]
- 63.Okholm TLH, Sathe S, Park SS, Kamstrup AB, Rasmussen AM, Shankar A, Chua ZM, Fristrup N, Nielsen MM, Vang S, et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 2020;12:112. doi: 10.1186/s13073-020-00812-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wong CH, Lou UK, Li Y, Chan SL, Tong JH, To KF, Chen Y. circFOXK2 promotes growth and metastasis of pancreatic ductal adenocarcinoma by complexing with RNA-binding proteins and sponging miR-942. Cancer Res. 2020;80:2138–2149. doi: 10.1158/0008-5472.CAN-19-3268. [DOI] [PubMed] [Google Scholar]
- 65.Liang Y, Wang H, Chen B, Mao Q, Xia W, Zhang T, Song X, Zhang Z, Xu L, Dong G, Jiang F. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol Ther Nucleic Acids. 2020;23:355–368. doi: 10.1016/j.omtn.2020.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Z, Wang Q, Wang X, Xu Z, Wei X, Li J. Circular RNA cIARS regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 2020;6:72. doi: 10.1038/s41420-020-00306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu Y, Liu X, Lin C, Jia X, Zhu H, Song J, Zhang Y. Noncoding RNAs regulate alternative splicing in cancer. J Exp Clin Cancer Res. 2021;40:11. doi: 10.1186/s13046-020-01798-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stagsted LVW, O'Leary ET, Ebbesen KK, Hansen TB. The RNA-binding protein SFPQ preserves long-intron splicing and regulates circRNA biogenesis in mammals. Elife. 2021;10:e63088. doi: 10.7554/eLife.63088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta C, Conn SJ. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat Plants. 2017;3:17053. doi: 10.1038/nplants.2017.53. [DOI] [PubMed] [Google Scholar]
- 70.Hu Q, Zhou T. EIciRNA-mediated gene expression: Tunability and bimodality. FEBS Lett. 2018;592:3460–3471. doi: 10.1002/1873-3468.13253. [DOI] [PubMed] [Google Scholar]
- 71.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 72.Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. doi: 10.1016/j.molcel.2013.08.017. [DOI] [PubMed] [Google Scholar]
- 73.Kong S, Tao M, Shen X, Ju S. Translatable circRNAs and lncRNAs: Driving mechanisms and functions of their translation products. Cancer Lett. 2020;483:59–65. doi: 10.1016/j.canlet.2020.04.006. [DOI] [PubMed] [Google Scholar]
- 74.Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19:30. doi: 10.1186/s12943-020-1135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wu X, Xiao S, Zhang M, Yang L, Zhong J, Li B, Li F, Xia X, Li X, Zhou H, et al. A novel protein encoded by circular SMO RNA is essential for Hedgehog signaling activation and glioblastoma tumorigenicity. Genome Biol. 2021;22:33. doi: 10.1186/s13059-020-02250-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, Wu C, Zhou Q, Hu W, Wu C, Jiang J. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, Huang N, Yang X, Zhao K, Zhou H, et al. Novel Role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–315. doi: 10.1093/jnci/djx166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, Huang N, Yang X, Xiao F, Liu D, et al. A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer. 2019;18:131. doi: 10.1186/s12943-019-1083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.El-Hashash AHK. Histone H3K27M mutation in brain tumors. Adv Exp Med Biol. 2021;1283:43–52. doi: 10.1007/978-981-15-8104-5_3. [DOI] [PubMed] [Google Scholar]
- 80.He Z, Yang C, He Y, Gong B, Yin C, Feng J, Chen L, Tang J, Chen Y. CAMTA1, a novel antitumor gene, regulates proliferation and the cell cycle in glioma by inhibiting AKT phosphorylation. Cell Signal. 2021;79:109882. doi: 10.1016/j.cellsig.2020.109882. [DOI] [PubMed] [Google Scholar]
- 81.Osama M, Mostafa MN, Alvi MA. Astrocyte elevated gene-1 as a novel therapeutic target in malignant gliomas and its interactions with oncogenes and tumor suppressor genes. Brain Res. 2020;1747:147034. doi: 10.1016/j.brainres.2020.147034. [DOI] [PubMed] [Google Scholar]
- 82.Zhang Y, Liu Q, Liao Q. CircHIPK3: A promising cancer-related circular RNA. Am J Transl Res. 2020;12:6694–6704. [PMC free article] [PubMed] [Google Scholar]
- 83.Smyth LCD, Rustenhoven J, Scotter EL, Schweder P, Faull RLM, Park TIH, Dragunow M. Markers for human brain pericytes and smooth muscle cells. J Chem Neuroanat. 2018;92:48–60. doi: 10.1016/j.jchemneu.2018.06.001. [DOI] [PubMed] [Google Scholar]
- 84.Jiang Y, Zhou J, Luo P, Gao H, Ma Y, Chen YS, Li L, Zou D, Zhang Y, Jing Z. Prosaposin promotes the proliferation and tumorigenesis of glioma through toll-like receptor 4 (TLR4)-mediated NF-κB signaling pathway. EBioMedicine. 2018;37:78–90. doi: 10.1016/j.ebiom.2018.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shi F, Shi Z, Zhao Y, Tian J. circRNA hsa-circ-0014359 promotes glioma progression by regulating miR-153/PI3K signaling. Biochem Biophys Res Commun. 2019;510:614–620. doi: 10.1016/j.bbrc.2019.02.019. [DOI] [PubMed] [Google Scholar]
- 86.Chen C, Deng L, Nie DK, Jia F, Fu LS, Wan ZQ, Lan Q. Circular RNA Pleiotrophin promotes carcinogenesis in glioma via regulation of microRNA-122/SRY-box transcription factor 6 axis. Eur J Cancer Prev. 2020;29:165–173. doi: 10.1097/CEJ.0000000000000535. [DOI] [PubMed] [Google Scholar]
- 87.Ye Z, Zhuo Q, Hu Q, Xu X, Liu M, Zhang Z, Xu W, Liu W, Fan G, Qin Y, et al. FBW7-NRA41-SCD1 axis synchronously regulates apoptosis and ferroptosis in pancreatic cancer cells. Redox Biol. 2021;38:101807. doi: 10.1016/j.redox.2020.101807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mohamad Anuar NN, Nor Hisam NS, Liew SL, Ugusman A. Clinical review: Navitoclax as a pro-apoptotic and anti-fibrotic agent. Front Pharmacol. 2020;11:564108. doi: 10.3389/fphar.2020.564108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hartman ML, Czyz M. BCL-w: Apoptotic and non-apoptotic role in health and disease. Cell Death Dis. 2020;11:260. doi: 10.1038/s41419-020-2417-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yan F, Fan B, Wang J, Wei W, Tang Q, Lu L, Luo Z, Pu J, Yang SS. circ_0008305-mediated miR-660/BAG5 axis contributes to hepatocellular carcinoma tumorigenesis. Cancer Med. 2021;10:833–842. doi: 10.1002/cam4.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lou J, Hao Y, Lin K, Lyu Y, Chen M, Wang H, Zou D, Jiang X, Wang R, Jin D, et al. Circular RNA CDR1as disrupts the p53/MDM2 complex to inhibit gliomagenesis. Mol Cancer. 2020;19:138. doi: 10.1186/s12943-020-01253-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He J, Huang Z, He M, Liao J, Zhang Q, Wang S, Xie L, Ouyang L, Koeffler HP, Yin D, Liu A. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol Cancer. 2020;19:17. doi: 10.1186/s12943-019-1120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cao Q, Shi Y, Wang X, Yang J, Mi Y, Zhai G, Zhang M. Circular METRN RNA hsa_circ_0037251 promotes glioma progression by sponging miR-1229-3p and regulating mTOR expression. Sci Rep. 2019;9:19791. doi: 10.1038/s41598-019-56417-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chi G, Xu D, Zhang B, Yang F. Matrine induces apoptosis and autophagy of glioma cell line U251 by regulation of circRNA-104075/BCL-9. Chem Biol Interact. 2019;308:198–205. doi: 10.1016/j.cbi.2019.05.030. [DOI] [PubMed] [Google Scholar]
- 95.Zheng X, Liu J, Li X, Tian R, Shang K, Dong X, Cao B. Angiogenesis is promoted by exosomal DPP4 derived from 5-fluorouracil-resistant colon cancer cells. Cancer Lett. 2021;497:190–201. doi: 10.1016/j.canlet.2020.10.009. [DOI] [PubMed] [Google Scholar]
- 96.Yetkin-Arik B, Kastelein AW, Klaassen I, Jansen CHJR, Latul YP, Vittori M, Biri A, Kahraman K, Griffioen AW, Amant F, et al. Angiogenesis in gynecological cancers and the options for anti-angiogenesis therapy. Biochim Biophys Acta Rev Cancer. 2021;1875:188446. doi: 10.1016/j.bbcan.2020.188446. [DOI] [PubMed] [Google Scholar]
- 97.Ahir BK, Engelhard HH, Lakka SS. Tumor development and angiogenesis in adult brain tumor: Glioblastoma. Mol Neurobiol. 2020;57:2461–2478. doi: 10.1007/s12035-020-01892-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Carlson JC, Cantu-Gutierrez M, Lozzi B, Huang-Hobbs E, Turner WD, Tepe B, Zhang Y, Herman AM, Rao G, Creighton CJ, et al. Identification of diverse tumor endothelial cell populations in malignant glioma. Neuro Oncol. 2021;23:932–944. doi: 10.1093/neuonc/noaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.He Z, Ruan X, Liu X, Zheng J, Liu Y, Liu L, Ma J, Shao L, Wang D, Shen S, et al. FUS/circ_002136/miR-138-5p/SOX13 feedback loop regulates angiogenesis in glioma. J Exp Clin Cancer Res. 2019;38:65. doi: 10.1186/s13046-019-1065-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.He Q, Zhao L, Liu X, Zheng J, Liu Y, Liu L, Ma J, Cai H, Li Z, Xue Y. MOV10 binding circ-DICER1 regulates the angiogenesis of glioma via miR-103a-3p/miR-382-5p mediated ZIC4 expression change. J Exp Clin Cancer Res. 2019;38:9. doi: 10.1186/s13046-018-0990-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu X, Shen S, Zhu L, Su R, Zheng J, Ruan X, Shao L, Wang D, Yang C, Liu Y. SRSF10 inhibits biogenesis of circ-ATXN1 to regulate glioma angiogenesis via miR-526b-3p/MMP2 pathway. J Exp Clin Cancer Res. 2020;39:121. doi: 10.1186/s13046-020-01625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Novikov NM, Zolotaryova SY, Gautreau AM, Denisov EV. Mutational drivers of cancer cell migration and invasion. Br J Cancer. 2021;124:102–114. doi: 10.1038/s41416-020-01149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yu-Ju Wu C, Chen CH, Lin CY, Feng LY, Lin YC, Wei KC, Huang CY, Fang JY, Chen PY. CCL5 of glioma-associated microglia/macrophages regulates glioma migration and invasion via calcium-dependent matrix metalloproteinase 2. Neuro Oncol. 2020;22:253–266. doi: 10.1093/neuonc/noz189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li X, Meng Y. Analyses of metastasis-associated genes in IDH wild-type glioma. BMC Cancer. 2020;20:1114. doi: 10.1186/s12885-020-07628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zuo CY, Qian W, Huang CJ, Lu J. Circular RNA circ-SMAD7 promoted glioma cell proliferation and metastasis by upregulating PCNA. Eur Rev Med Pharmacol Sci. 2019;23:10035–10041. doi: 10.26355/eurrev_201911_19570. [DOI] [PubMed] [Google Scholar]
- 106.Yi C, Li H, Li D, Qin X, Wang J, Liu Y, Liu Z, Zhang J. Upregulation of circular RNA circ_0034642 indicates unfavorable prognosis in glioma and facilitates cell proliferation and invasion via the miR-1205/BATF3 axis. J Cell Biochem. 2019;120:13737–13744. doi: 10.1002/jcb.28646. [DOI] [PubMed] [Google Scholar]
- 107.Su H, Zou D, Sun Y, Dai Y. Hypoxia-associated circDENND2A promotes glioma aggressiveness by sponging miR-625-5p. Cell Mol Biol Lett. 2019;24:24. doi: 10.1186/s11658-019-0149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Patil V, Mahalingam K. A four-protein expression prognostic signature predicts clinical outcome of lower-grade glioma. Gene. 2018;679:57–64. doi: 10.1016/j.gene.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 109.Choi J, Kim SH, Ahn SS, Choi HJ, Yoon HI, Cho JH, Roh TH, Kang SG, Chang JH, Suh CO. Extent of resection and molecular pathologic subtype are potent prognostic factors of adult WHO grade II glioma. Sci Rep. 2020;10:2086. doi: 10.1038/s41598-020-59089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zheng K, Xie H, Wu W, Wen X, Zeng Z, Shi Y. circRNA PIP5K1A promotes the progression of glioma through upregulation of the TCF12/PI3K/AKT pathway by sponging miR-515-5p. Cancer Cell Int. 2021;21:27. doi: 10.1186/s12935-020-01699-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Qu Y, Zhu J, Liu J, Qi L. Circular RNA circ_0079593 indicates a poor prognosis and facilitates cell growth and invasion by sponging miR-182 and miR-433 in glioma. J Cell Biochem. 2019;120:18005–18013. doi: 10.1002/jcb.29103. [DOI] [PubMed] [Google Scholar]
- 112.Ghiaseddin AP, Shin D, Melnick K, Tran DD. Tumor treating fields in the management of patients with malignant gliomas. Curr Treat Options Oncol. 2020;21:76. doi: 10.1007/s11864-020-00773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Oldrini B, Vaquero-Siguero N, Mu Q, Kroon P, Zhang Y, Galán-Ganga M, Bao Z, Wang Z, Liu H, Sa JK, et al. MGMT genomic rearrangements contribute to chemotherapy resistance in gliomas. Nat Commun. 2020;11:3883. doi: 10.1038/s41467-020-17717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Oprita A, Baloi SC, Staicu GA, Alexandru O, Tache DE, Danoiu S, Micu ES, Sevastre AS. Updated insights on EGFR signaling pathways in glioma. Int J Mol Sci. 2021;22:587. doi: 10.3390/ijms22020587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hua L, Huang L, Zhang X, Feng H, Shen B. Knockdown of circular RNA CEP128 suppresses proliferation and improves cytotoxic efficacy of temozolomide in glioma cells by regulating miR-145-5p. Neuroreport. 2019;30:1231–1238. doi: 10.1097/WNR.0000000000001326. [DOI] [PubMed] [Google Scholar]
- 116.Zhao M, Xu J, Zhong S, Liu Y, Xiao H, Geng L, Liu H. Expression profiles and potential functions of circular RNAs in extracellular vesicles isolated from radioresistant glioma cells. Oncol Rep. 2019;41:1893–1900. doi: 10.3892/or.2019.6972. [DOI] [PubMed] [Google Scholar]
- 117.Ding C, Wu Z, You H, Ge H, Zheng S, Lin Y, Wu X, Lin Z, Kang D. CircNFIX promotes progression of glioma through regulating miR-378e/RPN2 axis. J Exp Clin Cancer Res. 2019;38:506. doi: 10.1186/s13046-019-1483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Glazar P, Papavasileiou P, Rajewsky N. circBase: A database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Panda AC, Dudekula DB, Abdelmohsen K, Gorospe M. Analysis of circular RNAs using the web tool circinteractome. Methods Mol Biol. 2018;1724:43–56. doi: 10.1007/978-1-4939-7562-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang J, Cheng H, Yan J, Zhang S, Yang P, Zhao F. Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep. 2019;26:3444–3460.e5. doi: 10.1016/j.celrep.2019.02.078. [DOI] [PubMed] [Google Scholar]
- 121.Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al. The landscape of circular RNA in cancer. Cell. 2019;176:869–881.e13. doi: 10.1016/j.cell.2018.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fan C, Lei X, Fang Z, Jiang Q, Wu FX. CircR2Disease: A manually curated database for experimentally supported circular RNAs associated with various diseases. Database (Oxford) 2018;2018:bay044. doi: 10.1093/database/bay044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, Xiang Y, Liu L, Zhong S, Han L, He C. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984–992. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 124.Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, Jin Y, Gao Y, Xia L, Chang H, et al. CSCD: A database for cancer-specific circular RNAs. Nucleic Acids Res. 2018;46(D1):D925–D929. doi: 10.1093/nar/gkx863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.