Abstract
Despite signs of infection—including taste loss, dry mouth and mucosal lesions such as ulcerations, enanthema and macules— the involvement of the oral cavity in coronavirus disease 2019 (COVID-19) is poorly understood. To address this, we generated and analyzed two single-cell RNA sequencing datasets of the human minor salivary glands and gingiva (9 samples, 13,824 cells), identifying 50 cell clusters. Using integrated cell normalization and annotation, we classified 34 unique cell subpopulations between glands and gingiva. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) viral entry factors such as ACE2 and TMPRSS members were broadly enriched in epithelial cells of the glands and oral mucosae. Using orthogonal RNA and protein expression assessments, we confirmed SARS-CoV-2 infection in the glands and mucosae. Saliva from SARS-CoV-2-infected individuals harbored epithelial cells exhibiting ACE2 and TMPRSS expression and sustained SARS-CoV-2 infection. Acellular and cellular salivary fractions from asymptomatic individuals were found to transmit SARS-CoV-2 ex vivo. Matched nasopharyngeal and saliva samples displayed distinct viral shedding dynamics, and salivary viral burden correlated with COVID-19 symptoms, including taste loss. Upon recovery, this asymptomatic cohort exhibited sustained salivary IgG antibodies against SARS-CoV-2. Collectively, these data show that the oral cavity is an important site for SARS-CoV-2 infection and implicate saliva as a potential route of SARS-CoV-2 transmission.
SARS-CoV-2 is the causal agent for COVID-19, and the World Health Organization classifies this virus as an airborne pathogen transmitted by asymptomatic, pre-symptomatic and symptomatic individuals through close contact via exposure to infected droplets and aerosols1,2. Although SARS-CoV-2 transmission can occur by activities involving the oral cavity, such as speaking, breathing, coughing, sneezing and even singing3–5, most attention has focused on the nasal–lung axis of infection6. Oral manifestations, such as taste loss, dry mouth and oral lesions, are evident in about half of COVID-19 cases7–9, although it remains unknown whether SARS-CoV-2 can directly infect and replicate in oral tissues, such as the salivary glands (SGs) or mucosa. This is critical because, if these are sites of early infection, they could play an important role in transmitting the virus to the lungs or the gastrointestinal tract via saliva, as has been suggested for other microbial-associated diseases, such as pneumonia10 and inflammatory bowel diseases11,12 (Extended Data Fig. 1a).
SARS-CoV-2 uses host entry factors, such as ACE2 and TMPRSS family members (TMPRSS2 and TMPRSS4)13,14, and understanding the cell types that harbor these receptors is important for determining infection susceptibilities throughout the body15–17. ACE2 and TMPRSS2 expression has been reported in oral tissues18,19; however, there are no comprehensive descriptions of viral entry factor expression nor direct confirmation of SARS-CoV-2 infection in oral tissues. We hypothesized that SGs and barrier epithelia of the oral cavity and oropharynx can be infected by SARS-CoV-2 and contribute to the transmission of SARS-CoV-2. To test this, we generated two human oral single-cell RNA sequencing (scRNA-seq) atlases to predict cell-specific susceptibilities to SARS-CoV-2 infection. We confirmed ACE2 and TMPRSS expression in SGs and oral mucosa epithelia. SARS-CoV-2 infection was confirmed using autopsy and outpatient samples. Saliva from asymptomatic individuals with COVID-19 demonstrated the potential for viral transmission. In a prospective clinical cohort, we found a positive correlation between salivary viral load and taste loss; we also demonstrated persistent salivary antibody responses to SARS-CoV-2 nucleocapsid and spike proteins.
Results
Oral tissue atlases reveal resident immune cells and niche-specific epithelial diversity.
The SGs and the barrier mucosa of the oral cavity and oropharynx are likely gateways for viral infection, replication and transmission (Extended Data Fig. 1a). The oral cavity mucosa is lined by stratified squamous epithelia, divided into keratinized (gingiva and hard palate) and non-keratinized (buccal, labial, ventral tongue and oropharyngeal) mucosae20. The dorsal tongue mucosa is adapted for taste and contains specialized projections (papillae)21; moreover, the entirety of the oral cavity is bathed by saliva that lubricates and protects the underlying mucosa (Extended Data Fig. 1b).
Oral tissue niche heterogeneity is increasingly being recognized at the cellular level22–25; because of this, we predicted that SARS-CoV-2 infection would be non-uniform across oral sites. Using two unique sites—SGs (n = 5; labial minor) and oral mucosae (n = 4; gingiva/hard palate)—we generated and integrated two human oral scRNA-seq datasets to establish an initial database to explore viral infection susceptibilities (Extended Data Fig. 1c). Broad structural and immune cell heterogeneity was identified, including 22 and 28 cell types in the glands and mucosae, respectively (Fig. 1a,b and Supplementary Table 1).
Fig. 1 |. Single-cell atlases of distinct oral tissue niches reveal diverse immune and epithelial cells.

a,b, Using unpublished datasets, UMAPs of scRNA-seq data from dissociated (a) minor SGs and gingiva (b) delineate cell population contribution by sample and cell type annotation. We found 22 populations in the SG (7,141 cells) and 28 populations in the gingiva (6,683 cells). c–f, Distinct SG cell subpopulations were defined by a (c) dot plot expression matrix. Gene set (MSigDB) analysis for immune activation pathways (d) does not show clear enrichment, pointing to cells being in a resting/homeostatic state in the SGs. Gingival cell atlases were also defined by a dot plot expression matrix. f, Despite these biopsies coming from minorly inflamed gingival tissues (gingival index scoring an average of 1 out of 3), MSigDB analysis revealed minimal activation of T, natural killer and B cell populations. For gene lists related to a–d, see Supplementary Table 1. IFN, interferon; NK, natural killer; DC, dendritic cell.
The epithelial compartment of the SGs contained major cell types, including serous acini (LPO and ODAM), mucous acini (MUC5B and BPIFB2), ducts (S100A2 and WFDC2), a ductal/basal epithelial population (KRT15 and SOX2), myoepithelia (KRT14 and ACTA2) and neuroendocrine cells (CHGA and GFRA3) (Fig. 1c). Ionocytes (CFTR and FOXI1) were found in the SG in higher proportion than reports from the lung26; these cells regulate the ionic composition of secretions. The connective tissue compartment consisted of fibroblasts (DCN and LUM), endothelium—arterial (CLDN5), venules (AQP1) and capillaries (CA4)—smooth muscle (ITGA8) and pericytes (MYO1B). Eight distinct populations of immune cells were identified and consisted of a high proportion of IgA+ plasma cells, B lymphocytes (MS4A1), T helper (Th) cells (CD40LG) and two cytotoxic Tc subtypes (T cells (CD8A)—subtype 1 (EOMES and GMZA, exhausted type) and subtype 2 (SPRY1 and ITAG1, tissue-resident memory type). The myeloid-derived compartment included subpopulations of conventional DC2 (CD1C) and two types of macrophages—type 1 (CD163; M1) and type 2 (FCGR3A, IFNGR1 and CX3CR1; M2). Of note, M2 cells were CD86+TNF+>, and a minority of the Tc subtypes were IFNG+, although neither M2 nor Tc displayed activation signatures using gene set enrichment analysis (Molecular Signatures Database (MsigDB)) (Fig. 1d).
The gingiva can serve as an indicator of overall oral health12,27. Using healthy gingival biopsies, the mucosal atlas contributed to the identification of 15 immune subpopulations (Fig. 1e and Supplementary Table 1). Five populations of dendritic cells were identified, including conventional dendritic cell subtype 1 (XCR1) and 2 (CD300E), activated dendritic cells (CCR7+ and LAMP3 (ref. 28)), plasmacytoid dendritic cells (PACSIN1) and epithelial-resident Langerhans cells (CD207)29,30. Among T cells, cytotoxic T cells (Tc; CD8A), helper T cells (Th; CD40LG), regulatory T cells (Treg; LAIR2), γδ T cells (γδ; KLRC4), mucosal-associated invariant T (MAIT) cells (IL23R)) and a cycling subset (KI67) were cataloged. A fraction (approximately 20–30%) of MAIT cells expressed TNF; a fraction (~30%) of the cycling and Tc clusters were found to express IFNG and were CD95+ (Fas receptor). These cells also did not show immune activation profiles, pointing to a resting/homeostatic state (Fig. 1f)31,32.
Four basal epithelial cell populations (Basal 1–4) were discovered, defined by enriched expression of CXCL14, KRT19, COL7A1 and DSC2/3, respectively. KRT19-expressing cells define the non-keratinized gingiva ‘pocket’ epithelium33. Suprabasal cells were sparse in this dataset. In COVID-19, barrier epithelia are especially at risk for infection. These data reveal that, among many oral barrier cell populations, the diversity of oral epithelial cells provides multiple available targets for viral infection.
Oral single-cell atlases predict viral infection of epithelial subpopulations.
To predict the viral tropism of SARS-CoV-2, other coronaviruses (SARS-CoV-1, HCoV-NL63, MERS-CoV and HCoV-229E), influenza and rhinovirus C, we performed joint annotation of the SG and gingival scRNA-seq atlases (Fig. 2a, Extended Data Fig. 2a and Supplementary Table 1). Thirty-four unique cell types were identified, including 12 epithelial, 7 mesenchymal and 15 immune cell clusters (Fig. 2b). The cell type expression of known viral entry factor genes across cell clusters demonstrated broad potential susceptibilities to viral infection among the epithelial populations (Fig. 2c–e)16,17,34,35. Similarly to SARS-CoV-1 and HCoV-NL63, SARS-CoV-2 spike glycoprotein binds to ACE2 and is reported to be activated by tissue-specific proteases (TMPRSS2, TMPRSS4 and TMPRSS11D) and endosomal proteases (CTSB, CTSL, BSG and FURIN) to gain entry for replication36. These factors were broadly expressed across many clusters, but only epithelia commonly expressed ACE2 and TMPRSS members (Fig. 2c).
Fig. 2 |. An integrated oral cell atlas reveals broad epithelial infection susceptibilities.

a, To characterize the vulnerabilities of oral tissues to infection by SARS-CoV-2 and other common viruses, we integrated unpublished data from human oral gingiva and minor SGs. b, These 50 populations were jointly annotated before integration into 34 cell clusters to create the first human pan-oral cell atlas (see gene signatures in Extended Data Fig. 2a and Supplementary Table 2). These results illustrate that both shared and unique cell populations are represented in the gingiva and SG. c, Vulnerabilities to infection by coronaviruses, influenza and rhinovirus C can be predicted based on entry factor expression and visualized using expression matrices. Epithelia appear especially at risk for viral infection. d, When focused on the nine epithelial cell populations, vulnerabilities to SARS-CoV-2 were apparent in both SG and mucosa. These results strongly suggest that the oral cavity might be vulnerable to viral infection, especially for SARS-CoV-2. Expression matrices, including a low-frequency ACE2/TMPRSS2 co-expressing cells in Basal 1, ducts, mucous acini and myoepithelial clusters, further supporting SARS-CoV-2 infection susceptibilities. e, UMAPs demonstrate distinct cluster vulnerabilities, with ACE2 highest in most oral epithelia; however, expression of proteases demonstrated tissue-specific expression patterns with TMPRSS2 (enriched for SGs) and TMPRSS11D (enriched for mucosal cells). Endosomal proteases, CTSB and CTSL exhibited broad expression across vulnerable cell types. f, Normalized expression of epithelial clusters across oral, nasal and intestinal tissues demonstrate the relatively equivalent expression level of oral sites, especially in the minor SG. NK, natural killer; DC, dendritic cell; TA, transit amplifying.
SARS-CoV-2 entry factor expression analyses revealed that no single oral epithelial subpopulation appeared at singular risk for infection (Fig. 2d). ACE2 expression was detected in nine oral epithelial clusters, including Basal 1–3, basal cycling, SG ducts, SG serous and SG mucous acini clusters. These findings suggest that multiple oral epithelial cell subtypes are susceptible to infection. Uniform manifold approximation and projections (UMAPs) of putative SARS-CoV-2 entry factor expression revealed expression of ACE2 confined primarily to epithelial clusters of the SGs (Fig. 2e). Proteases were demonstrated to exhibit tissue-specific expression patterns, with TMPRSS2 enriched in the SG epithelia and TMPRSS11D enriched in mucosal keratinocytes. Endosomal proteases CTSB and CTSL exhibited broad expression patterns across epithelia.
Co-expression of the principal entry factors ACE2 and TMPRSS2 has been used to predict infection in the nasal cavity, lungs and gut. ACE2 expression was detected in our epithelial populations; however, ACE2 and TMPRSS2 co-expression in epithelial cells of the SGs and mucosa was generally rare, specifically in the SG ducts and acini and the Basal 1 population (~0.5% of cells) (Fig. 2d). To examine relative ACE2 and TMPRSS2 expression, we further integrated the oral epithelial cell atlas with nasal37 and intestinal38 scRNA-seq epithelial cell subsets (Fig. 2f). These normalized expression counts revealed that oral epithelia, especially the minor SGs, were similar to known sites of infection by SARS-CoV-2. Furthermore, using publicly available Genotype-Tissue Expression (GTEx) data39, similar viral entry factor expression levels for minor SGs were found across the gastrointestinal and respiratory sites (Extended Data Fig. 2b). When ACE2 and TMPRSS2 levels were plotted, the three consistently highest co-expressing sites were the lungs, intestines and glands (Extended Data Fig. 2c).
We validated both receptor (ACE2) and protease (TMPRSS2, TMPRSS4 and TMPRSS11D) expression in the SG and gingival mucosa (Fig. 3a,b) using in situ hybridization (ISH) and immunofluorescence (IF) microscopy (Fig. 3a,b and Extended Data Fig. 3a,b). These oral ISH expression levels are similar to the nasal cavity6. Available bulk RNA sequencing (RNA-seq) of the major and minor SGs was used to compare ACE2 and TMPRSS2 expression between different SGs, and minor SGs expressed higher ACE2 compared to parotid and submandibular SGs (Extended Data Fig. 3c–e). ISH of the gingiva demonstrated relatively higher expression of ACE2 and TMPRSS2 family proteins in suprabasal compared to basal cells (Fig. 3b).
Fig. 3 |. Oral and oropharyngeal ISH mapping supports oral infection by SARS-CoV-2.
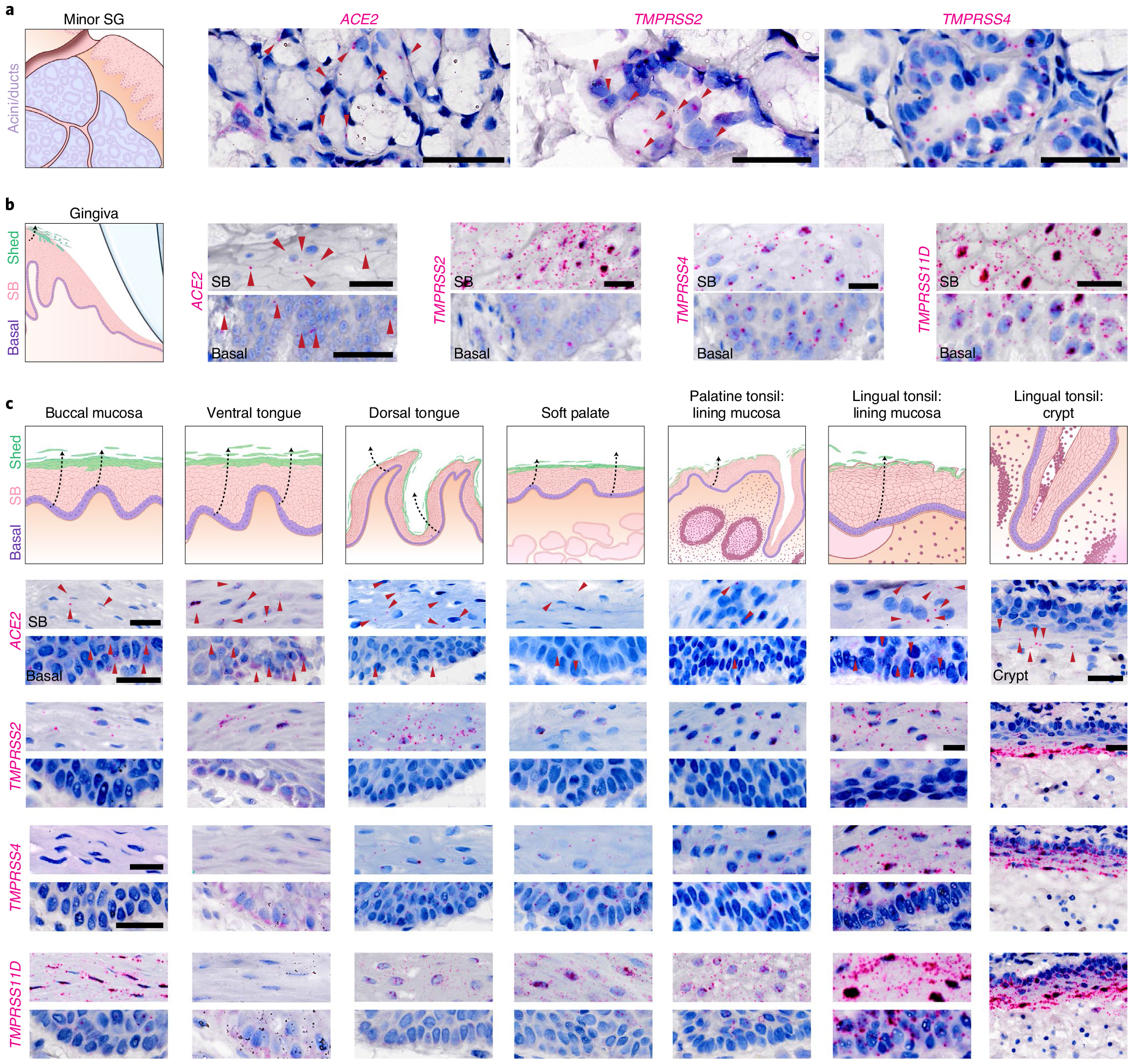
a,b, Using healthy volunteer (a) gland and (b) gingival tissue sections, mRNA expression was confirmed using RNAscope ISH for ACE2, TMPRSS2, TMPRSS4 and TMPRSS11D in gingiva; (see TMPRSS11D in SG: Extended Data Fig. 3b); three independent replications for each. b, Owing to the known shedding/sloughing of suprabasal epithelial cells (c, illustrations), we examined both basal and suprabasal (SB) expression, revealing enrichment of all examined entry factors in suprabasal over basal cells. c, Using ISH, we mapped ACE2 and TMPRSS2, TMPRSS4 and TMPRSS11D in diverse oral tissues (buccal mucosa, ventral tongue and the dorsal tongue) and the oropharynx (soft palate and tonsils); three independent replications for oral and two replications for oropharyngeal samples. ISH controls are included in Extended Data Fig. 3f–h. This again supported the heterogeneity that can be found in the oral cavity—not only considering suprabasal over basal enrichment but also across sites. This mapping also revealed that all sites are vulnerable to infection in suprabasal cells that are shed/sloughed into saliva. Arrowheads in a-c indicate high gene expression (red). Scale bars (a-c), 25 μm.
To further map ACE2/TMPRSS2 expression in oral sites, ISH assays were conducted using healthy adult tissue samples from the buccal mucosa, ventral/dorsal tongue, soft palate and palatine/lingual tonsils (Fig. 3c and Extended Data Fig. 3f–g). For each entry factor examined, increased suprabasal expression was observed when compared with the basal compartment (Fig. 3c and Extended Data Fig. 3h). Additionally, substantial ACE2 expression was observed in the tonsillar crypt. These results point to multiple sites in the oral cavity and oropharynx as potentially susceptible to infection by SARS-CoV-2.
SARS-CoV-2 can infect and replicate in the SGs and mucosae.
We assessed SARS-CoV-2 transcripts by droplet digital polymerase chain reaction (ddPCR) in a cohort of COVID-19 autopsy tissues from 28 SG sites and six mucosal sites from 18 patients (Supplementary Table 2). SARS-CoV-2 was detected in 57% of SGs (16/28; Fig. 4a) with a trend toward higher viral loads in the minor SGs than the paired parotid SGs (n = 8 pairs, P = 0.0625, Wilcoxon two-sided signed-rank test; Extended Data Fig. 4a). SARS-CoV-2 infection of the submandibular glands was also observed in two cases40. SARS-CoV-2 infection of lining cells of the oral mucosa was observed in two sets of tissues from autopsy patients (P11 and P23) where 5/6 of the available mucosal sites (dorsal tongue, tonsil and uvula) exhibited detectable SARS-CoV-2 by ddPCR (Supplementary Table 2).
Fig. 4 |. SGs are infected in patients with COVID-19 and exhibit robust immune responses.
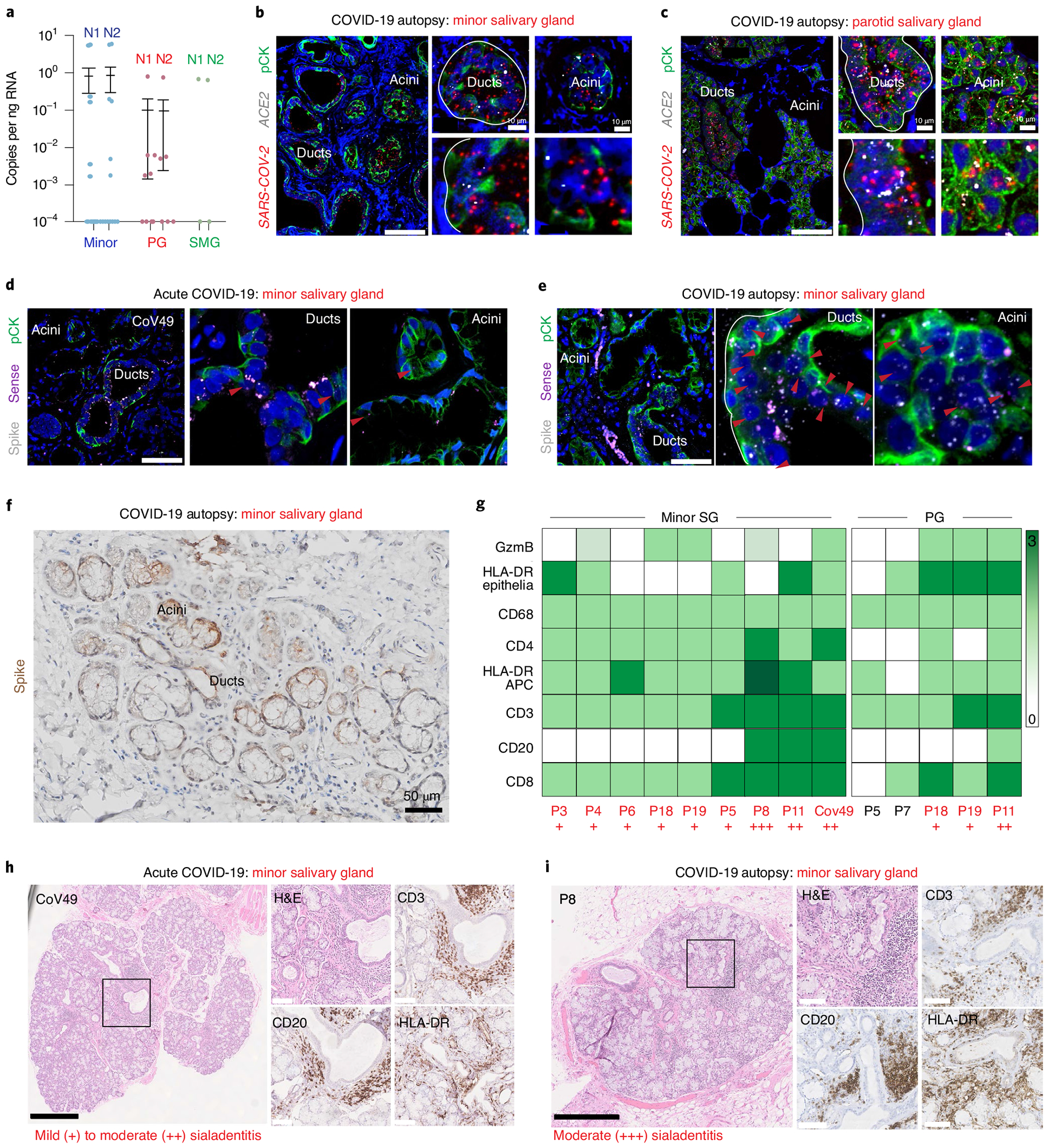
a, ddPCR was used to investigate SARS-CoV-2 RNA in SGs: (minor (n = 14), parotid (PG, n = 8) and submandibular (SMG, n = 2) (18 individuals and 28 glands) recovered from COVID-19 autopsies. N1 and N2 mean ± standard deviation are displayed as appropriate for minor: 0.82 ± 2.0 and 0.86 + 2.1; PG: 0.10 ± 0.28 and 0.10 ± 0.26; and SMG: 0.34 ± 0.48 and 0.32 ± 0.46. b, c, Infection was confirmed using RNAscope fluorescence in situ hybridization (FISH) and IHC. Both minor (b) and parotid (c) SGs demonstrated SARS-CoV-2 in ACE2-positive ducts/acini (one independent replication). d-f, FISH and IHC detected SARS-CoV-2 infection and replication in the minor SG from (d) an acutely infected and (e) an autopsy SG using the spike (V-nCoV2019-S) and sense (V-nCoV2019-orf1ab-sense) probes, respectively (one independent replication). f, Spike IHC confirms patchy regional expression (zero independent replications). g, Immunophenotyping of autopsied SG was completed for CD3, CD4, CD8, CD20, CD68, HLA-DR and granzyme B (GzmB); T cell responses dominate and are well correlated with sialadenitis. h, i, Representative immunophenotyping studies on (h) an acutely infected individual (CoV49) and (i) a COVID-19 autopsy (P8), demonstrating mild-to-moderate sialadenitis with focal lymphocytic sialadenitis and epithelial injury. Scale bars: b,c left, d and e, 25 μm; b,c right, 10 μm; f, 50 μm; h,i black, 500 μm; h,i white, 100 μm. A solid black box highlights the area enlarged in the inset. Experiments were completed on tissue sections from at least four separate individuals (five minor and four parotid). Where appropriate, summary data are presented as mean values ± s.e.m.
To confirm tissue tropism for SARS-CoV-2 in the SG, ACE2 and SARS-CoV-2 spike ISH and immunohistochemistry (IHC) were completed on tissue sections from minor and parotid SGs recovered from COVID-19 autopsies. SARS-CoV-2 messenger RNA (mRNA) was consistently detected in ACE2-expressing ducts and acini (Fig. 4b,c). However, SARS-CoV-2 infection was regional, with some acinar units exhibiting more viral transcript expression compared to others. We next evaluated whether ducts and/or acini support SARS-CoV-2 replication. Using minor SGs from an acutely infected individual with COVID-19 and from autopsies, we found infected acini and ducts harbored replicating SARS-CoV-2 (Fig. 4d,e and Extended Data Fig. 4b,c). To a lesser degree, this was also confirmed in parotid SGs (Extended Data Fig. 4c). Infection (spike) and replication (sense) counts were well correlated on a per-cell basis (Extended Data Fig. 4d nd Methods).
To characterize the heterogeneous inflammatory responses to the presence of viral infection in SGs (Fig. 4f), immunophenotyping and systematic microscopic analysis of acutely infected minor and autopsy SGs were performed (Fig. 4g–i, Extended Data Fig. 4e,f and Supplementary Table 2). The most common histological feature was chronic sialadenitis, including focal lymphocytic sialadenitis (FLS; Fig. 4h,i). Other findings included architectural distortion, atrophy and fibrosis, mucous inspissation and ductal rupture. Immunophenotyping revealed a predominance of T lymphocytic inflammation (CD3) with proportionally more B lymphocytes (CD20) in cases with FLS, reported in other tissues affected in COVID-19 (ref. 41). HLA-DR expression was adjacent to areas of inflammation and epithelial injury42. These results establish that the minor and major SGs are susceptible sites for infection, replication and local immune cell activation.
These findings raised two critical questions for SARS-CoV-2 pathogenesis. First, because principal entry factors are expressed in the oral mucosa suprabasal layers, SARS-CoV-2 might originally infect cells in situ and then be shed into saliva (Fig. 5a). Second, populations of shed infected cells might provide binding sites for SARS-CoV-2 and/or act as carriers to promote viral stability and transmissibility. To explore these possibilities, we again performed ISH on an acutely infected individual with COVID-19 (CoV49; Fig. 4) and found infection and replication in all layers of mucosa, primarily enriched in the differentiated epithelial cells (Fig. 5b). Mucosal scrapings suggested that these cells retained infection and replication capacity after shedding (Fig. 5c).
Fig. 5 |. Oral mucosal epithelial cells are infected by SARS-CoV-2 and shed into saliva.
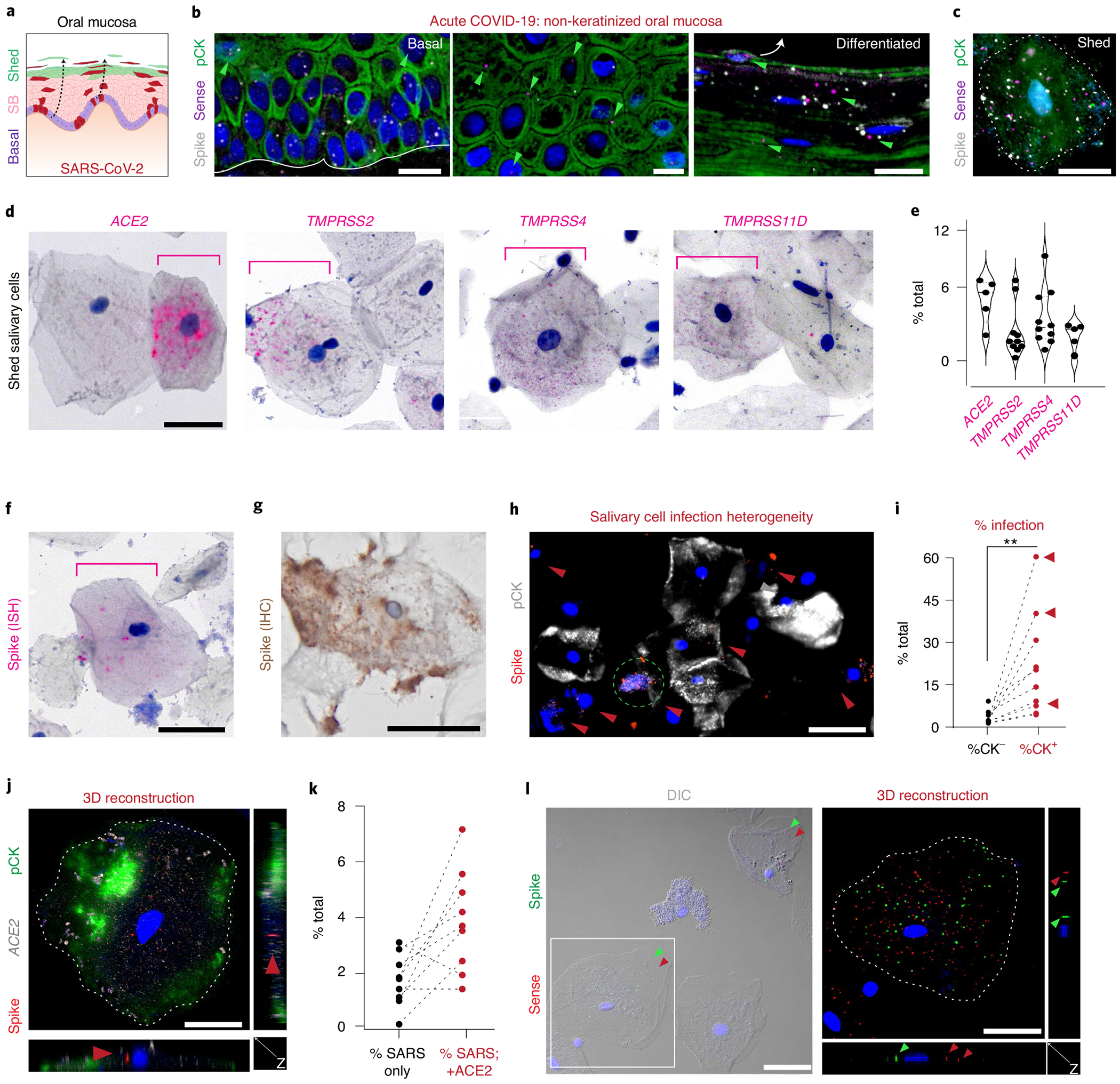
a, We hypothesized that oral mucosae would be infected by SARS-CoV-2. b, Using the overlying mucosa from SG biopsy of CoV49 (Fig. 4), we performed ISH using spike/sense probes to demonstrate infection in basal, suprabasal (SB), and differentiated cells. Like the expression of SARS-CoV-2 entry factors (Fig. 3), replication was found more often in suprabasal layers (one independent replication). c, Mucosal scrapings from CoV49 confirmed infection and replication in cells fated to be shed into saliva (no independent replication). d,e, Salivary epithelial cells were found to express (d) all SARS-CoV-2 entry factors in a minority of cells (n = 5 for ACE2 and TMPRSS11D; n = 10 for TMPRSS2 and TMPRSS4). e, Scatter violin plots highlight percentage of expressing cells per sample from d, with means represented by a thick solid line (5.4, 2.8, 3.9 and 2.4; respectively). f-l, Salivary epithelial cells can sustain infection by SARS-CoV-2 using (f) ISH (one independent replication) or (g) IHC (no independent replication). h, Across saliva cells, there is infection heterogeneity of pCK cells (one independent replication). i, Using ten samples collected from outpatients with COVID-19, we confirmed that pCK cells are the primary infected cell population; red arrows point to individuals with loss of taste (n = 10 with two independent replications; two-tailed paired t-test P=0.01). j, SARS-CoV-2 infection more often occurs in ACE2+ cells. Using 3D confocal microscopy, we demonstrated the virus inside of these cells, (k) most of which were ACE2+ (two independent replications). l, These epithelial cells were found to sustain viral replication inside the cells (two independent replications). The solid white line in b represents basement membrane; the white arrow in b represents differentiated cell trajectory. Dotted white lines (c, j and l) highlight cell membranes; the dotted green circle (h) indicates possibly CK+ cell undergoing cell death; red arrowheads (h, j and l) represent SARS-CoV-2 infection, and green arrowheads (b and l) represent SARS-CoV-2 replication. DIC, differential interference contrast microscopy. Scale bars: b, h and l left, 25 μm; c, d, f, g, j and l right, 10 μm. Statistical test (i): **P ≤ 0.001.
Saliva samples collected from mildly symptomatic individuals who were postitive for SARS-CoV-2 were analyzed for SARS-CoV-2 entry factor expression. ISH demonstrated heterogeneity of ACE2 in nucleated squamous keratinocytes; TMPRSS2, TMPRSS4 and TMPRSS11D were variably expressed (Fig. 5d,e and Extended Data Fig. 5a,b). Further analysis demonstrated detectable SARS-CoV-2 spike expression in these cells (Fig. 5f,g). Saliva was found to contain both epithelial (pan-cytokeratin positive (pCK+)) and immune (pCK−) populations. Using a ten-sample cohort (Supplementary Table 3), ISH revealed significantly more SARS-CoV-2 signals in pCK+ cells and further supported oral mucosal infection by SARS-CoV-2 (Fig. 5h,i). Using three-dimensional (3D) confocal microscopy, we confirmed that spike signals were found inside salivary-resident epithelial cells (Fig. 5j). In this cohort, ~5–10% of all salivary cells were found to be infected by SARS-CoV-2; using ISH, most infected cells were found to express ACE2 (Fig. 5k). Although typically found in the respiratory tract, a rare population of pCK+, ACE2-expressing SARS-CoV-2-positive ciliated cells was found in saliva, indicating that a very small fraction of saliva viral load reflects contributions from the respiratory tract (Extended Data Fig. 5c). Notably, SARS-CoV-2 spike independently localized both inside the shed epithelial cells and on their membrane surface. We also observed occasional co-localization of bacteria and SARS-CoV-2 on these epithelial cell membranes (Extended Data Fig. 5d).
The cell dynamics of shed oral epithelial cells have been poorly studied in health and disease12. Previous reports suggested that about 50% of shed cells in saliva from healthy individuals were epithelial43; however, our data suggest that only ~25% of shed cells in individuals with COVID-19 were epithelial (range, 4.0–39%). This reduction might reflect infection-related decreases in shed epithelial cell survival, leading to a relative enrichment of immune cell populations in saliva. Although rare, we observed pCKlow/SARS-CoV-2high cells that also presented with altered morphologies (Fig. 5h, green circle). Although some of these pCKlow/SARS-CoV-2high cells might be salivary immune populations, UMAPs of human salivary neutrophils do not express ACE2/TMPRSS2 (Extended Data Fig. 5e)44. Using ISH and 3D imaging, viral replication was detected inside of salivary epithelial cells (Fig. 5l). Concurrent analyses from National Institutes of Health (NIH) saliva samples confirmed our findings (Extended Data Fig. 5f–h).
Saliva from asymptomatic individuals contains infectious virus.
Our results pointed to two potential sources of SARS-CoV-2 in saliva: an acellular fraction from infected glands making de novo virus and a cellular fraction from shed, infected oral mucosa. To test the infectivity of both acellular and cellular salivary fractions, saliva samples with high viral load (n = 8; N1 Ct value 16–29) from individuals with COVID-19 were incubated with Vero cells to test for viral cytopathic effect (CPE) (Fig. 6a,b and Extended Data Fig. 6a,b). After confirming SARS-CoV-2 RNA in saliva from individuals who were entirely asymptomatic, specimens were processed to separate cell-free salivary fluid from epithelial cells (Methods). Acellular saliva induced CPE typical of coronavirus infection45 starting at Day 2 and becoming more pronounced on Day 6 in two of eight samples (N1 Ct 16 and 23, respectively). The cellular fraction induced CPE; however, this was observed only on Day 6 (Fig. 6a,b and Extended Data Fig. 6a,b). To confirm replication, supernatants were collected from monolayers exhibiting CPE and those without evidence of CPE and used to confirm infection in new Vero monolayers. These cultures followed the same time course as their parent cultures. These results demonstrate the infectiousness of saliva from asymptomatic/pre-symptomatic individuals with COVID-19. Notably, these results suggest the potential for expelled oral droplets containing infectious virus and infected cells to be a source of airborne transmission of SARS-CoV-2.
Fig. 6 |. Viral infection kinetics and the antibody response in saliva samples of patients with COVID-19.

a,b, To test the infectiousness of saliva, (a) acellular and (b) cellular fractions of saliva from individuals with high viral load (n = 8; Ct 16–29) were incubated in duplicate on Vero cells to observe viral CPE. Supernatants were incubated with new Vero cultures in duplicate to demonstrate replication of the virus. Both acellular and cellular fractions demonstrated CPE in some (n = 2) samples after 2 and 6 d, respectively; culture of supernatants elicited similar temporal dynamics as parent incubations (one independent replication). c,d, In a prospective, observational study, 39 NP-positive individuals with COVID-19 were grouped as having oral symptoms (with or without systemic symptoms), as having systemic symptoms only, or as being asymptomatic. Individuals demonstrated a heterogeneous disease course, with some requiring up to 13 weeks to clear detected virus in either the (c) nasopharynx or (d) saliva. Most asymptomatic individuals displayed only NP swab positivity, including two patients who developed symptoms during the prospective sampling (CoV12 and CoV13). e,f, Compared to healthy control saliva samples procured before the pandemic (n = 7), the LIPS assay shows that saliva from a cohort of individuals with COVID-19 (n = 32) contains antibodies to both (e) nucleocapsid and (f) spike viral antigens and occurred in 73% (22/30) and 54% (15/28) of samples, respectively. By week, the means ± s.e.m. are displayed to the right of each scatter plot as appropriate for nucleocapsid from weeks 1–7 (3.5 × 104 ± 1.2 × 104; 1.1 × 105 ± 4.8 × 104; 4.4 × 105 ± 1.8 × 105; 1.9 × 105 ± 8.4 × 104; 2.6 × 105 ± 9.0 × 104; 3.5 × 105 ± 9.1 × 104; 1.0 × 105± 2.5 × 104) and also for spike (1.8 × 104 ± 2.6 × 103; 2.2 × 104 ± 3.3 × 103; 3.7 × 104± 8.2×103; 4.2 × 104 ± 8.7 × 103; 4.7 × 104 ± 7.6 × 103; 5.7 × 104 ± 6.2 × 103; 3.3 × 104± 5.0 × 103). Fair to excellent agreement between presence of antibodies to viral antigens to nucleocapsid and spike was shown, respectively (Extended Data Fig. 6f). Horizontal dashed bar: (e and f) control light unit cutoff for positivity: mean plus 3 standard deviations. Scale bars (a and b), 100 μm. Ct, cycle threshold.
Levels of SARS-CoV-2 in saliva correlate with taste alterations.
To study SARS-CoV-2 kinetics in asymptomatic and symptomatic individuals with COVID-19, ambulatory individuals (n = 39) were enrolled from two groups: individuals with a positive nasopharyngeal (NP) swab (n = 30) or individuals with high-risk contact (cohabitation/occupational exposure, n = 9; Extended Data Fig. 6c, Supplementary Table 3 and Methods). Over a 5-week prospective interval, this trial evaluated SARS-CoV-2 RNA levels in saliva and NP swabs as well as the presence of antibodies against viral antigens in saliva. Based on the time from their first positive NP swab test, these data confirmed that salivary and NP swabs were generally well correlated over time (Fig. 6c,d), as has been previously shown46.
Some individuals cleared SARS-CoV-2 over very long periods (>2 months since the first test to negative NP and saliva tests), and not all truly asymptomatic individuals rapidly cleared the virus from their secretions. Of the 39 individuals enrolled in this study, 22% reported no symptoms at all, with a range of viral clearance from 0.5 weeks to 3.5 weeks (Extended Data Fig. 6c). This cohort’s average clearance rate was ~1 week less than in individuals with symptoms, suggesting that some asymptomatic individuals might carry SARS-CoV-2 in their nasopharynx and saliva over extended periods. Of eight asymptomatic participants, five were positive only in their NP swabs alone, including two who developed positive viral loads in the middle of the monitoring period. Two asymptomatic participants, CoV01 and CoV02, displayed only positive saliva when matched to NP viral loads 14 d after the positive NP test. These data highlight the possibility that the virus is cleared from the nasopharynx but can persist in saliva, suggesting sustained shedding of virus from SARS-CoV-2 infected oral sites.
In symptomatic individuals, the presence of SARS-CoV-2 RNA in saliva was positively associated with patient self-reported ‘loss of taste and smell’ (X2: 4.92, odds ratio (OR): 4.777 (1.137–19.977), P < 0.05; Fig. 6d and Supplementary Table 3). These data are corroborated by the saliva cell infection experiments (Fig. 6b and Extended Data Fig. 6b) showing that two patients with high viral load in saliva and reported taste changes displayed significant epithelial (pCK+) cell infection in ACE2-expressing cells (Extended Data Figs. 5g,h and 6d). Reports of ‘body ache/muscle pain’ was inversely correlated to saliva viral burden (X2: 4.74, OR: 0.171 (0.030–0.973), P < 0.05; Supplementary Table 3). Although a modest sample size, these data suggest that the presence of SARS-CoV-2 RNA in saliva positively correlates with taste alterations.
Public health measures, such as universal mask use and social distancing, are intended to reduce droplet and aerosol transmission. However, few studies have attempted to directly measure the change in saliva droplet ejection from individuals with COVID-19 by wearing a mask (Methods). The effectiveness of standard mask wearing to reduce droplet spread in these individuals was tested and demonstrated a more than ten-fold decrease in the detection of expelled salivary droplets (P < 0.005; Wilcoxon two-sided signed-rank test), including in some asymptomatic individuals with positive NP or saliva viral load (Extended Data Fig. 6e).
Virus-specific antibody kinetics in the saliva.
A previous study suggested rapid decay of IgA and IgM antibodies and stable IgG antibody titers in saliva samples up to 15 weeks after infection47. In our prospective cohort, salivary IgG antibodies were detected to the viral antigens nucleocapsid and spike in 73% (22/30) and 54% (15/28) of samples, respectively, in early recovered individuals with COVID-19 (Fig. 6e,f), with spike antibodies developing later than nucleocapsid antibodies. Nucleocapsid and spike antibodies in saliva and sera exhibited moderate (n = 10, kappa: 0.588, two-sided P = 0.0651) and strong (n = 9, kappa: 0.781, two-sided P = 0.0164) agreement, respectively (Extended Data Fig. 6f). Several of the individuals with both nucleocapsid and spike antibodies were those with protracted viral RNA positivity in nasopharynx and saliva, moderate symptoms and oral tissue infection (example: CoV23 in Extended Data Fig. 5g and CoV19 in Extended Data Fig. 6e). These data further support that SARS-CoV-2 infection can elicit sustained, local immune responses in saliva.
Discussion
Overall, the oral cavity represents a robust and underappreciated site for SARS-CoV-2 infection, and its direct role in viral transmission requires further exploration. To address this, we generated the first integrated adult human oral scRNA-seq atlas, and our data support a role for the oral axis in SARS-CoV-2 pathogenesis and transmission. Although principally focused on SARS-CoV-2, this atlas could be used to explore susceptibility to other pathogens and viruses. Our efforts were undertaken in parallel with the collaborative efforts led by the Human Cell Atlas that also used scRNA-seq to examine cell-specific SARS-CoV-2 tropism, leading to the COVID-19 Cell Atlas (https://www.covid19cellatlas.org/). Our and other atlases point to the permissiveness of the barrier epithelium to SARS-CoV-2 infection15,16.
There has been a historical focus on major SG viral infections48–50; however, a more detailed analysis of the minor SGs exhibit revealed enriched expression of principal entry factors in the glandular epithelia. Our atlases predicted rare expression of SARS-CoV-2 entry factors in the same cells of the minor SG glandular epithelia, but we found that this was more frequent using ISH. This demonstrated how both single-cell sequencing and ISH were important tools because subsequent experiments revealed that SARS-CoV-2 tropism was broadly identified for the ACE2/TMPRSS2-expressing acini and duct epithelial cells. Furthermore, minor SGs appear to be a relatively unexamined site for viral infection and replication, despite the detection of the virus throughout the functional unit for saliva production, which points to a mechanism whereby the acellular source of SARS-CoV-2 in saliva might be derived from these glands. The effect of this could be that SGs could propagate SARS-CoV-2 infection and sustain COVD-19 in other body sites51.
One limitation of our scRNA-seq approach was the underrepresentation of suprabasal epithelial cells relative to other epithelial cell subpopulations (Figs. 1 and 2). After ISH mapping of the major SARS-CoV-2 entry factor expression in basal and suprabasal epithelial cells (Fig. 3), it became imperative for us to look for infected epithelial cells in saliva. Suprabasal mucosal cells eventually shed from the most terminally differentiated tissue layers, estimated to occur once every 3 h52; they also maintained entry factor expression and evidence of SARS-CoV-2 infection (Fig. 5). Although the constant shedding of these cells might serve as a local protective mechanism for oral tissue infection52, this might come at the expense of other aerodigestive tissue niches deeper into the body if these shed oral epithelial cells are harboring infected cells11,12. However rare, these cell subpopulations were found to transmit the virus to Vero cells ex vivo from saliva with high viral load. In total, the connection of the oral cavity to the rest of the body via saliva remains to be fully understood in health and disease.
Our findings highlight how simultaneous testing of oral and NP sites might be required to fully understand SARS-CoV-2 spread. Moreover, our findings might partially explain some false-negative tests using only NP or saliva testing. However, these results raise new questions about COVID-19 pathogenesis, including 1) whether this is primarily a ‘nasal-first’ infection that spreads to the oral cavity, 2) the possibility of an ‘oral-first’ infection via droplet/aerosol inoculation or fomite ingestion, and/or 3) whether the pattern of infection affects disease severity and host immunological responses. To test whether oral transmission can precede nasal infection and/ or occur in the absence of nasal infection, it will be necessary to design studies that include daily surveillance using NP and salivary tests in an at-risk cohort.
Considering oral SARS-CoV-2 infection and the ease of saliva for transmission (Fig. 6), it remains critical to further understanding of the dominant modes of viral spread across the spectrum of asymptomatic, pre-symptomatic and symptomatic individuals. Asymptomatic transmission of SARS-CoV-2 remains the ‘Achilles’ Heel’ of this pandemic53, and, owing to the peripheral anatomical location and frequent exposure of oral tissues to the external environment, it remains possible that these tissues play a major role in the asymptomatic spread of SARS-CoV-2. Taken together, this raises the possibility that the oral cavity actively participates in SARS-CoV-2 transmission.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41591-021-01296-8.
Methods
All the research reported in this manuscript complies with all relevant ethical regulations, and informed consent was obtained from all human participants.
Ethical approval.
NIH.
The NIH single institutional review board (IRB) conducts ethical reviews for human research studies as required by Department of Health and Human Services regulations for the Protection of Human Subjects. All patients seen at the author’s (B.M.W.) institute (NIH/NIDCR) reported herein provided informed consent before participation in IRB-approved research protocols (NIH IRB: 20-D-0094, NCT04348240; NIH IRB: 15-D-0051, NCT02327884). Individuals on 20-D-0094 had the option to receive a $50 payment per visit ($300 total) to offset the cost of travel.
The University of North Carolina at Chapel Hill.
The Office of Human Research Ethics (OHRE) is responsible for ethical and regulatory oversight of research at the University of North Carolina at Chapel Hill (UNC-CH) that involves human subjects. The OHRE supports and oversees the work of the IRBs. IRB Boards A through F review research in a wide variety of areas, such as biomedical research conducted at the School of Medicine, School of Pharmacy, UNC Hospitals, the Adams School of Dentistry and other research in other units that involves biomedical interventions. Expertise is focused on medical, surgical, physiological or pharmacological studies and includes research with drugs, devices, counseling or other interventions. Studies that require dental expertise are reviewed by Biomedical Boards B and D. All patients seen at the author’s (K.M.B.) institute (UNC-CH) reported herein provided informed consent before participation in IRB-approved research protocols (UNC IRB: 19-0183, NCT04105569; UNC IRBs 20-1501 and 15-1814; UNC IRB: 20-0792). In the clinical protocol for human gingival single-cell sequencing, each individual received an electric toothbrush and $200 at the completion of the study; these individuals also received free parking vouchers for each appointment
Human gingival scRNA-seq.
Clinical protocol.
Five individuals were enrolled in this study conducted at the UNC Adams School of Dentistry, and five total healthy individuals (three females and two males, aged 20–30 years) completed the experimental gingivitis protocol in October 2019. One was used as a control and for future ISH studies. Each individual was determined to be systemically healthy. Individuals received initial prophylaxis (Day 0) with a 3-week, stent-induced, biofilm overgrowth phase to standardize inflammation induction temporally54,55. Gingivitis was experimentally induced over the upper left maxillary premolar and first and second molars only (Clinicaltrials.gov ID: NCT04105569; UNC-CH OHRE IRB approval no. 19-0183). The experimental protocol yielded mildly inflamed gingival mucosa (that is, gingivitis; gingival index scores56 ranging from 1.0 to 1.5; Extended Data Fig. 1c). Full-thickness gingival biopsies (0.3 μm3) were collected on the same day from each individual at the 3-week time point from the upper left maxillary lingual interproximal papilla (universal number 14–15). Biopsy samples were split for tissue dissociation and tissue fixation.
Cold protease dissociation/fluorescence-activated cell sorting.
Split tissues were placed immediately in ice-cold, calcium-free and magnesium-free 1× PBS for overnight shaking and incubation with 1:1,000 ROCK inhibitor (Sigma-Aldrich, Y27632). Cold protease dissociation cocktail was made using cold 1× PBS, 2× DNase (2.5 mg ml−1, Sigma-Aldrich, 9003-98-9) and 1:1,000 ROCK inhibitor. Protease (Sigma-Aldrich, P5380) was then added to a concentration of 100 mg ml−1. Sterilized dissecting forceps (Roboz) were used to mechanically separate gingival into several pieces. The enzymatic cocktail was incubated with gingival tissues for 35 min, but progress was checked every 10 min with 10-μL aliquots. Dissociation was completed when >75% of the solution was single cells, and then gingival single cells were pelleted at 1,800 relative centrifugal force and washed twice with an appreciable volume of a cocktail of cold culture medium, 1:1,000 Y-27632 and 10% FBS. After dissociation, viability was checked using Trypan blue (viability was confirmed >80%), and red blood cell (RBC) lysis was achieved using a 1× RBC buffer (BioLegend, 420301). A fluorescence-activated cell sorting (FACS) buffer (a mixture of PBS, 1% FBS and 1:1,000 ROCK inhibitor) was used throughout the resuspension and cell sorting process. Single cells were enriched using a 35-mm Falcon cell strainer (Thermo Fisher Scientific). Before FACS, the cell pellet was resuspended in the FACS buffer and stored on ice. To isolate living cells by FACS, rat anti-CD49f Alexa Fluor 647 (a6-Integrin, BioLegend clone GoH3) was added to cells and incubated for 60 min on ice. Sytox (Thermo Fisher Scientific, 1:1,000) was added 5 min before FACS to evaluate live/dead cells using a Sony SH800S Cell Sorter, with the following controls: anti-CD49f Alexa Fluor 647-negative and Sytox Dead Cell Stain-negative cell suspensions. Cells were sorted ~6 counts per s, and live cells were sorted into tubes directly for 10x capture, library preparation and scRNA-seq.
10x capture, library preparation and sequencing.
Cells were processed using the 10x Genomics Chromium Controller and the Chromium Single Cell 3’ GEM, Library & Gel Bead Kit v3 (PN-1000075) following the manufacturer’s user guide (https://tinyurl.com/y28bwe67). Aliquots of the sorted cells were stained with acridine orange and propidium iodide and assessed for viability and concentration using the LUNA-FL Dual Fluorescence Cell Counter (Logos Biosystems). Approximately 3,200 cells per sample were loaded onto the Chromium Chip B with a target recovery of 2,000 cells per sample for library preparation. Single cells, reverse transcription reagents and gel beads coated with barcoded oligos were encapsulated together in an oil droplet to produce gel beads in emulsion (GEMs). Reverse transcription was performed using a C1000 thermal cycler (Bio-Rad) to generate complementary DNA (cDNA) libraries tagged with a cell barcode and unique molecular index (UMI). GEMs were then broken, and the cDNA libraries were purified using Dynabeads MyOne SILANE (Invitrogen) before 12 amplification cycles. Amplified libraries were purified with SPRIselect magnetic beads (Beckman Coulter) and quantified using an Agilent Bioanalyzer High Sensitivity DNA chip (Agilent Technologies). Fragmentation, end repair, A-tailing and double-sided size selection using SPRIselect beads were then performed. Illumina-compatible adapters were ligated onto the size-selected cDNA fragments. Adapter-ligated cDNA was then purified using SPRIselect beads. Uniquely identifiable indexes were added during 12 amplification cycles. The finalized sequencing libraries were then purified using SPRIselect beads, visualized using the Bioanalyzer High Sensitivity DNA chip, quantified with the KAPA SYBR FAST Universal qPCR Kit for Illumina (Roche) and StepOnePlus Real-Time PCR System (Applied Biosystems), normalized to 4 nM and pooled. Pooled libraries were sequenced on a NextSeq 500 machine (Illumina). Libraries were denatured and diluted following standard Illumina protocol, spiked with 1% PhiX sequencing control (Illumina), loaded onto the flow cell at 1.8 pM and sequenced in paired-end format (Read 1: 28 bp, Read 2: 91 bp) to a total depth of 523 million read pairs passing quality filters. The 10x Genomics Cell Ranger analytical pipeline v3.1.0 and GRCh38 v3.0.0 reference package was used to analyze the data following default settings.
Human minor SG scRNA-seq.
Tissue dissociation.
Minor SGs were collected from healthy volunteers who provided informed consent on NIH protocols 15-D-0051 (PI: Warner) or 94-D-0094 (PI: Warner). De-identified submandibular and parotid glands were obtained fresh from the Human Cooperative Tissue Network and portioned into RNAlater for RNA-seq or were formalin fixed in 10% neutral buffered formalin overnight before being transferred to 70% PBS-buffered ethanol and then paraffin embedded for IHC and ISH studies. Five healthy individuals provided minor SG biopsies for scRNA-seq. Tissues were biopsied following standard methods and immediately placed in ice-cold RPMI. Tissues were sharply dissected into 1–2-mm pieces and then dissociated using the Miltenyi Multi-tissue Dissociation Kit A using the Multi_A01 in C-type tubes at 37 °C in an OctoMACS tissue disruptor using heated sleeves. Crude single-cell suspensions were serially filtered through 70- and 30-μm filters and rinsed with 1× Hanks’ buffered salt solution. Cells were centrifuged at 300g for 10 min at 4 °C and washed once with 1× Hanks’ buffered salt solution. Cell counting and viability were determined using a Trypan blue exclusion assay. Suspensions with greater than 35% viability were used for subsequent sequencing. Additional glands from these patients were submitted for histopathological assessment, including focus scoring.
Single-cell capture, library preparation and sequencing.
Single-cell suspensions targeting approximately 5,000 cells were prepared as described above and loaded onto a 10x Genomics Chromium Next GEM Chip B following the manufacturer’s recommendations. After cell capture, single-cell library preparation was performed following the instructions for the 10x Chromium Next GEM Single Cell 3’ kit v3 (10x Genomics). The libraries were pooled and sequenced on four lanes of a NextSeq500 sequencer (Illumina), adopting the read configuration indicated by the manufacturer.
scRNA-seq data processing and quality control.
Read processing was performed using the 10x Genomics workflow. Briefly, the Cell Ranger v3.0.1 Single-Cell Software Suite was used for demultiplexing, barcode assignment and UMI quantification (http://software.10xgenomics.com/single-cell/overview/welcome). Sequencing reads were aligned to the hg38 reference genome (Genome Reference Consortium Human Build 38) using a pre-built annotation package obtained from the 10x Genomics website (https://www.10xgenomics.com/). Samples were demultiplexed using the ‘cell ranger mkfastq’ function, and gene count matrices were generated using the ‘cellranger coun’ function. The single-cell data were analyzed in R (v3.5.0) using Seurat (v3.1.2). Filtering was performed using the standard quality control steps provided on the Satija Lab website (https://satijalab.org/). Cells containing more than 200 and fewer than 2,500 unique features were retained. From this set, cells with greater than 15% of read counts attributed to mitochondrial DNA were filtered out. We adjusted this value from 5% to 15% to increase the yield from each sample and did not observe substantive changes in our results after adjustment. Data were normalized using the ‘NormalizeData’ command (scale factor = 10,000).
Annotation of clusters.
SG cells from n = 5 non-Sjögren’s syndrome and otherwise healthy individuals were integrated into a single Seurat object containing 12 clusters at resolution = 0.1. The clusters were manually annotated with marker genes identified by Seura’s ‘FindAllMarkers’ function, using the receiver operating characteristic test. The ‘FeaturePlo‘ function was used to identify subpopulations within identified clusters and confirm the expression of known marker genes within clusters.
Immune cell activation analysis.
Immune activation signatures were extracted from MSigDB and scored using scanpy.tl.score_genes().
Pan-epithelial expression analysis for SARS-CoV-2 entry factors.
Expression of ACE2 and TMPRSS2 in epithelial cells from the human oral cavity were compared with a human nasal airway dataset37 and a human gut dataset38. The total UMI counts were downsampled using ‘log1p() in numpy’ -> ‘scanpy.pp.log1p()’ for each dataset so that the median number of UMIs per cell for epithelial cells of each dataset is equal to that of the lowest dataset. Datasets were then pooled, normalized by cell and log1p transformed, based on which fraction of cells expressing ACE2/TMPRSS2 and the respective level of expression were calculated.
Integrated single-cell analyses.
Clustering and annotation.
Oral atlases were retrieved from published and unpublished datasets (human SGs: Warner et al.; human gingiva/palatal mucosae: Byrd et al.). Raw expression matrices were filtered, normalized and log transformed for further processing using standard SCANPY (v1.4.3) procedure57. For the two unpublished human datasets, BBKNN was used for batch correction across samples, and multiple clustering rounds of different resolutions were performed to resolve subpopulations, and then manual annotations were made considering published human references58. For integration across datasets, each dataset was first randomly downsampled to having ≤500 cells per cell type, and then pooled raw expression values were re-processed following standard SCANPY procedure while using Harmony for correcting batch effects across datasets and samples59. Again, multiple clustering rounds of different resolutions were performed, and manual annotations were made. Illustration of the results was generated using Python code around SCANPY.
GTEx RNA-seq analysis.
Publicly available data were downloaded from the GTEx v7 Portal39 (http://www.gtexportal.org/home/datasets). Of the 2,964 samples corresponding to the esophagus, heart, lung, minor SG, small intestine and stomach, 2,544 had expression >0 transcripts per million (TPM) of any of the viral entry factors and were amenable to being fit into a dendrogram. Expression values were log10 transformed with an added pseudocount of 0.01 TPM, the lowest non-zero expression value in the dataset, and linkages were built using the UPGMA algorithm on Euclidean distances between samples. The heat map and dendrogram were visualized using the seaborn package v0.11.0 (https://seaborn.pydata.org) in Python 3.6.
IHC and ISH for human oral tissues and saliva.
Oral and oropharyngeal tissue acquisition.
Participant tissues included remnant oral specimens previously collected through the UNC Adams School of Dentistry Oral Pathology Biobank (UNC-CH OHRE IRB approval no. 20-1501) or the Center for Oral and Systemic Diseases Biorepository (UNC-CH OHRE IRB approval no. 15-1814). Lab and project personnel were double blinded and did not have access to protected health information (PHI) or any links to any PHI. Project members did not have interpersonal contact with patients who provided specimens. All de-identified tissues were previously fixed in 10% formalin and embedded in paraffin blocks.
Saliva cytospin.
Saliva samples were collected from patients with COVID-19 and fixed with 4% paraformaldehyde for 30 min. Then, the samples were centrifuged, and the cell pellet was dissolved into 5 mM EDTA in PBS. A portion of the samples was loaded into disposable sample chambers (double cytofunnel, Thermo Fisher Scientific) with slides and applied into a cytospin machine (Shandon Cytospin 4, Thermo Fisher Scientific) following the manufacturer’s instructions and centrifuged at 1,000 r.p.m. for 5 min. The cytospin slides were stored in 70% ethanol under 4 °C until IHC or RNA ISH. Saliva cell blocks: saliva samples were diluted in 10 volumes of 4% paraformaldehyde in 1× PBS. Cells were allowed to fix for 24 h at room temperature. After 5 min of centrifugation at 1,000g, cell pellets were transferred to 70% ethanol. Cell pellets were then ensnared by adding thrombin and fibrin to the cell pellet, which could be subsequently embedded for tissue sectioning.
ISH/IHC.
Formalin-fixed, paraffin-embedded (FFPE) tissue sections were baked overnight, followed by deparaffinization with xylene for 5 min twice and dehydration with 100% ethanol for 1 min twice. Then, the sections were incubated with hydrogen peroxide for 10 min, and antigen retrieval was performed with boiled water for 15 min and with protease at 40 °C for 15 min. Probes were hybridized at 40 °C for 2 h in an oven, followed by signal amplification and washing steps. The hybridized signals were visualized by Fast Red, followed by counterstaining with hematoxylin. The probe targeting the human housekeeping gene UBC was used as a positive control to test for RNA quality in the tissue sections. A bacterial gene, DapB, was used as a negative control target. The images were acquired using an Olympus VS200 slide scanner light microscope with a ×60 1.42 N.A. objective. All RNAscope ISHs were performed following the manufacturer’s recommended conditions, with the following exceptions: for ISH, pre-treatment conditions were modified as follows: protease plus digestion at 40 °C for 15 min; antigen retrieval in RNAscope target retrieval in slide steamer control temperature at 99 °C for 15 min.
UNC:
positive control probe: 320861, 310041; negative control probe: 320871, 310043; SARS-CoV-2 probe: ACD, 848561; ACE2 probe: 848151; TMPRSS2 probe: 470341; TMPRSS4 probe: 565301; TMPRSS11D probe: 870841. CSTL probe: 858611; ACE2 antibody: mouse monoclonal (protein tech no. 66699-1-IG), (ACE2 diluted 1/2,000, PBS 1% BSA, 1% donkey serum PBS); AQP5 antibody: goat antibody, Santa Cruz Biotechnology, C-19, sc9891 (dilution 1/300 PBS 1% BSA, 1% donkey serum, PBS); ACTA2 antibody: goat, Novus Biological, NB300–978 (dilution 1/200 PBS 1% BSA, 1% donkey serum, PBS); pCTK antibody: rabbit polyclonal (Abcam, ab234297); SARS2 antibody: rabbit polyclonal, Invitrogen, cat. no. PA1-41098 (1:500 dilution); cytokeratin antibody: mouse monoclonal, Dako cat. no. M3515; secondary antibodies: Al secondary antibodies were in donkey (D-anti-goat, D-anti-rabbit, diluted 1/300 in 1/300 PBS 1% BSA, 1% donkey serum, PBS). For the detection of anti-ACE2, we used TSA detection kit from Biotium no. 33000); mounting media. The slides were mounted in Prolong Gold anti-fade (with DAPI for the IF; without DAPI for the ISH).
NIH:ISH.
Positive control probe: 320861; negative control probe: 320871; ACE2 probe: 848151-C3; TMPRSS2 probe: 470341-C2. V-nCoV2019-S (848561) is an anti-sense probe specific to the spike protein sequence of the viral RNA and, thus, provides information about infection and viral load in tissue—that is, presence of the virus. V-nCoV2019-orf1ab-sense (859151) (40 probe pairs) detects replicating virus. Therefore, these can be used to detect viral replication in tissue. 848561 was used in the original set of ISH (Fig. 4) and, in the last set, was mixed with 859151 (Fig. 5) to detect both the virus and replication at the same time.
IHC studies.
ACE2 antibody: mouse monoclonal (protein tech no. 66699-1-IG), (ACE2 diluted 1/2,000, PBS 1% BSA, 1% donkey serum PBS); validated for immunofluorescence (IF), western blot (WB) and flow cytometry (FC) and on human tissues (colon). AQP5 antibody: goat antibody, Santa Cruz Biotechnology, C-19, sc9891 (dilution 1/300 PBS 1% BSA, 1% donkey serum, PBS); validated by WB, IHC on human tissues (human SG with primarily apical membrane expression on acini). ACTA2 antibody: goat, Novus Biological, NB300-978 (dilution 1/200 PBS 1% BSA, 1% donkey serum, PBS); validated using WB of mouse and human duodenum and frozen section IF of the mouse heart. Expected staining is demonstrated on the terminal acini of the human SGs. pCTK antibody: rabbit polyclonal (Abcam, ab234297) demonstrated reactivity with human epidermis. SARS-COV-2 spike antibody: mouse monoclonal (1A9) (GeneTex, GTX632604). HIER Citric Buffer pH 6.0. (dilution 1/50, in 10 FBS, 1% BSA, PBS).
Validation.
Not only are these antibodies verified by the manufacturer in each case, but we also used internal positive and negative controls to verify proper staining and hybridization on these tissues. For example, AQP5, ACE2 and ACTA2 proteins were co-stained on the same tissue sections. Expectedly, ACE2 resides in the membranes of ducts and acini (basolateral and apical) as well as in some endothelial cells. AQP5 co-localizes with some ACE2 signals (Fig. 4); however, ACTA2 does not. pCK exhibits staining only in epithelial cells in FFPE sections and also cell blocks. In our view, the antibodies used passed both the manufacturer’s validation and our internal validation. Proper ISH cell targeting using IF for ACE2 confirmed expected localization to positive cells as predicted by RNA-seq and as shown. All conditions were exhaustively optimized and repeated at least three times to capture representative images for publication.
Clinical studies.
UNC epidemiological and immunological aspects of COVID-19 saliva.
A subset of ten samples from outpatients with COVID-19 were collected from an IRB-approved study (UNC-CH OHRE IRB approval no. 20-0792). The purpose of this study was to describe the epidemiology, clinical features and immunological response to SARS-CoV-2 infection and develop new diagnostic tests focused on saliva. Study individuals were older than 18 years and were tested for COVID-19 (NP swab) at UNC Healthcare and contacted as outpatients for sample collection. The individuals in this study were classified as having ‘mild’ COVID-19. Outpatients were enrolled during diagnosis at the UNC-CH Respiratory Diagnostic Center (RDC) and were recruited after a positive clinical test. All participants were asked to come to the RDC for saliva collection, which occurred by the participant actively drooling into a 5-ml collection tube. For all samples, specimens were evaluated for volume, total cell count and cell differential before proceeding to fixation and ISH/IHC analyses. All samples were quantified using >200 cells per sample from multiple fields selected at random.
NIH transmissibility and viral load of SARS-CoV-2 in oral secretions.
The purpose of this NIH IRB-approved short-term longitudinal study (PI: Warner, 20-D-0094, Clinicaltrials.gov NCT04348240) was to examine the potential for transmission of COVID-19 via saliva in asymptomatic and mildly symptomatic individuals, to compare the utility of saliva as a diagnostic fluid with standard NP swabs, to examine the real-world effectiveness of standard mask use to reduce transmission via saliva and to collect biofluid and tissue samples for mechanistic and biomarker studies. A limited follow-up, prospective, observational study follows individuals from the initial diagnosis of COVID-19 or a high-risk exposure through the individual’s resolution of COVID-19. Individuals providing informed consent were principally recruited using NIH IRB-approved flyers handed out in a COVID-19 testing facility targeting high-risk individuals and those who very recently tested positive for COVID-19. Inclusion criteria included people 18 years or older without symptoms or with mild symptoms (for example, low-grade fever, mild malaise, sore throat, runny nose, cough and loss of taste or smell) who had been in close contact (for example, live in the same house or long-term occupational exposure) with someone who had tested positive for COVID-19 or individuals with or without symptoms who had recently tested positive for COVID-19. Exclusion criteria included individuals who tested positive for COVID-19 with active moderate or severe symptoms (for example, moderate or worse upper and lower respiratory symptoms) or currently requiring hospitalization at a hospital other than the NIH Clinical Center (recovering patients with COVID-19 can be enrolled). Individuals were scheduled to be seen in an outpatient setting every 7 d through day 15 if they were negative at day 15 or up to day 28 if they were not. Individuals were seen on study between the hours of 8:30 and 11:00 at the NIH COVID-19 Testing Center. Two individuals were seen in the NIH Special Clinical Studies Unit, including a single individual who provided lower lip minor SG biopsy at their day 1 appointment (CoV49). Before arriving at the testing facility, individuals were asked to refrain from eating 90 min before testing. First, individuals were asked to speak by reading Samantha Smith’s ‘Ambassador of Peace’ address (~2-min duration, 5th grade reading level) directly into an emesis bag shortened to a depth of 10 cm with a ¼ section of 3’ X 3’ Pro-Gauze Pads (Curad) dampened with 2 ml of 1× PBS. Gauze pads were recovered for saliva and viral burden analysis as described above. Individuals then repeated the speaking exercise with a disposable procedure mask on day 1, day 8 and/or 15 (or day 21 or day 28, when applicable). Whole unstimulated saliva was collected in sterile 50-ml conical vials by asking patients to refrain from swallowing, allowing the pooling of saliva under the tongue and in the vestibules of the mouth and then pushing the saliva into the open conical vial for 2–3 min. A synthetic Copan swab (NP swab) was used to transfer a standardized amount of saliva from the 50-ml tube to a new 15-ml conical tube containing 3 ml of viral transport media for Centers for Disease Control and Prevention (CDC) SARS-CoV-2 testing. Swabs of the nasopharynx were obtained and placed into 15-ml conical tubes containing 3 ml of viral transport media for CDC SARS-CoV-2 testing. The remaining samples were immediately placed on ice and transported to the NIH Department of Laboratory Medicine for testing and cryostorage. Symptom questionnaires were collected at consent and again every 2–3 d throughout the study, including the COVID-19 screening questionnaire and the Monell Smell and Taste Instruments, using an NIH IRB-approved online portal.
CDC qRT–PCR test for testing the NP swabs and saliva.
In response to the SARS-CoV-2 pandemic, the CDC 2019 novel Coronavirus (2019-nCoV) Real-Time PCR Diagnostic Panel received U.S. Food and Drug Administration approval and was adopted by the NIH Clinical Center, herein referred to as the SARS-CoV-2 RT–PCR assay. The SARS-CoV-2 RT–PCR assay is used to screen NP and oropharyngeal swab specimens in viral transport medium and bronchoalveolar lavage for SARS-CoV-2 from patients with respiratory symptoms. The SARS-CoV-2 RT–PCR assay implemented at the NIH Clinical Center uses the easyMAG automated nucleic acid extractor (bioMérieux). A Taqman assay using two primer/probe sets is used to detect two distinct regions of the N gene (nucleocapsid protein, referred to as N1 and N2) and an internal control to detect the human RNase P (RP1) gene present in all specimen collections. This assay was performed on the ABI 7500 Fast Real-Time PCR System (Thermo Fisher Scientific).
Procurement and testing of autopsy tissue specimens for SARS-COV-2.
Autopsy tissues.
Consent for autopsies of COVID-19 victims was coordinated by the NIH COVID-19 Autopsy Consortium and obtained from family members. Autopsies were performed in the National Cancer Institute (NCI) Laboratory of Pathology, and tissues were recovered per family wishes for histopathological and other downstream analyses.
Viral quantification of autopsy tissues.
Total RNA was extracted from RNAlater (Invitrogen)-preserved tissues collected at autopsy using the RNeasy Fibrous Tissue Mini Kit (Qiagen) according to the manufacturer’s protocol. Before extraction, tissues were mechanically homogenized on the GentleMACS Octo Dissociator with Heaters (Miltenyi Biotec) using the gentleMACS M tubes with the RNA_02 program. A NanoDrop ND-1000 Spectrophotometer (Thermo Fisher Scientific) was used to quantify RNA concentrations. The QX200 AutoDG Droplet Digital PCR System (Bio-Rad) was used to detect and quantify SARS-CoV-2 RNA using the SARS-CoV-2 Droplet Digital PCR Kit (Bio-Rad), which contains a triplex assay of primers/probes aligned to the CDC markers for SARS-CoV-2 N1 and N2 genes and human RPP30 gene. Ninety-six-well plates were prepared with technical replicates of up to 550 ng of RNA per well using the aforementioned kit according to the manufacturer’s instructions. The QX200 Automated Droplet Generator (Bio-Rad) provided microdroplet generation, and plates were sealed with the PX1 PCR Plate Sealer (Bio-Rad) before proceeding with RT–PCR on the C1000 Touch Thermal Cycler (Bio-Rad) according to the manufacturer’s instructions. Plates were read on the QX200 Droplet Reader (Bio-Rad) and analyzed using the freely available QuantaSoft Analysis Pro Software (Bio-Rad) to quantify copies of N1, N2 and RP genes per well, which was then normalized to RNA concentration input. For samples to be considered positive for SARS-CoV-2 N1 or N2 genes, they needed to average the manufacturer’s limit of detection of ≥ 0.1 copies per μl and two positive droplets per well. Additional ISH quantification: viral mRNA and replication strand (sense) RNA was quantified using the fully automated VIS DIA VisioMorph system (Visiopharm, v2020.05.17823). After ISH, slides were scanned at ×400 magnification using an Axioscan Z1 slide scanner (Zeiss Group) and imported into Visiopharm. A customized analysis protocol package was used to 1) subtract the background signal in all the channels used, 2) detect the probe signal base on the intensity/size/form of the signal in the channel corresponding to each probe set, 3) segment cells based on DAPI staining and 4) count the total number of dots and the number of dots per cell in the whole tissue area. All the slides were analyzed with the same conditions. The results obtained were processed, and the number of dots per cell in negative control was subtracted from the results obtained in the target samples. After this correction, the results were expressed as the number of dots (probe) per cell. The data were plotted in a colored matrix showing the number of cells for each combination of the number of probes per cell. The cells were colored based on the percentage of cells representing each section to the total number of positive cells.
Systematic microscopic assessment and immunophenotyping.
To conduct a systematic histological assessment of the available salivary and oral autopsy tissues, a convenience set of tissues were selected, and 5-μm serial sections were cut and immunohistochemical studies were performed (H&E, CD3, CD4, CD8, CD20, CD68, HLA-DR and GMZB) from the NIH COVID-19 Autopsy Consortium. H&E sections were evaluated at low and high magnification. Immunohistochemical stains were completed using the NIH Clinical Center clinical testing antibodies. Two board-certified pathologists (Oral and Maxillofacial Pathology, B.M.W.; Anatomic and Autopsy Pathology, D.E.K.) and one pathologist in training (Anatomic Pathology, B.G.) reviewed all the cases, recorded histopathological features and scored immunostains on a 0–3 scale (0, negative; 1, weak; 2, moderate; 3, strong). After consensus was achieved across the pathologists, data were summarized as a table, and an immunohistochemical scoring heat map was constructed.
Antibody validation.
All immunohistochemical studies used on anatomic pathology autopsy specimens were performed in the NCI Center for Cancer Research, Laboratory of Pathology, a laboratory approved by the College of American Pathologists Laboratory Accreditation Program. The included antibodies were clinically validated to the satisfaction of the College of American Pathologists checklists and inspectors. Positive and negative tissue controls for each antibody are regularly checked and maintained in the Laboratory of Pathology.
NCI Laboratory of Pathology clinically valid antibody information
| Antibody | Clone | Control | Source | Catalog no. | Dilution | Titer no. |
|---|---|---|---|---|---|---|
| CD20 | L26 | Tonsil | Roche | 760-2531 | Predilute | |
| CD3 | 2GV6 | Tonsil | Ventana | 790-4341 | Predilute | |
| CD4 | SP35 | Tonsil | Roche | 790-4423 | Predilute | |
| CD8 | SP57 | Tonsil | Roche | 790-4460 | Predilute | |
| CD68 | KP-1 | Tonsil | Roche | 790-2931 | Predilute | |
| HLA-DR | TAL.1B5 | Tonsil | Dako | M0746 | 1:200 | 956 |
| Granzyme B | GrB-7 + D170 | Tonsil | Millipore | MAB 3070 | 1:100 | 861 |
Salivary measurement of antibodies against the SARS-COV-2 proteins.
The luciferase immunoprecipitation systems (LIPS) immunoassay was used to study IgG antibody response against SARS-CoV-2 in saliva. Owing to the potential biohazard of infectious SARS-CoV-2 in saliva, a viral inactivation protocol involving heating saliva at 56 °C for 30 min was employed. For these SARS-CoV-2 antibody measurements, Renilla luciferase-nucleocapsid and Gaussia luciferase-spike fusion protein extracts were employed with protein A/G beads as the IgG capture reagent as previously described60. Owing to the lower levels of immunoglobulin present in saliva, 10 μl of each saliva sample was used in the LIPS assay as previously described61. Known SARS-CoV-2 serum samples for IgG antibodies against nucleocapsid and spike proteins and saliva from uninfected controls were used for assigning seropositive cutoff values and for standardization. The cutoffs for antibody-positive saliva were calculated as 3 standard deviations plus the mean of control saliva for nucleocapsid and spike: 63,000 and 47,000, respectively. The cutoffs for antibody-positive sera were calculated as 3 standard deviations plus the mean of control sera for nucleocapsid and spike are as follows: 125,000 and 45,000, respectively60,61.
Saliva infectivity and reinfection assay.
Saliva for SARS-CoV-2 infectivity testing. High viral load (n = 8; CDC SARS-CoV-2 N1 Ct value 16–29) for SARS-CoV-2 infectivity testing was collected on either NIH IRB-approved protocols (20-D-0094, PI: Warner; 20-CC-0128, PI: Frank) from individuals with COVID-19 or as de-identified and anonymized, discarded samples from the NIH Asymptomatic Testing Pool. Saliva samples from patients with COVID-19 were cultured with Vero cells (ATCC CL-81). Briefly, Vero cells were propagated in DMEM medium supplemented with GlutaMAX, sodium pyruvate, 10% heat-inactivated FBS, 100 U ml−1 of penicillin, 100 μg ml−1 of streptomycin and 25 ng ml−1 of amphotericin B. The day before culture initiation, 24-well plates (Corning) were seeded with 2 × 105 Vero cells per well in 1 ml and incubated at 37 °C and 5% CO2 overnight in duplicate. Frozen saliva samples were thawed at room temperature and then centrifuged for 5 min at 6,000 r.p.m. to separate the cellular fraction from the acellular fraction. Acellular fractions were collected, and 150 μl was used to directly infect the cell monolayer in duplicates for 1 h at 37 °C, after which 1 ml of virus isolation medium was added per well (VI medium: DMEM supplemented with 2% heat-inactivated FBS, GlutaMAX, sodium pyruvate, 100 U ml−1 of penicillin, 100 μg ml−1 of streptomycin and 275 ng ml−1 of amphotericin B). The cell pellet was resuspended in VI medium and centrifugated for 5 min at 6,000 r.p.m. The supernatant was discarded, and the cell pellet was resuspended in an initial volume of VI medium. Five-fold serial dilution was performed in VI medium. Co-cultures with Vero cells were initiated with 200 μl of 1/5 and 1/25 saliva cell dilution and 1 ml of VI medium. Cultures were monitored daily for the appearance of CPE using an EVOS 5000 digital microscope. Half of the culture medium was replaced on day 3, and cultures were terminated on day 6. Culture supernatants were collected on day 3 and day 6 and frozen at −80 °C until further use. For virus passages, Vero cells were seeded at 7.5 × 105 Vero cells per well in 3 ml in six-well plates the day before. Before infection, Vero cell monolayers were washed twice with VI medium and infected with culture supernatant from the initial culture diluted five-fold in VI medium. Cultures were monitored for the appearance of CPE using an EVOS 5000 digital microscope.
General statistical methods.
All non-sequencing-based data were analyzed in JMP(r) v14.0.0 and/or Prism 9. Appropriate statistical tests were selected based on data type and are described throughout the text, where applicable. Figures were constructed using Prism 9 when not otherwise described elsewhere.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1 |. A hypothetical oral infection and transmission axis for SARS-CoV-2.
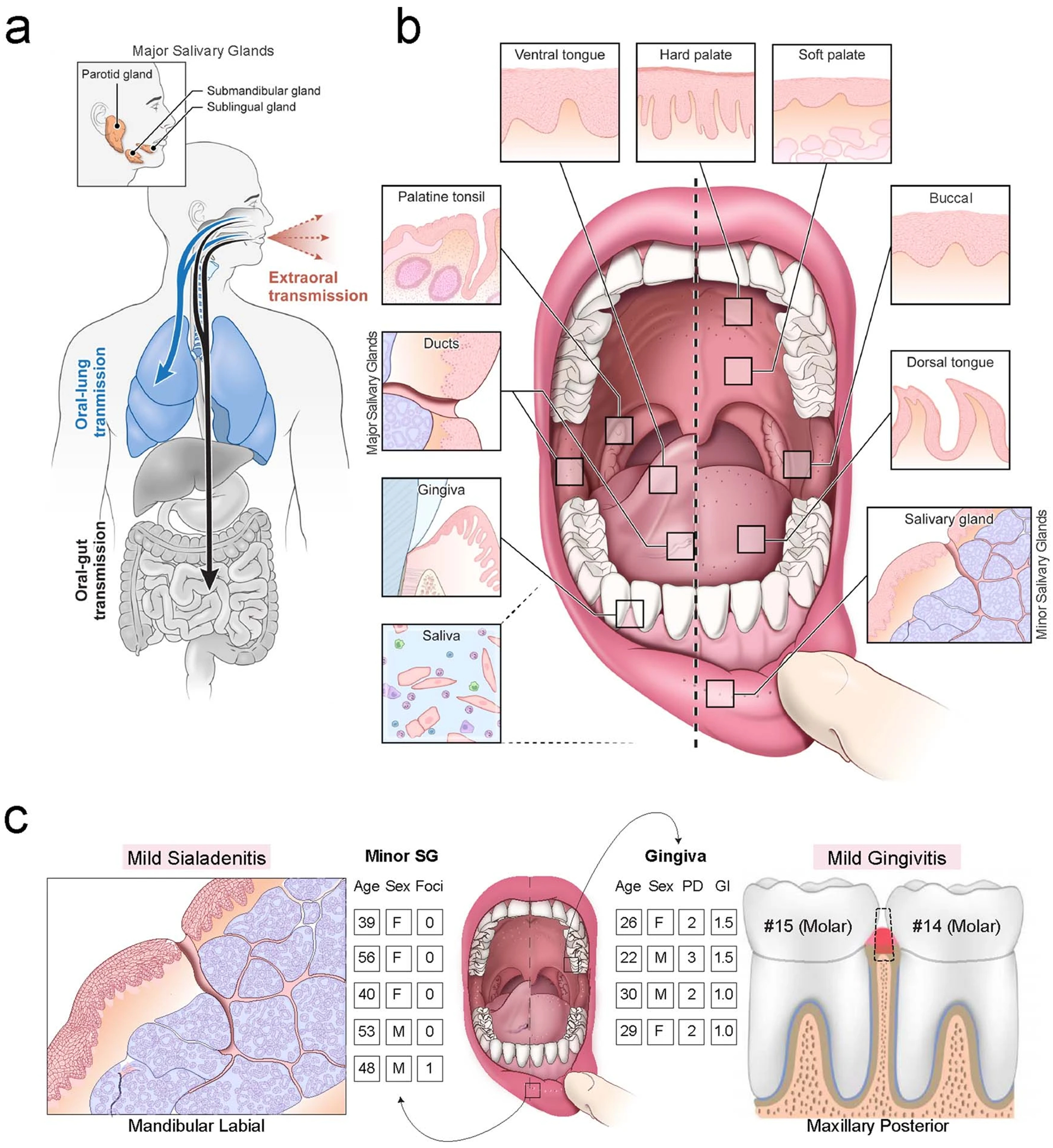
a, The contribution of the oral cavity to COVID-19 pathogenesis and transmission has been little explored. It is unknown whether SARS-CoV-2 can infect and replicate in the oral mucosae or glands. If the oral cavity is a site of early infection, this space may play important and underappreciated roles in transmitting SARS-CoV-2 ‘intermucosally’ to the lungs or gastrointestinal tract. Alternatively, saliva may also play a central role in transmitting the virus extraorally to others. b, The human oral cavity is a diverse collection of tissue niches with potentially unique vulnerabilities to viral infection (adapted from62). Additionally, saliva, a mixture of fluids, electrolytes, proteins, and cells (immune and sloughed mucosal epithelial cells) is made by the glands and empties into the oral cavity where it mixes with other fluids and cells). c, Labial minor salivary glands (SG) were procured from patients having biopsies for the clinical workup of Sjögren’s Syndrome who did not meet classification criteria and were otherwise healthy, including lacking SSA/SSB autoantibodies and without focal lymphocytic sialadenitis. Mucosal tissues including the gingiva and hard palate were harvested from four 20 to 30-year-old healthy subjects who were given a stent that prevented brushing only three maxillary teeth to induce localized gingivitis in subjects who never have progressed to periodontitis (Gingival Index; GI = avg. 1 out of 3; Probing depths <3 mm). For more annotated clinical characteristics, see Supplementary Table 1.
Extended Data Fig. 2 |. RNA sequencing reveals common viral infection susceptibilities.
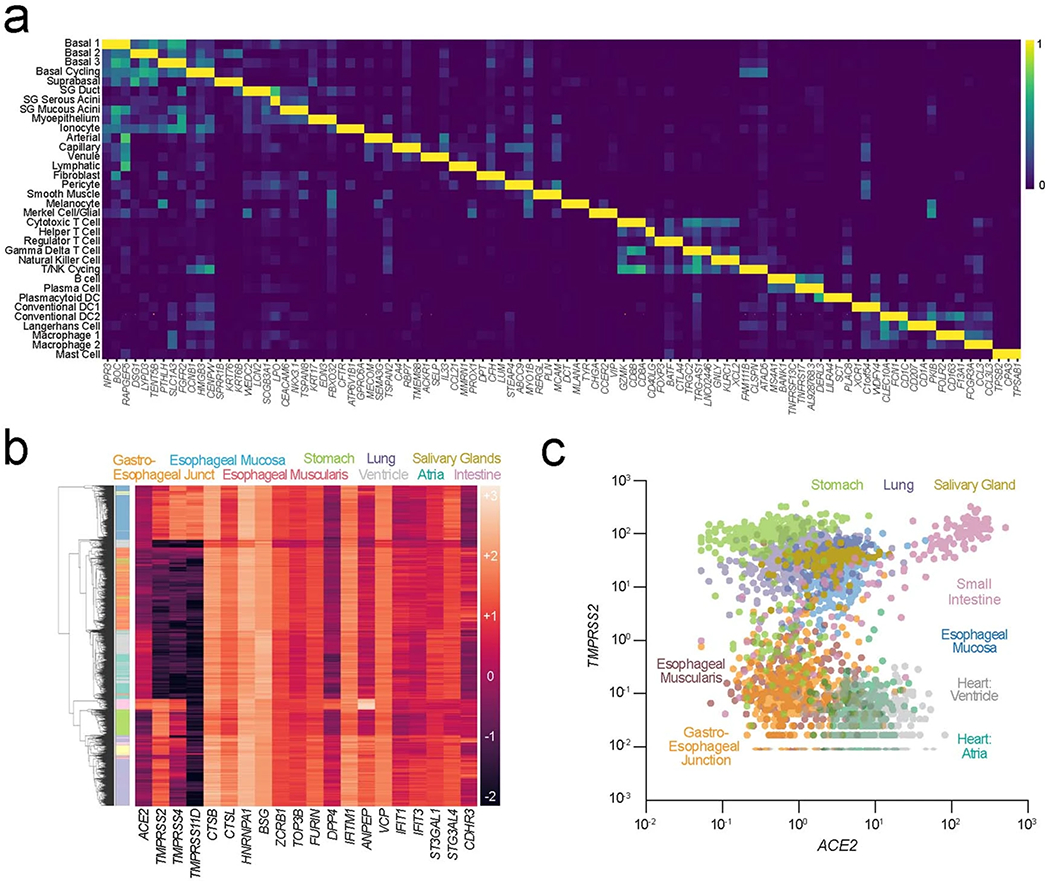
a, Expression matrix visualization of the integrated human oral atlas illustrates that cell types are distinguished by unique—and for some oral cell populations—as of yet, undescribed transcriptional signatures (defining list in Supplementary Table 1). b,c, (b) GTEx v7 RNA-seq data was used to construct a heatmap to visualize and compare the expression of viral entry factors across available tissue types including the esophagus, heart, lung, salivary gland, small intestine, and stomach. Salivary glands cluster with lung, small intestine, and stomach—all known tissues with tropism for SARS-CoV-2. c, Comparison of ACE2 and TMPRSS2 co-expression shows distinct clusters of tissues at risk of infection based on co-expression, including salivary glands clustering with the lungs and stomach—known sites of infection.
Extended Data Fig. 3 |. Validation of viral entry factor expression patterning across oral niches.

a, Immunofluorescence (IF) confocal microscopy demonstrated AQP5 (red, acini) colocalization with ACE2 (green) reveal that ACE2 is expressed on the apical (luminal) and basolateral membranes and is concentrated in the salivary gland (SG) acini/ducts (dotted/solid line, respectively; 1 independent replication). b-d, (b) Using healthy volunteer SG sections, minimal TMPRSS11D expression in ducts and acini was confirmed using RNAscope® in situ hybridization (1 independent replication). c, Analysis of available bulk RNA sequencing data suggests that the minor (minor) and parotid (PG) glands are vulnerable to SARS-CoV-2 infection compared to the submandibular glands (SMG); no independent replication. The minor SG express ~3x higher ACE2 compared to the major SG (violin plot highlights the mean with a solid line; minor: 1.42, 0.70; 0.53, respectively). For comparison, the glands express an equivalent amount of TMPRSS2 across all three SG samples (mean 63.60, 49.42; 64.78, respectively). d,e, To further confirm co-expression of ACE2 and TMPRSS2, RNAscope® fluorescent in situ hybridization and immunohistochemistry for pan-cytokeratin (pCK), shows that acini and ducts co-express ACE2 and TMPRSS2 further highlighting their vulnerability to infection in (d) minor and (e) parotid SG (1 independent replication). f, Examples of positive and negative controls for chromogenic assays in Fig. 3 (1 independent replication). g-h, (g) ISH mapping validation pipeline for discovering epithelial expression of SARS-CoV-2 entry factor expression in shedding suprabasal cells, first starting with H&E histochemical staining (1 independent replication). h, ISH was used with negative and positive probe controls. The dotted black box in (g) represents the zoomed-in areas. Experiments using immunofluorescence and in situ hybridization confocal microscopy were completed on tissue sections from 4 separate individuals (2 minor, 2 PG). Curved white arrows in (h) represent direction of differentiated epithelial cell shedding Scale bars: (f-h) 100μm, (a,b,d,e) 50μm.
Extended Data Fig. 4 |. Major and minor salivary glands are infected by SARS-CoV-2.

a, Viral load in minor salivary glands (SG) is generally higher when compared to parotid glands (PG) using ddPCR (n = 8 pairs, mean +/−s.e.m. = 1.38 +/−0.90 and 0.25 +/−0.16; respectively; p = 0.0625, Wilcoxon 2-sided signed-rank test). b, Infection and replication were tested using RNAscope® spike (V-nCoV2019-S), sense (V-nCoV2019-orf1ab-sense), and controls (See Fig. 4b–e, Fig. 5, Extended Data Fig. 5); 1 independent replication. Positive and negative controls for spike and sense probes are presented. c, Compared to minor SG (Fig. 4b–e), in situ hybridization (ISH) also reveals infection and replication in PG. d, Quantification of spike and sense signal per cell in P19 minor SG, P19 PG, and CoV49 minor SG reveals. The majority of cells in the minor SG expressed less than 7 positive signals per-cell that were well-correlated. e,f, (e) More summary immunophenotyping scoring (0-3) data from the submandibular SG and submucosal SG of the upper respiratory tract (minor and parotid gland data can be seen in Fig. 4g); 1 independent replication. f, Representative immunophenotyping studies on a parotid gland from COVID-19 autopsy (P19) demonstrating mild, non-specific sialadenitis with a predominant T cell infiltrate with the expression of HLA-DR in the ducts and acini associated with inflammation; 1 independent replication. Scale bars: (b) 25μm, (c) 10μm, (f) black: 500μm; white: 100μm. A solid black box highlights the area highlighted in the inset. Experiments using immunofluorescence and in situ hybridization confocal microscopy were completed on tissue sections from at least 4 separate individuals (5 minor, 4 PG). Immunophenotyping and was performed using autopsy sections (N =18 total tissue blocks) from individual specimens (N = 10). Three pathologists (BMW, DEK, and BG) independently scored the cases; disagreements were settled by consensus discussion.
Extended Data Fig. 5 |. Salivary cellular fractions are heterogeneous, but epithelia sustain infection.
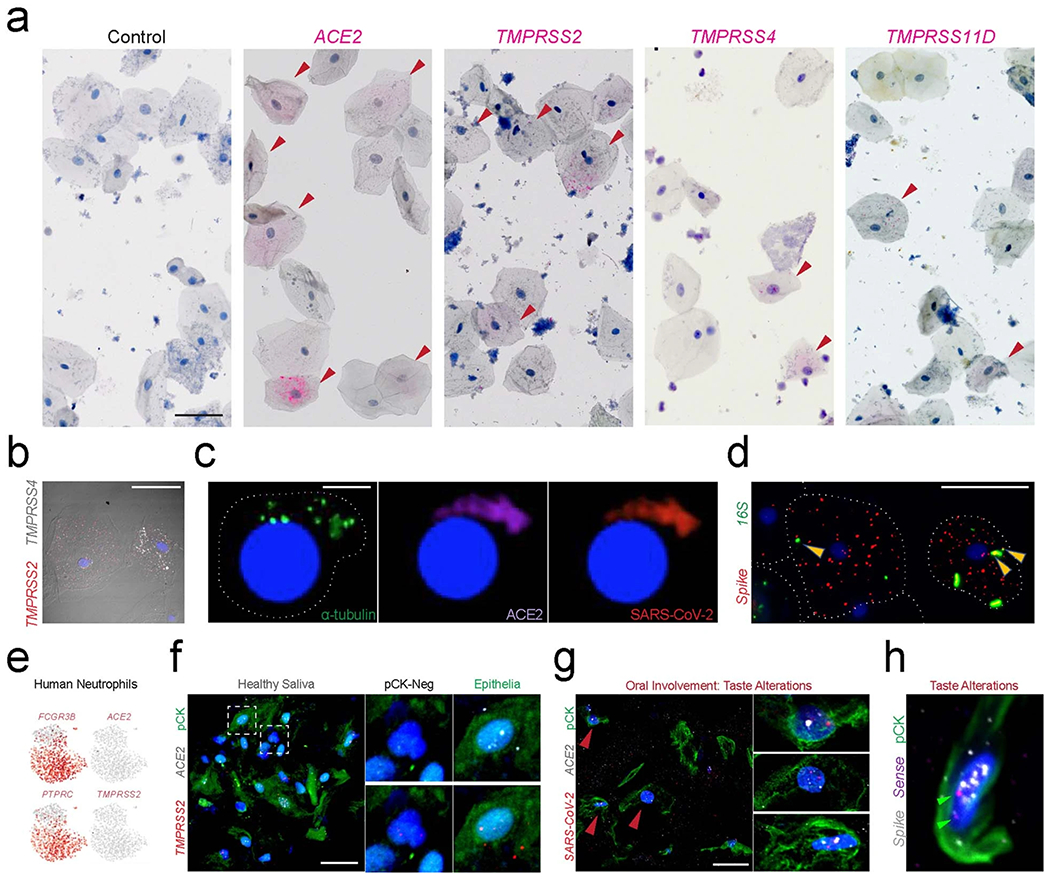
a, Salivary epithelial cells were found to heterogeneously express SARS-CoV-2 entry factors in a minority of cells; 1 independent replication. b, Shed cells in saliva display unique TMPRSS family heterogeneity, which we also observed in tissue-specific oral atlases for salivary glands (SG) and mucosa; 1 independent replication. SG express the highest TMPRSS2, and some shed epithelial cells express similar patterns, suggesting they may be shed from the SG ducts; no independent replication. Others express higher TMPRSS4, suggesting these cells may be from the suprabasal mucosa. c, Ciliated cells in saliva are exceedingly rare (α-tubulin + ) but can be present and infected; no independent replication. d, SARS-CoV-2 can be found to be associated with the diverse oral microbiome on shed epithelial cells; 1 independent replication. e, Using a salivary neutrophil atlas from healthy patients, we confirm that virtually no neutrophils express ACE2 or TMPRSS2. f-h, (f) Using cell blocks of heathy saliva from the NIH, we validate our findings that shed salivary epithelial (pan-cytokeratin positive; pCK + ) cells express both ACE2 and TMPRSS2; 1 independent replication. We see minimal to no expression of these SARS-CoV-2 entry factors in pCK−negative cells. g, These ACE2 + cells are also infected by SARS-CoV-2 and (h) able to replicate the virus (1 independent replication each). Red arrowheads in (a) indicate SARS-CoV-2 entry factors; in (g) SARS-CoV-2 infection; green arrowheads in (h) represent SARS-CoV-2 replication. Arrowheads in (h) indicate SARS-CoV-2 (red), universal 16 S probe (green) that are co-expressing (yellow); dotted lines (f) represent zoomed-in images. Scale bars: (a,b,d,g) 25 μm, (c,f,h) 10μm.
Extended Data Fig. 6 |. Saliva sampling supports a role for oral cavity in COVID-19 pathogenesis.
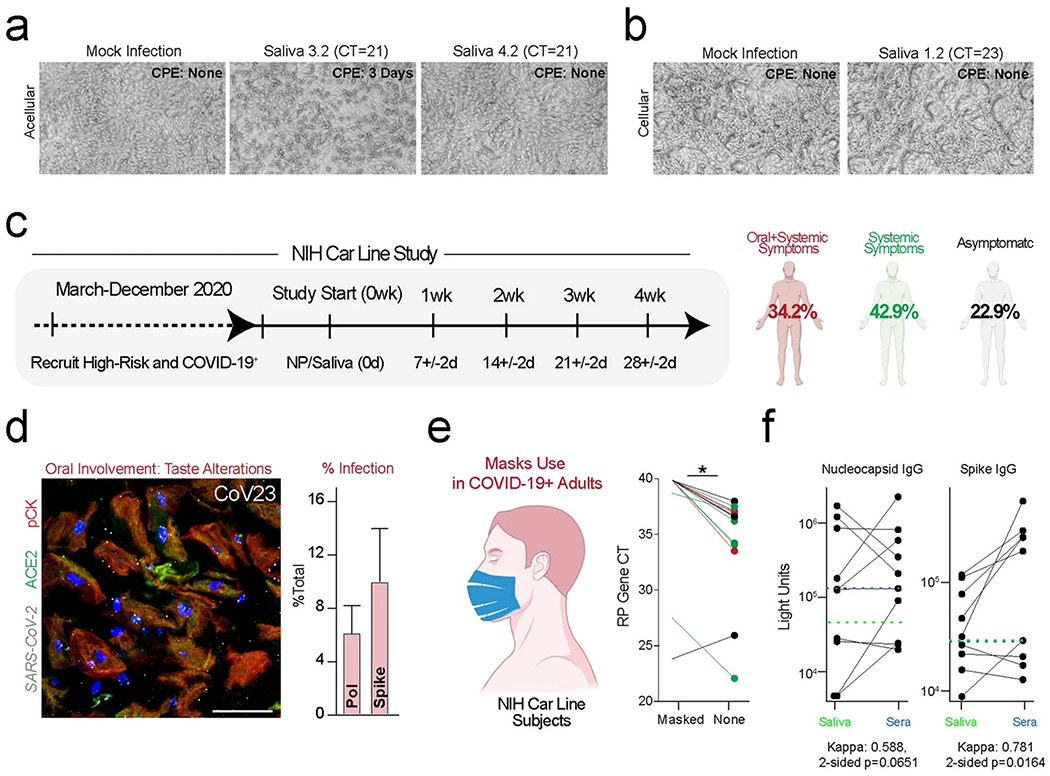
a,b, (a) Acellular (b) and cellular fractions (N = 8) were incubated with Vero cells in duplicate. Supernatants from the acellular saliva more often caused CPE (n = 2 of 8) compared to the cellular fraction (n = 1 of 8); 1 independent replication. c, A clinical study, NIH Transmissibility and Viral Load of SARS-CoV-2 Through Oral Secretions Study (AKA – ‘NIH Carline Study’), was used for symptom tracking and prospective sampling of nasopharyngeal (NP) swabs and saliva. d, Subjects with the highest saliva viral load and reported taste alterations (left); these individuals displayed infection of salivary epithelial (pCK+) cells using spike and polymerase probes--n = 3 (CoV19, CoV23, CoV25 where 2257, 28260, and 31804 cells, respectively, were counted). Bars are presented as mean +/− SEM., respectively with 1 independent replication). e, Infectious saliva fraction was measured with masks and unmasked in symptomatic (oral/systemic; red and systemic only; green). f, Sub-study of LIPS for nucleocapsid IgG and spike IgG levels comparing saliva and sera from the same subjects by calculating a two-sided agreement statistic (kappa) of antibody positivity without adjustment for multiple comparisons. Due to the limitations of the study design, samples were not temporally linked. In general, there was fair to excellent agreement between the presence of salivary and sera antibodies to nucleocapsid and spike, respectively. No pattern for salivary versus sera nucleocapsid IgG levels was observed, though this may reflect temporal differences. Sera trended toward relatively higher levels when assaying for spike IgG antibody levels. Green dotted lines in (f) represent saliva cutoff for nucleocapsid and spike; blue dotted lines, cutoffs for sera. Annotations: Scale bars: (a,b) 100μm, (d) 25μm. Statistical test (e): p< 0.005, two-sided Wilcoxon signed-rank test.
Supplementary Material
Acknowledgements
This work is dedicated to the more than 2.5 million worldwide fatalities and their families who have fallen victim to COVID-19. These experiments were principally supported by grants and awards to K.M.B. (NIH/NIDCR K08 DE026537; AAP Sunstar Innovation Grant) and to B.M.W. (NIH/NIDCR Division of Intramural Research Z01-DE000704). The following investigators are also supported by the NIH Intramural Research Program: J.A.C., L.A.T., D.E.K., S.H., B.A.L., D.C., S.P. and J.L. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH nor the ADA. Portions of the project were supported by the National Center for Advancing Translational Sciences, National Institutes of Health, through grant KL2TR002490 (A.J.K.). Gingival scRNA-seq was conducted by the UNC Advanced Analytics Core (Center for GI Biology and Disease, P30 DK034987; C.W.A.). Additional support is from the Senior Research Training Fellowship by the American Lung Association (RT-575362) and the Postdoctoral Research Fellowship by the Cystic Fibrosis Foundation (KATO20F0) to T.K., the Postdoctoral Research Fellowship by the Cystic Fibrosis Foundation (MIKAMI19XX0) to Y.M. and NIH/NIDCR (K01DE027087 to J.T.M.). The authors provide a special acknowledgment to the NIH Office of the Director and the NIH Principal Deputy Director, L. A. Tabak, for profound assistance in establishing the NIH COVID-19 Saliva Testing Study. We would like to acknowledge the NIDCR Office of the Clinical Director and the Office of the Scientific Director for their COVID-19-related support, including assistance in protocol development and research activity support. Special thanks as well to B. Brillante and C. Webb for their assistance in protocol navigation and project management. Healthy volunteer and COVID-19 clinical specimens were collected by members of the NIDCR Sjögren’s Syndrome Clinic. A special thanks for the coordinating efforts of the NIH COVID-19 Testing Facility, the NIH Occupational Medicine Service and NIH Medical Records for their support in executing the NIH Clinical Study. We are grateful to the NIAID Director of Intramural Research, S. M. Holland, for facilitating infectivity testing on SARS-CoV-2 saliva. scRNA-seq and bulk RNA-seq of the salivary glands were conducted by the NIDCR Genomics and Computational Biology Core, and a very deserving thanks to Z. Wei and D. Martin for their assistance in executing and processing the SG scRNAseq captures. We also thank W. Young for assistance in performing the COVID-19 autopsies. This publication is part of the Human Cell Atlas: http://www.humancellatlas.org/publications.
NIH COVID-19 Autopsy Consortium
David E. Kleiner4, Billel Gasmi2,4, Michelly Sampaio De Melo4, Esra Dikoglu4, Sabina Desar4, Stephen M. Hewitt4, Kris Ylaya4, Joon-Yong Chung4, Stefania Pittaluga4, Grace Smith4, Daniel S. Chertow5, Kevin M. Vannella5, Sydney Stein5, Marcos Ramos-Benitez5, Sabrina C. Ramelli5, Shelly J. Samet5, Ashley L. Babyak5, Luis Perez Valenica5, Mary E. Richert5, Alison Grazioli23, Nicole Hays28, Madeleine Purcell28, Shreya Singireddy28, Jocelyn Wu28, Jean Chung28, Amy Borth28, Kimberly Bowers28, Anne Weichold28, Douglas Tran29, Ronson J. Madathil29, Eric M. Krause30, Daniel L. Herr21, Joseph Rabin31, Joseph A. Herrold31, Ali Tabatabai31, Eric Hochberg31, Christopher Cornachione31, Andrea R. Levine31, Michael T. McCurdy31, Kapil K. Saharia32, Zack Chancer33, Michael A. Mazzeffi33, Justin E. Richards33, James W. Eagan Jr34 and Yashvir Sangwan35
28University of Maryland School of Medicine, Baltimore, MD, USA.
29Department of Surgery, Division of Cardiac Surgery, University of Maryland School of Medicine, Baltimore, MD, USA.
30Department of Surgery, Division of Thoracic Surgery, University of Maryland School of Medicine, Baltimore, MD, USA.
31Department of Medicine, Division of Pulmonary and Critical Care Medicine, University of Maryland School of Medicine, Baltimore, MD, USA.
32Department of Medicine, Division of Infectious Disease, University of Maryland School of Medicine, Baltimore, MD, USA.
33Department of Anesthesiology, Division of Critical Care Medicine, University of Maryland School of Medicine, Baltimore, MD, USA.
34Department of Pathology, University of Maryland, St. Joseph Medical Center, Towson, MD, USA.
35Department of Pulmonology, Tidal Health Peninsula Regional, Salisbury, MD, USA.
HCA Oral and Craniofacial Biological Network
Kevin M. Byrd11,27,38, Inês Sequeira36, Blake M. Warner2,38, Sarah A. Teichmann1,26, Marcelo Freire13,14, Adam J. Kimple3,20
36Institute of Dentistry, Barts and the London School of Medicine and Dentistry, Queen Mary University of London, London, UK.
Footnotes
Code availability
Analysis notebooks are available at https://github.com/Teichlab/covid19_oral.
Competing interests
Although the authors view each of these as non-competing financial interests, we report that, in the last year, K.M.B. has been a Scientific Advisor at Arcato Laboratories; K.M.B., B.M.W., M.O.F., A.J.K. and S.T. are active members of the Human Cell Atlas; for the past 6 months, M.O.F. has served as a Scientific Advisor for the PolyBio Research Foundation; and, for the last 3 years, S.A.T. has been remunerated for consulting by Roche and Genentech and is a member of Scientific Advisory Boards at Biogen, GlaxoSmithKline and Foresite Labs. All other authors declare no competing interests.
Extended data is available for this paper at https://doi.org/10.1038/s41591-021-01296-8.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s41591-021-01296-8.
Peer review information Nature Medicine thanks Joanne Konkel and the other, anonymous, reviewers for their contribution to the peer review of this work. Saheli Sadanand was the primary editor on this article and managed its editorial process and peer review in collaboration with the rest of the editorial team.
Data availability
Data were collected based on existing resources, and no additional software was generated for this manuscript. Analysis notebooks are available at http://github.com/Teichlab/covid19_oral. All of the published datasets and relevant data from unpublished sources in this study can be visualized and assessed through a website portal (https://www.covid19cellatlas.org/). Human nasal airway37 and human gut38 datasets for integrated pan-epithelial analysis can be found (PMID:31209336 and PMID:31348891, respectively). Human neutrophil data can be downloaded using the SRA Toolkit (SRP271375 (ref. 44)). Publicly available data can be downloaded from the GTEx v7 Portal39 (http://www.gtexportal.org/home/datasets). The Molecular Signatures Database can be accessed at http://www.gsea-msigdb.org/gsea/msigdb/index.jsp.
References
- 1.Zhu N et al. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med 382, 727–733 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang R, Li Y, Zhang AL, Wang Y & Molina MJ Identifying airborne transmission as the dominant route for the spread of COVID-19. Proc. Natl Acad. Sci. USA 117, 14857–14863 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghinai I et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet 382, 1137–1144 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pung R et al. Investigation of three clusters of COVID-19 in Singapore: implications for surveillance and response measures. Lancet 382, 1039–1046 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hamner L et al. High SARS-CoV-2 attack rate following exposure at a choir practice—Skagit County, Washington, March 2020. MMWR Morb. Mortal. Wkly. Rep 69, 606–610 (2020). [DOI] [PubMed] [Google Scholar]
- 6.Hou YJ et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell 382, 429–446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amorim dos Santos J et al. Oral manifestations in patients with COVID-19: a living systematic review. J. Dent. Res 382, 141–154 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Jimenez-Cauhe J et al. Enanthem in patients with COVID-19 and skin rash. JAMA Dermatol. 156, 1134–1136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.La Rosa GRM, Libra M, De Pasquale R, Ferlito S & Pedullà E Association of viral infections with oral cavity lesions: role of SARS-CoV-2 infection. Front. Med 7, 1059 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muhlebach MS et al. Initial acquisition and succession of the cystic fibrosis lung microbiome is associated with disease progression in infants and preschool children. PLoS Pathog. 14, e1006798 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kitamoto S et al. The intermucosal connection between the mouth and gut in commensal pathobiont-driven colitis. Cell 14, 447–462 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Byrd KM & Gulati AS The ‘Gum–Gut’ axis in inflammatory bowel diseases: a hypothesis-driven review of associations and advances. Front. Immunol 12, 39 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zang R et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol 5, eabc3582 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh M, Bansal V & Feschotte C A single-cell RNA expression map of human coronavirus entry factors. Cell Rep. 32, 108175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sungnak W et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med 26, 681–687 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brann DH et al. Non-neuronal expression of SARS-CoV-2 entry genes in the olfactory system suggests mechanisms underlying COVID-19-associated anosmia. Sci. Adv 5, eabc5801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong M et al. ACE2 and furin expressions in oral epithelial cells possibly facilitate COVID-19 infection via respiratory and fecal–oral routes. Front. Med 7, 580796 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L et al. Detection of 2019-nCoV in saliva and characterization of oral symptoms in COVID-19 patients. Cell Prolif. 12, e12923 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones KB & Klein OD Oral epithelial stem cells in tissue maintenance and disease: the first steps in a long journey. Int. J. Oral. Sci 5, 121–129 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Byrd KM et al. LGN plays distinct roles in oral epithelial stratification, filiform papilla morphogenesis and hair follicle development. Development 143, 2803–2817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krivanek J et al. Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nat. Commun 11, 1–18 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caetano AJ et al. Defining human mesenchymal and epithelial heterogeneity in response to oral inflammatory disease. eLife 10, e62810 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Byrd KM et al. Heterogeneity within stratified epithelial stem cell populations maintains the oral mucosa in response to physiological stress. Cell Stem Cell 25, 814–829 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones KB et al. Quantitative clonal analysis and single-cell transcriptomics reveal division kinetics, hierarchy, and fate of oral epithelial progenitor cells. Cell Stem Cell 24, 183–192 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plasschaert LW et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377–381 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamster IB, Rodrick ML, Sonis ST & Falchuk ZM An analysis of peripheral blood and salivary polymorphonuclear leukocyte function, circulating immune complex levels and oral status in patients with inflammatory bowel disease. J. Periodontol 53, 231–238 (1982). [DOI] [PubMed] [Google Scholar]
- 28.Fergusson JR et al. Maturing human CD127+ CCR7+ PDL1+ dendritic cells express AIRE in the absence of tissue restricted antigens. Front. Immunol 9, 2902 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dutzan N, Konkel JE, Greenwell-Wild T & Moutsopoulos NM Characterization of the human immune cell network at the gingival barrier. Mucosal Immunol. 9, 1163–1172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song L, Dong G, Guo L & Graves DT The function of dendritic cells in modulating the host response. Mol. Oral Microbiol 33, 13–21 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zemmour D et al. Single-cell gene expression reveals a landscape of regulatory T cell phenotypes shaped by the TCR. Nat. Immunol 19, 291–301 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.MacNeil AJ, Yang YJ & Lin TJ MAPK kinase 3 specifically regulates FcεRI-mediated IL-4 production by mast cells. J. Immunol 187, 3374–3382 (2011). [DOI] [PubMed] [Google Scholar]
- 33.Presland RB & Dale BA Epithelial structural proteins of the skin and oral cavity: function in health and disease. Crit. Rev. Oral Biol. Med 11, 383–408 (2000). [DOI] [PubMed] [Google Scholar]
- 34.Ziegler C et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is enriched in specific cell subsets across tissues. Cell 181, 1016–1035 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Muus C et al. Integrated analyses of single-cell atlases reveal age, gender, and smoking status associations with cell type-specific expression of mediators of SARS-CoV-2 viral entry and highlights inflammatory programs in putative target cells. Preprint at bioRxiv 10.1101/2020.04.19.049254 (2020). [DOI]
- 36.Singh M, Bansal V & Feschotte C A single-cell RNA expression map of human coronavirus entry factors. Cell Rep. 10.1016/j.celrep.2020.108175 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vieira Braga FA et al. A cellular census of human lungs identifies novel cell states in health and in asthma. Nat. Med 25, 1153–1163 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Smillie CS et al. Intra-and inter-cellular rewiring of the human colon during ulcerative colitis. Cell 178, 714–730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schurink B et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe 1, E290–E299 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karlsson AC, Humbert M & Buggert M The known unknowns of T cell immunity to COVID-19. Sci. Immunol 5, eabe8063 (2020). [DOI] [PubMed] [Google Scholar]
- 42.Speight PM, Cruchley A & Williams DM Epithelial HLA-DR expression in labial salivary glands in Sjögren’s syndrome and non-specific sialadenitis. J. Oral Pathol. Med 18, 178–183 (1989). [DOI] [PubMed] [Google Scholar]
- 43.Theda C et al. Quantitation of the cellular content of saliva and buccal swab samples. Sci. Rep 8, 1–8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choudhury SN et al. A protocol for revealing oral neutrophil heterogeneity by single-cell immune profiling in human saliva. Preprint at Protocol Exchange 10.21203/rs.3.pex-953/v1 (2020). [DOI]
- 45.Harcourt J et al. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis 26, 1266 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyllie AL et al. Saliva or nasopharyngeal swab specimens for detection of SARS-CoV-2. N. Engl. J. Med 383, 1283–1286 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Isho B et al. Persistence of serum and saliva antibody responses to SARS-CoV-2 spike antigens in COVID-19 patients. Sci Immunol. 5, eabe5511 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arrieta JJ et al. In situ detection of hepatitis C virus RNA in salivary glands. Am. J. Pathol 158, 259–264 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakamura H et al. Reevaluation for clinical manifestations of HTLV-I-seropositive patients with Sjögren’s syndrome. BMC Musculoskelet. Disord 16, 1–7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hviid A, Rubin S & Mühlemann K Mumps. Lancet 371, 932–944 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Silva J et al. Saliva viral load is a dynamic unifying correlate of COVID-19 severity and mortality. Preprint at medRxiv 10.1101/2021.01.04.21249236 (2021). [DOI] [Google Scholar]
- 52.Dawes C Estimates, from salivary analyses, of the turnover time of the oral mucosal epithelium in humans and the number of bacteria in an edentulous mouth. Arch. Oral Biol 48, 329–336 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Gandhi M, Yokoe DS & Havlir DV Asymptomatic transmission, the Achilles’ heel of current strategies to control Covid-19. N. Engl. J. Med 382, 2158–2160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Löe H, Theilade E & Jensen SB Experimental gingivitis in man. J. Periodontal 36, 177–187 (1965). [DOI] [PubMed] [Google Scholar]
- 55.Offenbacher S et al. Gingival transcriptome patterns during induction and resolution of experimental gingivitis in humans. J. Periodontal 80, 1963–1982 (2009). [DOI] [PubMed] [Google Scholar]
- 56.Löe H The gingival index, the plaque index and the retention index systems. J. Periodontol 38, 610–616 (1967). [DOI] [PubMed] [Google Scholar]
- 57.Wolf FA, Angerer P & Theis FJ SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Polański K et al. BBKNN: fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korsunsky I et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1–8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Burbelo PD et al. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J. Infect. Dis 222, 206–213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ching KH et al. Salivary anti-Ro60 and anti-Ro52 antibody profiles to diagnose Sjögren’s syndrome. J. Dent. Res 90, 445–449 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moutsopoulos NM & Konkel JE Tissue-specific immunity at the oral mucosal barrier. Trends Immunol. 39, 276–287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data were collected based on existing resources, and no additional software was generated for this manuscript. Analysis notebooks are available at http://github.com/Teichlab/covid19_oral. All of the published datasets and relevant data from unpublished sources in this study can be visualized and assessed through a website portal (https://www.covid19cellatlas.org/). Human nasal airway37 and human gut38 datasets for integrated pan-epithelial analysis can be found (PMID:31209336 and PMID:31348891, respectively). Human neutrophil data can be downloaded using the SRA Toolkit (SRP271375 (ref. 44)). Publicly available data can be downloaded from the GTEx v7 Portal39 (http://www.gtexportal.org/home/datasets). The Molecular Signatures Database can be accessed at http://www.gsea-msigdb.org/gsea/msigdb/index.jsp.


