Abstract
Wastewater monitoring for virus infections within communities can complement conventional clinical surveillance. Currently, most SARS‐CoV‐2 (severe acute respiratory syndrome coronavirus 2) clinical testing is voluntary and inconsistently available, except for a few occupational and educational settings, and therefore likely underrepresents actual population prevalence. Randomized testing on a regular basis to estimate accurate population‐level infection rates is prohibitively costly and is hampered by a range of limitations and barriers associated with participation in clinical research. In comparison, community‐level fecal monitoring can be performed through wastewater surveillance to effectively surveil communities. However, epidemiologically defined protocols for wastewater sample site selection are lacking. Herein, we describe methods for developing a geographically resolved population‐level wastewater sampling approach in Jefferson County, Kentucky, and present preliminary results. Utilizing this site selection protocol, samples (n = 237) were collected from 17 wastewater catchment areas, September 8 to October 30, 2020 from one to four times per week in each area and compared to concurrent clinical data aggregated to wastewater catchment areas and county level. SARS‐CoV‐2 RNA was consistently present in wastewater during the studied period, and varied by area. Data obtained using the site selection protocol showed variation in geographically resolved wastewater SARS‐CoV‐2 RNA concentration compared to clinical rates. These findings highlight the importance of neighborhood‐equivalent spatial scales and provide a promising approach for viral epidemic surveillance, thus better guiding spatially targeted public health mitigation strategies.
Keywords: COVID‐19, environmental surveillance, epidemiology, GIS, SARS‐CoV‐2, wastewater
Key Points
Epidemiologically defined protocols for wastewater sample site selection are lacking
Demographically distinct catchment areas were selected with high spatial resolution
Our protocol may inform disease surveillance for geographically targeted scales
1. Introduction
Wastewater monitoring can complement clinical surveillance for virus infections when fecal shedding has been established for the viral strain (Bisseux et al., 2020). Application of wastewater‐based epidemiology (WBE) has been used for monitoring public health risks, including illicit drug use, poliomyelitis, Hepatitis E, human immunodeficiency virus (HIV), adenoviruses, and coronavirus for low‐cost, real‐time estimates, and retrospective monitoring (Bibby & Peccia, 2013; Castiglioni et al., 2014; Masclaux et al., 2013; Pogka et al., 2017). Currently, most SARS‐CoV‐2 (severe acute respiratory syndrome coronavirus 2) testing is voluntary and inconsistently available, except for a few occupational and educational settings, and therefore likely underrepresents actual population prevalence (Moghadas et al., 2020), especially in low socioeconomic status subpopulations with substantially limited access to testing. This, and other limitations, skews estimated rates of infected individuals, making it difficult to accurately detect clusters of infections in a timely fashion in order to mount a targeted public health response.
Fecal shedding of SARS‐CoV‐2 has been documented in patients diagnosed with COVID‐19 (Wölfel et al., 2020) and is detectable in sewer systems. Thus, wastewater monitoring results could be an important early indicator of COVID‐19, and potential variants, in communities (Ahmed et al., 2020; Bivins et al., 2020; Gonzalez et al., 2020; Medema et al., 2020; Peccia et al., 2020; Sherchan et al., 2020; Randazzo et al., 2020). However, interpretation of a signal beyond presence/absence of SARS‐CoV‐2 RNA copies in wastewater continues to challenge broad application. However, while a close relationship between fecal and wastewater samples has been reported for poliomyelitis (Pogka et al., 2017), this is less well‐established for SARS‐CoV‐2 (Kitajima et al., 2020; Wölfel et al., 2020). Because viral RNA monitoring of wastewater is done anonymously, it may be an important surveillance option for community‐level monitoring with the added benefit that it includes pre‐symptomatic/asymptomatic individuals, underserved populations with less access to testing resources, and others who experience barriers to clinical testing (Bivins et al., 2020). Moreover, wastewater sampling may also indicate geographic locations of infection clusters (“hot spots”) that are more difficult to detect with conventional in‐person measurements or aggregations to large geographic areas.
Wastewater monitoring of SARS‐CoV‐2 RNA is available to complement community clinical diagnostic surveillance for infections at three units of analysis: (a) At an established centralized wastewater treatment facility, typically collecting wastewater from many neighborhoods or an entire city; (b) for neighborhood/multi‐neighborhood spatial scales, with samples collected at street sewer line manholes or pump stations; and (c) for congregate living facilities (e.g., ships, buildings), with samples collected at effluent access points. Much of the published literature on the monitoring of SARS‐CoV‐2 RNA in wastewater addresses samples collected at treatment facilities (Ahmed et al., 2020; Medema et al., 2020; Sherchan et al., 2020). Bisseux et al. (2020) report the usefulness of combining raw wastewater sampling with clinical encounters. From an epidemiological perspective, sampling derived from hundreds of thousands of households may produce useful aggregate infection prevalence information and may provide advance early warning about impending clinical facilities' burden or the need for adjusting broad social restrictions. However, this approach does not address the possibly important changes in infection dynamics in component areas of the community, which could be the focus of public health engagement and infection mitigation strategies. Examples of this could include public health intervention measures such as ad hoc “pop up” community testing or development of place‐ and culture‐specific health communications messaging.
Moving from centralized wastewater treatment facilities to neighborhood sewer lines introduces factors that preclude comparisons across sites. For example, sewer systems are not always accurately mapped, and sewer lines, in addition to household wastewater, may contain combined commercial, industrial, and often rain water runoff, which complicates the consistent and comparable sampling of feces due to inconsistent dilution of household waste and introduction of reactive agents. Furthermore, these sewer system lines often have complex technical and geographic characteristics that result from ad hoc expansion due to growing populations over many decades. Therefore, a key barrier to developing epidemiologically comparable and geographically resolved monitoring of SARS‐CoV‐2 RNA in wastewater is the lack of established methods for sample site selection specifically designed for spatially resolved community‐wide surveillance.
To the best of our knowledge, there is no published methodological framework for sample site selection to maximize the surveillance and response value of SARS‐CoV‐2 RNA wastewater testing within communities. Hence, we propose a pragmatic location‐based method for selection of raw wastewater sample sites, where wastewater testing results are representative of geographically resolved community prevalence of SARS‐CoV‐2 infection. We developed a protocol, which we describe here, in conjunction with a multidisciplinary team of public health scientists and practitioners, virologists, and wastewater professionals. Our main objectives were to describe the establishment of epidemiologically based and geographically resolved urban community surveillance for SARS‐CoV‐2 RNA in wastewater and to develop preliminary evidence to assess variation in SARS‐CoV‐2 across large aggregation sites and between smaller neighborhood/multi‐neighborhood scales. Such an approach could serve to inform public health officials and decision‐makers on spatial trends of SARS‐CoV‐2 infection prevalence and subsequently inform proactive place‐specific public health interventions.
2. Methods
With a population of approximately 770,000 residents over 380 square miles, Jefferson County, Kentucky is physically and demographically similar to many urban areas across the United States. There are five water quality treatment centers (WQTC) within the county, operated by Louisville/Jefferson County Metropolitan Sewer District (MSD), each treating wastewater of an average volume from 3.5 to 350 million gallons per day. While many WBE programs utilize sludge samples collected from WQTCs, we collected samples of raw wastewater from both WQTCs and pipes with a known wastewater catchment area. This approach enabled us to select sampling locations based on geographic location and scope, land use, and specific populations. Though the WQTCs are fixed, we have continuously added and removed sampling sites to refine our sampling approach during the pilot of this protocol.
2.1. Data
We collected existing sewer data in geographic information systems (GIS) format from MSD through the Louisville‐Jefferson County Information Consortium. These data included information on the location and attributes of all sewer lines and access points within the county, allowing for rapid design and deployment of this protocol and precise geographic definition of catchment areas represented by each sampling location. Countywide socioeconomic data was collected from the U.S. Census Bureau, including information on the distribution of income, race, ethnicity, and population characteristics, which was utilized to ensure representative sampling of minority and low socioeconomic status communities (U.S. Department of Commerce, 2020). Geocoded clinical case rate (per 100k) data was provided by Louisville Metro Public Health and Wellness under a data use agreement and are publicly available (https://covid-19-in-jefferson-county-ky-lojic.hub.arcgis.com). These case rates are an underestimate because we were unable to locate all case data due to errant records. We based demographic estimates on 2018 American Community Survey block group data and aggregated to the wastewater catchment areas with overlapping block group centroids.
2.2. Sample Size Justification
The recommendation for wastewater poliomyelitis monitoring is a sample population of 100,000 to 300,000 persons (Global Polio Eradication Initiative, 2015); however, our project required sample sensitivity to be directly comparable with our concurrent nasal diagnostic clinical study, justifying a smaller sample population at neighborhood/multi‐neighborhood resolution. Based on the physical layout of sewer lines in Jefferson County, there was a steep decline in informative value of additional sites beyond those selected. For example, complete spatially resolved monitoring of all residential areas would require many more sample locations, due to small sewage lines connecting directly to sewer mains with upstream sampling specifically placed to isolate populations between 5,000 and 100,000 residents.
2.3. Ethics
The University of Louisville Institutional Review Board classified this project as non‐human subjects research (reference #: 717950). For wastewater surveillance, we over‐selected sites that represent geographic areas that are often underrepresented in clinical testing, such as low‐income neighborhoods and communities of color. The research team has discussed this new approach to SARS‐CoV‐2 infection surveillance in community meetings and in local media interviews, and it is typically met with supportive interest. Wastewater samples cannot be used to identify individuals and are considered “community samples” by the sewer district.
2.4. Rationale for Choosing Sampling Locations and Field Sampling Limitations
Inclusion criteria for sample sites and corresponding catchment areas:
Residential coverage of areas with water toilets serviced by the municipal sanitary sewer system
Locations overlapping recruitment areas for randomized resident testing of the corresponding prevalence study; That is all of Jefferson County
Minimum population of 5,000 within the catchment area represented
Sample site accessibility to sewer lines with free‐flowing wastewater (street lines, pumping stations, and main treatment facilities)
Contextual information (flow rate, temperature)
Feasibility with field human resources
For manhole locations, free‐flowing wastewater at no more than 25 feet below the street level
Exclusion criteria for sample sites:
Catchment areas that include majority coverage by commercial, industrial, or medical sewer outflow, even if residential households are also present
Any sites not practical for field sampling, such as the area around a manhole that does not offer enough space or safety for field personnel
High temperature wastewater, usually due to industrial processes
Specific a priori additions:
Each WQTC
Higher sampling in the northwestern portion of the county to ensure inclusion of residents with disproportionate health risk
2.5. Site Selection
We identified ideal sample sites using a stepwise approach (Figure 1). Predominately residential areas were prioritized over large commercial areas due to predictable outflows from residential locations and the ability to quantify area population and demographics with census and property data. Because commercial areas may demonstrate greater day‐to‐day variability and workers commuting from many places, it is significantly more difficult to trace COVID‐19 in these sectors. Sampling of lines with effluent from hospitals or other facilities with high disinfectant effluent was also minimized via this approach. When dividing sewer lines to exclude non‐residential areas or capture more resolute spatial coverage areas, we typically placed sample sites upstream to points before two larger sewer lines joined. We additionally added sample sites at each treatment facility in the county, representing nearly complete coverage of residences within the county. The two major treatment facilities are downstream from some of the neighborhood sampling zones but also include areas which were not included in community sampling catchment areas. We then sent sample site recommendations to MSD, which reviewed these recommendations for field sampling logistics determined by MSD's occupational safety standards for field activities. Some practical obstacles included manholes that were not readily accessible for the sampling team (requiring 4‐wheel drive vehicles) and manholes that were located along high traffic roadways. If an ideal site was not logistically feasible, we typically moved the sampling point to the first feasible location upstream from the ideal location in order to capture most of the ideal catchment area. In addition to the diminishing returns of informative value from additional sites, the total number of sampling sites was also constrained by MSD staff availability, which in this case was two field teams that each required two full days to collect six to eight samples each. Additionally, the work needed to be completed in time to prepare the samples for delivery to and processing by the analysis laboratory at the University of Louisville. Based on this protocol and field sampling limitations, we selected a total 17 sample sites: Twelve community sites collecting from manholes or pump stations and five WQTCs.
Figure 1.
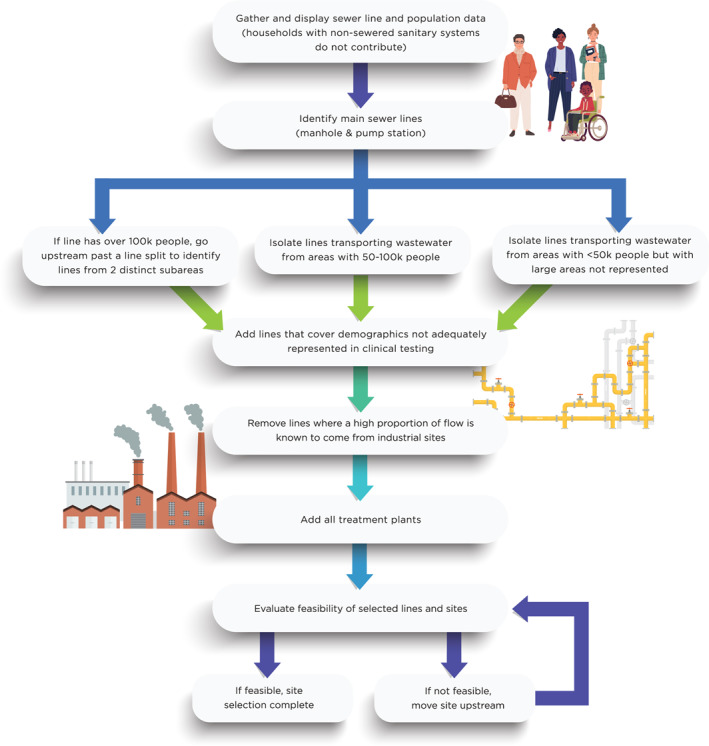
Flow diagram of iterative wastewater monitoring site selection process.
After our initial site selection, we performed an initial round of sampling. Upon field observations, we excluded one site with a pilot sample at an elevated temperature due to industrial discharges into the sewer system. Elevated temperatures have been reported to make wastewater samples vulnerable to over or under estimation due to possible degradation of constituents over time (Hart & Halden, 2020; Kitajima et al., 2020).
2.6. Wastewater Sample Collection, Transport, Preservation, and Storage
Wastewater samples were collected September 8 to October 30, 2020, one to four times per week. The samples consisted of 24‐h composite wastewater samples. We contracted with MSD for installation and extraction at sample sites, usually including at least one weekend sample to increase the likelihood of residential attribution. Sampling personnel from MSD initiated the portable composite autosampler (60‐2954‐001 Model GLS Sampler) approximately 24‐h proceeding sample collection. We used both 10‐L and 4‐L composite sample collection containers during the pilot. We determined the 4‐L containers to be preferable, to allow a higher proportion of ice between the cover and the collection container; ice was placed in the space between the cover and the collection jar as part of setting up the sampler. The lower volume 4‐L sample collection container is also preferable to reduce the weight of the equipment that is manually raised to ground level. The sampler collected 30 ml of wastewater into a larger container every 15 min. Samplers were placed based on site‐specific considerations, below a manhole and suspended with rope or on the ground adjacent to a sampling site. Our preferred set‐up had the composite sampler placed securely at street/ground level, but in several neighborhood sample locations, for safety and security reasons, the composite sampler was lowered into the manhole and suspended. Although composite samples are preferred, during our piloting, we found grab samples were required in instances of battery malfunction of the composite sampler or in cases where the composite sampler tubing was clogged by solid waste. To collect a grab sample, a sampling cup on a rope was used.
We homogenized each sample and utilized a portable pump to fill two sample containers (one 500‐ml and one 150‐ml). Samples were immediately placed on ice and, upon complete sample collection, transported to the University of Louisville, Center for Predictive Medicine laboratory. Transit time between sample sites was less than 1‐h, and the 17 samples were typically collected within a 5‐h time frame to minimize temperature variations and virus particle decay that may lead to artifactual differences in virus count measurements. After sampling, the portable pump and tubing were rinsed with bleach water and double rinsed with deionized water before reuse. Sampling personnel from MSD wore standard personal protective equipment (PPE) for wastewater sampling, including Tyvek coveralls, boots, hard hats, face shields, and gloves.
Upon arrival at the Center for Predictive Medicine laboratory, the samples were split, with half retained for archival. The further processing of these samples that followed ensured RNA extraction and detection within 24 h of collection. We pelleted each sample with centrifugation overnight, resuspended in Trizol™, and extracted with the Zymo MagBead RNA extraction kit followed by a Qiagen RNA clean‐up kit. Purified RNA was then evaluated with OD260/230/280. Samples resulting in RNA of sufficient quality and concentration were quantified with an Applied Biosystems QS3 RT PCR System for copy number of N1, in triplicate. Data were reported on unconcentrated sample basis (copies/ml of wastewater). We analyzed preliminary data in R version 3.6.1 (R Core Team, 2016) and figures were produced using the package ggplot2. Kruskal‐Wallis analysis was used to examine differences in total number of SARS‐CoV‐2 RNA detections and clinical cases, between weeks and wastewater catchment areas.
3. Results
We collected samples from manholes, pump stations, and treatment plants in Jefferson County over 8 weeks for a total of 237 samples from 17 wastewater catchment areas (Table 1; Figure 2). During the study period, clinical rate trends reported by Louisville Metro Public Health Department were increasing at the county level, while both the clinical case rate (per 100k) and SARS‐CoV‐2 RNA loads in neighborhood‐level wastewater showed wide variation (Figure 3). Between sampling locations, the wastewater SARS‐CoV‐2 RNA (Kruskal‐Wallis rank sum test p = 0.56) results were not significantly different. Similarly, between sampling locations, clinical rates (Kruskal‐Wallis rank sum test p = 0.058) were not significantly different. But, between the weeks sampled, both wastewater SARS‐CoV‐2 RNA (Kruskal‐Wallis rank sum p = <0.05) and clinical rates (Kruskal‐Wallis rank sum test p = <0.05) were significantly different. When deconstructed, an increasing trend only viewed at the county level loses data of localized outbreaks that may not be spreading uniformly in the wider county population.
Table 1.
Demographics of Study Sampling Site Catchment Areas in Jefferson County, Kentucky a
| ID | Sampling site | Household income (USD) (mean within catchment areas of reported block group median values) | Population | Race and hispanic origin | ||
|---|---|---|---|---|---|---|
| Non‐hispanic | Black (%) | Hispanic (%) | ||||
| White (%) | ||||||
| 1 | Treatment plant | $54,138 | 349,850 | 68 | 25 | 4 |
| 2 | Treatment plant | $53,577 | 295,910 | 72 | 21 | 7 |
| 3 | Treatment plant | $76,606 | 55,928 | 82 | 12 | 4 |
| 4 | Treatment plant | $113,699 | 32,460 | 87 | 8 | 3 |
| 5 | Treatment plant | $106,769 | 31,269 | 75 | 14 | 4 |
| 6 | Community | $27,695 | 10,739 | 9 | 88 | 1 |
| 7 | Community | $27,446 | 7,820 | 68 | 26 | 3 |
| 8 | Community | $103,304 | 11,203 | 92 | 3 | 2 |
| 9 | Community | $45,895 | 35,956 | 59 | 37 | 4 |
| 10 | Community | $51,656 | 25,073 | 83 | 12 | 5 |
| 11 | Community | $77,842 | 99,061 | 87 | 7 | 4 |
| 12 | Community | $68,259 | 139,251 | 79 | 13 | 5 |
| 13 | Community | $53,542 | 73,666 | 63 | 28 | 9 |
| 14 | Community | $61,837 | 46,659 | 75 | 18 | 8 |
| 15 | Community | $63,642 | 22,437 | 80 | 13 | 5 |
| 16 | Community | $49,031 | 8,071 | 90 | 6 | 3 |
| 17 | Community | $24,084 | 20,832 | 61 | 32 | 5 |
Based on 2018 U.S Census Bureau American Community Survey.
Figure 2.
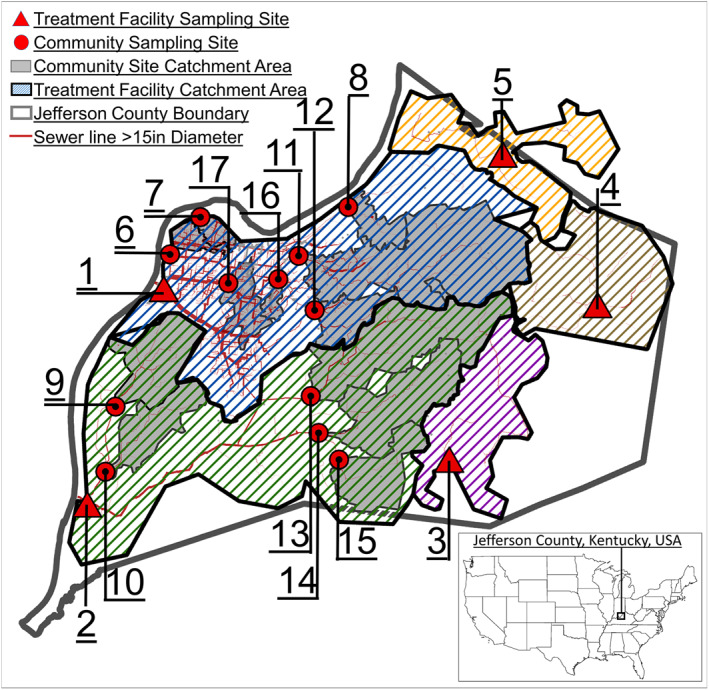
Location of wastewater sampling sites and corresponding wastewater catchment areas in Jefferson County, Kentucky. Underlying geographic information system data for sewer systems and Jefferson County was provided by the Louisville‐Jefferson County Information Consortium (LOJIC). Data are provided by permission of the LOJIC partners which include Louisville Metro Government, Louisville Water Company, the Louisville and Jefferson County Metropolitan Sewer District, and the Jefferson County Property Valuation Administrator.
Figure 3.
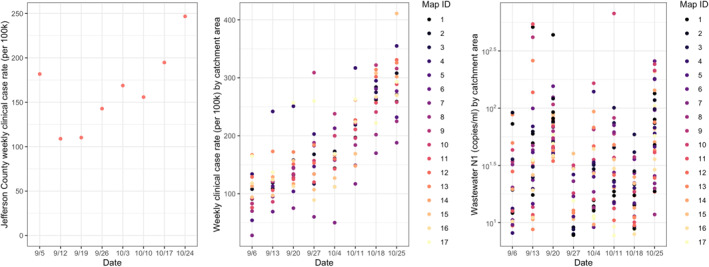
(a) Weekly Jefferson County reported clinical cases (per 100,000 people); (b) clinical cases reported based on wastewater catchment area scale of 17 sampling locations; (c) wastewater SARS‐CoV‐2 RNA concentration (N1 copies per ml) from the selected 17 sampling locations (n = 237).
4. Discussion
With this work, we propose a method to select wastewater sampling locations for the purpose of geographically resolved estimation of community prevalence of SARS‐CoV‐2 infection rates. The difference in week‐to‐week infection rates at the wastewater catchment area scale is important in demonstrating the value of this sampling method because it is clear that rates of infection are not spatially homogenous at the county level, with a high degree of spatial and temporal variance in prevalence. Furthermore, this WBE approach is not subject to substantial data biases of standard clinical testing. The development of an epidemiologically sound methodology for COVID‐19 surveillance depends upon clear definition of the spatiotemporal properties of monitoring methods and clarity regarding the assumptions about the correspondence between the amount of RNA detected in different sampling configurations and the estimation of infected persons in that service area. In Jefferson County, monitoring the five WQTCs allows for nearly complete population surveillance, but it would not allow for an understanding of SARS‐CoV‐2 infection rates with high spatial resolution. Hence, we selected additional community‐based wastewater sampling sites to allow for additional geographic resolution among WQTC catchment areas, utilizing a pragmatic approach to maximize the public health value of our monitoring resources. While previous work has similarly suggested increased spatial resolution of wastewater samples to identify small, localized COVID‐19 outbreaks (Gonzalez et al., 2020), that work was constrained to samples from treatment plants, rather than distributed throughout the sewer network as we describe.
Although wastewater cannot be a substitute for clinical testing data, it may be useful in estimating geographically resolved community‐level infection rates. Importantly, this approach could be used to objectively monitor rates of infection between communities with disproportionate testing rates–a major limitation of sub‐county level reporting of COVID‐19 rates. This approach could be implemented also to monitor emerging variants of SARS‐CoV‐2 and lead to new interventions within communities to contain emerging clusters of infection, making it a more feasible approach than regularly utilized widespread individual testing. The findings of this research strengthen the knowledge base to implement wastewater monitoring in conjunction with clinical surveillance to provide critical insight to isolating and quantifying community‐level SARS‐CoV‐2 infection prevalence and developing actionable public health responses.
This study contributes to advancement of knowledge by informing selection of sample sites for wastewater to correlate with spatial clinical data on infection rates from randomized testing. An important strength of this sampling approach is the provision of more accurate estimates of actual community‐level SARS‐CoV‐2 infections, independent of the proportion of symptomatic individuals, individual testing capacity, healthcare capacity, socioeconomic capacity, and other substantial biases that skew prevalence estimates based on individual‐level testing. Furthermore, sampling of wastewater closer to residences, as described here, allows for less time for virus concentrations from catchment areas to differentially degrade before sampling, as opposed to centralized facility testing. Thus, it allows for more specific prevalence estimates than WQTC samples alone. The results of this protocol provide a useful and cost‐effective scenario for public health surveillance to inform interventions and countermeasures to allow geographically targeted strategies to reduce transmission of SARS‐CoV‐2 infections. The preliminary data following this protocol attest to the value of the design by showing that analysis of catchment areas could be an opportunity to demonstrate geographic variation, which is advantageous when compared with homogenous county‐level data, for both clinical rates and wastewater SARS‐CoV‐2 RNA concentrations. There are several drawbacks of very large aggregations to informing public health understanding and response. Our work highlights the importance of neighborhood/multi‐neighborhood epidemiologically defined scales, which may be a better sampling approach than sampling only treatment plants.
There are several limitations to this protocol. The site selection is framed to consider physical variation in sample state (travel time, sample temperature, flow data, background chemistry, variation in shedding by infected persons), limitations that apply to any wastewater sampling. The rapidly evolving understanding of the decay of SARS‐CoV‐2 RNA in wastewater may further help refine sample location selection. The number of sites was not predetermined, but was based on spatial resolution of sewer catchment areas and on physical limitations of the system to select ideal population distributions. In some areas, MSD has a combined sanitary sewer and storm water system, possibly diluting concentrations of SARS‐CoV‐2 RNA. However, dilution may largely be accounted for by normalizing to population and flow rate. Furthermore, even though we can partially account for resulting dilution, it is not possible to fully exclude industrial and commercial outflow. It should be noted that clinical data from testing community residents potentially includes households not represented in the sewer system (e.g., septic tank users or individuals living in one area but spending the majority of their day in another area of the city).
Areas for future exploration include: (a) Sensitivity and temporal dynamics to determine if raw wastewater is a concurrent indicator of SARS‐CoV‐2 RNA in sludge; (b) quantification of field samples such as turbidity or another field proxy for SARS‐CoV‐2 RNA to allow expanded rapid indicator methods; (c) waste travel time in relation to sampling location selections; (d) attributes of physical sewer system infrastructure regarding age of sewage or opportunities for latent virus in the system; (e) the need for sewer sampling infrastructure that is “pandemic ready” and a better fit design for answering human health surveillance instead of only industrial surveillance and application of public health response protocols that integrate sewer sampling results; (f) protocols for ongoing monitoring for variants at wastewater catchment area scale; and (g) protocols for expanding to other cities.
5. Conclusions
Design of this protocol for epidemiologically defined population‐level sampling for SARS‐CoV‐2 RNA prevalence in wastewater can be used for geographically resolved community surveillance, enabling a low‐bias measure of population‐level infections and the ability to detect the presence and the progression of new variants‐of‐concern of the virus. The approach may therefore be a valuable guide for targeted and cost‐effective public health population level surveillance and mitigation strategies.
Conflict of Interest
The authors declare no conflicts of interest relevant to this study.
Supporting information
Data Set S1
Acknowledgments
We thank our field and analytical technicians Dwight Mitchell, David Hoetker, Rick Strehl, Brandon Ryan, and Ian Santisteban for their diligent commitment to this work. This work was supported in part by grants from the James Graham Brown Foundation, the Owsley Family Foundation and Louisville Metro Government Coronavirus Aid, Relief, and Economic Security (CARES) Act funds. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Yeager, R. , Holm, R. H. , Saurabh, K. , Fuqua, J. L. , Talley, D. , Bhatnagar, A. , & Smith, T. (2021). Wastewater sample site selection to estimate geographically resolved community prevalence of COVID‐19: A sampling protocol perspective. GeoHealth, 5, e2021GH000420. 10.1029/2021GH000420
R. Yeager, R. H. Holm, and T. Smith contributed equally to this work.
Data Availability Statement
R. Yeager (2021), “CatchmentAreas051821,” Mendeley Data, V1. Data set accessed 2021‐06‐02 at http://dx.doi.org/10.17632/t8mcw8f95x.1. Underlying GIS data for sewer systems and Jefferson County was provided by LOJIC, the Louisville‐Jefferson County Information Consortium and available at https://www.lojic.org. Data provided by permission of the LOJIC partners consisting of Louisville Metro Government, Louisville Water Company, the Louisville and Jefferson County Metropolitan Sewer District and the Jefferson County Property Valuation Administrator. Countywide socioeconomic data was collected from the U.S. Census Bureau and available at https://www.census.gov/.
References
- Ahmed, W. , Angel, N. , Edson, J. , Bibby, K. , Bivins, A. , O'Brien, J. W. , et al. (2020). First confirmed detection of SARS‐CoV‐2 in untreated wastewater in Australia: A proof of concept for the wastewater surveillance of COVID‐19 in the community. Science of the Total Environment, 728, 138764. 10.1016/j.scitotenv.2020.138764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibby, K. , & Peccia, J. (2013). Identification of viral pathogen diversity in sewage sludge by metagenome analysis. Environmental Science & Technology, 47, 1945–1951. 10.1021/es305181x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisseux, M. , Debroas, D. , Mirand, A. , Archimbaud, C. , Peigue‐Lafeuille, H. , Bailly, J.‐L. , & Henquell, C. (2020). Monitoring of enterovirus diversity in wastewater by ultra‐deep sequencing: An effective complementary tool for clinical enterovirus surveillance. Water Research, 169, 115246. 10.1016/j.watres.2019.115246 [DOI] [PubMed] [Google Scholar]
- Bivins, A. , North, D. , Ahmad, A. , Ahmed, W. , Alm, E. , Been, F. , et al. (2020). Wastewater‐based epidemiology: Global collaborative to maximize contributions in the fight against COVID‐19. Science of the Total Environment, 54, 7754–7757. 10.1021/acs.est.0c02388 [DOI] [PubMed] [Google Scholar]
- Castiglioni, S. , Thomas, K. V. , Kasprzyk‐Hordern, B. , Vandam, L. , & Griffiths, P. (2014). Testing wastewater to detect illicit drugs: State of the art, potential and research needs. Science of the Total Environment, 487, 613–620. 10.1016/j.scitotenv.2013.10.034 [DOI] [PubMed] [Google Scholar]
- Global Polio Eradication Initiative (2015). Guidelines on environmental surveillance for detection of polioviruses. Global Polio Eradication Initiative. Geneva, Switzerland. World Health Organization. http://polioeradication.org/wp-content/uploads/2016/07/GPLN_GuidelinesES_April2015.pdf [Google Scholar]
- Gonzalez, R. , Curtis, K. , Bivins, A. , Bibby, K. , Weir, M. H. , Yetka, K. , et al. (2020). COVID‐19 surveillance in Southeastern Virginia using wastewater‐based epidemiology. Water Research, 186, 116296. 10.1016/j.watres.2020.116296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, O. E. , & Halden, R. U. (2020). Computational analysis of SARS‐CoV‐2/COVID‐19 surveillance by wastewater‐based epidemiology locally and globally: Feasibility, economy, opportunities and challenges. Science of the Total Environment, 730, 138875. 10.1016/j.scitotenv.2020.138875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima, M. , Ahmed, W. , Bibby, K. , Carducci, A. , Gerba, C. P. , Hamilton, K. A. , et al. (2020). SARS‐CoV‐2 in wastewater: State of the knowledge and research needs. Science of the Total Environment, 739, 139076. 10.1016/j.scitotenv.2020.139076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux, F. G. , Hotz, P. , Friedli, D. , Savova‐Bianchi, D. , & Oppliger, A. (2013). High occurrence of hepatitis E virus in samples from wastewater treatment plants in Switzerland and comparison with other enteric viruses. Water Research, 47, 5101–5109. 10.1016/j.watres.2013.05.050 [DOI] [PubMed] [Google Scholar]
- Medema, G. , Heijnen, L. , Elsinga, G. , Italiaander, R. , & Brouwer, A. (2020). Presence of SARS‐Coronavirus‐2 RNA in sewage and correlation with reported COVID‐19 prevalence in the early stage of the epidemic in The Netherlands. Environmental Science & Technology, 7(7), 511–516. 10.1021/acs.estlett.0c00357 [DOI] [PubMed] [Google Scholar]
- Moghadas, S. M. , Fitzpatrick, M. C. , Pandey, A. , Shoukat, A. , Singer, B. H. , Galvani, A. P. , & Galvani, A. P. (2020). The implications of silent transmission for the control of COVID‐19 outbreaks. Proceedings of the National Academy of Sciences of the United States of America, 117, 17513–17515. 10.1073/pnas.2008373117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia, J. , Zulli, A. , Brackney, D. E. , Grubaugh, N. D. , Kaplan, E. H. , Casanovas‐Massana, A. , et al. (2020). Measurement of SARS‐CoV‐2 RNA in wastewater tracks community infection dynamics. Nature Biotechnology, 38, 1164–1167. 10.1038/s41587-020-0684-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogka, V. , Labropoulou, S. , Emmanouil, M. , Voulgari‐Kokota, A. , Vernardaki, A. , Georgakopoulou, T. , & Mentis, A. F. (2017). Laboratory surveillance of polio and other enteroviruses in high‐risk populations and environmental samples. Applied and Environmental Microbiology, 83(5), e02872–16. 10.1128/AEM.02872-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo, W. , Truchado, P. , Cuevas‐Ferrando, E. , Simón, P. , Allende, A. , & Sánchez, G. (2020). SARS‐CoV‐2 RNA in wastewater anticipated COVID‐19 occurrence in a low prevalence area. Water Research, 181, 115942. 10.1016/j.watres.2020.115942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherchan, S. P. , Shahin, S. , Ward, L. M. , Tandukar, S. , Aw, T. G. , Schmitz, B. , et al. (2020). First detection of SARS‐CoV‐2 RNA in wastewater in North America: A study in Louisiana, USA. Science of the Total Environment, 743, 140621. 10.1016/j.scitotenv.2020.140621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Commerce (2020). https://www.census.gov/
- Wölfel, R. , Corman, V. M. , Guggemos, W. , Seilmaier, M. , Zange, S. , Müller, M. A. , et al. (2020). Virological assessment of hospitalized patients with COVID‐2019. Nature, 581(7809), 465–469. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Set S1
Data Availability Statement
R. Yeager (2021), “CatchmentAreas051821,” Mendeley Data, V1. Data set accessed 2021‐06‐02 at http://dx.doi.org/10.17632/t8mcw8f95x.1. Underlying GIS data for sewer systems and Jefferson County was provided by LOJIC, the Louisville‐Jefferson County Information Consortium and available at https://www.lojic.org. Data provided by permission of the LOJIC partners consisting of Louisville Metro Government, Louisville Water Company, the Louisville and Jefferson County Metropolitan Sewer District and the Jefferson County Property Valuation Administrator. Countywide socioeconomic data was collected from the U.S. Census Bureau and available at https://www.census.gov/.


