Abstract
Transcriptomic profiling of tumor tissues introduces a large database, which has led to improvements in the ability of cancer diagnosis, treatment, and prevention. However, performing tumor transcriptomic profiling in the clinical setting is very challenging since the procurement of tumor tissues is inherently limited by invasive sampling procedures. Here, we demonstrated the feasibility of purifying hepatocellular carcinoma (HCC) circulating tumor cells (CTCs) from clinical patient samples with improved molecular integrity using Click Chips in conjunction with a multimarker antibody cocktail. The purified CTCs were then subjected to mRNA profiling by NanoString nCounter platform, targeting 64 HCC-specific genes, which were generated from an integrated data analysis framework with 8 tissue-based prognostic gene signatures from 7 publicly available HCC transcriptomic studies. After bioinformatics analysis and comparison, the HCC CTC-derived gene signatures showed high concordance with HCC tissue-derived gene signatures from TCGA database, suggesting that HCC CTCs purified by Click Chips could enable the translation of HCC tissue molecular profiling into a noninvasive setting.
Keywords: circulating tumor cells, click chemistry, nanosubstrate, hepatocellular carcinoma, transcriptome profiling
Graphical Abstract
Covalent chemistry-mediated multimarker purification of hepatocellular carcinoma circulating tumor cells (HCC CTCs) is developed to realize noninvasive detection of molecular signatures of HCC. The purification system enables improved purity and molecular integrity of the purified HCC CTCs for conducting transcriptomic profiling of a strictly selected HCC-specific gene panel.
Introduction
Over the past decade, integrative bioinformatics analysis[1] of large genomic and transcriptomic data sets has been leveraged to provide insights into the underlying cancer biology. For example, The Cancer Genome Atlas (TCGA) has generated over 2.5 petabytes of data including genomics, epigenomics, transcriptomics and proteomics, which play an important role in improving our ability on cancer diagnosis, treatment, and prevention and broadening our knowledge in underlying cancer biology governing disease progression.[2] These molecular data have relied exclusively on tissue specimens obtained from surgical excision or biopsy. However, the acquisition of specimens is inherently limited by availability, invasiveness, cost, and risk of morbidity that hampers clinical utility.[3] Translating these extensive cancer tissue-based molecular profiling data resources into clinical practice calls for a noninvasive sampling technology to obtain tumor cells.
Several groups have proposed circulating tumor cells (CTCs)[4–6] as the source material for “liquid biopsies” in solid tumors. CTCs can be recovered noninvasively throughout the natural history of disease. Compared to conventional diagnostic imaging and serum marker tests, CTC detection and enumeration offer a noninvasive systemic assessment of tumor that can aid with prognostication[7, 8] and evaluation of treatment response.[9, 10] However, identification, enumeration, and characterization of CTCs in clinical samples have been technically limited by their extremely low abundance compared to hematologic cells (i.e., white blood cells, WBCs). To obtain purified CTCs, numerous efforts[11–13] have been made in multiple fields including chemistry, materials science, and bioengineering, paving the way for realization of CTC-based molecular profiling.
Hepatocellular carcinoma (HCC), the fourth leading cause of cancer-related deaths worldwide,[14] is in dire need of diagnostic and prognostic biomarkers. CTCs have been considered to be of prognostic and predictive potential in HCC.[15, 16] For example, it has been reported that CTC enumeration, along with CTC analysis of biological characteristics and genomic heterogeneity, show clinical potential in predicting disease prognosis and monitoring treatment response in HCC patients.[17] Using NanoVelcro Chips[18], we have recently developed and validated a multi-marker capture cocktail[19] (targeting three HCC-specific surface markers, i.e., ASGPR1, GPC3, and EpCAM) for detecting HCC CTCs. By combining NanoVelcro Chips[18,20] with Laser Capture Microdissection technique,[21] we were able to harvest single HCC CTCs for whole-genome analysis, unveiling that somatic copy number alterations in HCC CTCs recapitulated those observed in the matching HCC tissues.[22] On the other hand, CTC-iChips[23] developed by the Massachusetts General Hospital team were employed to purify HCC CTCs. The purified HCC CTCs were subjected to mRNA analysis[24] by droplet digital polymerase chain reaction (ddPCR) to quantify 10 HCC-related genes, including AFP, AHSG, ALB, APOH, FABP1, FGB, FGG, GPC3, RBP4, and TF. The resulting CTC-derived mRNA signatures[24] can be used to specifically detect HCC CTCs, enabling dynamic monitoring of disease progression. These approaches demonstrated the potential for HCC CTC to serve as a tumor tissue surrogate.
In the last dozen years, our joint team pioneered a collection of nanostructure-embedded microchips, in which nanosubstrates[18,25] (e.g., silicon nanowire substrates, SiNWS) coated with capture agents exhibit superb “stickiness” to selectively capture CTCs in patients’ blood. Five generations of NanoVelcro Chips[13] have been developed to meet different clinical needs, i.e., 1st-gen chips for CTC enumeration[25], 2nd-gen chips for single-cell analysis[21, 26], 3rd-gen Thermoresponsive chips[27] and 4th-gen Sweet Chips[28] for purification of CTCs from patients’ blood. Challenges remain in the development of new-generation devices that are compatible with a multimarker capture cocktail and capable of efficient, specific, and rapid CTC purification in order to preserve RNA quality in CTCs. 5th-gen Click Chips[29] have been developed to achieve purification of CTCs with well-preserved RNA in conjunction with the use of antibody cocktails for different solid tumors.
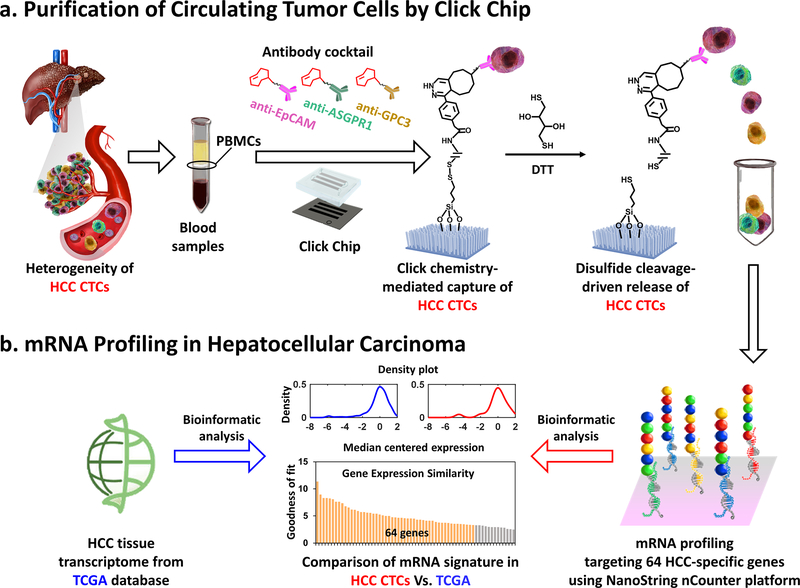
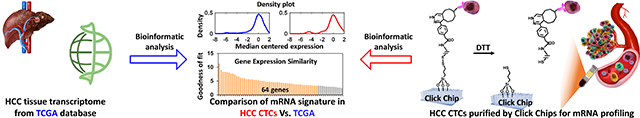
Here, we demonstrated the feasibility of purifying HCC CTCs from patients’ blood samples with improved purity and molecular integrity using Click Chips[29] in conjunction with a multimarker antibody cocktail[19] containing anti-EpCAM, anti-ASGPR1, and anti-GPC3 antibodies (Figure 1a). Such an HCC CTC purification approach combines two consecutive covalent chemistry-mediated reactions, i.e., 1) the click reaction-mediated HCC CTC capture from PBMC samples onto Click Chips, and 2) disulfide cleavage-driven HCC CTC release from Click Chips into phosphate-buffered saline (PBS) solution. Again, a pair of click chemistry motifs with high selectivity and reaction rates, i.e., tetrazine (Tz) and trans-cyclooctene (TCO), were grafted onto SiNWS (via chemical modification) and HCC CTCs (via antibody conjugation), respectively. By introducing a PBMC sample from an HCC patient into a Click Chip, click reaction between Tz and TCO enabled the rapid immobilization of HCC CTCs. Subsequently, 1,4-dithiothreitol (DTT) was employed to cleave the embedded disulfide bonds to release the captured HCC CTCs specifically. The released CTCs were then subjected to mRNA profiling by NanoString nCounter platform, targeting 64 HCC-specific genes, which were generated from an integrated data analysis framework with 8 tissue-based prognostic gene signatures from 7 publicly available HCC transcriptomic studies. After bioinformatics analysis and comparison, the HCC CTC-derived gene signatures in HCC CTCs showed high concordance with HCC tissue-derived gene signatures from TCGA database (Figure 1b), suggesting that HCC CTCs purified by Click Chips could enable the translation of HCC tissue molecular profiling into a noninvasive setting.
Figure 1. Multimarker purification of circulating tumor cells (CTCs) enables mRNA profiling of hepatocellular carcinoma (HCC).
a) Schematic illustration of the workflow employed for HCC CTC purification (capture/release) by Click Chips in conjunction with an HCC-specific antibody cocktail (i.e., anti-EpCAM, anti-ASGPR1, and anti-GPC3). The HCC CTC purification approach combines two consecutive covalent chemistry-mediated reactions, i.e., 1) the click reaction-mediated HCC CTC capture from peripheral blood mononuclear cell (PBMC) samples onto Click Chips, and 2) disulfide cleavage-driven HCC CTC release from Click Chips to phosphate-buffered saline (PBS) by 1,4-dithiothreitol (DTT). b) Quantification of the 64 HCC-specific genes in the purified HCC CTCs were performed by Nanostring nCounter platform. After bioinformatic analysis and comparison, the gene signatures in HCC CTCs showed high concordance with HCC tissue gene signatures from The Cancer Genome Atlas (TCGA) database.
2. Results and Discussions
2.1. The fabrication of a Click Chip
Based on the previous demonstration of Click Chips, where covalent chemistry-mediated capture and release of CTCs were applied for purification of CTCs in non-small cell lung cancer (NSCLC),[29] we explored the combined use of Click Chip’s device configuration with a multimarker cocktail for purification of HCC CTCs. A Click Chip (Figure 1a) is composed of two main functional components: 1) tetrazine (Tz)-grafted SiNWS (Figure S1, Supporting Information): a patterned SiNWS[30] covalently modified by terminal Tz motifs with disulfide bridges,[29] and 2) an overlaid polydimethylsiloxane (PDMS) chaotic mixer[25] on which a network of microchannels was designed to induce chaotic mixing.[31] They were housed in a custom-designed microfluidic chip holder to form a “Click Chip” device (Figure S2, Supporting Information). The Tz-grafted SiNWS and PDMS chaotic mixer were fabricated according to the previously published procedures.[29, 32]
2.2. General procedure for performing purification of HCC CTCs in Click Chips
The covalent chemistry-mediated HCC CTC purification approach[29] combines two consecutive steps, i.e., 1) the click reaction-mediated HCC CTC capture onto Click Chips and 2) disulfide cleavage-driven HCC CTC release from Click Chips. Prior to performing HCC CTC purification studies using Click Chips, trans-cyclooctene TCO motif was covalently conjugated onto the three antibody capture agents, i.e., anti-EpCAM, anti-ASGPR1, and anti-GPC3 (see Experimental Section/Methods). Single or combined antibodies were then incubated with the artificial or clinical HCC CTC samples. After washing off the excess antibody, the HCC CTC samples in 200-μL PBS were introduced into Click Chips, where click reaction-mediated capture led to the capture of HCC CTCs on SiNWS. After CTC capture, the DTT solution (50 mM) was introduced to cleave disulfide bonds for specific CTC release.
2.3. Optimization of Click Chips for HCC CTC capture and release using artificial HCC CTC samples
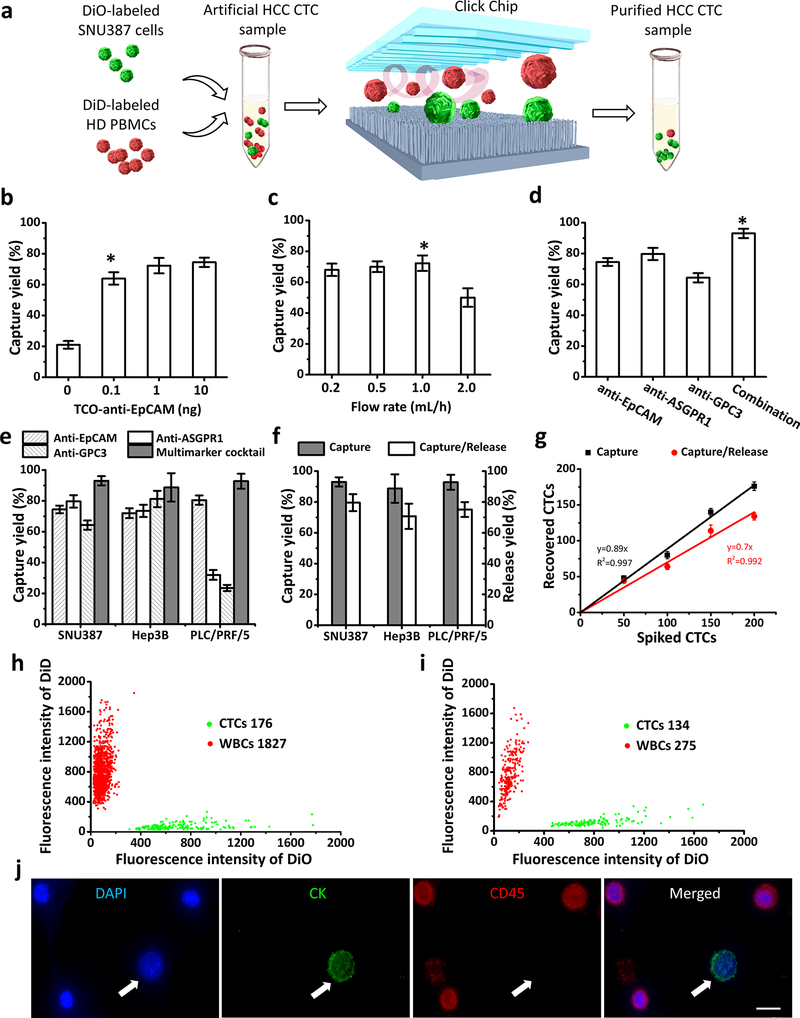
To optimize the capture performance of Click Chips, artificial HCC CTC samples (Figure 2a) were prepared by spiking 200 DiO-labeled HCC cell lines (green color) into the DiD-labeled PBMCs (red color, 5 × 106 cells mL−1) isolated from a healthy donor’s whole blood. After incubating with single or combined antibody capture agents, the artificial HCC CTC samples were introduced into Click Chips for click chemistry-mediated capture of HCC CTCs. After staining with 4’, 6-diamidino-2-phenylindole (DAPI), DiO-labeled HCC cells and background DiD-labeled WBCs were counted under a fluorescence microscope (Nikon 90i). The HCC CTC capture yields were calculated by dividing the counts of CTCs captured on the Click Chips by the counts of target cells that were initially spiked to the artificial HCC CTC samples. We first evaluated the effect of different quantities of TCO-labeled anti-EpCAM (i.e., 0 ng, 0.1 ng, 1.0 ng, and 10 ng) on HCC CTC capture yields (Figure 2b). The capture yield of Click Chips was up to 72% when 1 ng of TCO-anti-EpCAM was used for HCC CTC capture at a flow rate of 1.0 mL h−1. HCC CTC capture yields reached the plateau even with a higher quantity of TCO-labeled anti-EpCAM. Next, the effects of different flow rates (0.2, 0.5, 1.0, and 2.0 mL h−1) on HCC CTC capture yields of Click Chips were evaluated (Figure 2c), and an optimal flow rate of 1.0 mL h−1 was identified, which was consistent with our works.[29] Our group has previously demonstrated the combined use of NanoVelcro Assay with the multimarker antibody cocktail (i.e., anti-EpCAM, anti-ASGPR1, and anti-GPC3 antibodies) for enumeration of HCC CTCs.[19] Under the optimal flow rate of 1.0 mL h−1, we examined the capture yields (Figure 2d) using either single or antibody cocktails. Similarly, an optimal capture yield (93.02 ± 3.00 %) was achieved when the multimarker cocktail was used. We further validated the single antibodies versus the multimarker cocktail study using two additional HCC cell lines (i.e., Hep3B and PLC/PRF/5). As shown in Figure 2e, the HCC CTC capture performance observed for the multimarker cocktail group outperformed those from any single antibody groups.
Figure 2. Click Chip optimization for purifying hepatocellular carcinoma (HCC) circulating tumor cells (CTCs) using artificial HCC CTC samples.
a) DiO-labeled SNU387 HCC cells (green) and DiD-labeled peripheral blood mononuclear cells (PBMCs) (red) from a healthy donor (HD) were mixed to prepare the artificial HCC CTC samples for Click Chip optimization. b) The capture yields of Click Chips at different quantity of TCO-labeled anti-EpCAM. c) The capture yields of Click Chips at different flow rates. d) The capture yields acheived by using single and combined antibody, i.e., anti-EpCAM, anti-ASGPR1, anti-GPC3, and combination of the three capture agents. e) The capture yields achieved by using single and combined antibody capture agents. Different artificial HCC CTC samples containing three different HCC cells (i.e., SNU387, Hep3B, PLC/PRF/5) were validated. f) Capture/release yields of Click Chips observed for different artificial HCC CTC samples. g) Dynamic ranges of HCC CTC capture and capture/release yields of Click Chips. Here, artificial HCC CTC samples containing 0 to 200 SNU387 cells were tested. (b-g, The means ± SD of three independent assays are presented). h) A 2-parameter scatter plot showed the cell distribution of SNU387/White blood cell (WBC) observed for CTC capture in a Click Chip. i) A 2-parameter scatter plot showed the cell distribution of SNU387/WBC after CTC release, following the CTC capture (h). j) The representative fluorescent images of SNU387 (DAPI+/CK+/CD45-; white arrow)/WBC cells (DAPI+/CK-/CD45+) captured on the Click Chips. bar, 10 μm. *denotes the optimal condition.
After determining the optimal condition for click creaction-mediated capture of HCC CTCs, we tested the performance of HCC CTC release in Click Chips, in which 200 μL of DTT (50 mM) soluiton was flowed through[29] at a rate of 1.0 mL h−1 to release the immobilized HCC CTCs. The HCC CTC release yield was evaluated by dividing the CTC counts released from the Click Chips by the counts of target cells that were initially spiked to the artificial HCC CTC samples. Furthermore, the universal applicability of Click Chips for purification (capture/release) of HCC CTCs was evaluated using three artificial HCC CTC samples spiking with three HCC cell lines, i.e., SNU387, Hep3B, or PLC/PRF/5. Figure 2f showed that the HCC CTC capture/release yields across three HCC cell lines were ranging from 88.75% to 93.02% (capture yields), and 70.74% to 79.57% (release yields), respectively. The dynamic range of Click Chips was also tested using artificial HCC CTC samples by spiking 0 to 200 SNU387 cells into a healthy donor’s PBMCs. Consistent capture yields (y = 0.89x, R2 = 0.997) and release yields (y = 0.70x, R2 = 0.992) were observed for Click Chips that are sufficient for testing clinical samples (Figure 2g). A representative study using an artificial sample spiked with 200 SNU387 cells demonstrated (Figure 2h,i) a minimal background (1,827 WBC-capture and 275 WBC-release) of Click Chips in CTC-capture (176/200) and release (134/176). Figure 2j depicted the representative fluorescent images of SNU387 (DAPI+/CK+/CD45-)/WBC cells (DAPI+/CK-/CD45+) captured on the Click Chips for cell enumeration and capture yield calculation. The CTC purification data indicate that the disulfide cleavage-driven release mechanism can further improvethe purity of the purified CTCs (ca. one order of magnitude reduction in WBC contamination), providing robust and reproducible samples with high purity for downstream molecular characterization. Overall, these results suggest that the HCC CTCs can be purified by Click Chips effectively (click chemistry-driven CTC capture followed by disulfide cleavage-driven CTC release).
2.4. Development of HCC-specific gene panel with 64 genes
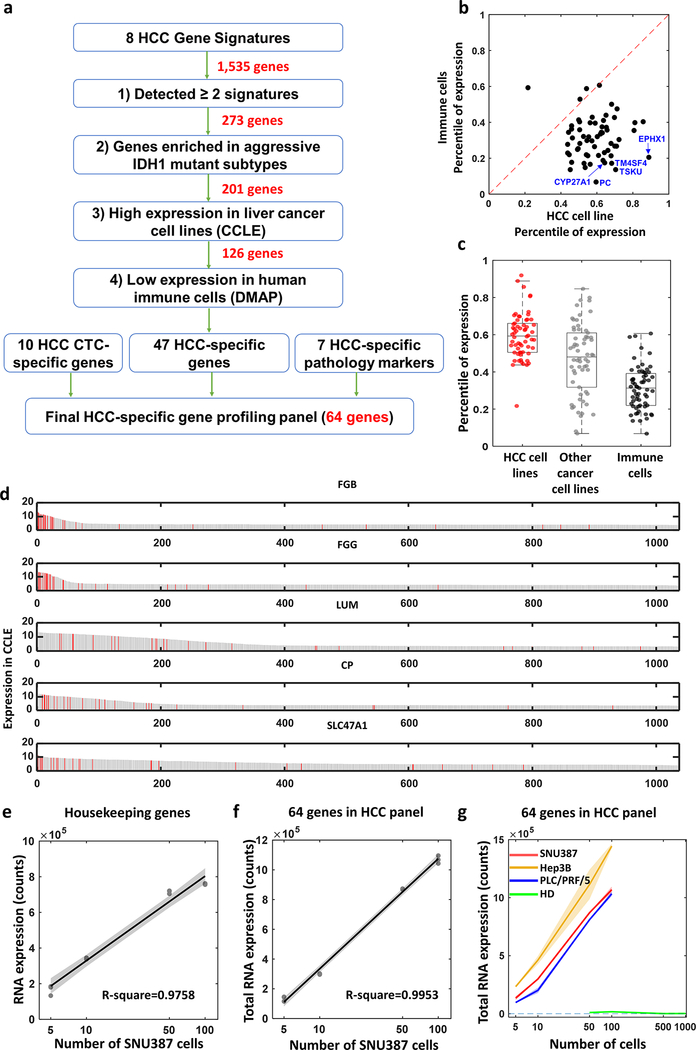
Before performing mRNA profiling on the HCC CTCs purified by Click Chips, we adopted an integrated data analysis framework to select and develop an HCC-specific gene panel, capable of detecting HCC CTCs in the presence of non-specifically purified WBCs. We started HCC-specific gene panel selection by assembling 8 tissue-based prognostic gene signatures (Table S1, Supporting Information) from 7 publicly available HCC transcriptomic studies,[33–39] including Hoshida’s HCC subtype gene signature,[33] cholangiocarcinoma-like (CCL) HCC gene signature,[34] Hippo pathway inactivation associated gene signature,[36] risk score classifier based on 65 genes for HCC survival prediction,[37] NCI proliferation signature,[38] hepatoblastoma-like tumor signature,[39] iCluster1signature, and IDH1-like signature. A total of 1,535 candidate genes were collected, and serially and selectively pared down in a step-wise fashion to enrich for genes with high expression in HCC CTCs and low expression in immune cells in order to avoid the signals from non-specifically trapped WBCs in the Click Chips.[40] To this end, an integrated data analysis framework (Figure 3a) was applied for the selection of the HCC-specific gene panel. This framework consists of four major steps: 1) Select genes involved in at least two independent tissue-based HCC prognostic signatures to further support their role in HCC tumor biology, this selection step results in 273 candidate genes; 2) Select the subset of genes also included in IDH1-like signature, known to be associated with aggressive HCC subtypes, resulting in 201 genes; 3) Select genes that are highly expressed in HCC cell lines from the Cancer Cell Line Encyclopedia (CCLE),[41] further enriching genes specific to HCC compared to other cancer cell lines, resulting in 126 genes; and 4) Select genes with low expression in various immune cells using Differentiation MAP dataset (DMAP),[42] to minimize the signal from non-specifically captured WBCs. This yielded 47 genes that are specific for detecting HCC. Finally, we incorporated the 10 HCC-specific genes[24] (i.e., AFP, AHSG, ALB, APOH, FABP1, FGB, FGG, GPC3, RBP4, and TF), that were used to specifically detect HCC CTCs and monitor disease progression, and 7 markers (i.e., ARG1, ASGR1, ASGR2, CPS1, KRT8, KRT18, and ERRFI1) that are commonly used in clinical pathology diagnosis,[43, 44] resulting in a 64-gene HCC-specific gene panel (Table S2, Supporting Information).
Figure 3. Development and validation of of hepatocellular carcinoma (HCC)-specific gene panel with 64 genes.
a) An integrated data analysis framework adopted for selecting and developing the 64 genes of HCC-specific gene panel. b) Scatter plot shows expression of the 64 genes comparing HCC cell lines to humane immune cells. Dotted line indicates the same expression between HCC cells and other cancer cells which is y=x axis. c) Box plot depicts differential expression of 64 genes between HCC cell lines (red dots), other cancer cell lines (gray dots) from Cancer Cell Line Encyclopedia (CCLE), and humane immune cells from Differentiation MAP dataset (DMAP) (black dots). d) Bar charts show distribution of all the cells in CCLE based on the expression level of the genes (i.e., FGB, FGG, LUM, CP, SLC47A1). Red bar indicates HCC cell lines. e, f) Dots with regression line depict expression dynamics of housekeeping genes (e, R2 = 0.976) and 64 genes (f, R2 = 0.995) was assessed with difference SNU387 cell counts from 0 to 100. g) Line plots with distinct colors represent dynamic changes of the 64 gene expression counts from SNU387, Hep3B, PLC/PRF/5 and healthy donor peripheral blood mononuclear cells (PBMCs) with different cell numbers. Slopes of the curve- SNU387: 9,829.5 counts/cell, Hep3B: 12,744.9 counts/cell, PLC/PRF/5: 9,862.5 counts/cell, healthy donor PBMCs: −8.8 counts/cell.
2.5. Analytical validation of HCC-specific gene panel
We first confirmed that the 64 genes of the HCC-specific gene panel are HCC cell-specific by performing bioinformatics analysis using the CCLE and DMAP. As shown in Figure 3b, 61 out of 64 genes (95%) are below the regression line, indicating that these genes are highly expressed in HCC cell lines and lowly expressed in immune cells. We then compared expression ranks (percentiles) of the 64 genes in HCC and other cancer cell lines from the CCLE and immune cells from the DMAP. Expression ranks of the 64 genes are significantly higher in HCC cells compared to other cancer cell lines (p < 0.001) or immune cells (p < 0.001) (Figure 3c). Comparison of the most differentially expressed top 5 representative genes between HCC cell lines versus humane immune cells, and HCC cell lines versus other cancer cell lines are depicted in Figure S3 and Figure S4, respectively (Supporting Information). The top 5 genes (i.e., FGB, FGG, LUM, CP, SLC47A1) with the highest expression in HCC cell lines was assessed in all CCLE cancer cell lines (Figure 3d). This result demonstrates the predominant expression of the 64 genes in HCC cell lines.
To validate the feasibility of applying Nanostring nCounter platform for profiling the 64 genes of the HCC-specific gene panel, we then prepared the cell mixture by spiking different numbers of SNU387 cells with serial dilution (i.e., 5, 10, 50, 100 cells) into PBMCs (5 × 106 cells mL−1) isolated from a healthy donor’s blood. The cell mixtures were then subjected to RNA extraction and Nanostring nCounter platform for the 64 gene expression analysis. Five housekeeping genes (i.e., DYNLL1, GAPDH, RPL13A, RPS11, and RPS16) were tested simultaneously as an internal control of the samples. The dynamic ranges of mRNA expression detected for housekeeping genes (Figure 3e, R2 = 0.976) and the 64 genes (Figure 3f, R2 = 0.995) showed excellent linearity of the Nanostring nCounter platform. We then tested the general applicability of the Nanostring nCounter platform for the 64 gene expression analysis using three cell mixtures spiked with three HCC cell lines, i.e., SNU387, Hep3B, or PLC/PRF/5. PBMCs (5 × 106 cells mL−1) isolated from a healthy donor’s blood served as the negative control. Results summarized in Figure 3g showed that the Nanostring nCounter platform for the 64 gene expression analysis exhibited consistent performances across all three cell mixtures spiked with three HCC cell lines.
2.6. Comparative analysis of the 64-gene expressions between HCC CTCs purified by Click Chips and HCC tissues from TCGA
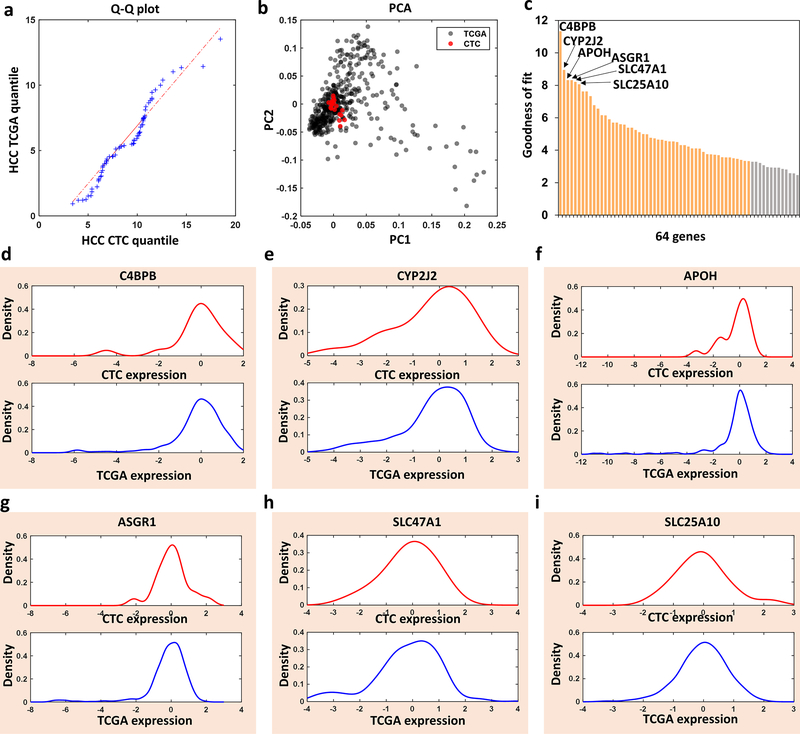
We performed HCC CTC purification by Click Chips using 20 patient blood samples with HCC across all clinical stages (Table 1). Subsequently, the purified HCC CTCs were subjected to analyze the 64 genes of the HCC-specific gene panel using the Nanostring nCounter platform. The 64-gene expressions from HCC CTCs were compared with the ones from HCC tissues from TCGA. We first examined whether the two 64-gene expression profiles (one from HCC CTCs and the other from HCC tissues) came from populations with a common distribution. The quantile-quantile plot (Q-Q plot) suggests that both are from HCC patient populations with a common gene expression distribution (Figure 4a). In addition, the principal component analysis (PCA) score plot spanned by the first two principal components based on the 64-gene expression data showed that 20 patient samples of HCC CTCs (red dots) were overlapped with HCC tissues from TCGA (gray dots), supporting the high concordance of expression variance between the HCC CTCs and HCC tissues from TCGA (Figure 4b). Further assessment of expression concordance was performed using the goodness of fit measure based on the Kolmogorov-Smirnov (KS) distance between the HCC CTCs and HCC tissues from TCGA, and 51 out of the 64 genes (80%) have a significantly high level of expression concordance (KS distance<0.3, Figure 4c). Empirical cumulative distribution function (CDF) analysis of 64 gene expression from the HCC CTCs and HCC tissue from TCGA reveals the expression concordance of 64 genes between HCC CTCs (red) and HCC tissues from TCGA (blue). The numbers on the individual plots indicate KS distance between HCC CTCs and HCC tissues from TCGA (Figure S5, Supporting Information). The density plots of the 6 most representative genes having the best concordance expression between HCC CTCs and HCC tissues from TCGA were shown in Figures 4d-i. Collectively, these results demonstrate the high expression concordance of 64 genes of HCC CTCs by Click Chips with HCC tissues.
Table 1.
Clinical information of hepatocellular carcinoma (HCC) patients enrolled in this study.
| Sample ID | Age (Y) | Gender | Cirrhosis | Etiology | Tumor number | Cumulative tumor size (cm) | BCLC Stage | AJCC Stage | Within Milan Criteria |
|---|---|---|---|---|---|---|---|---|---|
| HCC01 | 84 | Male | No | idiopathic | 1 | 7.1 | B | 1b | No |
| HCC02 | 82 | Female | Yes | HCV | 1 | 3.6 | A | 1b | Yes |
| HCC03 | 73 | Male | Yes | HCV | 1 | 2.8 | A | 1b | Yes |
| HCC04 | 63 | Male | Yes | NAFLD/HCV | 1 | 2.4 | C | 4b | No |
| HCC05 | 60 | Female | Yes | HCV | 1 | 5.4 | A | 1b | No |
| HCC06 | 70 | Male | Yes | HCV | 1 | 2.2 | A | 1b | Yes |
| HCC07 | 61 | Male | Yes | HCV | No record | N/A | C | 4b | No |
| HCC08 | 75 | Male | Yes | NASH | 2 | 5.5 | B | 3b | Yes |
| HCC09 | 53 | Male | Yes | EtOH | 1 | 3.4 | A | 1b | Yes |
| HCC10 | 73 | Male | No | NASH | 1 | 13.5 | A | 1b | No |
| HCC11 | 73 | Male | Yes | NASH | 4 | 13.5 | C | 4b | No |
| HCC12 | 63 | Female | Yes | HBV | 1 | 7.9 | B | 3a | No |
| HCC13 | 61 | Male | Yes | HBV | infiltrative | 5.5 | C | 3b | No |
| HCC14 | 71 | Male | Yes | HCV | 1 | 2.1 | A | 1b | Yes |
| HCC15 | 53 | Male | No | HBV | Multiple >10 | 38.3 | C | 3b | No |
| HCC16 | 63 | Male | Yes | HCV | 3 | 7.5 | D | 4b | No |
| HCC17 | 78 | Male | No | idiopathic | 1 | 3.7 | A | 1b | Yes |
| HCC18 | 73 | Male | Yes | HCV | 1 | 4.2 | A | 1b | Yes |
| HCC19 | 56 | Female | Yes | HCV | 2 | 6.8 | A | 2 | No |
| HCC20 | 59 | Male | Yes | HCV/EtOH | 1 | 6.2 | C | 2 | No |
ALD, alcoholic liver disease; AJCC, American Joint Committee on Cancer; BCLC, Barcelona clinic liver cancer; HBV, hepatitis B virus; HCV, Hepatitis C virus; NAFLD, nonalcoholic fatty liver disease.
Figure 4. Bioinformatic analysis and comparison of the 64-gene mRNA signatures of hepatocellular carcinoma (HCC) circulating tumor cells (CTCs) purified by Click Chips from HCC patients and HCC tissue mRNA signatures from The Cancer Genome Atlas (TCGA) database.
a) Q-Q Plot showed that the 64-gene expression profiles from the HCC CTCs and HCC TCGA came from populations with a common distribution. b) PCA plot based on the 64-gene expression from the HCC CTCs from 20 patients (red dots) and HCC TCGA (gray dots). c) Bar charts showed the goodness of fit (inverse KS distance) of the 64 genes between the HCC CTCs and HCC TCGA. d-i) Kernel density plot of the 6 representative genes with the highest expression concordance between HCC CTCs and HCC TCGA.
3. Conclusion
We demonstrated the feasibility of purifying HCC CTCs from patients’ blood samples with improved purity and molecular integrity using Click Chips in conjunction with a multimarker cocktail. Compared with other immunoaffinity-mediated CTC purification system, Click Chips exhibit several unique advantages: 1) Click Chips can be optimized to achieve desired HCC CTC purification performance with dramatically reduced consumption of capture agents. This improvement is attributed to its rapid, irreversible, and biorthogonal click reaction-mediated cell capture mechanism, as well as the significantly increased number of click reaction sites between TCO moieties grafted on CTCs and Tz moieties functionalized on the embedded SiNWS; 2) Click Chips are compatible with the multimarker cocktail, which improves the efficiency of HCC CTC enrichment significantly and overcomes the high heterogeneity of HCC CTCs. 3) The mild DTT-mediated disulfide cleavage enables the specific release of HCC CTCs effectively. In parallel with the development of Click Chips for purification of HCC CTCs, we adopted an integrated data analysis framework with high-dimensional transcriptomics data from public resource databases. While many HCC gene signatures[33–39] have been proposed, they do not necessarily identify the genes for CTC gene expression detection. In addition, very few studies in the literature have investigated the degree of similarity/dissimilarity between CTCs and tissues in the context of biomarker discovery. We thus start with 7 publicly available HCC transcriptomic studies to identify the signature genes specific to HCC. Such an HCC-specific gene panel allows for the detection of HCC CTCs in the presence of non-specifically trapped WBCs. We purified HCC CTCs from 20 HCC blood samples, and conducted the downstream mRNA profiling using NanoString nCounter platform which targeted the 64 genes. In a systematic way, the 64 gene expressions in HCC CTCs showed high concordance with those in HCC tissues from TCGA, suggesting that biomarker discovery framework in this study can provide a window for identifying common gene expression characteristics between tissue and blood.
4. Experimental Section/Methods
The fabrication and surface characterization of Tz-grafted SiNWS, and molding of PDMS chaotic mixer can be referred to our previously published procedures.[29]
Preparation of trans-cyclooctene-antibody conjugate:
Trans-cyclooctene (TCO)-antibody conjugate was prepared by incubating TCO-PEG4-NHS ester (0.5 mM, Click Chemistry Tools) with human EpCAM/TROP-1 antibody (Goat IgG, 0.1 μg mL−1, R&D Systems, Inc.), human ASGPR1 antibody (Rabbit IgG, 0.5 μg mL−1, LifeSpan BioSciences, Inc.,), and human GPC3 antibody (Sheep IgG, 0.1 μg mL−1, R&D Systems, Inc.) in PBS for 30 min at room temperature. The TCO-antibody conjugate was freshly prepared before use.
HCC cell lines:
Three HCC cell lines (i.e., SNU387, Hep3B, and PLC/PRF/5) were purchased from American Type Culture Collection (ATCC) and cultured in RPMI-1640 medium (SNU387) or Eagle’s Minimum Essential Medium (Hep 3B and PLC/PRF/5) with 10% fetal bovine serum (FBS), 1% GlutaMAX-I and 100 U mL−1 penicillin-streptomycin (Thermo Fisher Scientific) in a humidified incubator with 5% CO2.
Artificial HCC CTC sample studies:
Artificial CTC samples were prepared by spiking 200 HCC cells, pre-stained with Vybrant™ DiO green fluorescent dye (Invitrogen), into the freshly isolated PBMCs (5 × 106 cells mL−1), pre-stained with Vybrant™ DiD red fluorescent dye (Invitrogen), in 200 μL of RPMI medium. These artificial CTC samples were incubated with TCO-labeled antibodies in RPMI medium (200 μL) for 30 min at room temperature, followed by a centrifugation at 300g to remove the surplus TCO-labeled antibody and non-reactive TCO-PEG4-NHS ester. Then, the TCO-grafted CTC samples were purified using Click Chips. Finally, these captured cells were imaged and counted under a fluorescence microscope (Nikon 90i, W1, 325–375 nm; W2, 465–495 nm; W3, 590–650 nm) after staining with DAPI, cytokeratin (CK), and CD45. WBCs were defined as 4’,6-diamidino-2-phenylindole (DAPI)+/cytokeratin (CK)-/CD45+ round/ovoid cells. HCC cells were defined as DAPI+/CK+/CD45- round/ovoid cells.
CTC capture and release from Click Chips:
For CTC capture, the TCO-labeled artificial samples and HCC patient PBMC samples (200 μL) were processed in Click Chips at the flow rate of 1 mL h−1. The captured CTCs in Click Chips were then fixed with 2.0% formaldehyde (PFA in PBS, 200 μL) for CTC staining and enumeration. For CTC release, 200 μL of DTT (50 mM) solution was injected into Click Chips at a flow rate of 1 mL h−1, followed by injecting100 μL of PBS to collect the released CTCs. The released CTCs were subjected to either glass smear for fluorescence imaging or transferred into RNase-free tubes for the following molecular analysis.
HCC patients:
Twenty HCC (Table 1) patients from October 2018 to June 2019 at Ronald Reagan UCLA Medical Center were enrolled in this study. Patients who had severe mental disease, or other uncontrolled malignant tumors, uncontrolled infection of mycobacterium, tuberculosis were excluded. All participants were elder than 18 years old. Treatment-naïve HCC patients across all clinical stages were enrolled in this study. The written informed consents according to the IRB protocol (IRB #14–000197) at UCLA were provided by each patient and healthy donor for this study. None of the enrolled patients was enrolled in any clinical trial.
Collection and treatment of HCC patients’ blood samples:
Peripheral venous blood samples were collected with the written informed consent of these patients. Each 10 mL blood sample was collected in a BD Vacutainer glass tube (BD Medical, Fisher Cat. #02–684-26) with acid citrate dextrose. For each HCC patient sample, peripheral blood mononuclear cells (PBMCs) were isolated from one 2.0 mL vial of blood, and then incubated with TCO-labeled antibody in PBS for 30 min at room temperature before CTC purification by Click Chips.
RNA extraction and preamplification:
The released CTCs were lysed by TRI Reagent, and CTC-derived RNA was extracted using a Direct-zol™ RNA MicroPrep Kit according to the manufacturer’s protocol (ZYMO Research Corp.). The extracted RNA was collected in 12 μL of RNase-free water, and then subjected to cDNA Synthesis. The internal quality control requires that the RNA amount must be more than 2 ng/μL for each sample.Prior to runing on the NanoString nCounter platform (NanoString Technologies, Inc., WA, USA), the whole transcriptome amplification was performed using the nCounter Low RNA Input Amplification Kit (NanoString Technologies, Inc., WA, USA) according to the manufacturer’s protocol.
Data Analysis:
The 8 HCC gene expression signatures were obtained from the literature except iCluster1 and IDH1-like signatures. We selected the top 100 genes by comparing iCluster1 tumors with other tumors in TCGA HCC cohort using an integrated hypothesis testing method.[45] TCGA HCC transcriptome data were obtained from the NCI GDC data portal (https://portal.gdc.cancer.gov/). To identify the IDH1-like signature, the same approach was performed, and top 100 genes were selected using TCGA HCC cohort data. With the iCluster1 and IDH1-like signature, we found 622 genes in cholangiocarcinoma-like HCC[34], 496 genes reflecting Hippo pathway inactivation[36], 65 genes from risk score classifier based on Cox hazard model[37], 347 genes from NCI proliferation signature[38], 16 genes in hepatoblastoma-like tumors[39], and 115 genes from Hoshida’s HCC subtyping study[33]. NanoStringNorm R package (1.2.0) was used for expression data normalization. The housekeeping geometric mean option was used for sample contents normalization and quantile method with adopting log2 was applied to entire data to reduce systematic variance. We performed Wilcoxon Rank Sum Test for comparison of the expression ranks (percentiles) of the 64 genes in HCC and other cancer cell lines from the CCLE and immune cells from the DMAP. Linear regression analysis was performed to assess the linearity of the mRNA expression readout in HCC cell lines (SNU-387 and PLC/PRF/5) and healthy donor PBMCs. The quantile-quantile plot, PCA analysis, and the Kolmogorov-Smirnov (KS) distance were used to show expression concordance between the HCC CTCs and HCC tissues from TCGA. All the bioinformatical and statistical analysis in this study were performed using the R statistical software version 3.5 (http://www.r-project.org/) and MATLAB (MathWorks). All statistical tests are two-sided and P* < 0.05 is considered statistically significant.
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health (U01CA198900, U01EB026421, R01CA218356, R21CA235340, and R21CA240887).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Contributor Information
Na Sun, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles (UCLA), 570 Westwood Plaza, Los Angeles, CA 90095, USA; Key Laboratory for Nano-Bio Interface, Suzhou Institute of Nano-Tech and Nano-Bionics, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Suzhou 215123, P.R. China.
Yi-Te Lee, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles (UCLA), 570 Westwood Plaza, Los Angeles, CA 90095, USA.
Minhyung Kim, Cedars-Sinai Cancer, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA..
Jasmine J. Wang, Cedars-Sinai Cancer, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Ceng Zhang, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles (UCLA), 570 Westwood Plaza, Los Angeles, CA 90095, USA; Department of Pathology, Nanfang Hospital, Southern Medical University, Guangzhou, 510515, Guangdong Province, P.R. China.
Pai-Chi Teng, Cedars-Sinai Cancer, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA..
Dongping Qi, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles (UCLA), 570 Westwood Plaza, Los Angeles, CA 90095, USA.
Ryan Y. Zhang, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles (UCLA), 570 Westwood Plaza, Los Angeles, CA 90095, USA
Benjamin V. Tran, Department of Surgery, UCLA, 200 Medical Plaza, Los Angeles, CA, 90024, USA
Yue Tung Lee, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles (UCLA), 570 Westwood Plaza, Los Angeles, CA 90095, USA.
Jinglei Ye, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles (UCLA), 570 Westwood Plaza, Los Angeles, CA 90095, USA.
Juvelyn Palomique, Comprehensive Transplant Center, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA..
Nicholas N. Nissen, Comprehensive Transplant Center, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Steven-Huy B. Han, Department of Surgery, UCLA, 200 Medical Plaza, Los Angeles, CA, 90024, USA
Saeed Sadeghi, Department of Surgery, UCLA, 200 Medical Plaza, Los Angeles, CA, 90024, USA.
Richard S. Finn, Department of Surgery, UCLA, 200 Medical Plaza, Los Angeles, CA, 90024, USA
Sammy Saab, Department of Surgery, UCLA, 200 Medical Plaza, Los Angeles, CA, 90024, USA.
Ronald W. Busuttil, Department of Surgery, UCLA, 200 Medical Plaza, Los Angeles, CA, 90024, USA
Edwin M. Posadas, Cedars-Sinai Cancer, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA.
Li Liang, Department of Pathology, Nanfang Hospital, Southern Medical University, Guangzhou, 510515, Guangdong Province, P.R. China.
Renjun Pei, Key Laboratory for Nano-Bio Interface, Suzhou Institute of Nano-Tech and Nano-Bionics, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Suzhou 215123, P.R. China.
Ju Dong Yang, Comprehensive Transplant Center, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA..
Sungyong You, Cedars-Sinai Cancer, Cedars-Sinai Medical Center, Los Angeles, CA 90048, USA..
Vatche G. Agopian, Department of Surgery, UCLA, 200 Medical Plaza, Los Angeles, CA, 90024, USA
Hsian-Rong Tseng, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles (UCLA), 570 Westwood Plaza, Los Angeles, CA 90095, USA.
Yazhen Zhu, California NanoSystems Institute, Crump Institute for Molecular Imaging, Department of Molecular and Medical Pharmacology, University of California, Los Angeles (UCLA), 570 Westwood Plaza, Los Angeles, CA 90095, USA.
References
- [1].Manzoni C, Kia DA, Vandrovcova J, Hardy J, Wood NW, Lewis PA, Ferrari R, Brief. Bioinform. 2018, 19, 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Gao GF, Parker JS, Reynolds SM, Silva TC, Wang LB, Zhou W, Akbani R, Bailey M, Balu S, Berman BP, Brooks D, Chen H, Cherniack AD, Demchok JA, Ding L, Felau I, Gaheen S, Gerhard DS, Heiman DI, Hernandez KM, Hoadley KA, Jayasinghe R, Kemal A, Knijnenburg TA, Laird PW, Mensah MKA, Mungall AJ, Robertson AG, Shen H, Tarnuzzer R, Wang Z, Wyczalkowski M, Yang L, Zenklusen JC, Zhang Z, Genomic Data Analysis N, Liang H, Noble MS, Cell Syst. 2019, 9, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rockey DC, Caldwell SH, Goodman ZD, Nelson RC, Smith AD, American D Association for the Study of Liver, Hepatology 2009, 49, 1017. [DOI] [PubMed] [Google Scholar]
- [4].Krebs MG, Metcalf RL, Carter L, Brady G, Blackhall FH, Dive C, Nat. Rev. Clin. Oncol. 2014, 11, 129. [DOI] [PubMed] [Google Scholar]
- [5].Alix-Panabières C, Pantel K, Nat. Rev. Cancer 2014, 14, 623. [DOI] [PubMed] [Google Scholar]
- [6].Speicher MR, Pantel K, Nat. Biotechnol. 2014, 32, 441. [DOI] [PubMed] [Google Scholar]
- [7].Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, Hayes DF, Engl N. J. Med. 2004, 351, 781. [DOI] [PubMed] [Google Scholar]
- [8].de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, Doyle GV, Terstappen LW, Pienta KJ, Raghavan D, Clin. Cancer Res. 2008, 14, 6302. [DOI] [PubMed] [Google Scholar]
- [9].Heller G, McCormack R, Kheoh T, Molina A, Smith MR, Dreicer R, Saad F, de Wit R, Aftab DT, Hirmand M, Limon A, Fizazi K, Fleisher M, de Bono JS, Scher HI, J. Clin. Oncol. 2018, 36, 572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Hong X, Sullivan RJ, Kalinich M, Kwan TT, Giobbie-Hurder A, Pan S, LiCausi JA, Milner JD, Nieman LT, Wittner BS, Ho U, Chen T, Kapur R, Lawrence DP, Flaherty KT, Sequist LV, Ramaswamy S, Miyamoto DT, Lawrence M, Toner M, Isselbacher KJ, Maheswaran S, Haber DA, Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Shen Z, Wu A, Chen X, Chem. Soc. Rev. 2017, 46, 2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yoon HJ, Kozminsky M, Nagrath S, ACS nano 2014, 8, 1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Dong J, Chen JF, Smalley M, Zhao M, Ke Z, Zhu Y, Tseng HR, 2020, 32, e1903663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A, CA Cancer J. Clin. 2018, 68, 394. [DOI] [PubMed] [Google Scholar]
- [15].Xu W, Cao L, Chen L, Li J, Zhang XF, Qian HH, Kang XY, Zhang Y, Liao J, Shi LH, Yang YF, Wu MC, Yin ZF, Clin Cancer Res 2011, 17, 3783. [DOI] [PubMed] [Google Scholar]
- [16].Fan JL, Yang YF, Yuan CH, Chen H, Wang FB, Cell Physiol. Biochem. 2015, 37, 629. [DOI] [PubMed] [Google Scholar]
- [17].Ahn JC, Teng PC, Chen PJ, Posadas E, Tseng HR, Lu SC, Yang JD, Hepatology 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Jan YJ, Chen JF, Zhu Y, Lu YT, Chen SH, Chung H, Smalley M, Huang YW, Dong J, Chen LC, Yu HH, Tomlinson JS, Hou S, Agopian VG, Posadas EM, Tseng HR, Adv. Drug Deliv. Rev. 2018, 125, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Court CM, Hou S, Winograd P, Segel NH, Li QW, Zhu Y, Sadeghi S, Finn RS, Ganapathy E, Song M, French SW, Naini BV, Sho S, Kaldas FM, Busuttil RW, Tomlinson JS, Tseng HR, Agopian VG, Liver Transpl. 2018, 24, 946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lin M, Chen JF, Lu YT, Zhang Y, Song J, Hou S, Ke Z, Tseng HR, Acc. Chem. Res. 2014, 47, 2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hou S, Zhao L, Shen Q, Yu J, Ng C, Kong X, Wu D, Song M, Shi X, Xu X, OuYang WH, He R, Zhao XZ, Lee T, Brunicardi FC, Garcia MA, Ribas A, Lo RS, Tseng HR, Angewandte Chemie. 2013, 52, 3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Court CM, Hou S, Liu L, Winograd P, DiPardo BJ, Liu SX, Chen PJ, Zhu Y, Smalley M, Zhang R, Sadeghi S, Finn RS, Kaldas FM, Busuttil RW, Zhou XJ, Tseng HR, Tomlinson JS, Graeber TG, Agopian VG, NPJ Precis. Oncol. 2020, 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, Kimura A, Sengupta S, Stott SL, Karabacak NM, Barber TA, Walsh JR, Smith K, Spuhler PS, Sullivan JP, Lee RJ, Ting DT, Luo X, Shaw AT, Bardia A, Sequist LV, Louis DN, Maheswaran S, Kapur R, Haber DA, Toner M, Sci. Transl. Med. 2013, 5, 179ra47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kalinich M, Bhan I, Kwan TT, Miyamoto DT, Javaid S, LiCausi JA, Milner JD, Hong X, Goyal L, Sil S, Choz M, Ho U, Kapur R, Muzikansky A, Zhang H, Weitz DA, Sequist LV, Ryan DP, Chung RT, Zhu AX, Isselbacher KJ, Ting DT, Toner M, Maheswaran S, Haber DA, Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Wang S, Liu K, Liu J, Yu ZT, Xu X, Zhao L, Lee T, Lee EK, Reiss J, Lee YK, Chung LW, Huang J, Rettig M, Seligson D, Duraiswamy KN, Shen CK, Tseng HR, Angewandte Chemie. 2011, 50, 3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Zhao L, Lu YT, Li F, Wu K, Hou S, Yu J, Shen Q, Wu D, Song M, OuYang WH, Luo Z, Lee T, Fang X, Shao C, Xu X, Garcia MA, Chung LW, Rettig M, Tseng HR, Posadas EM, Adv. Mater. 2013, 25, 2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Ke Z, Lin M, Chen JF, Choi JS, Zhang Y, Fong A, Liang AJ, Chen SF, Li Q, Fang W, Zhang P, Garcia MA, Lee T, Song M, Lin HA, Zhao H, Luo SC, Hou S, Yu HH, Tseng HR, ACS nano 2015, 9, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Shen MY, Chen JF, Luo CH, Lee S, Li CH, Yang YL, Tsai YH, Ho BC, Bao LR, Lee TJ, Jan YJ, Zhu YZ, Cheng S, Feng FY, Chen P, Hou S, Agopian V, Hsiao YS, Tseng HR, Posadas EM, Yu HH, Adv. Healthc. Mater. 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dong J, Jan YJ, Cheng J, Zhang RY, Meng M, Smalley M, Chen PJ, Tang X, Tseng P, Bao L, Huang TY, Zhou D, Liu Y, Chai X, Zhang H, Zhou A, Agopian VG, Posadas EM, Shyue JJ, Jonas SJ, Weiss PS, Li M, Zheng G, Yu HH, Zhao M, Tseng HR, Zhu Y, Sci. Adv. 2019, 5, eaav9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang S, Wang H, Jiao J, Chen KJ, Owens GE, Kamei K, Sun J, Sherman DJ, Behrenbruch CP, Wu H, Tseng HR, Angew. Chem. Int. Ed. 2009, 48, 8970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Stroock AD, Dertinger SKW, Ajdari A, Mezić I, Stone HA, Whitesides GM, Science 2002, 295, 647. [DOI] [PubMed] [Google Scholar]
- [32].Sun N, Lee YT, Zhang RY, Kao R, Teng PC, Yang Y, Yang P, Wang JJ, Smalley M, Chen PJ, Kim M, Chou SJ, Bao L, Wang J, Zhang X, Qi D, Palomique J, Nissen N, Han SB, Sadeghi S, Finn RS, Saab S, Busuttil RW, Markovic D, Elashoff D, Yu HH, Li H, Heaney AP, Posadas E, You S, Yang JD, Pei R, Agopian VG, Tseng HR, Zhu Y, Nat. Commun. 2020, 11, 4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, Watanabe G, Gabriel S, Friedman SL, Kumada H, Llovet JM, Golub TR, Cancer Res. 2009, 69, 7385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Woo HG, Lee JH, Yoon JH, Kim CY, Lee HS, Jang JJ, Yi NJ, Suh KS, Lee KU, Park ES, Thorgeirsson SS, Kim YJ, Cancer. Res. 2010, 70, 3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].w. b. e. Cancer Genome Atlas Research Network. Electronic address, N. Cancer Genome Atlas Research, Cell 2017, 169, 1327.28622513 [Google Scholar]
- [36].Sohn BH, Shim JJ, Kim SB, Jang KY, Kim SM, Kim JH, Hwang JE, Jang HJ, Lee HS, Kim SC, Jeong W, Kim SS, Park ES, Heo J, Kim YJ, Kim DG, Leem SH, Kaseb A, Hassan MM, Cha M, Chu IS, Johnson RL, Park YY, Lee JS, Clin. Cancer Res. 2016, 22, 1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kim SM, Leem SH, Chu IS, Park YY, Kim SC, Kim SB, Park ES, Lim JY, Heo J, Kim YJ, Kim DG, Kaseb A, Park YN, Wang XW, Thorgeirsson SS, Lee JS, Hepatology 2012, 55, 1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lee JS, Chu IS, Heo J, Calvisi DF, Sun Z, Roskams T, Durnez A, Demetris AJ, Thorgeirsson SS, Hepatology 2004, 40, 667. [DOI] [PubMed] [Google Scholar]
- [39].Cairo S, Armengol C, De Reynies A, Wei Y, Thomas E, Renard CA, Goga A, Balakrishnan A, Semeraro M, Gresh L, Pontoglio M, Strick-Marchand H, Levillayer F, Nouet Y, Rickman D, Gauthier F, Branchereau S, Brugieres L, Laithier V, Bouvier R, Boman F, Basso G, Michiels JF, Hofman P, Arbez-Gindre F, Jouan H, Rousselet-Chapeau MC, Berrebi D, Marcellin L, Plenat F, Zachar D, Joubert M, Selves J, Pasquier D, Bioulac-Sage P, Grotzer M, Childs M, Fabre M, Buendia MA, Cancer Cell 2008, 14, 471. [DOI] [PubMed] [Google Scholar]
- [40].Jan YJ, Yoon J, Chen JF, Teng PC, Yao N, Cheng S, Lozano A, Chu GCY, Chung H, Lu YT, Chen PJ, Wang JJ, Lee YT, Kim M, Zhu Y, Knudsen BS, Feng FY, Garraway IP, Gao AC, Chung LWK, Freeman MR, You S, Tseng HR, Posadas EM, Theranostics 2019, 9, 2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehar J, Kryukov GV, Sonkin D, Reddy A, Liu M, Murray L, Berger MF, Monahan JE, Morais P, Meltzer J, Korejwa A, Jane-Valbuena J, Mapa FA, Thibault J, Bric-Furlong E, Raman P, Shipway A, Engels IH, Cheng J, Yu GK, Yu J, Aspesi P Jr., de Silva M, Jagtap K, Jones MD, Wang L, Hatton C, Palescandolo E, Gupta S, Mahan S, Sougnez C, Onofrio RC, Liefeld T, MacConaill L, Winckler W, Reich M, Li N, Mesirov JP, Gabriel SB, Getz G, Ardlie K, Chan V, Myer VE, Weber BL, Porter J, Warmuth M, Finan P, Harris JL, Meyerson M, Golub TR, Morrissey MP, Sellers WR, Schlegel R, Garraway LA, Nature 2012, 483, 603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, Frampton GM, Drake AC, Leskov I, Nilsson B, Preffer F, Dombkowski D, Evans JW, Liefeld T, Smutko JS, Chen J, Friedman N, Young RA, Golub TR, Regev A, Ebert BL, Cell 2011, 144, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Choi WT, Ramachandran R, Kakar S, Hum Pathol.. 2017, 63, 1. [DOI] [PubMed] [Google Scholar]
- [44].Choi WT, Kakar S, Gastroenterol Clin. North Am. 2017, 46, 311. [DOI] [PubMed] [Google Scholar]
- [45].Hwang D, Rust AG, Ramsey S, Smith JJ, Leslie DM, Weston AD, de Atauri P, Aitchison JD, Hood L, Siegel AF, Bolouri H, Proc. Natl. Acad. Sci. U. S. A. 2005, 102, 17296. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.