Abstract
3-dimensional (3D) laboratory tissue cultures have emerged as an alternative to traditional 2-dimensional (2D) culture systems that do not recapitulate native cell behavior. The discrepancy between in vivo and in vitro tissue-cell-molecular responses impedes understanding of human physiology in general and creates roadblocks for the discovery of therapeutic solutions. Two parallel approaches have emerged for the design of 3D culture systems. The first is biomedical engineering methodology, including bioengineered materials, bioprinting, microfluidics and bioreactors, used alone or in combination, to mimic the microenvironments of native tissues. The second approach is organoid technology, in which stem cells are exposed to chemical and/or biological cues to activate differentiation programs that are reminiscent of human (prenatal) development. This review article describes recent technological advances in engineering 3D cultures that more closely resemble the human brain. The contributions of in vitro 3D tissue culture systems to new insights in neurophysiology, neurological diseases and regenerative medicine are highlighted. Perspectives on designing improved tissue models of the human brain are offered, focusing on an integrative approach merging biomedical engineering tools with organoid biology.
Keywords: bioengineering, organoids, stem cells, neurodevelopment, neurodegenerative and psychiatric diseases
Graphical Abstract
This review outlines current bioengineering and organoid technology approaches to developing 3-dimensional (3D) in vitro models of the human brain. Advances in generating representative microenvironments to observe and manipulate cellular behavior are described, with emphasis on opportunities to combine these approaches to generate advanced tissue models that are suitable for modeling human brain development, disease, and injury.
1. Introduction
To understand human brain physiology, the human condition must be studied in a laboratory setting. Animal models, such as rodents, primates, worms and experimental systems such as yeast, have yielded great insights into development, function and diseases of the brain. However, these model systems do not fully recapitulate the complexity of the human brain related to neural circuit architecture, genetics, gene expression, cell-type diversification, lipidomics and proteomics. The cerebral cortex, the part of the brain involved in motor and sensory function and processing information, as well as associated with psychiatric diseases, is less complex in rodents in comparison to primates in general, and to humans in particular.[1–5] Evolution also had its impact on the divergence of genetic make-up of humans compared to other mammals, notably the expression of non-coding RNAs.[6–7] Species-specific gene expression patterns are found in the context of neural circuit function and specific cell-types, and they may be related to the organization of cis-regulatory elements in DNA or stress conditions such as aging.[8–15] Ultimately, they may lead to changes in interactome at the mesoscale, such as lipid profiles or the composition of the synaptic proteome.[16–17] In light of this broad spectrum of species-specific differences on the level of DNA, molecular pathways, cells and brain networks, it should not come as a surprise that the modeling in rodents of complex neurodegenerative diseases and psychiatric disorders pose challenges, in particular for drug and target discovery.[3, 5, 18] Unlike rodent models, access to living human test subjects and primates, or ex vivo brain slices derived thereof, is surrounded with ethical issues. Moreover, only a limited number of scientists and clinicians master the necessary expertise in, or have access to, brain imaging techniques such as non-invasive functional magnetic resonance imaging (fMRI) and electroencephalography (EEG), or minimally invasive methods such as the implantation of recording and stimulating electrodes in the brain. These methods also suffer from low spatial and/or temporal resolution, as well as the need for large patient cohorts and consortia to identify the features of the human brain connectome related to normal aging or disease.[19–22]
The advent of stem cell engineering, both embryonic (ESC) and induced pluripotent stem cells (iPSC) has been a game changer in the study of the human brain. These advancements in cell biology made possible, for the first time, the generation of a renewable source (vis-à-vis terminal explant studies) and propagation of in vitro human culture models of the human brain that are scalable and available to the scientific community at large.[23] iPSCs, which are generated from healthy individuals or patient fibroblasts or their peripheral blood monocytes, have demonstrated to be excellent tools to study human neurological diseases. The literature is now replete with examples from Alzheimer’s and Parkinson’s disease, schizophrenia, autism, epilepsy, pain and other fields.[23] Genome engineering approaches, in particular based on clustered regularly interspaced short palindromic repeats (CRISPR) technology (other examples are Zinc-fingers and transcription activator-like effector nucleases (TALENs)), have made a similar transformative impact. Genetic variants associated with increased (or decreased) risk of disease can now be introduced, with relative ease, in the germ-line of healthy cells to replicate human disease. They can also be removed from the cells as a strategy to correct brain illnesses.[24–28] These cells may be used in traditional in vitro mono-cultures grown on flat, 2-dimensional (2D) surfaces, but these experimental cell systems do not replicate the complexity and biology of the human brain. Salient examples of this fact can be found in studies of gene expression signatures, extracellular matrix microenvironment, neural network firing and biomarkers of brain development and disease.[29–34] This realization has spurred the development of 3-dimensional (3D) laboratory cell and tissue models that aim to better replicate the cellular composition, microenvironment and architecture of the brain.
We will discuss two different techniques that have become popular for designing 3D brain-like tissue structures. The first is the use of organoids (grown in bioreactors or otherwise), dense multicellular tissues differentiated from stem cells following their innate developmental cues.[35–37] The other is bioengineering, a term collectively used to describe methods that make use of a bottom up strategy, involving seeding (multiple) cells (types) in biomaterial scaffolds and/or hydrogel environments, including those generated by 3-dimensional (3D) printing, and the use of bioreactors such as microfluidic devices or perfusion devices.[38–39] These two complementary approaches have matured to the stage that we now see bioengineering techniques being used in a complementary fashion to improve on organoid cultures.[40] We will conclude with future perspectives on generating more complex in vitro tissue models of the human brain that integrate organoid and bioengineering technology, with a vision towards high-throughput screening systems for biomarker and therapeutic discovery.
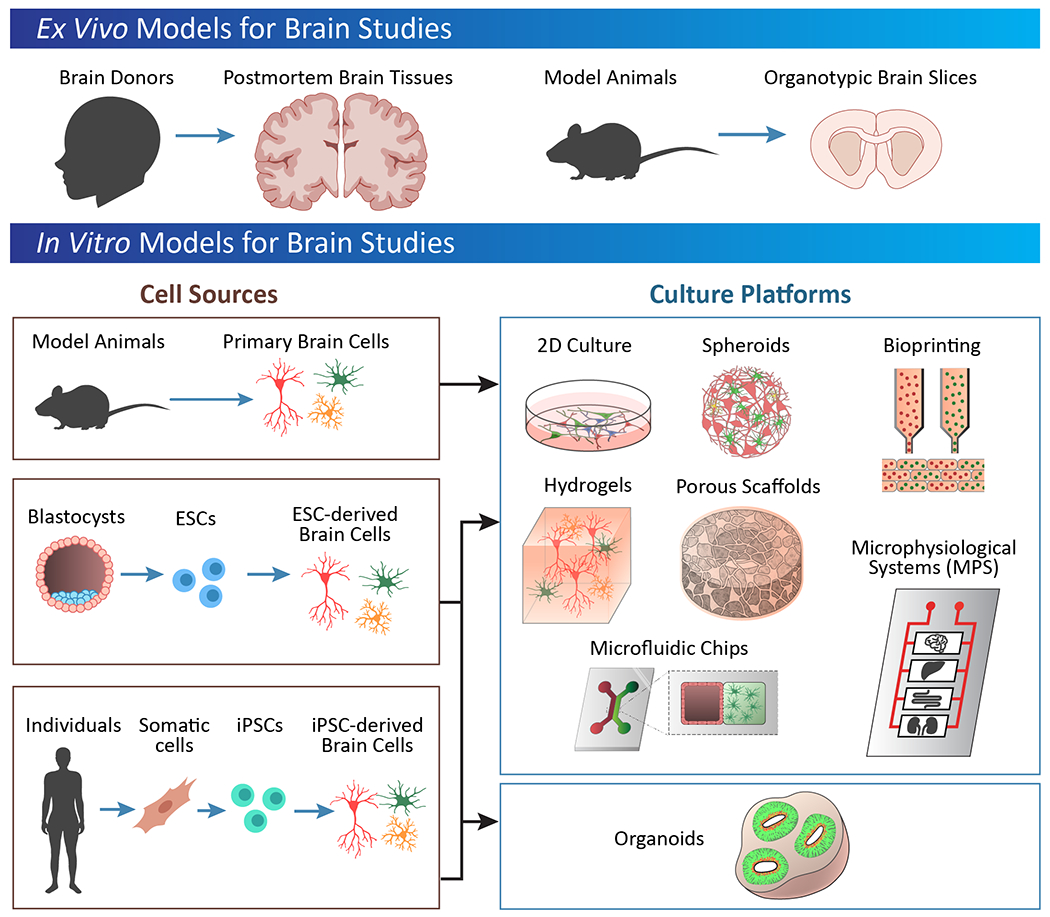
2. Ex Vivo and 2D In Vitro Platforms to Study the Brain
Much of our fundamental understanding of the brain is derived from traditional in vitro culture systems. Through extensive research studies of postmortem brain tissues as well as isolated cell cultures in 2D, many key features of brain physiology and mechanisms of disease have been identified. This knowledge serves as the basis for generating 3D models of the brain, and thus we begin by reviewing the literature on traditional in vitro systems to provide context to the sections describing 3D brain models.
2.1. Human postmortem brain tissues
Historically, preliminary attempts to understand the structure and function of the brain, the most complex organ in the body, have relied on the use of postmortem tissues. In a classic example from the 1860’s, the French physician Pierre Paul Broca found lesions in the same area of the postmortem brains of twelve patients with impaired language skills, providing the first evidence linking particular brain regions to specific functions.[41] Since then, many other key discoveries have been made by examining and identifying the hallmark anatomical changes in postmortem brains associated with patients who suffer from neurological dysfunctions such as Alzheimer’s disease (AD; loss of neurons in cortex and subcortical regions) and Parkinson’s disease (PD; loss of dopamine neurons in the substantia nigra of the midbrain). Due to advances in technological capabilities, postmortem brain studies continue to be an indispensable tool to study human brain function and disorders through the ability to observe and analyze tissue structures, cellular morphologies, and protein and gene expression.[42] As an example, the examination of AD patient brain tissues has been used to identify what are now described as the two classical molecular hallmarks of the disease - extracellular accumulation of beta-amyloid (Aβ) plaques and intracellular aggregation of hyperphosphorylated tau proteins in neurofibrillary tangles.[43] More recently, transcriptomics analysis and single cell-RNA sequencing from AD brain tissues have identified region-specific and cell-type-specific expression patterns.[43] By taking advantage of new technologies such as single-cell RNA sequencing, a reduction in excitatory neurons in the AD-gene carrier but not in unaffected family members or sporadic cases has been found, indicating differences in disease mechanisms between familiar and sporadic forms of the disease.[44] Similarly, combinations of postmortem studies detailed the molecular changes in the PD midbrain, including abnormal levels of α-synuclein production and aggregation, reduction in dopamine synthesis, increased oxidative stress, mitochondrial dysfunction, and reduction in neurotrophic factors essential for cell survival.[45] A recent study closely examined a small number of postmortem brain tissues of patients who received deep brain stimulation therapy, an approved surgical intervention alternative to medication. The results suggested recovery of mitochondria numbers and volume as underlying mechanisms of neuroprotection.[46]
In a recent and high-profile example of identifying neurodegenerative disease through examination of postmortem tissues, the study of brains subjected to repeated injuries, particularly from American football players, has led to the identification of chronic traumatic encephalopathy (CTE). Using postmortem brain tissues, researchers were able to define the distinguishable molecular feature of perivascular tau protein pathology. A postmortem study on four teenage athletes’ brains during the acute and subacute (1 day – 4 months) phase post brain trauma showed microvascular damage, neuroinflammation, axonal injuries, and tau pathology – indication of early stage of CTE.[47]
Postmortem brains have similarly been used to study neurodevelopmental and neuropsychiatric disorders, such as autism spectrum disorder (ASD), schizophrenia, and bipolar disorder. These disorders are particularly challenging as the majority of cases have no known genetic cause. To begin to address this issue, hundreds of postmortem brains from ASD patients and control cohorts were recently investigated, with allelic imbalance (expression favors one allele) reported in two specific regions associated with small nucleolar RNA.[48] More recent studies examining large numbers of tissue samples and data sets have identified the association of neuroinflammation with mental illnesses, such as schizophrenia and bipolar disorder.[49–51]
As evidenced above, the precise genetics, cell types involved, and pathogenesis are not well understood for most brain disorders. Phenotype heterogeneity in presentations and confounding factors such as sex, age, medications, and environmental factors add to the difficulties in investigating brain disorders.[52]. Also limiting is the availability of postmortem brain samples. Recently, successful efforts to increase awareness for brain donations has made possible the building of networks of brain tissue repositories and databases. The NIH NeuroBioBank (NBB) networks six brain tissue repositories, with the largest one storing over two-thousand brain samples, in order to facilitate the collection and redistribution of high quality brain tissues in the United States.[53]
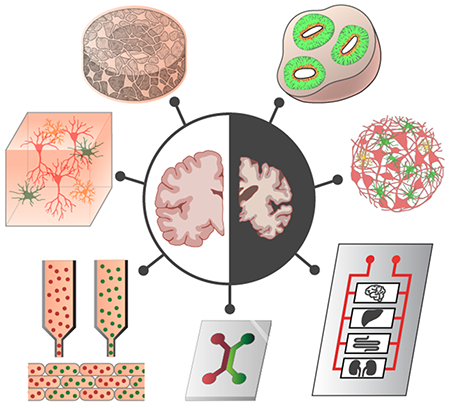
Despite the high level of relevance to human brain disorders, postmortem brain studies are limited in their ability to provide information on dynamic, temporal changes of disease progression. Furthermore, from the perspective of disease prevention or treatment, molecular and cellular changes may occur years prior to clinical presentation of symptoms of the decline in cognitive or motor skills[54] and researchers cannot control the genetic background or the external insults to the study objects. Thus, the need for well-controlled laboratory models are essential for probing the human brain (Figure 1).
Figure 1.
Overview of commonly used model systems for studying the brain.
2.2. Organotypic slice culture from animals
In contrast to human post mortem tissues, in vitro organotypic slice cultures from animals allow for the control of genetics, sex, age, external insults, and reduction of confounding factors. The major advantage of in vitro organotypic slice cultures is that they provide investigators access to the cells while preserving native cell types, ECM, cell organization, tissue architecture, and local synaptic connections. In addition, multiple tissue samples are generated from each brain, reducing the number of animals needed. In typical organotypic slice studies, brain slices are prepared with a thickness of 100-400 µm from acutely sacrificed animals.[55] These slices can be used in acute experiments, in which they are analyzed immediately after dissection to provide information from a near-in vivo state, or cultured in media for longer term studies (days to months) for the purpose of studying cell behavior over time after in vitro stimuli. A recent study compared the level of membrane associated tau protein vs. tau protein released into the media in brain slice cultures from wild type and AD model mice over the duration of 1 month.[56] A higher level of tau protein was released from the AD slices and, unlike in wild type slices, the release did not respond to neuronal stimulation. These results suggested differences in the mechanisms in tau protein release and spreading in AD.[56]
Organotypic slices are particularly useful when the tissue structure and organization are critical to the study outcome, such as in traumatic brain injury (TBI), cancer biology or neural circuit structure and functional activity. The tearing and shearing of the soft, connected brain tissue as a consequence of TBI, is not sufficiently recapitulated by single cells cultured on conventional 2D hard plastic surface, but can be mimicked by mechanically stretching the brain slices. Complementary to in vivo mouse models, slice culture allows for the tracking of cell death, tissue injury, inflammation and changes in neural network activity over time.[57–58] Brain slice models of TBI also provide a useful tool for screening therapeutic strategies, and several candidates have already been identified, such as hypothermia,[59] inert gases,[60] sodium calcium exchanger blockers,[61–62] and glutamate receptor blockers.[63] Although, hypothermia has been used in the treatment for traumatic brain injury, a recent review did not identify improvement in clinical outcomes.[64]
Because tumor metastasis to the brain and brain tumor invasiveness directly correlates to poor prognosis, many researchers study tumor cell migration in vitro. The majority of these studies, however, utilize 2D scratch or transwell assays that capture few of the actual in vivo interactions between tumor cells, host cells and ECM, or the cellular heterogeneity of the cancer. Moreover, the majority of organotypic slice studies use rodent brains, and therefore they face the same drawbacks of lacking proper human-equivalent models as other brain disorders. There are slice culture models that attempt to address these critical issues by employing the implantation of human tumor cell lines in the form of single cells or spheroids on rodent brain slices. This made it possible to examine in detail the interactions between glioblastoma cells and the vasculature, identifying surface protein targets and small molecule candidates to reduce tumor invasion or migration. Further, this approach provided insight into changes in gene expression in reactive astrocytes, turning the microenvironment to become pro-metastasis.[65–67]
In any slice culture, one of the key limitations is the age-dependency of slice survival and the resulting impact on study design. Slices have high viability when prepared from embryonic animal brains, but at the same time contain immature cell and phenotype. Slices from postnatal animals provide a higher level of cellular and tissue maturation than from embryonic animals and are more resistant to the mechanical trauma from slice preparation (up to months).[68] Adult brain slices, including from animals and from human biopsy and postmortem tissues, historically have a poor survival rate, but improvements have been made by adding neurotrophic factors, the addition of cerebrospinal fluid in the medium, or media flow systems to enhance nutrient and oxygen exchange.[69–72] However, it should be noted that slice preparation induces significant mechanical trauma to the tissue, which can cause the tissue to have injury-like responses that may affect the outcome of the experiment and introduce artifacts to interpretations into brain functions.
2.3. Two-dimensional cell culture systems
Culturing dissociated cells in vitro offers the advantages of optical transparency, substrate consistency, easy handling, robustness, reproducibility, relatively low cost, compatibility with electrophysiology techniques and high-throughput screening. In conventional 2D culture, dissociated cells are cultured on plastic or glass surfaces coated with laminin, fibronectin, Matrigel, poly-lysine, and/or poly-L-ornithine for adhesion.[73] In comparison to the abovementioned models, cell culture allows for controlled co-cultures with different cell types and/or cells with different genetic backgrounds (Table 1).
Table 1.
Summary of strengths and limitations of ex vivo and 2D in vitro platforms for brain studies.
| Strengths | Limitations | |
|---|---|---|
| Human Postmortem Tissues[41–54] | Complexity of brain preserved, including cell composition and spatial organization | Tissue not living, no evaluation of function or development over time |
| Identification of disease markers | Limited reproducibility or scalability for high-throughput screening | |
| Imaging of brain structure | ||
| Rodent Slice Cultures[55–72] | Complexity of brain preserved, including cell composition and spatial organization | Limited capability for evaluating function or development over time |
| Analysis of neural network anatomy and function | Limited reproducibility or scalability | |
| Not suitable for high-throughput screening | ||
| 2D Cell Culture[73–86] | Reproducible and scalable: high-throughput screening | Limited culture longevity, particularly for primary cells as opposed to stem cells |
| Suitable for non-invasive imaging and electrophysiological recordings | Low complexity, not representative of native cell environment | |
| Modeling of human biology through stem cell technology |
To date, investigating the human brain in vitro has been hindered by the inaccessibility to live human brain tissue and the extreme difficulty of isolating and culturing primary human neurons. Historically, rodent embryonic or postnatal neuronal culture has been used for studying neuronal functions and to model diseases. For these studies, the neurons are often genetically modified or cultured in the presence of toxins or disease-associated protein, such as Aβ for Alzheimer’s disease. Given that the underlying etiologies are not well understood for many human neurodevelopmental, neuropsychiatric, and neurodegenerative disorders, there is a lack of appropriate rodent models and the subsequent results from rodent cell culture often fail to translate to the human condition. A major breakthrough towards this goal was the method of reprogramming somatic cells to iPSCs, developed by Takahashi and Yamanaka in 2006, achieved by overexpressing transcription factors Oct3/4, Sox2, Klf4, and c-Myc in fibroblasts. Since then, many protocols have been developed for the direct differentiation of iPSCs or the transdifferentiation into relevant brain cell types, including cortical neurons, striatal dopamine neurons, astrocytes, microglia, oligodendrocytes, pericytes, and brain microvasculature endothelial cells.[74–75] The use of iPSCs from individuals affected by neurological disorders presents an unprecedented tool to unveil the underlying genetics, mechanisms, and pathogenesis associated with these disorders. Within the past decade, iPSC-derived neurons have provided novel insights on the etiology and demonstrated opportunities to screen for therapeutic interventions.
Due in part to the fact that AD is the most prevalent neurodegenerative disorder, many iPSC lines have been derived from AD patients, and their healthy family members, for the study of monogenetic and sporadic forms of the disease, as well as novel candidate genetic risk factors identified in genetic analysis of patients.[75–78] Studies have shown that human iPSC (hiPSC)-derived cortical neurons carrying familial Alzheimer’s disease (fAD) mutations presented elevated levels of Aβ42 – the main component of Aβ plaque, as well as pathological tau protein phenotype and neuroinflammation as seen in patients, demonstrating the utility of iPSCs as a preclinical model of the disease.[79–81] Treatment with pharmacological compounds targeting pathways related to Aβ production and cleavage reduced AD pathology, supporting the feasibility of iPSC models as in vitro platforms for identifying therapeutic interventions.
These approaches have been used to study aspects of other neurodegenerative disorders such as Parkinson’s or Huntington diseases. One of the hallmarks of Parkinson’s disease is death of midbrain dopamine neurons and the formation of α-synuclein protein aggregates. Several different types of PD models have been established, either by treating healthy iPSC-derived dopamine neurons with α-synuclein aggregates or by generating DA neurons from familial PD (fPD) and sporadic PD (sPD) patients. iPSC lines derived from rare fPD cases have helped in the validation of novel risk genes.[82] One study compared neurons derived from sPD and fPD iPSCs, interestingly finding epigenetic dysregulation in both groups with DNA methylation patterns differing from healthy dopamine neurons and hypermethylation in regions relevant to PD.[83] For the study of Huntington disease, a hereditary, monogenic, neurodegenerative movement disorder caused by an expansion of CAG repeats in the huntingtin (HTT) gene, iPSC models have been used to identify the genetic and cellular mechanisms of disease progression. A recent study of patient iPSC-derived cortical neurons found modifications in transcriptomics, morphology, and maturation, suggesting altered corticogenesis.[84] Another study detected astrocyte dysfunction in a HD-iPSC co-culture model, suggesting contribution of glial-neuron interactions in neuronal death and the importance of studying multiple cell types in a disease model.
Since the timeline for iPSC differentiation and maturation into neurons or other cell-types follows the same trajectory as in the developing embryo, these cells are particularly useful tools to study neurodevelopmental disorders such as autism spectrum disorders (ASD). Several iPSC lines have been established from monogenic ASD-iPSC (e.g., Fragile X syndrome, Rett syndrome, Timothy syndrome, Angelman syndrome, and Phelan-McDermid syndrome) as well as from idiopathic ASD patients. Investigators use these ASD-iPSC-neurons to study cell proliferation, excitatory and inhibitory balance, synapse connectivity, neural networks, epigenetics, and proteomics.[85] Many of them showed upregulation of inhibitory neuron genes, deficits in excitatory synaptic transmission, reduction in neurite length and synaptogenesis, indicating the relevance of abnormal network formation in ASD.[85]
The development of cells and 2D cell culture models has led to tremendous discoveries in the field. Despite this insight, the main drawback of 2D cell culture remains that they are oversimplified and lack the complexity and organization of the in vivo microenvironment (Figure 2). As an example, the architecture of the original neural networks is lost when neurons are dissociated into single cells. Furthermore, cell density in 2D culture is typically much lower than that found in vivo, with cell-cell interactions limited to the XY directions. The local cellular environment is also altered in 2D as typical cell culture substrates are several orders of magnitude more rigid than brain tissues, and the dynamics of nutrient, oxygen, and secreted factor transport are much different in open media than in 3D tissues. As an example, an AD model with iPSC-neurons cultured in 2D were unable to generate Aβ plaques as efficiently as in 3D because the secreted Aβ freely diffused away.[86] Because many environmental factors can significantly impact cell behavior and thus disease mechanisms, the development of 3D tissue model systems are actively pursued.
Figure 2.
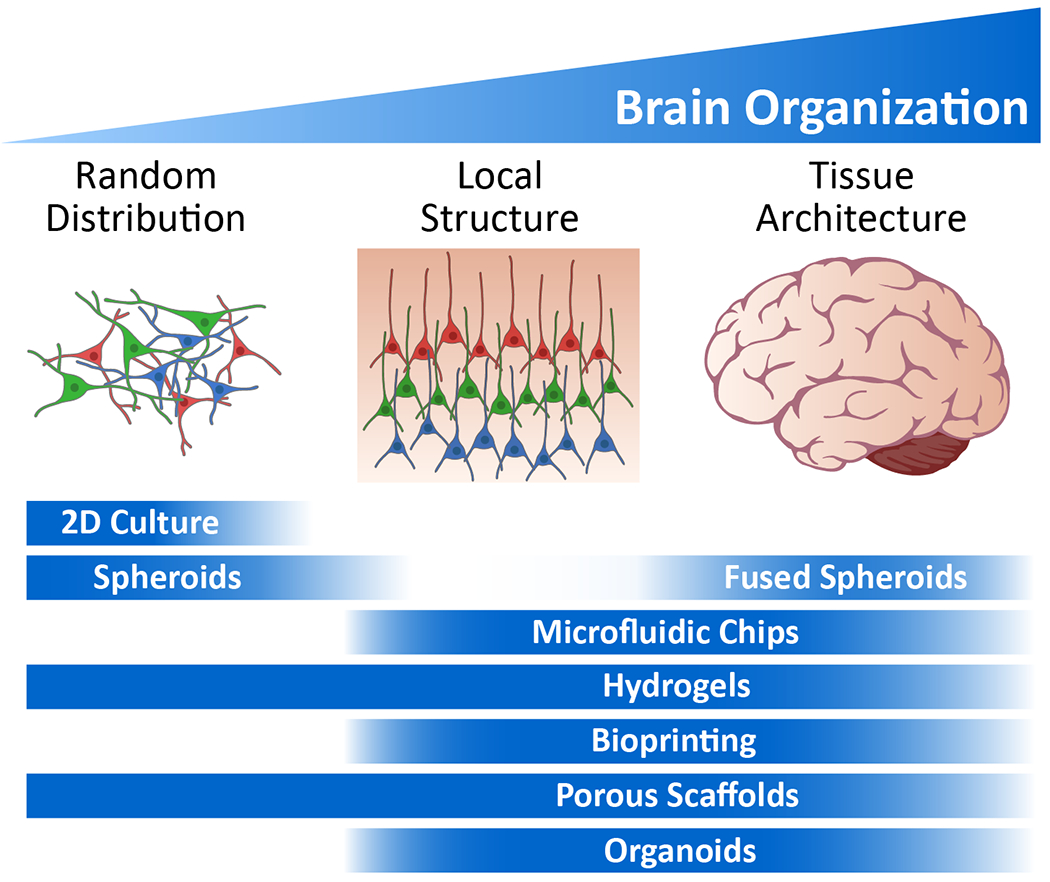
Overview of the applicability of in vitro platforms to emulate brain physiology at different levels of brain organization. Bars represent the optimal application of each technology.
3. 3D Brain Organoid Models
3.1. Organoids as an emerging technology for analyzing the human brain
All methods for generating organoids rely on the innate properties of stem cells to develop and self-organize into 3-dimensional architectures that share similarities with native tissues. Stem cells are uniquely endowed with the capacity to self-renew, differentiate and mature into virtually any type of cell.[23] As such, ESCs and iPSCs are ideal substrates for bioengineering of organoids. The first organoid systems generated modelled peripheral (i.e., not the central nervous system) tissues using mouse, and then human, cells. Salient examples derive from the retina[87–88] and the anterior pituitary,[89] two tissue types composed of neural and non-neural cell-types, and the intestine.[90–92]
The first protocols for engineering brain organoids made use of embryoid bodies generated from hiPCSs and grown on mouse embryonic fibroblasts. These cells were then embedded in Matrigel droplets and cultured for prolonged time (up to 10 months) in a spinning bioreactor. The resulting cerebral organoid resembled the characteristics of the human cortex, including the multilayered structure and inner and outer sub-ventricular zones, which house progenitor cells and (outer) radial glia. This type of glia plays an important role in human evolution, driving the expansion of the human brain compared to other primates. Generated from hiPSCs, the emerging 3D systems were heterogenous, also showing characteristics of meninges, choroid plexus and retina structures.[35, 93] A related approach relied on low-adhesion plates instead of bioreactors, and embryonic stem cells rather than iPSCs.[37]
Subsequent large-scale RNA sequence analysis of thousands of single cells have expanded on the notion that organoids give rise to a broad range of cell-types, such as neuroepithelial cells, photoreceptors and dopamine neurons.[94] Gene expression, epigenetic and image analysis studies further indicate that the maturing organoids follow the normal pace of development of the human fetal brain,[95–98] adhering to species specific rules of evolution.[99–101] As such, organoids have become exciting tools to examine the evolutionary cues that shape development of the brain, in particular for studies inspired by genetic analysis comparing humans to Neanderthals, chimpanzee or other animals.[102–105]
3.2. Optimizing differentiation protocols for organoid cultures
Organoid technology is often associated with limitations related to reproducibility and heterogeneity between organoids from the same or different batches as well as necrosis. As a consequence, the implementation of organoid cultures to study brain development and function is challenging. To overcome these hurdles, several groups have developed the next generation of protocols for culturing organoids that are more homogenous. Commonly referred to as ‘directed differentiation’, these methods make use of cocktails of chemicals and trophic factors, either alone[106] or in combination with low-adhesion plates, and custom-built miniaturized bioreactors or 3D scaffold environments.[107] With these techniques, it is now possible to guide the development of iPSCs towards specific sublineages, such as forebrain, midbrain or hypothalamus.[98, 108–117] The success of these efforts in achieving more reliable organoid differentiation was demonstrated recently in the testing of multiple iPSC lines, independent differentiation, and analyzing hundreds of thousands of single cells and in 96-well plate format assays.[115, 118–119]
3.3. Organoids as a tool to study neurodevelopment, disease and evolution
The power of organoid technology for characterization of neurological disease signatures was first demonstrated in a patient population stratified based on clinical record but with no knowledge of the genetic underpinnings. Focusing on idiopathic forms of autism spectrum disorder (ASD), differences in cell proliferation, neural differentiation and synapse biology were observed in organoids of iPSC selected from patients with macrocephaly phenotypes compared to healthy controls. With ‘omics’ analyses, changes in a forkhead box G1 (FOXG1) transcription factor dependent signaling was found associated with an imbalance between gamma-aminobutyric acid (GABA)-ergic and glutamatergic neurons,[108] often cited as one of the underlying culprits of ASD.[120] Interestingly, aberrant FOXG1 signaling is also implicated in Rett syndrome, a rare neurodevelopmental disorder mainly affecting girls.[121–123] Organoid studies link microRNA pathways to altered extracellular signal-regulated kinase (ERK) and serine/threonine-protein kinase (AKT) signaling pathways involved in neurogenesis in patients with Rett syndrome.[124]
A unique approach to the study of human (brain) development is the comparison of our genome to closely related species, such as Neanderthals and the chimpanzee. Such comparative analysis led to an intriguing organoid study of NOTCH signaling and regulators of the PI3K-AKT-mTOR pathway in radial glia, implicating the rapid evolution of the larger human cortex to loss of genome stability and ensuing neurodevelopmental disorders, such as 1q21.1 deletion and duplication syndrome.[103, 125] Related studies point to the importance of specific sets of chromatin enzymes, gene networks regulated by long non-coding RNAs and enhancers involved with brain development,[126] with a potential link to autism.[105, 126–128]
Organoids have been used to successfully study a wide range of neurological disease such as hereditary spastic paraplegia, periventricular heterotopia, a disease associated with intellectual disability and dysfunction of cadherin adhesion molecules[129] and autoimmune diseases caused by neuro-inflammation.[130] Organoid studies of neurodevelopmental diseases successfully highlighted defects in a broad range of cell-molecular functions, such as oxidative stress,[131] vesicular traffic,[132] calcium channels,[114] the expansion of DNA nucleotide repeats in the context of Huntingtin disease,[133] chromatin remodeling enzymes,[134–135] mRNA chemical modifications,[136] and lipid homeostasis.[137] A particular intriguing application that is otherwise hard to conduct in actual living humans is the real-time tracking of dynamic phenotypes in organoids, such as neuronal migration and cytoskeleton dynamics.[35, 97, 114, 138–143]
3.4. Organoids as a tool to study neurodegenerative diseases
Despite their typical fetal or early post-natal state,[95, 99, 144–145] it has been possible to successfully model phenotypes in organoids that are typically associated with older age human brains. Alterations in cortical structure, neurogenesis, cell proliferation and cell signaling related to schizophrenia, which emerges during adolescence, have been replicated in ESC and iPSC type organoids.[131, 146–147] Organoids from patients suffering from early-onset (EO) fAD or Down Syndrome (DS), and the feeding of pathogenic Aβ peptides, recapitulate the progressive neuropathology of patients, with the appearance first of Aβ and subsequently Tau pathology[148–150] and inflammatory and extracellular matrix responses.[151] Intriguingly, DS derived organoids also show an increase in subclasses of GABAergic interneurons through overexpression of OLIG2, providing one potential explanation for impaired cognitive ability of patients.[152]
Further mirroring clinical observations, the development of AD molecular pathology was shown to be delayed in organoids expressing the late-onset (LO) E4 risk variant of a gene called apolipoprotein E (APOE) relative to organoids from early-onset patients.[153] Related studies have highlighted a disease pathway shared between EO-fAD and LO-fAD involving gene networks regulated by the repressor element-1 silencing transcription factor (REST).[154] In one of the first forays in using organoids to profile responses to therapeutic drugs, treatment with beta and gamma-secretase inhibitors reduced amyloid and tau phenotypes associated with Alzheimer’s disease.[148]
In PD, midbrain organoids from patients expressing the G2019S mutation in the leucine rich repeat kinase 2 (LRRK2), one of the prominent genetic causes of the disease, faithfully reproduced hallmarks of the disease, such as reduced dopamine neuron content, an increase in the pathogenic alpha-synuclein protein and changes in gene expression that implicate the lysosome.[154] In an interesting contrast with conventional 2D cultures, only mid-brain style organoids produce neuromelanin in vitro, a biomarker for aging and PD that is produced by the substantia nigra, the part of the brain affected early in PD.[31]
3.5. The impact of environmental insults and infection on human biology in brain organoids
Brain organoids are also useful tools for examining brain function and related pathology that does not depend on a genetic component per se. Organoids have been used to model changes in neurodevelopment caused by hypoxia (and rescue thereof)[155–156] and exposure to trace elements,[157] anti-cancer agents,[158] substances of abuse,[159–161] therapeutic drugs,[162] and natural products that purportedly improve brain function.[163] Brain organoids have similarly proven to be a useful tool in elucidating the pathology underlying prion[164] and viral infection, such as Herpes Simplex,[165] Dengue,[166] and, in particular, Zika virus (see below). Studies indicate that Japanese encephalitis virus appears to have a preference for astrocytes and neuroprogenitor cells, in particular outer radial glial cells, inducing cell death, and immune responses when telencephalic organoids mature.[167]
In the face of the global public health crises that ensued after the re-emergence of the virus in the Western world in 2015, organoids quickly became a popular model to study Zika virus (ZIKV) related pathology. Organoid models were instrumental in finding determinants of viral tropism and identifying how the virus may negatively impact brain development by impeding neurogenesis and cortical layer development processes.[101, 109, 168–172] An interesting connection between ZIKV infection and the genesis of other brain pathologies such as schizophrenia and intellectual disability was established, pointing to changes in the DNA methylome of ZIKV-infected neuroprogenitors, neurons and astrocytes with associated alterations in gene expression.[173] Organoids have also been used in the search for therapeutic solutions for Zika virus, leading to the discovery of key players in cellular entry of ZIKV[174–175] and small molecules with potential clinical benefits.[175–179] ZIKV triggers the endogenous RNAi-based anti-viral response, and broad-spectrum antibiotics such as enoxacin, which stimulates RNAi, abolishes viral infections and the ensuing microcephalic morphology of cerebral organoids.[180]
3.6. Modeling interactions between different cell-subtypes and different brain regions
The circuitry of the brain is complex. Connections form between local and distant sub-networks, involving the communication between many neuronal subtypes and support cells.[1–2] The functional maturation of developing organoids is now well established through computational, fluorescence imaging, and electrophysiology techniques.[181–182] Recent data suggests that the activity patterns emerging in organoids are more irregular in time and space than conventional 2D-cultures, in a way more akin to that of human brain.[33]
It is possible to generate functional organoids and use them as a source to generate, isolate and study a broad range of cell subtypes, and to model the interactions between different types of cells, including cancer cells,[183–185] tissue elements such as the blood-brain barrier,[186] or even disparate brain regions. Protocols for glutamatergic, midbrain and dopaminergic neurons, inhibitory neurons, astrocytes and oligodendrocytes, thalamic neuron and organoid models have been published.[31, 98, 108, 110, 114, 143, 145, 175, 187–190] Long-term (20 month) cultivation of organoids created an environment supportive of the transition of astrocytes from fetal to a more mature stage.[145] Microglia-like cells, the brain resident immune cells that are derived from the mesoderm lineage, were successfully grafted onto organoids.[191] Recent literature suggest that it is possible that functional microglia arise naturally in brain organoids, which are ectoderm derived, possibly from a small pool of mesodermal cells.[192] When derived from Alzheimer’s patients carrying defined early-onset mutations, microglia exacerbated AD phenotypes when grafted onto organoid cultures.[153] Furthermore, within a single organoid, developing cells may specialize into regions resembling the make-up of specific subregions of the brain. Organoids can be fused to create 3D structures that emulate connections between ventral-rostral and dorsal areas of the cortex and with the medial ganglionic eminence and thalamus.[110–111, 114, 143, 187, 193] In one of the first steps towards regenerative medicine applications, human organoids grafted in mouse brains showed enhanced functional maturation and integration with the host tissue compared to the implantation of neuroprogenitor cells.[194–195]
4. Engineering Approaches to 3D Brain Models
In order to address the limitations of the systems described earlier, microfluidics and bioreactor systems are increasingly being used as enabling technologies for precise manipulation of the cellular environment (Table 2). These microscale systems, now commonly referred to as “tissue-on-a-chip,” are used to recapitulate the physiological function of tissues in vitro, with noteworthy success in creating tissue models as diverse as the human lung,[196–197] intestine[198] and liver,[199] among others. Recently, these systems have incorporated multiple tissue types into microphysiological systems (MPS), or “body-on-a-chip” devices, comprised of individual chambers that mimic the physiological function of different tissues all connected to mimic inter-tissue communication.[200–201] Here we focus on brain-specific microfluidic systems, but a thorough examination of all advances in tissue- and body-on-a-chip systems can be found expertly reviewed elsewhere.[202–203]
Table 2.
Summary of strengths and limitations of 3D in vitro platforms for brain studies.
| Strengths | Limitations | |
|---|---|---|
| Organoids[35–37, 87–195] |
Complexity of brain achieved through cell-guided tissue organization | Limited control of cell composition and spatial organization |
| Models of neurodevelopment, neurodegenerative disease, injury, infection, evolution, etc. | Necrotic core limits tissue size | |
| Long-term culture capability and establishment of functional networks | Low reproducibility and capability for high-throughput screening | |
| Microfluidic Chips[196–253] | Control of spatial organization and delivery of nutrients/growth factors via flow | Not suitable for long-term cultures to evaluate function or development |
| Capability for multi-tissue systems | Limited capability for high-throughput screening applications | |
| Integration with imaging or electrophysiological modalities | Binding of drugs and biomolecules to polymer surfaces | |
| Hydrogels[283–320, 344–345, 348–350, 359–361] | Ease of preparation for reproducible and scalable cultures | Limited culture longevity due to hydrogel degradation |
| Suitable for non-invasive imaging and electrophysiological recordings | Variable extracellular matrix composition | |
| Some control of spatial organization and size of tissues | ||
| Bioprinting[289, 321–329, 353–356] | Control of spatial organization and size of tissues | Requires optimization of cells and solution printing parameters |
| Suitable for non-invasive imaging and electrophysiological recordings | Limited culture longevity due to hydrogel degradation | |
| Limited capability for high-throughput screening applications | ||
| Porous Scaffolds[330–342, 346, 351–352, 358] | Long-term culture capability of larger scale tissue models | Challenging to scale-up for high-throughput screening applications |
| Greater control of tissue mechanics, particularly useful for injury models | Matrix needs to provide appropriate cellular microenvironment | |
| Suitable for non-invasive imaging and multi-tissue constructs |
4.1. Microfluidic culture devices
The production of a tissue-on-a-chip system requires control over pattern and flow in a microscale environment.[202] Borrowing concepts from the photolithographic etching methods used to make computer chips, the innovative use of elastomeric materials such as poly(dimethylsiloxane) (PDMS) allowed for the design and fabrication of devices with micro- to nano-scale control of feature shapes and sizes.[204] Networks of microfluidic channels and/or wells designed using computer-assisted design (CAD) software are used to create photomasks which are used to generate a master pattern etched into a photosensitive material (photoresist). This master pattern is then used as a mold for casting liquid PDMS, curing the polymer, and peeling the resultant replica with a negative relief from the master. When oxidized in oxygen plasma, the surface of the replica PDMS can be sealed using other materials, typically PDMS (patterned or slabs) or glass.[204] This soft lithography approach creates hollow channels for the perfusion of fluids, with applications in controlling fluid flow and cell patterning in biological systems, which offer advantages as compared to traditional 2D cultures. These devices can also be integrated with microsensors to provide feedback on culture conditions, including tissue barrier functions. To date, microfluidic brain-on-a-chip systems have been designed to model the blood-brain barrier, neural differentiation and brain development, and brain cancer.[205]
4.2. Blood-brain barrier-on-a-chip
Barrier functions at tissue interfaces can be modeled and modulated by seeding cells in different compartments or channels of a microfluidic chip. By incorporating a porous membrane between two microfluidic compartments and seeding with the appropriate cell types, the microfluidic components can be used to control barrier function biochemically or mechanically through exposure to fluid shear stress. The fidelity of the in vitro model to in vivo physiology is governed by the ability to replicate key characteristics of the BBB including tight junctions between endothelial cells, co-culture with astrocytes, and integration of fluid shear stress as well as functional outcomes such as high electrical resistance and selective permeability.[206] Early models of the blood-brain barrier (BBB) used murine cells such as mouse endothelial and astrocyte cell lines seeded on both sides of a porous membrane to establish a micro-BBB that mimics the cerebrovascular environment including expression of tight junction component zonula occludens-1 (ZO-1) and increased transendothelial electrical resistance (TEER) in response to fluid flow. Experimental measures such as a transient drop in TEER in response to histamine exposure as well as selective permeability to fluorescent tracers as a function of molecular size were used to evaluate barrier function.[206] In another model system, tight junction molecules were shown to be upregulated by culturing an immortalized rat brain endothelial cell line (RBE4) with a perfusate of astrocyte conditioned medium in a synthetic BBB model.[207]
While these studies demonstrate the utility of murine cell models of the BBB, they do not wholly recapitulate human biology. Similar results were observed using human cell lines, as a microfluidic system was used to study the barrier function of immortalized human brain endothelial cells (hCMEC/D3), demonstrating that shear stress increased TEER, while the addition of tumor necrosis factor alpha (TNF-α) had the opposite effect.[208] This further demonstrated the importance of a shear force component in mimicking barrier function in BBB models, particularly over prior approaches using human cells that were largely focused on static transwells. A hybrid model using rat cortical neurons and astrocytes, along with human endothelial cells, was used to form a 3D neurovascular microfluidic model of the BBB.[209] These systems demonstrated cell-type specific markers, endothelial barrier and selective permeability, and also showed neural functionality and could be used in the future as a platform for assessing therapeutic effects on neurons as well as BBB permeability.
As in murine models, the contribution of multiple cell types (endothelial cells, pericytes, smooth muscle cells, astrocytes, microglia, and neurons) is critical for BBB formation. The increased availability of hiPSCs, including differentiation protocols for the various cell types, has facilitated the development of human cell-based models. This has been accomplished by interfacing human brain-derived microvascular endothelial cells (BMECs) with primary human astrocytes and pericytes to provide critical intercellular communication for the establishment and maintenance of the high level barrier function of the in vivo human BBB.[210] The further incorporation of neurons, differentiated from hiPSCs, into a microfluidic BBB system provides a mechanism for investigating drug effects on neuronal function in addition to drug permeability and effects on barrier function.[211] More recent efforts have sought to create a fully iPSC-derived BBB model using hiPSC-derived BMECs and neural cells (neurons and astrocytes) that can be personalized to detect individual variability in BBB function.[212]
Importantly, these systems can be used to examine the delivery of therapeutics across the BBB. By co-culturing BMECs derived from hiPSCs with rat primary astrocytes, the drug permeability of large molecules (fluorescein isothiocyanate (FITC)-dextrans) and model drugs (caffeine, cimetidine, and doxorubicin) were measured and correlated with in vivo data.[213] Recent efforts have focused on developing high-throughput microfluidic platforms using human immortalized cell lines of brain microvascular endothelial cells, pericytes, and astrocytes suitable for drug screening applications.[214] These systems represent a promising alternative to current options for preclinical studies such as in vitro rodent or human cell culture or in vivo animal models.[215] In fact, microfluidic systems mimicking the BBB are now being used to probe disease mechanisms, such as the role of BBB dysfunction in AD and may be a platform for screening new drugs.[216] The development of microfluidic platforms, combined with the advancement in stem cell technology, will continue to improve understanding of the BBB and neurodegenerative disorders, with the ultimate goal of discovering effective therapies.[217]
4.3. Platforms for enhancing brain organoid and neural spheroid culture
Engineering tools and techniques have similarly been used to overcome the limitations of brain organoid and neural spheroid cultures (e.g., reproducibility, heterogeneity, and necrosis). In contrast to traditional well plates, these systems offer spatiotemporal control of culture conditions, which can be used to enhance cell viability and to direct neural tissue engineering.[218] With the help of microfluidic and microwell arrays, researchers have uncovered mechanisms of neural differentiation, brain development, and neurological disease. Early studies used microfluidic systems to maintain a gradient of cytokines to control differentiation of neural progenitors derived from human embryonic stem cells into neurons.[219] More recent work has focused on immobilizing embryoid bodies (EBs) within an extracellular matrix (e.g., Matrigel) in a microfluidic channel and cultured under static or perfused conditions to induce differentiation into brain organoids.[220] These systems have been used to model neurodevelopmental disorders such as fetal brain dysfunction due to prenatal nicotine exposure, demonstrating premature neuronal differentiation and disrupted cortical development as a function of nicotine exposure.[160] Other studies of brain development have used microfabricated compartments to model the physics of brain wrinkling during the development of human brain organoids.[221]
In addition to using microfluidic chips to control the stem cell environment, microwell arrays have been engineered to control sphere size and shape and aid in homogeneous formation of 3D neural spheroids tissues. Neural spheroids derived from cell lines, primary rodent brain cells, or human stem cell-derived neural cells have been used to recapitulate key features of the brain such as cellular microenvironment and diversity.[222–225] However, because typical 3D neural spheroid fabrication methods such as hanging drop or spinner flask culture do not provide adequate size control and reproducibility, non-adhesive molds, made of agarose or PDMS, or ultra-low attachment plates are used to allow cellular self-assembly into spheroids in a controlled manner.[226–227] Using a micromold technique to control size and composition, spheroids prepared from primary rat cortical cells demonstrated formation of synaptic connections between neurons, electrical activity, brain-native ECM, and brain-like mechanical properties.[227] Similarly, neurospheres comprised of rat cortical neurons cultured in microwell arrays demonstrated controllable size distribution, enhanced viability, neural architecture and network formation, as well as decreased cell viability upon exposure to Aβ, a hallmark of AD.[228] Additional physiological relevance was achieved by using a concave microarray system and osmotic pump to culture neurospheroids under continuous low flow rates to mimic the interstitial fluid responsible for delivering nutrients and clearing waste in brain tissue in vivo. In addition to investigating flow effects in static versus dynamic cultures on neural differentiation and network formation, neurospheroids were cultured with Aβ to examine neurotoxicity in an in vitro model of AD.[229]
One of the critical remaining technical obstacles for organoid and spheroid cultures remains overcoming limitations of the distribution of oxygen and nutrients as organoids develop into larger and more mature tissues. The high cell density, in combination with the absence of active perfusion, can lead to a necrotic core in larger (>500 μm) spheroids or organoids. Miniaturized spinning bioreactors were 3D printed and used to optimize forebrain organoid cultures derived from human iPSCs.[109] These systems provided enhanced cell viability compared to stationary cultures and demonstrated cortical neurogenesis. Exposure of the cultured organoids to Zika virus led to characteristic features of microencephaly, offering potential utility of the bioreactor platform for disease modeling of brain development with subsequent compound testing during drug screening.[109] Another future option is to implement microfluidic perfusion networks to drive fluid flow or to grow organoids directly onto microfluidic channels seeded with endothelial cells to mimic vasculature.[230]
4.4. Chip-based models of neurodegenerative disease
While the use of brain organoid-on-a-chip systems for studying neurodegenerative diseases is in the early stages of development, other neural cultures have used microfluidic systems to help model Alzheimer’s and other CNS diseases due to their ability to control fluid delivery, incorporate micro-chambers for protein accumulation, and establish gradients to study chemotaxis.[231] Compared to traditional 2D cultures, these are more representative systems for studying specific pathologies of neurodegenerative diseases such as propagation of Aβ or tau protein through neuronal networks, chemotaxis of microglia, and/or synapse activity. Compartmentalized microchannels separating soma from axons have demonstrated early-stage AD pathological events in response to Aβ application such as transmission of Aβ to cell bodies via axons,[232] impaired axonal transport of brain-derived neurotrophic factor (BDNF)[233] or synaptic dysfunction in a cortico-hippocampal neuronal network.[234] In addition to Aβ transport, microfluidic platforms can be used to study other AD pathologies such as the accumulation of microglia near Aβ plaques by observing directional migration of human microglial in response to concentrations of monomer and oligomers.[235]
Mechanisms behind other hallmarks of AD pathology, such as the formation of tau protein aggregates, have been similarly been studied using microfluidic systems. As with Aβ, a microfluidic device with separate somatodendritic and axonal compartments was used to demonstrate that human wild type tau protein could be transferred between primary rat neuronal cells.[236] Using a tripartite microfluidic chamber device, it was further shown that tau aggregate formation could be induced in downstream neurons through cell to cell propagation as well as through extracellular medium.[237] More recently, 3D tissue models have been developed that incorporate multiple human cell types (neurons, astrocytes, and microglia) and hallmarks of AD (Aβ aggregation, phosphorylated tau accumulation, and glial cell activation), thereby offering a more complete representation of neurodegeneration and neuroinflammation in AD.[238] By incorporating many of the advances described in prior work, this microfluidic 3D triculture advances understanding of pathogenic mechanisms, particularly as it relates to glial activation and inflammatory processes.
Similar approaches have also been applied to the study of other neurodegenerative disorders such as Parkinson’s disease by examining the role of alpha-synuclein (aSyn) spreading and aggregation as well as neuroinflammation in disease pathology. A microfluidic system was used to study intercellular communication between human neuroglioma cells and murine microglial cells by controlling the fluid exchange of soluble factors between two isolated cell culture chambers.[239] More recent efforts have focused on using patient-derived iPSCs to generate midbrain-specific dopaminergic neurons to study PD in 3D microfluidic devices. 3D cultures of neurons carrying the LRRK2-G2019S mutation demonstrated time-dependent dopaminergic degeneration as well as the preceding mitochondrial abnormalities. Interestingly, treatment with LRRK2 inhibitor 2 rescued dopaminergic phenotype, suggesting that this system may be useful for drug screening purposes.[240] High-content imaging systems utilizing automated fluorescent microscopy and analysis algorithms allow for real-time assessment of these cultures with other recent efforts aimed at constructing fully automated workstations for differentiating PD patient-derived stem cells into dopaminergic neurons, with cultures maintained for over 100 days and monitored for electrophysiological activity in real-time.[241]
4.5. Brain cancer-on-a-chip
Microfluidic technologies have been used in brain cancer research for diagnostic applications such as circulating tumor cell isolation and biomarker detection, as well as to create in vitro models to study drug efficacy and tumor progression.[242] As with models of neurodevelopment or neurodegeneration, brain cancer-on-a-chip models rely on precise control of cellular microenvironment and capability for real-time outputs (e.g., high-resolution imaging, biochemical detection) to understand disease pathology. To date, researchers have focused on controlling variations in extracellular matrix (ECM) composition and mechanics, vascular integration, and cellular interactions as a means of studying glioblastoma multiforme (GBM) tumor progression. As an example, single cell microchips can be used to quantitate the distance dependence of signaling between two GBM cancer cells, providing insight into the cell-cell interactions that may govern diffuse nature of GBM tumor architecture.[243] Tumor progression can be predicted using a microfluidic platform for single-cell proteomics of human glioblastoma cell lines and brain tumor specimens.[244] For understanding tumor metastasis, microfabricated polyacrylamide channels of defined wall stiffness and width were used to study the effects of matrix composition and geometry on tumor cell migration, with narrow channels enhancing migration.[245] Other microfluidic cell culture devices have been developed to investigate the role of hypoxia in maintaining a stem cell niche in glioblastoma spheroids[246] or to examine clinical hallmarks of glioblastoma such as the formation of migratory hypercellular regions induced by oxygen and nutrient depletion by controlling the flow of medium using microfluidics.[247] Each of these systems demonstrate the utility of 3D microfluidic systems by providing insights due to the enhanced control over cell environment and experimental variables that are not otherwise achievable using standard 2D cultures.
Recent microfluidic models have examined the role of vascular cells in mediating glioma stem cell invasion. Using a microchannel co-culture system of human endothelial cells and glioma stem cells (GSC) separated by a collagen gel, migration of the glioma stem cells was promoted by endothelial cells through the genetic upregulation of cell-ECM adhesion-associated proteins and down-regulation of cell-cell adhesion proteins.[248] A more representative 3D microfluidic model of GSC and vascular cell interactions was established with a microvascular network in a fibrin hydrogel prior to introducing patient-derived GSCs in Matrigel to the microfluidic device. Using a receptor antagonist, this system demonstrated enhanced migration of GSCs in the presence of endothelial cells via the chemokine (CXC motif) ligand 12/chemokine (CXC motif) receptor 4 axis (CXCL12-CXCR4) signaling pathway.[249]
Brain cancer-on-a-chip platforms are often utilized as more representative model systems for preclinical drug screening as compared to traditional monolayer cultures due to their ability to recapitulate drug pharmacokinetics in a 3D environment. Brain cancer spheroids, cultured from glioblastoma cells within microwells in a 3D chip, were treated with anti-cancer drugs, pitavastatin and irinotecan, as a combinatorial treatment at varied concentrations in a high-throughput manner to determine the optimal treatment to reduce cell viability.[250] A modified version of this chip was able culture primary GBM cancer cells derived from primary human brain tumors and treat with combinations of chemotherapy drugs, temozolmide and bevacizumab to determine the optimal concentrations for personalized medicine.[251] Importantly, since these microchips are prepared from a polymer that blocks non-specific protein adsorption, poly(ethylene) glycol diacrylate (PEGDA), this system avoids the pitfalls of PDMS-based microfluidics which can adsorb hydrophobic molecules, an important consideration for drug screening applications. [250–251] In addition to drug efficacy, other personalized approaches to drug screening have included predicting drug resistivity of GBM cell lines by measuring mRNA markers of temozolmide resistance[252] as well as determining the drug sensitivity of live slice cultures to chemotherapies in a dose-dependent manner.[253]
4.6. Technology and systems integration in microfluidic platforms
The development of advanced in vitro 3D models of human brain physiology and disease through brain-on-a-chip, tissue engineering or other approaches, has necessitated the integration of technologies to monitor real-time changes in cellular microenvironment and activity, particularly in response to pharmaceutical intervention. As outlined in many of the systems described above, interfacing microfluidic platforms with optical, electrical and biochemical capabilities is critical to identifying biological changes in real time. Due to the optical clarity of PDMS and other polymers used in microfluidic systems, conventional microscopy techniques (bright field, fluorescence, phase contrast) have been used to observe cells and tissues in these devices. As microfluidic devices have trended towards thicker 3D tissues, the emergence of scanning-based imaging techniques such as confocal microscopy have allowed for increased depth resolution.[254] Future integration of light sheet microscopy in microdevices may allow for even greater spatial and temporal resolution, with lower phototoxicity than confocal microscopy.[255] While standard microscopes are still the most frequently used, continued development of compact microscope-on-a-chip imaging systems will reduce costs and increase capabilities for automation and portability.[256] Future integration of smartphones, utilizing their high-resolution complementary metal oxide semiconductor (CMOS) cameras, offers another alternative for optical biosensors,[257] with proof-of-concept already demonstrated in a microfluidic chip system.[258]
For neurological systems, the ability to monitor electrical signaling in real-time is critically important for understanding information processing in neural circuits. For microfluidic systems, this is typically achieved through the integration of multi-electrode arrays (MEA) that are capable of recording electrical activity with high spatial and temporal resolution. Because these are recorded non-invasively, repeated measurements can be achieved over time in either brain slices or tissue culture preparations.[259] Based on the design of the microfluidic system and MEA, research can range from investigating intracellular dynamics in a single axon[260] to propagation between neuronal sub-populations (e.g., cortical-thalmic) in a co-culture system.[261] Although not part of a microfluidic chip, MEAs have also shown utility in drug screening applications such as the evaluation of neurotoxic compounds.[262–265] Importantly, electrical recordings by MEAs can be combined with optical methods for a more comprehensive assessment of neuronal function. As an example, by loading fluorescent calcium sensitive dye (Fluo-4) in the sample medium, simultaneous recordings of extracellular calcium and electrical traces can be resolved to show a propagating wave in response to electrical or chemical stimulation of murine brain slices.[266] Though not explicitly for brain organoids, the recent development of a non-invasive, automated platform for physical, biochemical and optical monitoring represents the latest achievement in real-time multisensory feedback in a microfluidic device.[267]
In addition to advances in the hardware for MEA and imaging techniques, protein and genetic engineering approaches, such as genetically encoded calcium indicators (GECIs) or optogenetic techniques, are increasingly being used as alternative methods to voltage-sensitive fluorescent dyes to observe, analyze, and control neuronal activity. The development of a high affinity, green fluorescent protein (GFP)-based calcium (Ca2+) probe (GCaMP) allows for chronic imaging and quantification of changes in intracellular calcium concentration as a function of firing action potentials and synaptic input.[268–269] By transfecting neurons with GCaMP and culturing them in 3D hydrogel matrices, multi-planar images provide spatial specificity and the functional activity of the developed 3D neural networks, particularly in response to drug input.[270–271] Additional fluorescent protein-based probes such as red GECIs (RCaMP) provide the option of using different colors within the same cell or to distinguish cell populations (e.g., neurons and astrocytes) and may be integrated with optogenetic approaches.[272]
The emergence of optogenetics, starting from the use of a light-sensitive ion channel protein (Channelrhodopsin-2, ChR2), as a means of optically controlling the activity of neuronal populations has provided researchers with another tool to activate or inhibit the activity of specific neurons non-invasively in culture with high temporal and spatial resolution.[273–275] While the majority of development is focused on in vivo applications, such as neural probes for brain stimulation, microfluidic systems have used optogenetics and calcium imaging to observe synaptic communication in vitro.[276] 3D in vitro tissue models have demonstrated the ability to record changes in fluorescent intensity upon photostimulation of neural cells that are transfected with both a ChR2 and a GECI and cultured in a collagen gel on a CMOS chip.[277] As optogenetic tools continue to be developed, additional methods of cellular control and/or control of small molecule delivery in 3D systems are available for researchers to study different mechanisms of neuromodulation.[278] As a key example, the ability to combine optogenetic actuators (channelrhodopsins) with archaerhodopsin-based voltage indicators in hiPSC-derived neurons allows for non-invasive, all-optical electrophysiology that is well-suited for high-throughput drug screening.[279]
5. Materials-Based Approaches to 3D Brain Models
Whether in a chip-based or stand-alone system, the choice of materials for a 3D brain model is multifaceted, with considerations for biocompatibility, mechanics, porosity and imaging compatibility, among the key concerns. Neural cells must be able to proliferate, differentiate, and form viable networks within materials that also need to provide the appropriate porosity, structure and mechanical support over time. Additional biophysical cues such as electrical conductivity and architectural patterning are increasingly being integrated into material designs to guide cell growth, development and organization.[280] In addition to support structure, materials must also incorporate domains for cell attachment as well as appropriate bioactive signaling molecules (e.g., immobilized growth factors or chemical functional groups). By carefully designing and engineering the appropriate biochemical and biophysical cues, researchers can direct cellular behavior by recapitulating the natural cellular microenvironment in a 3D brain model.[281] For many researchers, particularly in the organoid field, Matrigel is the chosen support material as it offers a complex mixture of structural, functional and signaling molecules suitable for 3D culture, but remains a poorly defined and heterogeneous mixture of extracellular matrix proteins that is subject to batch-to-batch variability.[282] To address these deficiencies, a variety of material-based approaches have been used to bioengineer 3D scaffolds using a bottom-up approach to support in vitro neural tissue development and disease modeling.
5.1. Natural hydrogel systems
Hydrogels are commonly used in bioengineered neuronal culture systems as their chemical and mechanical properties can be tailored to resemble those naturally occurring in brain.[283] Derived from natural or synthetic polymers, they are cross-linked to form hydrated networks of tunable mechanical stiffness and may be functionalized with bioactive components.[283] In contrast to Matrigel systems, hydrogels prepared from fully-defined source materials can be formulated with specific control over ECM components and signaling cues, using in vivo conditions as a guide. Because brain ECM is composed of a mixture of proteoglycans, hyaluronic acid, tenascins, collagens, fibronectin, vitronectin, and laminin,[284] combinations of these materials have been used as a starting point to engineer hydrogel scaffolds for neural tissue engineering.[282] Importantly, hydrogels must be formulated to mimic the mechanical properties of the brain (elastic modulus of <1 kPa), providing an appropriately soft matrix to direct neuronal cell behavior.[285] This is typically achieved by preparing hydrogels that are cross-linked with sufficiently low density to achieve this mechanical stiffness while also allowing for exchange of oxygen and soluble factors to support neuronal culture.
Due to its wide availability, biocompatibility, and biodegradability, as well as its well-documented historical use as a hydrogel scaffold for tissue engineering, the most commonly used natural biomaterial for neural cell culture applications is collagen type I. It has been demonstrated that rodent-derived neural stem and progenitor cells (NSPCs) in type I collagen gels can expand and differentiate into neurons with functional synapse and neuronal circuit formation.[286] These gels have also shown the ability to support the differentiation into multiple cell types (neurons, astrocytes, and oligodendrocytes) under optimal conditions of cell density and collagen concentration.[287] In order to further increase relevancy to native brain tissue, additional natural polymers such as hyaluronan have been combined with collagen to enhance neuronal differentiation of NSPCs.[288] Similarly, the incorporation of soluble growth factors in engineered hydrogels have been used to direct cell proliferation, migration, and fate. Using a layer-by-layer bioprinting approach, collagen-fibrin composite hydrogels were used to promote the migration of neural stem cells toward hydrogel layers containing vascular endothelial growth factor (VEGF).[289] Combinations of neurotrophic factors such as nerve growth factor (NGF), glial cell-line derived neurotrophic factor (GDNF) and ciliary neurotrophic factor (CNTF) have been explored in dorsal root ganglion (DRG)-seeded collagen gels to optimize neurite outgrowth.[290] Mechanical cues such as gradients of stiffness in 3D collagen gels or bio-acoustic levitation in fibrin gels have also been utilized to drive neurite growth or generate multilayered 3D brain-like constructs from hESC-derived neuroprogenitors, respectively.[291–292]
Due to the prevalence of hyaluronic acid (HA) in native brain tissue, this polymer has been extensively studied as a substrate for central neural tissue engineering.[293] However, since unmodified HA does not support cell attachment, HA hydrogels must be modified with cell adhesion molecules or blended with other ECM components such as collagen.[294] As an example, a Nogo-66 receptor antagonist was used to modify HA hydrogels to improve the neuronal adherence and survival of DRGs in vitro as well as induce neurite outgrowth.[295] HA-poly-D-lysine (PDL) copolymer hydrogels have also been used to control neuronal cell adhesion and network formation based on HA:PDL ratios.[296] Similarly, ECM components such as laminin or fibronectin- or laminin-derived peptides (RGD, IKVAV) have been used to facilitate cell attachment to HA.[294] A combination of HA, collagen, and laminin was used as a mimic of natural ECM composition to support the co-culture of Schwann cells and neurons in 3D hydrogels for 2 weeks, offering a potential platform for studying mechanisms of interactions between the two cell types with applications in nerve regeneration.[297] Interestingly, as with collagen hydrogels, mechanical properties of HA gels influenced neural progenitor cell (NPC) differentiation towards neuronal subtype in softer hydrogels versus an astrocytic subtype in stiffer hydrogels.[298]
In addition to HA, other naturally-derived polysaccharides such as alginate, agarose, chitosan, and composites thereof have been used as substrates for 3D neural tissue engineering. As with HA, these polymers are inert biomaterials that can be tailored for 3D neural cell culture by varying composition and crosslink density. These polymers are similarly non-adhesive for cultured neurons and must incorporate cell attachment factors, although there is some evidence that unmodified soft gels can support neurite growth.[299] As an example, alginate hydrogels functionalized with cell-specific attachment ligands (laminin, IKVAV, or RGD) were used to study co-cultures of neurons, glia and endothelial cells, observing neural functions such as gap junction formation, synaptic vesicle release, and electrophysiological activity.[300] Another approach to incorporating cell binding domains has been to mix in collagen fibrils, with this collagen/alginate blended hydrogel used to culture neurons generated from human iPSCs.[301] Other researchers have used defined mixtures of nutrients in agarose and alginate hydrogels to generate 3D multi-layered scaffolds that mimic cortical layers using primary rat cortical neurons in a microfluidic device.[302]
Though the vast majority of neuronal tissue engineering has been achieved using rodent-derived neurons, combinatorial studies on matrix composition and architecture have sought to provide a more comprehensive understanding of how cellular microenvironment (ECM and signaling molecules) can influence cell fate in 3D cultures. This includes efforts to use these parameters to direct human stem cell differentiation toward neural and glial lineages in natural polymers such as Matrigel, collagen type I, gelatin, or HA.[303] As the field progresses towards high-throughput, human cell-based neuronal models for developmental/disease biology, regenerative medicine, or drug/toxicity screening applications, researchers aim to overcome limitations on reproducibility and culture duration that is inherent for natural materials. One approach is the use of novel biomaterials such as silk fibroin protein to generate hydrogels with tunable stiffness in the range of native neural tissues, capability for functionalization with growth factors or ECM molecules, as well as the ability to maintain structural integrity for much longer duration (months to years) than typical hydrogel materials.[304] Others approaches center around the development of synthetic materials, including polymers and self-assembling peptides, to address these concerns.
5.2. Synthetic hydrogel systems
In contrast to naturally-derived polymers which offer physiological relevance but also greater variability in chemical composition, synthetic hydrogels derived from polymeric materials such as polyethylene glycol (PEG) and methacrylates, among others, offer the advantage of tight control over physical and mechanical properties based on chemistry and polymerization.[283] However, these hydrogels are generally biologically inert and with minimal cell adhesion and protein adsorption to support tissue formation.[305] This requires chemical modifications to the polymer hydrogels for neural tissue engineering applications that are similar to those used for polysaccharide-derived hydrogels (e.g., RGD, IKVAV) or mixtures with other ECM proteins (e.g., collagen). As with chemically-defined natural hydrogels, the use of synthetic polymers is an alternative to Matrigel, replacing the inherent variability of a poorly defined mixture of bioactive components with a well-defined and controlled matrix for neural tissue engineering applications.
To date, PEG is the most extensively used synthetic polymer for neural cell culture in 3D hydrogels. Preliminary studies incorporated rat neurons in photopolymerizable PEG hydrogels, modified with poly(lactic acid) (PLA) and poly(glycolic acid) (PGA) to control degradation rate and supplemented with basic fibroblast growth factor 2 (bFGF-2) to support cell growth.[306] These tissues demonstrated neural tissue development and functionality, with the time-scale of neurite outgrowth a function of gel degradation rate. In another study, peripheral nerve regeneration was enhanced by covalently attaching cell adhesion ligands (RGD, IKVAV, and YIGSR) to PEG hydrogels.[307] Adhesion ligands conjugated to PEG hydrogels have similarly been used to study human iPSC-derived NPCs attachment and migration induced by astrocytes.[308] Recent advancement in PEG hydrogels has been achieved by incorporating various functional groups, including polymer-peptide conjugates, for cell attachment, signaling, and cleavage sites to tune physical and mechanical properties. Using star-shaped PEG (starPEG) and heparin peptide conjugates, hydrogel compositions were optimized to embed human vascular endothelial cells and DRGs and stimulated to control cell phenotype (angiogenic or neurite outgrowth).[309]
Synthetic polymers have been used to modify natural polymers, preserving the beneficial properties of the natural material while offering additional functionality. One notable example is the synthesis of methacrylamide chitosan (MAC), a synthetic polymer that can be modified with peptides for cell adhesion and neurite outgrowth, formed into a transparent gel via chemical crosslinking, and seeded on the surface with rat neurons.[310] This polymer has subsequently been modified to enhance oxygen diffusion within the hydrogel through the incorporation of D-mannitol[311] or conjugation of perfluorocarbons to MAC[312] to enhance the neuronal differentiation of rat neural stem cells (NSCs)/progenitor cells (NPCs) in 3D. Methacrylate has also been used to modify hyaluronic-acid, with researchers using this material to create layered 3D hydrogels of variable mechanical properties to study the differentiation and migration of human iPSC-derived NPCs.[308]
Synthetic, self-assembling peptides (SAPs) have been developed as a new class of biologically-inspired materials for neuronal culture. Oligopeptides of varied sequences of arginine-alanine-aspartate (RAD) are able to self-assemble under physiological conditions and support neuronal cell attachment, neurite outgrowth, and synapse formation,[313] with the most commonly used peptide, RADA16-I, commercially available as PuraMatrix. Neural stem cell proliferation, migration, and differentiation may be further promoted by modifying these SAPs with a functional cell adhesion motif (RADA16-IKVAV) in a 3D hydrogel scaffold using both rodent and human cells.[314–316] Despite these successes, these formulations suffer from low pH impacting cell viability. To address this issue, oppositely charged SAPs with different cell adhesion epitopes (RADA16-IKVAV and RADA16-RGD) were combined to form a 3D hydrogel at neutral pH that was capable of encapsulating NPCs/NSCs and differentiating into neurons and astrocytes.[317] Because SAPs often suffer from poor mechanical properties, linear or branched (LDLK)3 SAPs were recently used as an alternative to form hydrogels of controlled mechanical properties, demonstrating that neuronal differentiation is enhanced when scaffold stiffness is comparable to that of the native brain.[318–319] These constructs have also demonstrated the ability to support in vitro, serum-free differentiation of human NSCs into GABAergic, glutamatergic, and cholinergic neurons that give rise to the formation of electrically active networks.[320]
5.3. Bioprinting
The emergence of bioprinting as a technique to control the spatial distribution of cells, biomaterials, and signaling molecules offers new possibilities for building 3D neural tissues in vitro using a bottom-up approach. Using hydrogel-based bioinks to precisely arrange cells in 3D, heterogeneous tissues can be composed with the appropriate physiological cues and resultant functionality that is similar to that of native brain tissue. This approach further improves reproducibility in model development and potential for automation, critical elements downstream high-throughput screening applications.[222]
Many of the hydrogel materials described above have been investigated as potential bioinks for neural tissue engineering applications, with the choice dependent on a number of considerations including the printing technique (e.g., inkjet, extrusion-based, laser-guided), cytocompatibility, curing time and mechanical properties, among others.[321–322] In one of the first examples of bioprinting neurons, inkjet printing was used to pattern rat hippocampal and cortical cells in a single layer, with these cells demonstrating electrophysiological activity. Researchers in this study used the same technique to fabricate multi-layer constructs of human NT2 neurons and fibrin gels via a layer-by-layer printing approach to create 3D neural sheets.[323] A similar approach using multilayered collagen type I gels and rat embryonic neurons and astrocytes was used to form multicellular constructs with defined spatial patterning to examine cell-to-cell interactions.[324] Layer-by-layer inkjet printing has also been used to study cell migration through the controlled spatial distribution of VEGF-containing fibrin gel and murine NSCs within a 3D collagen scaffolds.[289] Subsequent work has focused on exploring tunable polymers as inks for 3D bioprinting such as thermoresponsive polyurethane hydrogels[325] or stereolithography-based printing of functionalized materials such as dopamine-modified gelatin methacrylate[326] to improve NSC proliferation and differentiation.
Improvements in printing techniques and bioink material properties have supported the fabrication of more complex structures, such as cortical layers, that mimic the native brain. Through the use of a bioink (gellan gum-RGD) that forms a porous structure suitable for oxygen and nutrient exchange, primary rat neural cells were printed in a layered, brain-like cortical construct.[327] More recently, human NSCs and iPSCs in a polysaccharide-based (alginate, carboxymethyl-chitosan, agarose) bioink have been used to print mini-tissues that differentiate into neurons, form neural networks, and demonstrate electrical activity. Using an extrusion-based printing technique, cells are co-printed with the bioink before gelation to encapsulate them in a 3D matrix for in situ expansion and differentiation into functional neurons and neuroglia.[328–329]
5.4. Solid porous scaffolds
While hydrogel-based techniques offer the benefit of soft mechanical properties for neurite outgrowth, they generally lack the ability for long-term culture (greater than 1 month) due to fast degradation rates, uncontrolled contraction, or the presence of necrotic cores. As an alternative, solid porous scaffolds prepared from synthetic or natural polymers offer mechanical and morphological stability and pore interconnectivity to facilitate nutrient transport required in long-term neural tissue models. Furthermore, these scaffolds may be modified with topographical cues for cell attachment and directed neurite outgrowth.[222].
As an example, researchers have used polystyrene as a synthetic polymer for generating porous scaffolds for 3D culture of human neural progenitor cells. Scaffolds were prepared by a gaseous salt leaching method where polystyrene is dissolved in chloroform, mixed with salt particles of known size, and cast into wells before curing in an oven.[330] When using these scaffolds to culture human neural progenitors, the cells more accurately emulated the gene expression profiles of in vivo surrogates (3D neurospheres) in comparison to standard 2D culture.[331] Porous polystyrene scaffolds cultured with primary rodent neural cells also demonstrated more representative calcium channel functionality versus standard 2D cultures in comparison to freshly dissected intact tissues.[332] More recently, graphene foam has been pursued as a biocompatible and conductive scaffold for 3D neural culture that represents an alternative to the inert, non-biodegradable polymers such as polystyrene. These scaffolds, prepared via a chemical vapor deposition method, showed that they can enhance NSC differentiation into neurons/astrocytes as well as serve as a conductive platform for electrical stimulation.[333] As an alternative to porous scaffolds, electrospinning techniques have been used to prepare fibrous scaffolds from synthetic polymers that can be functionalized to support neural cell culture and neurite outgrowth. However, because standard methods typically produce dense fiber meshes that function more as a 2D surface, recent efforts have used an array of needles as a collector to form lower density electrospun fiber scaffolds that support 3D culture of human NPCs and their formation of highly integrated, functional networks.[334]
Similar to synthetic polymers, multiple methods have been used to create porous or fibrous 3D scaffolds from natural polymers. Freeze-drying, in particular, has been commonly used to form porous scaffolds from biological materials such as collagen or HA. The freeze drying process can be manipulated to control the mechanical properties and to determine pore size of collagen-glycosaminoglycan scaffolds.[335–336] Collagen and HA were mixed at different ratios, freeze dried, and cross-linked to tailor the resultant scaffolds to have moduli and porosity similar to those of native brain tissue.[337] This provides the appropriate conditions to support neural progenitor cells by allowing for the exchange of nutrients, the outgrowth of neurites, or the infiltration of cells in in vivo applications.[338] Collagen-based scaffolds can be tailored to include cell adhesion molecules as in the modification of 3D freeze-dried collagen matrices with IKVAV and/or RGD peptide to enhance the attachment of rat DRGs.[339] Cryogels from gelatin-laminin were used as porous scaffolds for differentiating human cord blood-derived stem cells into artificial neural tissue with mature networks of neurons and glia, suitable for implantation in rat brain tissue or high-throughput personalized medicine applications.[340] Recently, porous silk scaffolds have been used in combination with collagen hydrogels to create functional brain-like cortical tissue, where the compartmentalization of neuronal cell bodies and axons recapitulates grey and white matter.[341] This combination of porous scaffold and hydrogel matrix was able to mimic the mechanics of the native rodent brain while allowing for a robust neural network formation and electrophysiological activity. The composite system (silk and collagen type I) was further used as a model of traumatic brain injury, monitoring changes in cellular damage, biochemical signaling, and electrical activity following a controlled impact.[341] These versatile systems have also demonstrated utility as engineered neural tissue implants for replacing large brain cavities.[342]
5.5. Materials-based models of neurological disorders, damage and dysfunction
In addition to supporting 3D cultures to study neurite outgrowth and neural network formation, materials-based approaches have been utilized to create models of neurological disease. To date, work has primarily focused on the development of 3D in vitro models of Alzheimer’s disease and glioblastoma as well as platforms for studying traumatic brain injury or neurotoxicity. By controlling material properties as well as cell type and genetic composition, particularly using human iPSCs or neural progenitors, key features of the disease may be recapitulated and used to understand mechanisms and/or to screen therapeutic interventions.
Initial 3D models of Alzheimer’s disease have sought to recapitulate the pathological hallmarks of the disease, Aβ accumulation and plaque formation as well as aggregation of phosphorylated tau protein. In one example, fAD mutations in Aβ precursor protein (APP) and presenilin 1 (PS1) genes were overexpressed in human NPCs. These cells were cultured in Matrigel as a 3D support matrix, which limited diffusion and accelerated β deposition and tau protein aggregation. This model system recapitulated, for the first time, these key features of AD in a 3D model and was subsequently used to screen beta- or gamma-secretase inhibitors to decrease Aβ pathology.[86] In another study, Aβ aggregates were added to conventional 2D cultures and 3D cultures of human stem cell-derived neurons in a PuraMatrix hydrogel. Cellular response to the Aβ aggregates, as mediated by p21-activated kinase, was only observed in the 3D cultures demonstrating the critical role of the 3D environment in modeling AD mechanisms.[343] Subsequent 3D models of AD have used collagen or PEG hydrogels to study Aβ-induced toxicity of neural cell lines or plasticity loss of human neural stem cells, respectively.[344–345] Others have used a composite silk scaffold-collagen gel system to support long-term (>8 month) 3D human neural network models using iPSCs derived from healthy individuals and Alzheimer’s disease patients, offering a potential platform for studying development, plasticity, and degeneration over time.[346] The continued development of 3D models that incorporate multiple cell types (neurons, astrocytes, microglia, and oligodendrocytes) will be critical in understanding the mechanisms of neurodegeneration and neuroinflammation that are key features of neurodegenerative disorders such as AD or PD.[347]
In addition to AD, the development of in vitro 3D brain cancer models has received a significant amount of attention, with specific focus on materials-based approaches to mimic the tumor microenvironment in vitro that drives glioblastoma (GBM) progression. Currently used bioengineered models are primarily based on the encapsulation of glioblastoma cells in 3D biomaterial microenvironments that recapitulate the hypoxic conditions observed in vivo and allow for cell migration within the scaffold.[348] Through the use of natural ECM-derived materials, primarily HA, researchers are able to mimic the composition and structure of the native brain. In fact, glioma cell invasion into HA hydrogels has been shown to be mediated by controlling the stiffness of HA-methacrylate hydrogels and functionalization with RGD peptides for cell adhesion.[349] One of the major drawbacks of using these systems is that by increasing polymer concentration or cross-linking density, biochemical signaling is affected. Others have sought to independently control matrix stiffness using synthetic polymer-based hydrogels such as polyacrylamide or PEG to isolate the effects of matrix stiffness and confinement on human glioma cell migration.[245, 350] In addition to hydrogels, solid porous scaffolds of chitosan and HA have been developed to study glioblastoma microenvironment, demonstrating enhanced invasiveness of human GBM cells and chemotherapy drug resistance.[351] Using porous silk scaffolds infused with ECM hydrogels as a brain-mimicking microenvironment, researchers have further showed that pediatric and adult brain tumor progression is mediated via ECM-dependent alterations in transcriptomic and metabolic profiles.[352] Additional spatial control may be achieved by using bioprinting approaches to control the deposition of different cells types as well as ECM and signaling molecules in complex 3D models of the tumor microenvironment.[353] As an example, a 3D brain tumor model was developed from glioma stem cells bioprinted in a gelatin/alginate/fibrinogen hydrogel. Similar to conditions found in vivo, these cells formed spheroids, remodeled ECM to occupy a larger space, and demonstrated chemotherapy drug resistance.[354] Spatial control of cells, matrix, signaling molecules, and oxygen content may also be controlled using microfluidic mixing as demonstrated in a HA-gelatin-methacrylate hydrogel brain tumor model.[355] More recently, bioprinting techniques have been used to create 3D mini-brains that include GBM cells as well as macrophages to study cellular interactions and therapeutic control of those interactions.[356] With these 3D systems, the goal is to not only understand the mechanisms of tumor progression, but also serve as a platform for preclinical screening of therapeutics and/or radiotherapy.[357] Using GBM cell lines seeded in a 3D Matrigel-coated polystyrene scaffold, researchers showed that the in vitro cellular response to radiotherapy in combination with clinically-used agents (temozolomide, bevacizumab, erlotinib) strongly correlated with clinical observations.[358]
In addition to studying neurological disease, recent work has sought to develop preclinical models for studying traumatic brain injury or neurotoxicity. These models are used correlate clinical observations with in vitro results to better understand mechanisms and/or to more accurately predict in vivo outcomes than traditional 2D cultures or animal models. For traumatic brain injury, collagen gels seeded with rat cortical neurons were subjected to compressive deformation, providing insight on how neuron viability and function is dependent on strain magnitude and rate.[359] Others have used a weight drop method to inflict mechanical damage in a 3D model of cortical brain-like tissue formed from a silk-collagen composite scaffold, observing enhanced glutamate release and transient hyperactivity that is consistent with in vivo results.[341] For neurotoxicity studies, 3D in vitro models using human cells represent an alternative to animal models that may be more predictive of chemical safety and reduce the number of drug failures in clinical trials. Recently, human embryonic stem cell-derived neural progenitor cells, endothelial cells, mesenchymal stem cells, and microglia/macrophage precursors formed neural tissue constructs following seeding on top of PEG hydrogels functionalized with biodegradable peptide cross-links and RGD peptides to promote cell adhesion. Leveraging the reproducible and controllable intracellular interactions achievable via this method, these constructs were then used for neurotoxicity screening in vitro by treating the tissues with toxic and non-toxic chemicals, collecting RNA sequencing data, and then using machine learning to build a predictive model capable of correctly predicting the toxicity of additional compounds.[360]
6. Future Perspectives for 3D Brain Models
Over the past decade, there has been tremendous progress in the development of 3D models of the human brain. Through technological advances and innovation in the key components of engineered tissues, cells, scaffolds, and signaling molecules, otherwise known as the tissue engineering triad, researchers can now create models that offer greater fidelity to neurological development and disease. These models offer the ability to better understand human physiology and the potential to identify therapeutic or regenerative medicine approaches to treat disorders of the human brain. As outlined in this review, there have been two generally parallel approaches to creating 3D brain culture systems, organoid technology and bioengineering methodologies. The fundamental difference between these approaches is that organoid technology is largely a cell-driven, bottom-up approach whereas the bioengineering methodology is a materials-templating bottom-up approach for generating 3D brain models. Looking forward, we see tremendous potential for integrating these two approaches, leveraging the strengths of each. Furthermore, we see potential for integrating other tools and techniques such as microfluidics and bioreactors for improving cell culture duration or electrode arrays and imaging modalities for non-invasive monitoring of the models in real time. Lastly, as the potential for 3D models as a screening tool is currently limited by reproducibility issues, we highlight prospective solutions for generating high-throughput 3D brain models for biomarker or drug screening applications (Figure 3).
Figure 3.
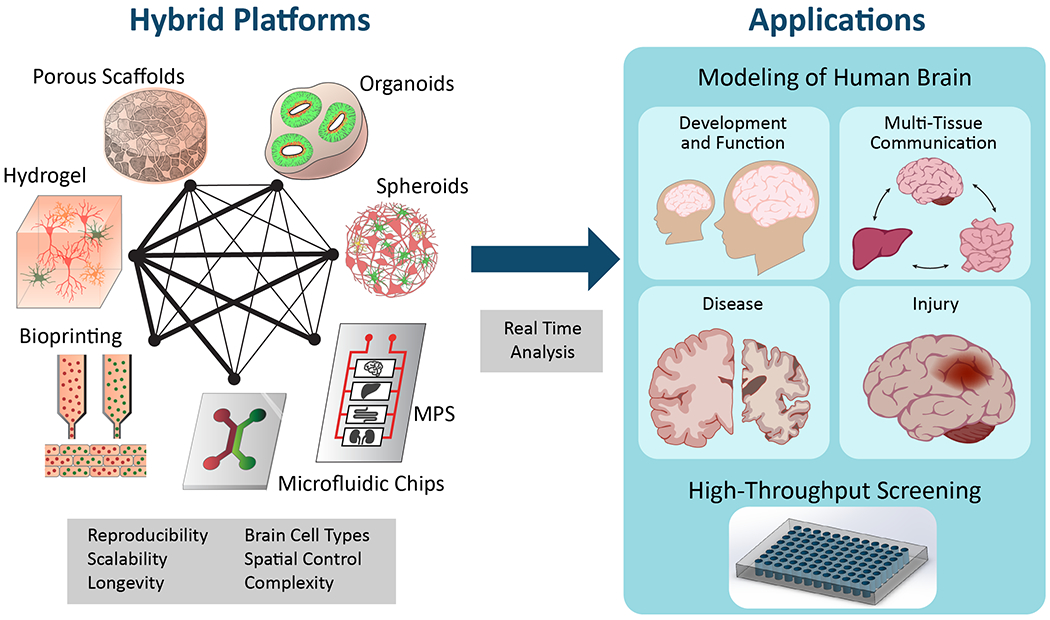
Proposed integration of technology platforms for optimizing 3D tissue systems for modeling human brain physiology and high-throughput screening. Thicker lines between platforms indicate current areas of investigation whereas thinner lines show areas for further consideration. Gray boxes identify some of the key stem cell and bioengineering technology parameters to be addressed.
As described in previous sections, organoids have proven to be an effective developmental model of the human brain with additional utility as a tool for studying neurological function or modeling neurodegenerative, psychiatric and infectious disease. The drawbacks associated with organoid cultures, including issues with scalability, reproducibility, heterogeneity and longevity due to necrotic cores, remain active areas of pursuit in the field. While microfluidic or bioreactor approaches have most commonly been used to improve the reproducibility of organoid cultures with regard to overall size distribution and neural cell viability and differentiation, a less explored option is the use of materials-based approaches to improving brain organoid cultures. In one example, bioengineered materials were used to help developing organoids adopt 3D architectures similar to the native brain as culturing embryoid bodies on floating scaffolds of synthetic polymer (PLGA) filaments caused self-organization in an elongated morphology and the formation of a polarized cortical plate and radial units.[40] Others have used defined, modular synthetic matrices composed of PEG-based engineered hydrogels to understand the role of biochemical and biophysical cues in regulating early neurogenesis in organoid cultures derived from mouse ESCs. By controlling the mechanical stiffness, degradability, ECM components, and soluble factors of the engineered hydrogels, they were able to identify the specific material properties that promoted neuroepithelial differentiation, apical-basal polarity and dorsal-ventral patterning, while also generating more homogenous cultures in these defined synthetic matrices in comparison to ESC cultures in Matrigel.[361] These pioneering studies demonstrate the great potential for integrating organoid and bioengineering technology to support brain organoid cultures, but with significant insight to be gained by studying bioengineering approaches used in other in vitro organoid-based tissue models.
To date, engineered materials for supporting organoid cultures have mainly focused on solutions for intestinal organoid tissue models. Through these studies, the importance of providing a reproducible, chemically-defined matrix with controllable mechanical properties and defined signaling cues for specific stem cell niches has been established. The knowledge gained from these studies can be transferred to guide the optimization of materials for 3D brain organoid culture.[362] Among the key goals of an engineered material for this application would be the spatiotemporal delivery of biochemical cues to direct neural differentiation. In fact, placing morphogen gradients within organoids has been shown to facilitate tissue self-organization.[363–364] This could be further optimized through the patterning of a bioengineered scaffold with growth factors known to direct neural progenitor differentiation to specific lineages. Cell migration and self-organization within the organoid could similarly be directed by controlling the spatial organization of cell-adhesive ligands, biochemical cues (including gradients), and/or mechanical properties.[362]
Controlling these aspects of cell-material interactions have been explored using synthetic or biological materials that otherwise do not possess cell instructive cues, notably HA or silk fibroin, and using the materials as a blank slate to design cell-matrix interfaces.[365–366] Additional spatial control may be achieved through continued improvement in 3D printing technology such as the printing of gradients of signaling molecules to direct neural stem cell differentiation.[367] Other recent efforts using biocompatible electroconductive materials, such as graphene or polypyrrole, offer another potential option for guiding the formation of neural networks in a 3D system through the use of electrical signaling.[366] This includes combinations of these materials to create novel matrices for neural tissue engineering applications.[368–369] In each of these examples, the engineered materials would serve to improve on the commonly used matrices, which are often undefined (e.g., Matrigel) or with limited complexity (i.e., build of a single polymer, such as collagen). Such an approach would offer the potential to improve the consistency of organoid cultures through the use of tunable and reproducible matrices that also provide the versatility of incorporating multiple signaling cues (biochemical, mechanical, spatial, electrical) as points of control.
To address the formation of necrotic cores during 3D brain tissue development, a key limitation of organoid cultures, engineering approaches may be used to control scaffold architecture to allow the proper exchange of oxygen, nutrients and waste products. One approach has been to expose cerebral organoids in liquid culture to an air interface which augments the survival and axonal outgrowth of neurons, presumably through an increased nutrient supply.[370] For a more sophisticated bioengineering approach, porous scaffolds have been used to support 3D neural tissue models, but the continued development of new fabrication methods using computer-aided design and 3D printing offer unprecedented levels of control for preparing porous scaffolds.[38, 371] Specialized architecture may be used to design suitable implants for regenerative medicine purposes in vivo or to support the long-term in vitro culture of neural tissue engineered constructs, which may be critical for the examination of neurodegenerative diseases such as AD or PD. In addition to scaffold porosity, the other commonly used engineering approaches to support chronic cultures is the use of microfluidic or bioreactor systems. Bioengineered scaffolds, particularly hydrogels, are often limited in culture duration due to concerns over relatively fast degradation rate or restricted oxygen/nutrient diffusion for dense cultures. Early bioreactor systems such as the rotary wall vessel bioreactor have been utilized to extend grow cortex-like tissue constructs without necrotic cores in collagen gels.[372] More recently, microfluidic or microarray technologies have been used, but many cultures are still short-term (less than 1 month). We expect the continued development of bioreactor systems to support longer-term culture will help in recapitulating the pathophysiology of neurological disorders.[205] Furthermore, given the recent interest in the association of brain disease with dysfunction in other physiological systems (e.g., the gut-brain axis), the incorporation of multiple systems in multi-compartment chips or bioreactors will help identify the impact of peripheral tissues in brain neurodegeneration.[373]
The combinatorial nature of neurological function and dysfunction has necessitated innovations in the ability to model interactions between different cell subtypes, brain regions, and other tissues. Brain tissue models with higher complexity have been achieved by fusing different types of spheroids together to study neural circuitry that spans multiple brain regions, such as cortical-hippocampal[374] or dorsal-ventral.[114] Furthermore, as described above, there are now published protocols for differentiating iPSCs into glutamatergic, midbrain and dopaminergic neurons, inhibitory neurons, astrocytes and oligodendrocytes. Through the use of bioengineering approaches, cells can be combined into 3D scaffolds or patterned networks to form circuit connections that can be used to examine functional communication between neuronal subtypes and support cells.[375–376] Furthermore, through the use of cells isolated from healthy or patients afflicted with the disease, researchers can probe mechanisms of circuit dysfunction at the scale of actual tissues in the native 3D environment, and examine genetic or therapeutic approaches to rescuing activity. Another pressing issue in the development of 3D brain models is the use of iPSCs as representative models of neurodegenerative disease. iPSCs have been ‘aged’ through genetic and chemical approaches interfering with specific pathways, such as mitochondria or DNA replication, and by performing direct differentiation of aged fibroblasts to neurons, however these approaches remains artificial, reduce the aging phenotype to a single factor.[377–387]
For each of the technologies described, one of the main goals is to improve the reproducibility of 3D brain models for biomarker or drug screening purposes. In considering systems that will be useful for this application, some key issues include ease of fabrication and use and confounding factors such as non-specific adsorption of therapeutics to materials used in the system. As an example of a reproducible scaffold system, researchers used an automated process to incorporate 3D polystyrene scaffolds into standard 96-well plates and were able to create a high-throughput capable platform for culturing neural stem cells and evaluating outputs in growth, gene expression, and functionality via a calcium assay.[330, 388] Other material-based approaches for high-throughput applications will similarly need to demonstrate the capability for automated production of reproducible scaffolds. Another consideration for drug screening applications, particularly for microfluidic or chip-based approaches, is the non-specific adsorption of therapeutics. Because hydrophobic molecules adsorb to PDMS, alternative materials such as silk, polyurethane or others have been proposed as the starting material for microfluidic systems. As an example, polyurethane elastomers demonstrated similar beneficial optical and mechanical properties to PDMS but offered improved resistance to adsorption of a hydrophobic small molecule dye.[389] Other researchers used poly(ethylene) glycol diacrylate (PEGDA), a synthetic polymer that blocks non-specific protein adsorption, to develop a brain cancer chip for high-throughput drug screening. Using this system to culture glioblastoma cells in a 3D brain cancer model, they were able to examine combinatorial treatments of chemotherapy drugs pitavastatin and irinotecan.[250] Subsequent systems should similarly seek to achieve a similar ease of use and applicability in developing a commercially and clinically promising high-throughput platform.
Another important consideration for commercial and clinical applicability is the integration of technological innovations in electrical or optical interfaces for monitoring 3D brain tissue models in real-time for high-throughput screening applications. While automated fluorescence image-based methods for quantifying brain tissue to classify cell types (neurons, astrocytes, microglia, and endothelial cells) and analyze their associations in cellular networks have been developed,[390] the use of label-free methods for real-time monitoring of cells is generally preferred. Using MEA-based impedance spectroscopy methods, researchers were able to monitor cellular pathology in real-time through the degradation of neurites in response to tau hyperphosphorylation in slice cultures.[391] However, because MEAs generally offer a 2-dimensional measurement of a 3D tissue, recent efforts have sought to use optical methods such as multiphoton microscopy for non-contact and non-invasive functional imaging of bioengineered neural tissues.[392] Wide-volume imaging demonstrated single cell resolution of neuronal activity via the expression of a calcium sensor (GCaMP6s). By using single and layered co-cultures of hippocampal and cortical neurons, researchers were able to examine cell-type specific and interactive neural network activity.[270] Optogenetic techniques are similarly being optimized non-invasive stimulation and recording of electrophysiological activity. The further optimization of technologies such as these for volumetric functional imaging with high resolution will continue to drive neuroscience discovery, particularly for high-throughput screening applications.
7. Conclusion
The emergence of 3-dimensional (3D) in vitro models of the human brain as an alternative to traditional 2-dimensonal (2D) culture systems offers a more representative environment to observe and manipulate cellular behavior. To date, organoid technology or bioengineering approaches have been the primary tools and techniques used to prepare these 3D brain models. Though only in existence for a few years, organoids have proven their utility as a tool to model human development, function, disease and evolution, with early examples of therapeutic applications in neurological and infectious disease and cancer biology. Despite this success, issues with reproducibility, heterogeneity, scalability and necrosis continue to plague these cultures given that their formation and development are largely driven by self-assembly. Bioengineering methodology, on the other hand, has been used to mimic microenvironments of the brain via cell-instructive hydrogels or scaffolding systems and/or connect with other tissues through the use of chip-based microfluidic systems and bioreactors. While these approaches provide for a greater level of control and reproducibility, they often lack the inherent complexity and physiological characteristics of the native microenvironment. Given the state of the field, there is a clear opportunity to combine these approaches to generate advanced 3D in vitro models with greater fidelity to the human brain and enhanced utility in disease modeling, biomarker identification, drug screening, and regenerative medicine. Through the convergence of interdisciplinary fields including biology, neuroscience, biomedical engineering and materials science, among others, we expect that future research will lead to better 3D models of the human brain that are suitable for high-throughput screening and compatible with technology for real-time functional monitoring. Through these efforts, researchers may finally be able to address the current lack of treatment options and offer solutions for patients currently suffering from neurological disease.
Acknowledgments
We thank the NIH (P41EB002520, R01NS092847, R01NS094218) for their support of this work.
Biographies
Thomas Nieland is a Research Associate Professor at The Department of Biomedical Engineering at Tufts University. His research interest is to understand the interactions between different brain cell-types and neural circuits that may lead to neurodegenerative and psychiatric diseases. Bringing unique academic and industry experiences, his ultimate goal is drug discovery using a combination of bioengineering with stem cell technology and systems biology.

David Kaplan is the Stern Family Endowed Professor of Engineering at Tufts University and a Distinguished University Professor. He is Professor and Chair of the Department of Biomedical Engineering. His research focus is on biopolymer engineering to understand structure-function relationships for biomaterials, tissue engineering and regenerative medicine. He is the editor-in-chief of ACS Biomaterials Science and Engineering and he serves on many editorial boards and programs for journals and universities.

References
- [1].Buckner RL, Krienen FM, Trends Cognit. Sci. 2013, 17, 648. [DOI] [PubMed] [Google Scholar]
- [2].Defelipe J, Front. Neuroanat. 2011, 5, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Nestler EJ, Hyman SE, Nat. Neurosci. 2010, 13, 1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pinson A, Namba T, Huttner WB, Front. Cell. Neurosci. 2019, 13, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Laubach M, Amarante LM, Swanson K, White SR, eNeuro 2018, 5, e.0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Andersen RE, Lim DA, Cell Tissue Res. 2018, 371, 55. [DOI] [PubMed] [Google Scholar]
- [7].Bartha I, di Iulio J, Venter JC, Telenti A, Nat. Rev. Genet. 2018, 19, 51. [DOI] [PubMed] [Google Scholar]
- [8].Burns TC, Li MD, Mehta S, Awad AJ, Morgan AA, Eur. J. Pharmacol. 2015, 759, 101. [DOI] [PubMed] [Google Scholar]
- [9].Galatro TF, Holtman IR, Lerario AM, Vainchtein ID, Brouwer N, Sola PR, Veras MM, Pereira TF, Leite REP, Moller T, Wes PD, Sogayar MC, Laman JD, den Dunnen W, Pasqualucci CA, Oba-Shinjo SM, Boddeke E, Marie SKN, Eggen BJL, Nat. Neurosci. 2017, 20, 1162. [DOI] [PubMed] [Google Scholar]
- [10].Gosselin D, Skola D, Coufal NG, Holtman IR, Schlachetzki JCM, Sajti E, Jaeger BN, O’Connor C, Fitzpatrick C, Pasillas MP, Pena M, Adair A, Gonda DD, Levy ML, Ransohoff RM, Gage FH, Glass CK, Science 2017, 356, eaal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL, Jegga AG, Aronow BJ, Lee CK, Bernard A, Glasser MF, Dierker DL, Menche J, Szafer A, Collman F, Grange P, Berman KA, Mihalas S, Yao Z, Stewart L, Barabasi AL, Schulkin J, Phillips J, Ng L, Dang C, Haynor DR, Jones A, Van Essen DC, Koch C, Lein E, Nat. Neurosci. 2015, 18, 1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hodge RD, Bakken TE, Miller JA, Smith KA, Barkan ER, Graybuck LT, Close JL, Long B, Johansen N, Penn O, Yao Z, Eggermont J, Hollt T, Levi BP, Shehata SI, Aevermann B, Beller A, Bertagnolli D, Brouner K, Casper T, Cobbs C, Dalley R, Dee N, Ding SL, Ellenbogen RG, Fong O, Garren E, Goldy J, Gwinn RP, Hirschstein D, Keene CD, Keshk M, Ko AL, Lathia K, Mahfouz A, Maltzer Z, McGraw M, Nguyen TN, Nyhus J, Ojemann JG, Oldre A, Parry S, Reynolds S, Rimorin C, Shapovalova NV, Somasundaram S, Szafer A, Thomsen ER, Tieu M, Quon G, Scheuermann RH, Yuste R, Sunkin SM, Lelieveldt B, Feng D, Ng L, Bernard A, Hawrylycz M, Phillips JW, Tasic B, Zeng H, Jones AR, Koch C, Lein ES, Nature 2019, 573, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, Lonnerberg P, Ryge J, Barker RA, Arenas E, Linnarsson S, Cell 2016, 167, 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Masuda T, Sankowski R, Staszewski O, Bottcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, Coenen VA, Reinacher PC, Michel A, Sure U, Gold R, Grun D, Priller J, Stadelmann C, Prinz M, Nature 2019, 566, 388. [DOI] [PubMed] [Google Scholar]
- [15].Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, Yang N, Danko T, Chen L, Wernig M, Sudhof TC, Neuron 2013, 78, 785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bayes A, Collins MO, Croning MD, van de Lagemaat LN, Choudhary JS, Grant SG, PLoS One 2012, 7, e46683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Bozek K, Wei Y, Yan Z, Liu X, Xiong J, Sugimoto M, Tomita M, Paabo S, Sherwood CC, Hof PR, Ely JJ, Li Y, Steinhauser D, Willmitzer L, Giavalisco P, Khaitovich P, Neuron 2015, 85, 695. [DOI] [PubMed] [Google Scholar]
- [18].Ransohoff RM, J. Exp. Med. 2018, 215, 2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Luft CD, Pereda E, Banissy MJ, Bhattacharya J, Front. Syst. Neurosci. 2014, 8, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Michel CM, Koenig T, NeuroImage 2018, 180, 577. [DOI] [PubMed] [Google Scholar]
- [21].Youngerman BE, Khan FA, McKhann GM, Neuropsychiatr. Dis. Treat. 2019, 15, 1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kaufmann T, van der Meer D, Doan NT, Schwarz E, Lund MJ, Agartz I, Alnaes D, Barch DM, Baur-Streubel R, Bertolino A, Bettella F, Beyer MK, Boen E, Borgwardt S, Brandt CL, Buitelaar J, Celius EG, Cervenka S, Conzelmann A, Cordova-Palomera A, Dale AM, de Quervain DJF, Carlo P, Djurovic S, Dorum ES, Eisenacher S, Elvsashagen T, Espeseth T, Fatouros-Bergman H, Flyckt L, Franke B, Frei O, Haatveit B, Haberg AK, Harbo HF, Hartman CA, Heslenfeld D, Hoekstra PJ, Hogestol EA, Jernigan TL, Jonassen R, Jonsson EG, Karolinska Schizophrenia P, Kirsch P, Kloszewska I, Kolskar KK, Landro NI, Hellard S, Lesch KP, Lovestone S, Lundervold A, Lundervold AJ, Maglanoc LA, Malt UF, Mecocci P, Melle I, Meyer-Lindenberg A, Moberget T, Norbom LB, Nordvik JE, Nyberg L, Oosterlaan J, Papalino M, Papassotiropoulos A, Pauli P, Pergola G, Persson K, Richard G, Rokicki J, Sanders AM, Selbaek G, Shadrin AA, Smeland OB, Soininen H, Sowa P, Steen VM, Tsolaki M, Ulrichsen KM, Vellas B, Wang L, Westman E, Ziegler GC, Zink M, Andreassen OA, Westlye LT, Nat. Neurosci. 2019, 22, 1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rowe RG, Daley GQ, Nat. Rev. Genet. 2019, 20, 377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F, Science 2013, 339, 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA, Cell 2013, 152, 1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E, Science 2012, 337, 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Engreitz J, Abudayyeh O, Gootenberg J, Zhang F, Cold Spring Harbor Perspect. Biol. 2019, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gaj T, Gersbach CA, Barbas CF 3rd, Trends Biotechnol. 2013, 31, 397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Frega M, Tedesco M, Massobrio P, Pesce M, Martinoia S, Sci. Rep. 2014, 4, 5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tekin H, Simmons S, Cummings B, Gao L, Adiconis X, Hession CC, Ghoshal A, Dionne D, Choudhury SR, Yesilyurt V, Sanjana NE, Shi X, Lu C, Heidenreich M, Pan JQ, Levin JZ, Zhang F, Nat. Biomed. Eng. 2018, 2, 540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jo J, Xiao Y, Sun AX, Cukuroglu E, Tran HD, Goke J, Tan ZY, Saw TY, Tan CP, Lokman H, Lee Y, Kim D, Ko HS, Kim SO, Park JH, Cho NJ, Hyde TM, Kleinman JE, Shin JH, Weinberger DR, Tan EK, Je HS, Ng HH, Cell Stem Cell 2016, 19, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim YH, Choi SH, D’Avanzo C, Hebisch M, Sliwinski C, Bylykbashi E, Washicosky KJ, Klee JB, Brustle O, Tanzi RE, Kim DY, Nat. Protoc. 2015, 10, 985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Trujillo CA, Gao R, Negraes PD, Gu J, Buchanan J, Preissl S, Wang A, Wu W, Haddad GG, Chaim IA, Domissy A, Vandenberghe M, Devor A, Yeo GW, Voytek B, Muotri AR, Cell Stem Cell 2019, 558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wen TH, Binder DK, Ethell IM, Razak KA, Front. Mol. Neurosci. 2018, 11, 270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA, Nature 2013, 501, 373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Arlotta P, Pasca SP, Curr. Opin. Neurobiol. 2019, 56, 194. [DOI] [PubMed] [Google Scholar]
- [37].Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, Sasai Y, Proc. Natl. Acad. Sci. U. S. A. 2013, 110, 20284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mahumane GD, Kumar P, du Toit LC, Choonara YE, Pillay V, Biomater. Sci. 2018, 6, 2812. [DOI] [PubMed] [Google Scholar]
- [39].Shafiee A, Atala A, Annu. Rev. Med. 2017, 68, 29. [DOI] [PubMed] [Google Scholar]
- [40].Lancaster MA, Corsini NS, Wolfinger S, Gustafson EH, Phillips AW, Burkard TR, Otani T, Livesey FJ, Knoblich JA, Nat. Biotechnol. 2017, 35, 659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Genon S, Reid A, Langner R, Amunts K, Eickhoff SB, Trends Cognit. Sci. 2018, 22, 350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Schumann CM, Nordahl CW, Brain Res. 2011, 1380, 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Verheijen J, Sleegers K, Trends in genetics : TIG 2018, 34, 434. [DOI] [PubMed] [Google Scholar]
- [44].Del-Aguila JL, Li Z, Dube U, Mihindukulasuriya KA, Budde JP, Fernandez MV, Ibanez L, Bradley J, Wang F, Bergmann K, Davenport R, Morris JC, Holtzman DM, Perrin RJ, Benitez BA, Dougherty J, Cruchaga C, Harari O, Alzheimer’s Res. Ther. 2019, 11, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Toulorge D, Schapira AH, Hajj R, Neurochem J. 2016, 139 Suppl 1, 27. [DOI] [PubMed] [Google Scholar]
- [46].Mallach A, Weinert M, Arthur J, Gveric D, Tierney TS, Alavian KN, FASEB J. 2019, 33, 6957. [DOI] [PubMed] [Google Scholar]
- [47].Tagge CA, Fisher AM, Minaeva OV, Gaudreau-Balderrama A, Moncaster JA, Zhang XL, Wojnarowicz MW, Casey N, Lu H, Kokiko-Cochran ON, Saman S, Ericsson M, Onos KD, Veksler R, Senatorov VV Jr., Kondo A, Zhou XZ, Miry O, Vose LR, Gopaul KR, Upreti C, Nowinski CJ, Cantu RC, Alvarez VE, Hildebrandt AM, Franz ES, Konrad J, Hamilton JA, Hua N, Tripodis Y, Anderson AT, Howell GR, Kaufer D, Hall GF, Lu KP, Ransohoff RM, Cleveland RO, Kowall NW, Stein TD, Lamb BT, Huber BR, Moss WC, Friedman A, Stanton PK, McKee AC, Goldstein LE, Brain : a journal of neurology 2018, 141, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Lee C, Kang EY, Gandal MJ, Eskin E, Geschwind DH, Nat. Neurosci. 2019, 22, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Breen MS, Dobbyn A, Li Q, Roussos P, Hoffman GE, Stahl E, Chess A, Sklar P, Li JB, Devlin B, Buxbaum JD, Nat. Neurosci. 2019, 22, 1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Giridharan VV, Sayana P, Pinjari OF, Ahmad N, da Rosa MI, Quevedo J, Barichello T, Mol. Psychiatry 2020, 25, 94. [DOI] [PubMed] [Google Scholar]
- [51].van Kesteren CF, Gremmels H, de Witte LD, Hol EM, Van Gool AR, Falkai PG, Kahn RS, Sommer IE, Transl. Psychiatry 2017, 7, e1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].McCullumsmith RE, Hammond JH, Shan D, Meador-Woodruff JH, Neuropsychopharmacology 2014, 39, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Freund M, Taylor A, Ng C, Little AR, Handb. Clin. Neurol. 2018, 150, 41. [DOI] [PubMed] [Google Scholar]
- [54].Howieson DB, Dame A, Camicioli R, Sexton G, Payami H, Kaye JA, J. Am. Geriatr. Soc. 1997, 45, 584. [DOI] [PubMed] [Google Scholar]
- [55].Humpel C, Neuroscience 2015, 305, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Croft CL, Wade MA, Kurbatskaya K, Mastrandreas P, Hughes MM, Phillips EC, Pooler AM, Perkinton MS, Hanger DP, Noble W, Cell Death Dis. 2017, 8, e2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Yu Z, Graudejus O, Tsay C, Lacour SP, Wagner S, Morrison B 3rd, Neurotrauma J 2009, 26, 1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Li Q, Han X, Wang J, Mol. Neurobiol. 2016, 53, 4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ooba S, Hasuo H, Shigemori M, Yamashita S, Akasu T, Kurume Med. J. 2009, 56, 49. [DOI] [PubMed] [Google Scholar]
- [60].Harris K, Armstrong SP, Campos-Pires R, Kiru L, Franks NP, Dickinson R, Anesthesiology 2013, 119, 1137. [DOI] [PubMed] [Google Scholar]
- [61].Breder J, Sabelhaus CF, Opitz T, Reymann KG, Schroder UH, Neuropharmacology 2000, 39, 1779. [DOI] [PubMed] [Google Scholar]
- [62].Schroder UH, Breder J, Sabelhaus CF, Reymann KG, Neuropharmacology 1999, 38, 319. [DOI] [PubMed] [Google Scholar]
- [63].Lamprecht MR, Morrison B 3rd, Neurotrauma J 2015, 32, 1361. [DOI] [PubMed] [Google Scholar]
- [64].Shaefi S, Mittel AM, Hyam JA, Boone MD, Chen CC, Kasper EM, Surg. Neurol. Int. 2016, 7, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Marques-Torrejon MA, Gangoso E, Pollard SM, Dis. Models Mech. 2018, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Priego N, Zhu L, Monteiro C, Mulders M, Wasilewski D, Bindeman W, Doglio L, Martinez L, Martinez-Saez E, Ramon YCS, Megias D, Hernandez-Encinas E, Blanco-Aparicio C, Martinez L, Zarzuela E, Munoz J, Fustero-Torre C, Pineiro-Yanez E, Hernandez-Lain A, Bertero L, Poli V, Sanchez-Martinez M, Menendez JA, Soffietti R, Bosch-Barrera J, Valiente M, Nat. Med. 2018, 24, 1024. [DOI] [PubMed] [Google Scholar]
- [67].Eisemann T, Costa B, Strelau J, Mittelbronn M, Angel P, Peterziel H, BMC cancer 2018, 18, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Marksteiner J, Humpel C, Mol. Psychiatry 2008, 13, 939. [DOI] [PubMed] [Google Scholar]
- [69].Romero-Leguizamon CR, Elnagar MR, Kristiansen U, Kohlmeier KA, Sci. Rep. 2019, 9, 1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Schwarz N, Hedrich UBS, Schwarz H, H. P A, Dammeier N, Auffenberg E, Bedogni F, Honegger JB, Lerche H, Wuttke TV, Koch H, Sci. Rep. 2017, 7, 12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Rakotomamonjy J, Ghoumari AM, Int. J. Mol. Sci. 2019, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Jang S, Kim H, Kim HJ, Lee SK, Kim EW, Namkoong K, Kim E, Psychiatry Invest. 2018, 15, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Centeno EGZ, Cimarosti H, Bithell A, Mol. Neurodegener. 2018, 13, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Li L, Chao J, Shi Y, Cell Tissue Res. 2018, 371, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Penney J, Ralvenius WT, Tsai LH, Mol. Psychiatry 2020, 25, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Marioni RE, Harris SE, Zhang Q, McRae AF, Hagenaars SP, Hill WD, Davies G, Ritchie CW, Gale CR, Starr JM, Goate AM, Porteous DJ, Yang J, Evans KL, Deary IJ, Wray NR, Visscher PM, Transl. Psychiatry 2018, 8, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, Sealock J, Karlsson IK, Hagg S, Athanasiu L, Voyle N, Proitsi P, Witoelar A, Stringer S, Aarsland D, Almdahl IS, Andersen F, Bergh S, Bettella F, Bjornsson S, Braekhus A, Brathen G, de Leeuw C, Desikan RS, Djurovic S, Dumitrescu L, Fladby T, Hohman TJ, Jonsson PV, Kiddle SJ, Rongve A, Saltvedt I, Sando SB, Selbaek G, Shoai M, Skene NG, Snaedal J, Stordal E, Ulstein ID, Wang Y, White LR, Hardy J, Hjerling-Leffler J, Sullivan PF, van der Flier WM, Dobson R, Davis LK, Stefansson H, Stefansson K, Pedersen NL, Ripke S, Andreassen OA, Posthuma D, Nat. Genet. 2019, 51, 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, Boland A, Vronskaya M, van der Lee SJ, Amlie-Wolf A, Bellenguez C, Frizatti A, Chouraki V, Martin ER, Sleegers K, Badarinarayan N, Jakobsdottir J, Hamilton-Nelson KL, Moreno-Grau S, Olaso R, Raybould R, Chen Y, Kuzma AB, Hiltunen M, Morgan T, Ahmad S, Vardarajan BN, Epelbaum J, Hoffmann P, Boada M, Beecham GW, Garnier JG, Harold D, Fitzpatrick AL, Valladares O, Moutet ML, Gerrish A, Smith AV, Qu L, Bacq D, Denning N, Jian X, Zhao Y, Del Zompo M, Fox NC, Choi SH, Mateo I, Hughes JT, Adams HH, Malamon J, Sanchez-Garcia F, Patel Y, Brody JA, Dombroski BA, Naranjo MCD, Daniilidou M, Eiriksdottir G, Mukherjee S, Wallon D, Uphill J, Aspelund T, Cantwell LB, Garzia F, Galimberti D, Hofer E, Butkiewicz M, Fin B, Scarpini E, Sarnowski C, Bush WS, Meslage S, Kornhuber J, White CC, Song Y, Barber RC, Engelborghs S, Sordon S, Voijnovic D, Adams PM, Vandenberghe R, Mayhaus M, Cupples LA, Albert MS, De Deyn PP, Gu W, Himali JJ, Beekly D, Squassina A, Hartmann AM, Orellana A, Blacker D, Rodriguez-Rodriguez E, Lovestone S, Garcia ME, Doody RS, Munoz-Fernadez C, Sussams R, Lin H, Fairchild TJ, Benito YA, Holmes C, Karamujic-Comic H, Frosch MP, Thonberg H, Maier W, Roshchupkin G, Ghetti B, Giedraitis V, Kawalia A, Li S, Huebinger RM, Kilander L, Moebus S, Hernandez I, Kamboh MI, Brundin R, Turton J, Yang Q, Katz MJ, Concari L, Lord J, Beiser AS, Keene CD, Helisalmi S, Kloszewska I, Kukull WA, Koivisto AM, Lynch A, Tarraga L, Larson EB, Haapasalo A, Lawlor B, Mosley TH, Lipton RB, Solfrizzi V, Gill M, Longstreth WT Jr., Montine TJ, Frisardi V, Diez-Fairen M, Rivadeneira F, Petersen RC, Deramecourt V, Alvarez I, Salani F, Ciaramella A, Boerwinkle E, Reiman EM, Fievet N, Rotter JI, Reisch JS, Hanon O, Cupidi C, Andre Uitterlinden AG, Royall DR, Dufouil C, Maletta RG, de Rojas I, Sano M, Brice A, Cecchetti R, George-Hyslop PS, Ritchie K, Tsolaki M, Tsuang DW, Dubois B, Craig D, Wu CK, Soininen H, Avramidou D, Albin RL, Fratiglioni L, Germanou A, Apostolova LG, Keller L, Koutroumani M, Arnold SE, Panza F, Gkatzima O, Asthana S, Hannequin D, Whitehead P, Atwood CS, Caffarra P, Hampel H, Quintela I, Carracedo A, Lannfelt L, Rubinsztein DC, Barnes LL, Pasquier F, Frolich L, Barral S, McGuinness B, Beach TG, Johnston JA, Becker JT, Passmore P, Bigio EH, Schott JM, Bird TD, Warren JD, Boeve BF, Lupton MK, Bowen JD, Proitsi P, Boxer A, Powell JF, Burke JR, Kauwe JSK, Burns JM, Mancuso M, Buxbaum JD, Bonuccelli U, Cairns NJ, McQuillin A, Cao C, Livingston G, Carlson CS, Bass NJ, Carlsson CM, Hardy J, Carney RM, Bras J, Carrasquillo MM, Guerreiro R, Allen M, Chui HC, Fisher E, Masullo C, Crocco EA, DeCarli C, Bisceglio G, Dick M, Ma L, Duara R, Graff-Radford NR, Evans DA, Hodges A, Faber KM, Scherer M, Fallon KB, Riemenschneider M, Fardo DW, Heun R, Farlow MR, Kolsch H, Ferris S, Leber M, Foroud TM, Heuser I, Galasko DR, Giegling I, Gearing M, Hull M, Geschwind DH, Gilbert JR, Morris J, Green RC, Mayo K, Growdon JH, Feulner T, Hamilton RL, Harrell LE, Drichel D, Honig LS, Cushion TD, Huentelman MJ, Hollingworth P, Hulette CM, Hyman BT, Marshall R, Jarvik GP, Meggy A, Abner E, Menzies GE, Jin LW, Leonenko G, Real LM, Jun GR, Baldwin CT, Grozeva D, Karydas A, Russo G, Kaye JA, Kim R, Jessen F, Kowall NW, Vellas B, Kramer JH, Vardy E, LaFerla FM, Jockel KH, Lah JJ, Dichgans M, Leverenz JB, Mann D, Levey AI, Pickering-Brown S, Lieberman AP, Klopp N, Lunetta KL, Wichmann HE, Lyketsos CG, Morgan K, Marson DC, Brown K, Martiniuk F, Medway C, Mash DC, Nothen MM, Masliah E, Hooper NM, McCormick WC, Daniele A, McCurry SM, Bayer A, McDavid AN, Gallacher J, McKee AC, van den Bussche H, Mesulam M, Brayne C, Miller BL, Riedel-Heller S, Miller CA, Miller JW, Al-Chalabi A, Morris JC, Shaw CE, Myers AJ, Wiltfang J, O’Bryant S, Olichney JM, Alvarez V, Parisi JE, Singleton AB, Paulson HL, Collinge J, Perry WR, Mead S, Peskind E, Cribbs DH, Rossor M, Pierce A, Ryan NS, Poon WW, Nacmias B, Potter H, Sorbi S, Quinn JF, Sacchinelli E, Raj A, Spalletta G, Raskind M, Caltagirone C, Bossu P, Orfei MD, Reisberg B, Clarke R, Reitz C, Smith AD, Ringman JM, Warden D, Roberson ED, Wilcock G, Rogaeva E, Bruni AC, Rosen HJ, Gallo M, Rosenberg RN, Ben-Shlomo Y, Sager MA, Mecocci P, Saykin AJ, Pastor P, Cuccaro ML, Vance JM, Schneider JA, Schneider LS, Slifer S, Seeley WW, Smith AG, Sonnen JA, Spina S, Stern RA, Swerdlow RH, Tang M, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Van Eldik LJ, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Wilhelmsen KC, Williamson J, Wingo TS, Woltjer RL, Wright CB, Yu CE, Yu L, Saba Y, Pilotto A, Bullido MJ, Peters O, Crane PK, Bennett D, Bosco P, Coto E, Boccardi V, De Jager PL, Lleo A, Warner N, Lopez OL, Ingelsson M, Deloukas P, Cruchaga C, Graff C, Gwilliam R, Fornage M, Goate AM, Sanchez-Juan P, Kehoe PG, Amin N, Ertekin-Taner N, Berr C, Debette S, Love S, Launer LJ, Younkin SG, Dartigues JF, Corcoran C, Ikram MA, Dickson DW, Nicolas G, Campion D, Tschanz J, Schmidt H, Hakonarson H, Clarimon J, Munger R, Schmidt R, Farrer LA, Van Broeckhoven C, C. O. D. M, DeStefano AL, Jones L, Haines JL, Deleuze JF, Owen MJ, Gudnason V, Mayeux R, Escott-Price V, Psaty BM, Ramirez A, Wang LS, Ruiz A, van Duijn CM, Holmans PA, Seshadri S, Williams J, Amouyel P, Schellenberg GD, Lambert JC, Pericak-Vance MA, Nat. Genet. 2019, 51, 414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lee HK, Velazquez Sanchez C, Chen M, Morin PJ, Wells JM, Hanlon EB, Xia W, PLoS One 2016, 11, e0163072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Sproul AA, Jacob S, Pre D, Kim SH, Nestor MW, Navarro-Sobrino M, Santa-Maria I, Zimmer M, Aubry S, Steele JW, Kahler DJ, Dranovsky A, Arancio O, Crary JF, Gandy S, Noggle SA, PLoS One 2014, 9, e84547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N, Hum. Mol. Genet. 2011, 20, 4530. [DOI] [PubMed] [Google Scholar]
- [82].Deng HX, Shi Y, Yang Y, Ahmeti KB, Miller N, Huang C, Cheng L, Zhai H, Deng S, Nuytemans K, Corbett NJ, Kim MJ, Deng H, Tang B, Yang Z, Xu Y, Chan P, Huang B, Gao XP, Song Z, Liu Z, Fecto F, Siddique N, Foroud T, Jankovic J, Ghetti B, Nicholson DA, Krainc D, Melen O, Vance JM, Pericak-Vance MA, Ma YC, Rajput AH, Siddique T, Nat. Genet. 2016, 48, 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Fernandez-Santiago R, Carballo-Carbajal I, Castellano G, Torrent R, Richaud Y, Sanchez-Danes A, Vilarrasa-Blasi R, Sanchez-Pla A, Mosquera JL, Soriano J, Lopez-Barneo J, Canals JM, Alberch J, Raya A, Vila M, Consiglio A, Martin-Subero JI, Ezquerra M, Tolosa E, EMBO Mol. Med. 2015, 7, 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Mehta SR, Tom CM, Wang Y, Bresee C, Rushton D, Mathkar PP, Tang J, Mattis VB, Cell Rep. 2018, 25, 1081. [DOI] [PubMed] [Google Scholar]
- [85].Russo FB, Brito A, de Freitas AM, Castanha A, de Freitas BC, Beltrao-Braga PCB, Neurobiol. Dis. 2019, 130, 104483. [DOI] [PubMed] [Google Scholar]
- [86].Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D’Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, Zhang C, Wainger BJ, Peitz M, Kovacs DM, Woolf CJ, Wagner SL, Tanzi RE, Kim DY, Nature 2014, 515, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y, Cell Stem Cell 2012, 10, 771. [DOI] [PubMed] [Google Scholar]
- [88].Eiraku M, Takata N, Ishibashi H, Kawada M, Sakakura E, Okuda S, Sekiguchi K, Adachi T, Sasai Y, Nature 2011, 472, 51. [DOI] [PubMed] [Google Scholar]
- [89].Suga H, Kadoshima T, Minaguchi M, Ohgushi M, Soen M, Nakano T, Takata N, Wataya T, Muguruma K, Miyoshi H, Yonemura S, Oiso Y, Sasai Y, Nature 2011, 480, 57. [DOI] [PubMed] [Google Scholar]
- [90].Sato T, Clevers H, Science 2013, 340, 1190. [DOI] [PubMed] [Google Scholar]
- [91].Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, Clevers H, Nature 2009, 459, 262. [DOI] [PubMed] [Google Scholar]
- [92].Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, Shroyer NF, Wells JM, Nature 2011, 470, 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Lancaster MA, Knoblich JA, Nat. Protoc. 2014, 9, 2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, Boyden ES, Lichtman JW, Williams ZM, McCarroll SA, Arlotta P, Nature 2017, 545, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Brauninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, Knoblich JA, Lachmann R, Paabo S, Huttner WB, Treutlein B, Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 15672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Luo C, Lancaster MA, Castanon R, Nery JR, Knoblich JA, Ecker JR, Cell Rep. 2016, 17, 3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Subramanian L, Bershteyn M, Paredes MF, Kriegstein AR, Nat. Commun. 2017, 8, 14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park JY, O’Rourke NA, Nguyen KD, Smith SJ, Huguenard JR, Geschwind DH, Barres BA, Pasca SP, Nat. Methods 2015, 12, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Otani T, Marchetto MC, Gage FH, Simons BD, Livesey FJ, Cell Stem Cell 2016, 18, 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Mora-Bermudez F, Badsha F, Kanton S, Camp JG, Vernot B, Kohler K, Voigt B, Okita K, Maricic T, He Z, Lachmann R, Paabo S, Treutlein B, Huttner WB, eLife 2016, 5, e18683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Li Y, Muffat J, Omer A, Bosch I, Lancaster MA, Sur M, Gehrke L, Knoblich JA, Jaenisch R, Cell Stem Cell 2017, 20, 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Cardenas A, Villalba A, de Juan Romero C, Pico E, Kyrousi C, Tzika AC, Tessier-Lavigne M, Ma L, Drukker M, Cappello S, Borrell V, Cell 2018, 174, 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Fiddes IT, Lodewijk GA, Mooring M, Bosworth CM, Ewing AD, Mantalas GL, Novak AM, van den Bout A, Bishara A, Rosenkrantz JL, Lorig-Roach R, Field AR, Haeussler M, Russo L, Bhaduri A, Nowakowski TJ, Pollen AA, Dougherty ML, Nuttle X, Addor MC, Zwolinski S, Katzman S, Kriegstein A, Eichler EE, Salama SR, Jacobs FMJ, Haussler D, Cell 2018, 173, 1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].O’Neill AC, Kyrousi C, Klaus J, Leventer RJ, Kirk EP, Fry A, Pilz DT, Morgan T, Jenkins ZA, Drukker M, Berkovic SF, Scheffer IE, Guerrini R, Markie DM, Gotz M, Cappello S, Robertson SP, Cell Rep. 2018, 25, 2729. [DOI] [PubMed] [Google Scholar]
- [105].Amiri A, Coppola G, Scuderi S, Wu F, Roychowdhury T, Liu F, Pochareddy S, Shin Y, Safi A, Song L, Zhu Y, Sousa AMM, Psych EC, Gerstein M, Crawford GE, Sestan N, Abyzov A, Vaccarino FM, Science 2018, 362, eaat6720.30545853 [Google Scholar]
- [106].Wang L, Hou S, Han YG, Nat. Neurosci. 2016, 19, 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Lindborg BA, Brekke JH, Vegoe AL, Ulrich CB, Haider KT, Subramaniam S, Venhuizen SL, Eide CR, Orchard PJ, Chen W, Wang Q, Pelaez F, Scott CM, Kokkoli E, Keirstead SA, Dutton JR, Tolar J, O’Brien TD, Stem Cells Transl. Med. 2016, 5, 970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Mariani J, Coppola G, Zhang P, Abyzov A, Provini L, Tomasini L, Amenduni M, Szekely A, Palejev D, Wilson M, Gerstein M, Grigorenko EL, Chawarska K, Pelphrey KA, Howe JR, Vaccarino FM, Cell 2015, 162, 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, Yoon KJ, Jeang W, Lin L, Li Y, Thakor J, Berg DA, Zhang C, Kang E, Chickering M, Nauen D, Ho CY, Wen Z, Christian KM, Shi PY, Maher BJ, Wu H, Jin P, Tang H, Song H, Ming GL, Cell 2016, 165, 1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Monzel AS, Smits LM, Hemmer K, Hachi S, Moreno EL, van Wuellen T, Jarazo J, Walter J, Bruggemann I, Boussaad I, Berger E, Fleming RMT, Bolognin S, Schwamborn JC, Stem Cell Rep. 2017, 8, 1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Bagley JA, Reumann D, Bian S, Levi-Strauss J, Knoblich JA, Nat. Methods 2017, 14, 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Qian X, Jacob F, Song MM, Nguyen HN, Song H, Ming GL, Nat. Protoc. 2018, 13, 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Yan Y, Song L, Madinya J, Ma T, Li Y, Tissue Eng., Part A 2018, 24, 418. [DOI] [PubMed] [Google Scholar]
- [114].Birey F, Andersen J, Makinson CD, Islam S, Wei W, Huber N, Fan HC, Metzler KRC, Panagiotakos G, Thom N, O’Rourke NA, Steinmetz LM, Bernstein JA, Hallmayer J, Huguenard JR, Pasca SP, Nature 2017, 545, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Yoon SJ, Elahi LS, Pasca AM, Marton RM, Gordon A, Revah O, Miura Y, Walczak EM, Holdgate GM, Fan HC, Huguenard JR, Geschwind DH, Pasca SP, Nat. Methods 2019, 16, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Sloan SA, Andersen J, Pasca AM, Birey F, Pasca SP, Nat. Protoc. 2018, 13, 2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Tieng V, Stoppini L, Villy S, Fathi M, Dubois-Dauphin M, Krause KH, Stem Cells Dev. 2014, 23, 1535. [DOI] [PubMed] [Google Scholar]
- [118].Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, Levin JZ, Arlotta P, Nature 2019, 570, 523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Schukking M, Miranda HC, Trujillo CA, Negraes PD, Muotri AR, Stem Cells Dev. 2018, 27, 1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Sohal VS, Rubenstein JLR, Mol. Psychiatry 2019, 24, 1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ariani F, Hayek G, Rondinella D, Artuso R, Mencarelli MA, Spanhol-Rosseto A, Pollazzon M, Buoni S, Spiga O, Ricciardi S, Meloni I, Longo I, Mari F, Broccoli V, Zappella M, Renieri A, Am. J. Hum. Genet. 2008, 83, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Bahi-Buisson N, Nectoux J, Girard B, Van Esch H, De Ravel T, Boddaert N, Plouin P, Rio M, Fichou Y, Chelly J, Bienvenu T, Neurogenetics 2010, 11, 241. [DOI] [PubMed] [Google Scholar]
- [123].Shoichet SA, Kunde SA, Viertel P, Schell-Apacik C, von Voss H, Tommerup N, Ropers HH, Kalscheuer VM, Hum. Genet. 2005, 117, 536. [DOI] [PubMed] [Google Scholar]
- [124].Mellios N, Feldman DA, Sheridan SD, Ip JPK, Kwok S, Amoah SK, Rosen B, Rodriguez BA, Crawford B, Swaminathan R, Chou S, Li Y, Ziats M, Ernst C, Jaenisch R, Haggarty SJ, Sur M, Mol. Psychiatry 2018, 23, 1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, Di Lullo E, Alvarado B, Bedolli M, Dougherty ML, Fiddes IT, Kronenberg ZN, Shuga J, Leyrat AA, West JA, Bershteyn M, Lowe CB, Pavlovic BJ, Salama SR, Haussler D, Eichler EE, Kriegstein AR, Cell 2019, 176, 743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Field AR, Jacobs FMJ, Fiddes IT, Phillips APR, Reyes-Ortiz AM, LaMontagne E, Whitehead L, Meng V, Rosenkrantz JL, Olsen M, Hauessler M, Katzman S, Salama SR, Haussler D, Stem Cell Rep. 2019, 12, 245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Yang X, Xu B, Mulvey B, Evans M, Jordan S, Wang YD, Pagala V, Peng J, Fan Y, Patel A, Peng JC, Nat. Neurosci. 2019, 22, 362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Perez-Branguli F, Buchsbaum IY, Pozner T, Regensburger M, Fan W, Schray A, Borstler T, Mishra H, Graf D, Kohl Z, Winkler J, Berninger B, Cappello S, Winner B, Hum. Mol. Genet. 2019, 28, 961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Klaus J, Kanton S, Kyrousi C, Ayo-Martin AC, Di Giaimo R, Riesenberg S, O’Neill AC, Camp JG, Tocco C, Santel M, Rusha E, Drukker M, Schroeder M, Gotz M, Robertson SP, Treutlein B, Cappello S, Nat. Med. 2019, 25, 561. [DOI] [PubMed] [Google Scholar]
- [130].Thomas CA, Tejwani L, Trujillo CA, Negraes PD, Herai RH, Mesci P, Macia A, Crow YJ, Muotri AR, Cell Stem Cell 2017, 21, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Stachowiak EK, Benson CA, Narla ST, Dimitri A, Chuye LEB, Dhiman S, Harikrishnan K, Elahi S, Freedman D, Brennand KJ, Sarder P, Stachowiak MK, Transl. Psychiatry 2017, 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Coulter ME, Dorobantu CM, Lodewijk GA, Delalande F, Cianferani S, Ganesh VS, Smith RS, Lim ET, Xu CS, Pang S, Wong ET, Lidov HGW, Calicchio ML, Yang E, Gonzalez DM, Schlaeger TM, Mochida GH, Hess H, Lee WA, Lehtinen MK, Kirchhausen T, Haussler D, Jacobs FMJ, Gaudin R, Walsh CA, Cell Rep. 2018, 24, 973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Conforti P, Besusso D, Bocchi VD, Faedo A, Cesana E, Rossetti G, Ranzani V, Svendsen CN, Thompson LM, Toselli M, Biella G, Pagani M, Cattaneo E, Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Wang P, Mokhtari R, Pedrosa E, Kirschenbaum M, Bayrak C, Zheng D, Lachman HM, Mol. Autism 2017, 8, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Siu MT, Butcher DT, Turinsky AL, Cytrynbaum C, Stavropoulos DJ, Walker S, Caluseriu O, Carter M, Lou Y, Nicolson R, Georgiades S, Szatmari P, Anagnostou E, Scherer SW, Choufani S, Brudno M, Weksberg R, Clin. Epigenet. 2019, 11, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Yoon KJ, Ringeling FR, Vissers C, Jacob F, Pokrass M, Jimenez-Cyrus D, Su Y, Kim NS, Zhu Y, Zheng L, Kim S, Wang X, Dore LC, Jin P, Regot S, Zhuang X, Canzar S, He C, Ming GL, Song H, Cell 2017, 171, 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Allende ML, Cook EK, Larman BC, Nugent A, Brady JM, Golebiowski D, Sena-Esteves M, Tifft CJ, Proia RL, J. Lipid Res. 2018, 59, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Jin M, Pomp O, Shinoda T, Toba S, Torisawa T, Furuta K, Oiwa K, Yasunaga T, Kitagawa D, Matsumura S, Miyata T, Tan TT, Reversade B, Hirotsune S, Sci. Rep. 2017, 7, 39902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Iefremova V, Manikakis G, Krefft O, Jabali A, Weynans K, Wilkens R, Marsoner F, Brandl B, Muller FJ, Koch P, Ladewig J, Cell Rep. 2017, 19, 50. [DOI] [PubMed] [Google Scholar]
- [140].Bershteyn M, Nowakowski TJ, Pollen AA, Di Lullo E, Nene A, Wynshaw-Boris A, Kriegstein AR, Cell Stem Cell 2017, 20, 435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Li R, Sun L, Fang A, Li P, Wu Q, Wang X, Protein Cell 2017, 8, 823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Matsui T, Nieto-Estevez V, Kyrychenko S, Schneider JW, Hsieh J, Development 2017, 144, 1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Xiang Y, Tanaka Y, Patterson B, Kang YJ, Govindaiah G, Roselaar N, Cakir B, Kim KY, Lombroso AP, Hwang SM, Zhong M, Stanley EG, Elefanty AG, Naegele JR, Lee SH, Weissman SM, Park IH, Cell Stem Cell 2017, 21, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Tejero R, Huang Y, Katsyv I, Kluge M, Lin JY, Tome-Garcia J, Daviaud N, Wang Y, Zhang B, Tsankova NM, Friedel CC, Zou H, Friedel RH, EBioMedicine 2019, 42, 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA, Pasca SP, Neuron 2017, 95, 779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Ye F, Kang E, Yu C, Qian X, Jacob F, Yu C, Mao M, Poon RYC, Kim J, Song H, Ming GL, Zhang M, Neuron 2017, 96, 1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Srikanth P, Lagomarsino VN, Muratore CR, Ryu SC, He A, Taylor WM, Zhou C, Arellano M, Young-Pearse TL, Transl. Psychiatry 2018, 8, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Raja WK, Mungenast AE, Lin YT, Ko T, Abdurrob F, Seo J, Tsai LH, PLoS One 2016, 11, e0161969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Gonzalez C, Armijo E, Bravo-Alegria J, Becerra-Calixto A, Mays CE, Soto C, Mol. Psychiatry 2018, 23, 2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Pavoni S, Jarray R, Nassor F, Guyot AC, Cottin S, Rontard J, Mikol J, Mabondzo A, Deslys JP, Yates F, PLoS One 2018, 13, e0209150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Yan Y, Song L, Bejoy J, Zhao J, Kanekiyo T, Bu G, Zhou Y, Li Y, Tissue Eng., Part A 2018, 24, 1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Xu R, Brawner AT, Li S, Liu JJ, Kim H, Xue H, Pang ZP, Kim WY, Hart RP, Liu Y, Jiang P, Cell Stem Cell 2019, 24, 908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Lin YT, Seo J, Gao F, Feldman HM, Wen HL, Penney J, Cam HP, Gjoneska E, Raja WK, Cheng J, Rueda R, Kritskiy O, Abdurrob F, Peng Z, Milo B, Yu CJ, Elmsaouri S, Dey D, Ko T, Yankner BA, Tsai LH, Neuron 2018, 98, 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Meyer K, Feldman HM, Lu T, Drake D, Lim ET, Ling KH, Bishop NA, Pan Y, Seo J, Lin YT, Su SC, Church GM, Tsai LH, Yankner BA, Cell Rep. 2019, 26, 1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Boisvert EM, Means RE, Michaud M, Madri JA, Katz SG, Cell Death Dis. 2019, 10, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Pasca AM, Park JY, Shin HW, Qi Q, Revah O, Krasnoff R, O’Hara R, Willsey AJ, Palmer TD, Pasca SP, Nat. Med. 2019, 25, 784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Sartore RC, Cardoso SC, Lages YV, Paraguassu JM, Stelling MP, Madeiro da Costa RF, Guimaraes MZ, Perez CA, Rehen SK, PeerJ 2017, 5, e2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Liu F, Huang J, Liu Z, Neuroscience 2019, 404, 530. [DOI] [PubMed] [Google Scholar]
- [159].Lee CT, Chen J, Kindberg AA, Bendriem RM, Spivak CE, Williams MP, Richie CT, Handreck A, Mallon BS, Lupica CR, Lin DT, Harvey BK, Mash DC, Freed WJ, Neuropsychopharmacology 2017, 42, 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Wang Y, Wang L, Zhu Y, Qin J, Lab Chip 2018, 18, 851. [DOI] [PubMed] [Google Scholar]
- [161].Zhu Y, Wang L, Yin F, Yu Y, Wang Y, Shepard MJ, Zhuang Z, Qin J, Integr. Biol. 2017, 9, 968. [DOI] [PubMed] [Google Scholar]
- [162].Huang J, Liu F, Tang H, Wu H, Li L, Wu R, Zhao J, Wu Y, Liu Z, Chen J, Front. Neurol. 2017, 8, 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Dakic V, Minardi Nascimento J, Costa Sartore R, Maciel RM, de Araujo DB, Ribeiro S, Martins-de-Souza D, Rehen SK, Sci. Rep. 2017, 7, 12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Groveman BR, Foliaki ST, Orru CD, Zanusso G, Carroll JA, Race B, Haigh CL, Acta Neuropathol. Commun. 2019, 7, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].D’Aiuto L, Bloom DC, Naciri JN, Smith A, Edwards TG, McClain L, Callio JA, Jessup M, Wood J, Chowdari K, Demers M, Abrahamson EE, Ikonomovic MD, Viggiano L, De Zio R, Watkins S, Kinchington PR, Nimgaonkar VL, J. Virol. 2019, 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Muffat J, Li Y, Omer A, Durbin A, Bosch I, Bakiasi G, Richards E, Meyer A, Gehrke L, Jaenisch R, Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Zhang B, He Y, Xu Y, Mo F, Mi T, Shen QS, Li C, Li Y, Liu J, Wu Y, Chen G, Zhu W, Qin C, Hu B, Zhou G, Cell Death Dis. 2018, 9, 719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK, Science 2016, 352, 816. [DOI] [PubMed] [Google Scholar]
- [169].Yoon KJ, Song G, Qian X, Pan J, Xu D, Rho HS, Kim NS, Habela C, Zheng L, Jacob F, Zhang F, Lee EM, Huang WK, Ringeling FR, Vissers C, Li C, Yuan L, Kang K, Kim S, Yeo J, Cheng Y, Liu S, Wen Z, Qin CF, Wu Q, Christian KM, Tang H, Jin P, Xu Z, Qian J, Zhu H, Song H, Ming GL, Cell Stem Cell 2017, 21, 349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, Polonio CM, Cunha I, Freitas CL, Brandao WN, Rossato C, Andrade DG, Faria Dde P, Garcez AT, Buchpigel CA, Braconi CT, Mendes E, Sall AA, Zanotto PM, Peron JP, Muotri AR, Beltrao-Braga PC, Nature 2016, 534, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, Kuivanen S, Korhonen E, Smura T, Vapalahti O, Papantonis A, Schmidt-Chanasit J, Riparbelli M, Callaini G, Kronke M, Utermohlen O, Gopalakrishnan J, Cell Stem Cell 2017, 20, 397. [DOI] [PubMed] [Google Scholar]
- [172].Setoh YX, Amarilla AA, Peng NYG, Griffiths RE, Carrera J, Freney ME, Nakayama E, Ogawa S, Watterson D, Modhiran N, Nanyonga FE, Torres FJ, Slonchak A, Periasamy P, Prow NA, Tang B, Harrison J, Hobson-Peters J, Cuddihy T, Cooper-White J, Hall RA, Young PR, Mackenzie JM, Wolvetang E, Bloom JD, Suhrbier A, Khromykh AA, Nat. Microbiol. 2019, 4, 876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Janssens S, Schotsaert M, Karnik R, Balasubramaniam V, Dejosez M, Meissner A, Garcia-Sastre A, Zwaka TP, mSystems 2018, 3, e00219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Wells MF, Salick MR, Wiskow O, Ho DJ, Worringer KA, Ihry RJ, Kommineni S, Bilican B, Klim JR, Hill EJ, Kane LT, Ye C, Kaykas A, Eggan K, Cell Stem Cell 2016, 19, 703. [DOI] [PubMed] [Google Scholar]
- [175].Watanabe M, Buth JE, Vishlaghi N, de la Torre-Ubieta L, Taxidis J, Khakh BS, Coppola G, Pearson CA, Yamauchi K, Gong D, Dai X, Damoiseaux R, Aliyari R, Liebscher S, Schenke-Layland K, Caneda C, Huang EJ, Zhang Y, Cheng G, Geschwind DH, Golshani P, Sun R, Novitch BG, Cell Rep. 2017, 21, 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, Jacob F, Nguyen HN, Itkin M, Hanna C, Shinn P, Allen C, Michael SG, Simeonov A, Huang W, Christian KM, Goate A, Brennand KJ, Huang R, Xia M, Ming GL, Zheng W, Song H, Tang H, Nat. Med. 2016, 22, 1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Sacramento CQ, de Melo GR, de Freitas CS, Rocha N, Hoelz LV, Miranda M, Fintelman-Rodrigues N, Marttorelli A, Ferreira AC, Barbosa-Lima G, Abrantes JL, Vieira YR, Bastos MM, de Mello Volotao E, Nunes EP, Tschoeke DA, Leomil L, Loiola EC, Trindade P, Rehen SK, Bozza FA, Bozza PT, Boechat N, Thompson FL, de Filippis AM, Bruning K, Souza TM, Sci. Rep. 2017, 7, 40920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Li C, Deng YQ, Wang S, Ma F, Aliyari R, Huang XY, Zhang NN, Watanabe M, Dong HL, Liu P, Li XF, Ye Q, Tian M, Hong S, Fan J, Zhao H, Li L, Vishlaghi N, Buth JE, Au C, Liu Y, Lu N, Du P, Qin FX, Zhang B, Gong D, Dai X, Sun R, Novitch BG, Xu Z, Qin CF, Cheng G, Immunity 2017, 46, 446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Zhou T, Tan L, Cederquist GY, Fan Y, Hartley BJ, Mukherjee S, Tomishima M, Brennand KJ, Zhang Q, Schwartz RE, Evans T, Studer L, Chen S, Cell Stem Cell 2017, 21, 274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Xu YP, Qiu Y, Zhang B, Chen G, Chen Q, Wang M, Mo F, Xu J, Wu J, Zhang RR, Cheng ML, Zhang NN, Lyu B, Zhu WL, Wu MH, Ye Q, Zhang D, Man JH, Li XF, Cui J, Xu Z, Hu B, Zhou X, Qin CF, Cell Res. 2019, 29, 265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Sakaguchi H, Ozaki Y, Ashida T, Matsubara T, Oishi N, Kihara S, Takahashi J, Stem Cell Rep. 2019, 13, 458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Poli D, Magliaro C, Ahluwalia A, Front. Neurosci. 2019, 13, 162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Zhao Y, Liu P, Zhang N, Chen J, Landegger LD, Wu L, Zhao F, Zhao Y, Zhang Y, Zhang J, Fujita T, Stemmer-Rachamimov A, Ferraro GB, Liu H, Muzikansky A, Plotkin SR, Stankovic KM, Jain RK, Xu L, Proc. Natl. Acad. Sci. U. S. A. 2018, 115, E2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Bian S, Repic M, Guo Z, Kavirayani A, Burkard T, Bagley JA, Krauditsch C, Knoblich JA, Nat. Methods 2018, 15, 631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Decaesteker B, Denecker G, Van Neste C, Dolman EM, Van Loocke W, Gartlgruber M, Nunes C, De Vloed F, Depuydt P, Verboom K, Rombaut D, Loontiens S, De Wyn J, Kholosy WM, Koopmans B, Essing AHW, Herrmann C, Dreidax D, Durinck K, Deforce D, Van Nieuwerburgh F, Henssen A, Versteeg R, Boeva V, Schleiermacher G, van Nes J, Mestdagh P, Vanhauwaert S, Schulte JH, Westermann F, Molenaar JJ, De Preter K, Speleman F, Nat. Commun. 2018, 9, 4866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Bergmann S, Lawler SE, Qu Y, Fadzen CM, Wolfe JM, Regan MS, Pentelute BL, Agar NYR, Cho CF, Nat. Protoc. 2018, 13, 2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Xiang Y, Yoshiaki T, Patterson B, Cakir B, Kim KY, Cho YS, Park IH, Curr. Protoc. Stem Cell Biol. 2018, 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Marton RM, Miura Y, Sloan SA, Li Q, Revah O, Levy RJ, Huguenard JR, Pasca SP, Nat. Neurosci. 2019, 22, 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Kim H, Xu R, Padmashri R, Dunaevsky A, Liu Y, Dreyfus CF, Jiang P, Stem Cell Rep. 2019, 12, 890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Carter BS, Bova GS, Beaty TH, Steinberg GD, Childs B, Isaacs WB, Walsh PC, J. Urol. 1993, 150, 797. [DOI] [PubMed] [Google Scholar]
- [191].Abud EM, Ramirez RN, Martinez ES, Healy LM, Nguyen CHH, Newman SA, Yeromin AV, Scarfone VM, Marsh SE, Fimbres C, Caraway CA, Fote GM, Madany AM, Agrawal A, Kayed R, Gylys KH, Cahalan MD, Cummings BJ, Antel JP, Mortazavi A, Carson MJ, Poon WW, Blurton-Jones M, Neuron 2017, 94, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Ormel PR, Vieira de Sa R, van Bodegraven EJ, Karst H, Harschnitz O, Sneeboer MAM, Johansen LE, van Dijk RE, Scheefhals N, Berdenis van Berlekom A, Ribes Martinez E, Kling S, MacGillavry HD, van den Berg LH, Kahn RS, Hol EM, de Witte LD, Pasterkamp RJ, Nat. Commun. 2018, 9, 4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Xiang Y, Tanaka Y, Cakir B, Patterson B, Kim KY, Sun P, Kang YJ, Zhong M, Liu X, Patra P, Lee SH, Weissman SM, Park IH, Cell Stem Cell 2019, 24, 487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Mansour AA, Goncalves JT, Bloyd CW, Li H, Fernandes S, Quang D, Johnston S, Parylak SL, Jin X, Gage FH, Nat. Biotechnol. 2018, 36, 432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Daviaud N, Friedel RH, Zou H, eNeuro 2018, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Huh D, Fujioka H, Tung YC, Futai N, Paine R 3rd, Grotberg JB, Takayama S, Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 18886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Huh D, Matthews BD, Mammoto A, Montoya-Zavala M, Hsin HY, Ingber DE, Science 2010, 328, 1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Kimura H, Yamamoto T, Sakai H, Sakai Y, Fujii T, Lab Chip 2008, 8, 741. [DOI] [PubMed] [Google Scholar]
- [199].Khetani SR, Bhatia SN, Nat. Biotechnol. 2008, 26, 120. [DOI] [PubMed] [Google Scholar]
- [200].Miller PG, Shuler ML, Biotechnol. Bioeng. 2016, 113, 2213. [DOI] [PubMed] [Google Scholar]
- [201].Edington CD, Chen WLK, Geishecker E, Kassis T, Soenksen LR, Bhushan BM, Freake D, Kirschner J, Maass C, Tsamandouras N, Valdez J, Cook CD, Parent T, Snyder S, Yu J, Suter E, Shockley M, Velazquez J, Velazquez JJ, Stockdale L, Papps JP, Lee I, Vann N, Gamboa M, LaBarge ME, Zhong Z, Wang X, Boyer LA, Lauffenburger DA, Carrier RL, Communal C, Tannenbaum SR, Stokes CL, Hughes DJ, Rohatgi G, Trumper DL, Cirit M, Griffith LG, Sci. Rep. 2018, 8, 4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Zhang B, Korolj A, Lai BFL, Radisic M, Nat. Rev. Mater. 2018, 3, 257. [Google Scholar]
- [203].Bhatia SN, Ingber DE, Nat. Biotechnol. 2014, 32, 760. [DOI] [PubMed] [Google Scholar]
- [204].Duffy DC, McDonald JC, Schueller OJA, Whitesides GM, Anal. Chem. 1998, 70, 4974. [DOI] [PubMed] [Google Scholar]
- [205].Mofazzal Jahromi MA, Abdoli A, Rahmanian M, Bardania H, Bayandori M, Moosavi Basri SM, Kalbasi A, Aref AR, Karimi M, Hamblin MR, Mol. Neurobiol. 2019, 56, 8489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Booth R, Kim H, Lab Chip 2012, 12, 1784. [DOI] [PubMed] [Google Scholar]
- [207].Prabhakarpandian B, Shen MC, Nichols JB, Mills IR, Sidoryk-Wegrzynowicz M, Aschner M, Pant K, Lab Chip 2013, 13, 1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Griep LM, Wolbers F, De Wagenaar B, Ter Braak PM, Weksler BB, Romero IA, Couraud PO, Vermes I, Van Der Meer AD, Van Den Berg A, Biomed. Microdevices 2013, 15, 145. [DOI] [PubMed] [Google Scholar]
- [209].Adriani G, Ma D, Pavesi A, Kamm RD, Goh EL, Lab Chip 2017, 17, 448. [DOI] [PubMed] [Google Scholar]
- [210].Park TE, Mustafaoglu N, Herland A, Hasselkus R, Mannix R, FitzGerald EA, Prantil-Baun R, Watters A, Henry O, Benz M, Sanchez H, McCrea HJ, Goumnerova LC, Song HW, Palecek SP, Shusta E, Ingber DE, Nat. Commun. 2019, 10, 2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Brown JA, Pensabene V, Markov DA, Allwardt V, Neely MD, Shi M, Britt CM, Hoilett OS, Yang Q, Brewer BM, Samson PC, McCawley LJ, May JM, Webb DJ, Li D, Bowman AB, Reiserer RS, Wikswo JP, Biomicrofluidics 2015, 9, 054124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Vatine GD, Barrile R, Workman MJ, Sances S, Barriga BK, Rahnama M, Barthakur S, Kasendra M, Lucchesi C, Kerns J, Wen N, Spivia WR, Chen Z, Van Eyk J, Svendsen CN, Cell Stem Cell 2019, 24, 995. [DOI] [PubMed] [Google Scholar]
- [213].Wang YI, Abaci HE, Shuler ML, Biotechnol. Bioeng. 2017, 114, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Wevers NR, Kasi DG, Gray T, Wilschut KJ, Smith B, van Vught R, Shimizu F, Sano Y, Kanda T, Marsh G, Trietsch SJ, Vulto P, Lanz HL, Obermeier B, Fluids and barriers of the CNS 2018, 15, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Ronaldson-Bouchard K, Vunjak-Novakovic G, Cell Stem Cell 2018, 22, 310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Shin Y, Choi SH, Kim E, Bylykbashi E, Kim JA, Chung S, Kim DY, Kamm RD, Tanzi RE, Adv. Sci. 2019, 6, 1900962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Fabre KM, Delsing L, Hicks R, Colclough N, Crowther DC, Ewart L, Adv. Drug Delivery Rev. 2019, 140, 129. [DOI] [PubMed] [Google Scholar]
- [218].Karimi M, Bahrami S, Mirshekari H, Basri SMM, Nik AB, Aref AR, Akbari M, Hamblin MR, Lab Chip 2016, 16, 2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Park JY, Kim SK, Woo DH, Lee EJ, Kim JH, Lee SH, Stem cells (Dayton, Ohio) 2009, 27, 2646. [DOI] [PubMed] [Google Scholar]
- [220].Wang Y, Wang L, Guo Y, Zhu Y, Qin J, RSC Adv. 2018, 8, 1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Karzbrun E, Kshirsagar A, Cohen SR, Hanna JH, Reiner O, Nat. Phys. 2018, 14, 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Zhuang P, Sun AX, An J, Chua CK, Chew SY, Biomaterials 2018, 154, 113. [DOI] [PubMed] [Google Scholar]
- [223].Simao D, Silva MM, Terrasso AP, Arez F, Sousa MFQ, Mehrjardi NZ, Saric T, Gomes-Alves P, Raimundo N, Alves PM, Brito C, Stem Cell Rep. 2018, 11, 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Boutin ME, Kramer LL, Livi LL, Brown T, Moore C, Hoffman-Kim D, Neurosci J. Methods 2018, 299, 55. [DOI] [PubMed] [Google Scholar]
- [225].Nzou G, Wicks RT, Wicks EE, Seale SA, Sane CH, Chen A, Murphy SV, Jackson JD, Atala AJ, Sci. Rep. 2018, 8, 7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Choi YY, Chung BG, Lee DH, Khademhosseini A, Kim JH, Lee SH, Biomaterials 2010, 31, 4296. [DOI] [PubMed] [Google Scholar]
- [227].Dingle YT, Boutin ME, Chirila AM, Livi LL, Labriola NR, Jakubek LM, Morgan JR, Darling EM, Kauer JA, Hoffman-Kim D, Tissue Eng., Part C 2015, 21, 1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Choi YJ, Park J, Lee SH, Biomaterials 2013, 34, 2938. [DOI] [PubMed] [Google Scholar]
- [229].Park J, Lee BK, Jeong GS, Hyun JK, Lee CJ, Lee SH, Lab Chip 2015, 15, 141. [DOI] [PubMed] [Google Scholar]
- [230].Yin X, Mead BE, Safaee H, Langer R, Karp JM, Levy O, Cell Stem Cell 2016, 18, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Yi Y, Park J, Lim J, Lee CJ, Lee SH, Trends Biotechnol. 2015, 33, 762. [DOI] [PubMed] [Google Scholar]
- [232].Song HL, Shim S, Kim DH, Won SH, Joo S, Kim S, Jeon NL, Yoon SY, Ann. Neurol. 2014, 75, 88. [DOI] [PubMed] [Google Scholar]
- [233].Poon WW, Blurton-Jones M, Tu CH, Feinberg LM, Chabrier MA, Harris JW, Jeon NL, Cotman CW, Neurobiol. Aging 2011, 32, 821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Deleglise B, Magnifico S, Duplus E, Vaur P, Soubeyre V, Belle M, Vignes M, Viovy JL, Jacotot E, Peyrin JM, Brugg B, Acta Neuropathol. Commun. 2014, 2, 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Cho H, Hashimoto T, Wong E, Hori Y, Wood LB, Zhao L, Haigis KM, Hyman BT, Irimia D, Sci. Rep. 2013, 3, 1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Dujardin S, Lecolle K, Caillierez R, Begard S, Zommer N, Lachaud C, Carrier S, Dufour N, Auregan G, Winderickx J, Hantraye P, Deglon N, Colin M, Buee L, Acta Neuropathol. Commun. 2014, 2, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Wu JW, Hussaini SA, Bastille IM, Rodriguez GA, Mrejeru A, Rilett K, Sanders DW, Cook C, Fu H, Boonen RA, Herman M, Nahmani E, Emrani S, Figueroa YH, Diamond MI, Clelland CL, Wray S, Duff KE, Nat. Neurosci. 2016, 19, 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Park J, Wetzel I, Marriott I, Dreau D, D’Avanzo C, Kim DY, Tanzi RE, Cho H, Nat. Neurosci. 2018, 21, 941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Fernandes JT, Chutna O, Chu V, Conde JP, Outeiro TF, Front. Neurosci. 2016, 10, 511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Bolognin S, Fossepre M, Qing X, Jarazo J, Scancar J, Moreno EL, Nickels SL, Wasner K, Ouzren N, Walter J, Grunewald A, Glaab E, Salamanca L, Fleming RMT, Antony PMA, Schwamborn JC, Adv. Sci. 2019, 6, 1800927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Kane KIW, Moreno EL, Hachi S, Walter M, Jarazo J, Oliveira MAP, Hankemeier T, Vulto P, Schwamborn JC, Thoma M, Fleming RMT, Sci. Rep. 2019, 9, 1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Logun M, Zhao W, Mao L, Karumbaiah L, Adv. Biosyst. 2018, 2, 1700221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Wang J, Tham D, Wei W, Shin YS, Ma C, Ahmad H, Shi Q, Yu J, Levine RD, Heath JR, Nano Lett. 2012, 12, 6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Sun J, Masterman-Smith MD, Graham NA, Jiao J, Mottahedeh J, Laks DR, Ohashi M, DeJesus J, Kamei K, Lee KB, Wang H, Yu ZT, Lu YT, Hou S, Li K, Liu M, Zhang N, Wang S, Angenieux B, Panosyan E, Samuels ER, Park J, Williams D, Konkankit V, Nathanson D, van Dam RM, Phelps ME, Wu H, Liau LM, Mischel PS, Lazareff JA, Kornblum HI, Yong WH, Graeber TG, Tseng HR, Cancer Res. 2010, 70, 6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Pathak A, Kumar S, Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Han J, Oh S, Hoang HH, Nguyen DTT, Lim W, Shin TH, Lee G, Park S, J. Ind. Eng. Chem. 2018, 62, 352. [Google Scholar]
- [247].Ayuso JM, Monge R, Martinez-Gonzalez A, Virumbrales-Munoz M, Llamazares GA, Berganzo J, Hernandez-Lain A, Santolaria J, Doblare M, Hubert C, Rich JN, Sanchez-Gomez P, Perez-Garcia VM, Ochoa I, Fernandez LJ, Neuro-oncology 2017, 19, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Chonan Y, Taki S, Sampetrean O, Saya H, Sudo R, Integr. Biol. 2017, 9, 762. [DOI] [PubMed] [Google Scholar]
- [249].Truong D, Fiorelli R, Barrientos ES, Melendez EL, Sanai N, Mehta S, Nikkhah M, Biomaterials 2019, 198, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Fan Y, Nguyen DT, Akay Y, Xu F, Akay M, Sci. Rep. 2016, 6, 25062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Akay M, Hite J, Avci NG, Fan Y, Akay Y, Lu G, Zhu JJ, Sci. Rep. 2018, 8, 15423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R, Nat. Commun. 2015, 6, 6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Chang TC, Mikheev AM, Huynh W, Monnat RJ, Rostomily RC, Folch A, Lab Chip 2014, 14, 4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Wu J, Zheng G, Lee LM, Lab Chip 2012, 12, 3566. [DOI] [PubMed] [Google Scholar]
- [255].Albert-Smet I, Marcos-Vidal A, Vaquero JJ, Desco M, Munoz-Barrutia A, Ripoll J, Front. Neuroanat. 2019, 13, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Paie P, Martinez Vazquez R, Osellame R, Bragheri F, Bassi A, Cytometry, Part A 2018, 93, 987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Geng Z, Zhang X, Fan Z, Lv X, Su Y, Chen H, Sensors 2017, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Cho S, Islas-Robles A, Nicolini AM, Monks TJ, Yoon JY, Biosens. Bioelectron. 2016, 86, 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Hofmann F, Bading H, J. Physiol. (Paris) 2006, 99, 125. [DOI] [PubMed] [Google Scholar]
- [260].Moutaux E, Charlot B, Genoux A, Saudou F, Cazorla M, Lab Chip 2018, 18, 3425. [DOI] [PubMed] [Google Scholar]
- [261].Kanagasabapathi TT, Franco M, Barone RA, Martinoia S, Wadman WJ, Decre MM, Neurosci J. Methods 2013, 214, 1. [DOI] [PubMed] [Google Scholar]
- [262].Hogberg HT, Sobanski T, Novellino A, Whelan M, Weiss DG, Bal-Price AK, Neurotoxicology 2011, 32, 158. [DOI] [PubMed] [Google Scholar]
- [263].Scelfo B, Politi M, Reniero F, Palosaari T, Whelan M, Zaldivar JM, Toxicology 2012, 299, 172. [DOI] [PubMed] [Google Scholar]
- [264].Yla-Outinen L, Heikkila J, Skottman H, Suuronen R, Aanismaa R, Narkilahti S, Front. Neuroeng. 2010, 3, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Defranchi E, Novellino A, Whelan M, Vogel S, Ramirez T, van Ravenzwaay B, Landsiedel R, Front. Neuroeng. 2011, 4, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Scott A, Weir K, Easton C, Huynh W, Moody WJ, Folch A, Lab Chip 2013, 13, 527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Zhang YS, Aleman J, Shin SR, Kilic T, Kim D, Mousavi Shaegh SA, Massa S, Riahi R, Chae S, Hu N, Avci H, Zhang W, Silvestri A, Sanati Nezhad A, Manbohi A, De Ferrari F, Polini A, Calzone G, Shaikh N, Alerasool P, Budina E, Kang J, Bhise N, Ribas J, Pourmand A, Skardal A, Shupe T, Bishop CE, Dokmeci MR, Atala A, Khademhosseini A, Proc. Natl. Acad. Sci. U. S. A. 2017, 114, E2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Nakai J, Ohkura M, Imoto K, Nat. Biotechnol. 2001, 19, 137. [DOI] [PubMed] [Google Scholar]
- [269].Akerboom J, Chen TW, Wardill TJ, Tian L, Marvin JS, Mutlu S, Calderon NC, Esposti F, Borghuis BG, Sun XR, Gordus A, Orger MB, Portugues R, Engert F, Macklin JJ, Filosa A, Aggarwal A, Kerr RA, Takagi R, Kracun S, Shigetomi E, Khakh BS, Baier H, Lagnado L, Wang SS, Bargmann CI, Kimmel BE, Jayaraman V, Svoboda K, Kim DS, Schreiter ER, Looger LL, J. Neurosci. 2012, 32, 13819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Palazzolo G, Moroni M, Soloperto A, Aletti G, Naldi G, Vassalli M, Nieus T, Difato F, Sci. Rep. 2017, 7, 8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Marom A, Mahto SK, Shor E, Tenenbaum-Katan J, Sznitman J, Shoham S, Adv. Healthcare Mater. 2015, 4, 1478. [DOI] [PubMed] [Google Scholar]
- [272].Akerboom J, Carreras Calderon N, Tian L, Wabnig S, Prigge M, Tolo J, Gordus A, Orger MB, Severi KE, Macklin JJ, Patel R, Pulver SR, Wardill TJ, Fischer E, Schuler C, Chen TW, Sarkisyan KS, Marvin JS, Bargmann CI, Kim DS, Kugler S, Lagnado L, Hegemann P, Gottschalk A, Schreiter ER, Looger LL, Front. Mol. Neurosci. 2013, 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K, Nat. Neurosci. 2005, 8, 1263. [DOI] [PubMed] [Google Scholar]
- [274].Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K, Nature 2007, 446, 633. [DOI] [PubMed] [Google Scholar]
- [275].Han X, Boyden ES, PLoS One 2007, 2, e299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Renault R, Sukenik N, Descroix S, Malaquin L, Viovy JL, Peyrin JM, Bottani S, Monceau P, Moses E, Vignes M, PLoS One 2015, 10, e0120680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Kobayashi T, Haruta M, Sasagawa K, Matsumata M, Eizumi K, Kitsumoto C, Motoyama M, Maezawa Y, Ohta Y, Noda T, Tokuda T, Ishikawa Y, Ohta J, Sci. Rep. 2016, 6, 21247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Delbeke J, Hoffman L, Mols K, Braeken D, Prodanov D, Front. Neurosci. 2017, 11, 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Hochbaum DR, Zhao Y, Farhi SL, Klapoetke N, Werley CA, Kapoor V, Zou P, Kralj JM, Maclaurin D, Smedemark-Margulies N, Saulnier JL, Boulting GL, Straub C, Cho YK, Melkonian M, Wong GK, Harrison DJ, Murthy VN, Sabatini BL, Boyden ES, Campbell RE, Cohen AE, Nat. Methods 2014, 11, 825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Huang G, Li F, Zhao X, Ma Y, Li Y, Lin M, Jin G, Lu TJ, Genin GM, Xu F, Chem. Rev. 2017, 117, 12764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Yildirimer L, Zhang Q, Kuang S, Cheung CJ, Chu KA, He Y, Yang M, Zhao X, Biofabrication 2019, 11, 032003. [DOI] [PubMed] [Google Scholar]
- [282].Murphy AR, Laslett A, O’Brien CM, Cameron NR, Acta Biomater. 2017, 54, 1. [DOI] [PubMed] [Google Scholar]
- [283].Aurand ER, Lampe KJ, Bjugstad KB, Neurosci. Res. 2012, 72, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [284].Ruoslahti E, Glycobiology 1996, 6, 489. [DOI] [PubMed] [Google Scholar]
- [285].Georges PC, Miller WJ, Meaney DF, Sawyer ES, Janmey PA, Biophys. J. 2006, 90, 3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Ma W, Fitzgerald W, Liu QY, O’Shaughnessy TJ, Maric D, Lin HJ, Alkon DL, Barker JL, Exp. Neurol. 2004, 190, 276. [DOI] [PubMed] [Google Scholar]
- [287].Watanabe K, Nakamura M, Okano H, Toyama Y, Restor. Neurol. Neurosci. 2007, 25, 109. [PubMed] [Google Scholar]
- [288].Brannvall K, Bergman K, Wallenquist U, Svahn S, Bowden T, Hilborn J, Forsberg-Nilsson K, J. Neurosci. Res. 2007, 85, 2138. [DOI] [PubMed] [Google Scholar]
- [289].Lee YB, Polio S, Lee W, Dai G, Menon L, Carroll RS, Yoo SS, Exp. Neurol. 2010, 223, 645. [DOI] [PubMed] [Google Scholar]
- [290].Deister C, Schmidt CE, J. Neural Eng. 2006, 3, 172. [DOI] [PubMed] [Google Scholar]
- [291].Sundararaghavan HG, Monteiro GA, Firestein BL, Shreiber DI, Biotechnol. Bioeng. 2009, 102, 632. [DOI] [PubMed] [Google Scholar]
- [292].Bouyer C, Chen P, Guven S, Demirtas TT, Nieland TJ, Padilla F, Demirci U, Adv. Mater. 2016, 28, 161. [DOI] [PubMed] [Google Scholar]
- [293].Kornev VA, Grebenik EA, Solovieva AB, Dmitriev RI, Timashev PS, Comput. Struct. Biotechnol. J. 2018, 16, 488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Wang X, He J, Wang Y, Cui FZ, Interface focus 2012, 2, 278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Hou S, Tian W, Xu Q, Cui F, Zhang J, Lu Q, Zhao C, Neuroscience 2006, 137, 519. [DOI] [PubMed] [Google Scholar]
- [296].Tian WM, Hou SP, Ma J, Zhang CL, Xu QY, Lee IS, Li HD, Spector M, Cui FZ, Tissue Eng. 2005, 11, 513. [DOI] [PubMed] [Google Scholar]
- [297].Suri S, Schmidt CE, Tissue Eng., Part A 2010, 16, 1703. [DOI] [PubMed] [Google Scholar]
- [298].Seidlits SK, Khaing ZZ, Petersen RR, Nickels JD, Vanscoy JE, Shear JB, Schmidt CE, Biomaterials 2010, 31, 3930. [DOI] [PubMed] [Google Scholar]
- [299].Matyash M, Despang F, Mandal R, Fiore D, Gelinsky M, Ikonomidou C, Tissue Eng., Part A 2012, 18, 55. [DOI] [PubMed] [Google Scholar]
- [300].Frampton JP, Hynd MR, Shuler ML, Shain W, Biomed. Mater. 2011, 6, 015002. [DOI] [PubMed] [Google Scholar]
- [301].Moxon SR, Corbett NJ, Fisher K, Potjewyd G, Domingos M, Hooper NM, Mater. Sci. Eng., C 2019, 104, 109904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Kunze A, Giugliano M, Valero A, Renaud P, Biomaterials 2011, 32, 2088. [DOI] [PubMed] [Google Scholar]
- [303].Kothapalli CR, Kamm RD, Biomaterials 2013, 34, 5995. [DOI] [PubMed] [Google Scholar]
- [304].Hopkins AM, De Laporte L, Tortelli F, Spedden E, Staii C, Atherton TJ, Hubbell JA, Kaplan DL, Adv. Funct. Mater. 2013, 23, 5140. [Google Scholar]
- [305].Li X, Katsanevakis E, Liu X, Zhang N, Wen X, Prog. Polym. Sci. 2012, 37, 1105. [Google Scholar]
- [306].Mahoney MJ, Anseth KS, Biomaterials 2006, 27, 2265. [DOI] [PubMed] [Google Scholar]
- [307].Gunn JW, Turner SD, Mann BK, J. Biomed. Mater. Res., Part A 2005, 72, 91. [DOI] [PubMed] [Google Scholar]
- [308].Zhang ZN, Freitas BC, Qian H, Lux J, Acab A, Trujillo CA, Herai RH, Nguyen Huu VA, Wen JH, Joshi-Barr S, Karpiak JV, Engler AJ, Fu XD, Muotri AR, Almutairi A, Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Tsurkan MV, Chwalek K, Prokoph S, Zieris A, Levental KR, Freudenberg U, Werner C, Adv. Mater. 2013, 25, 2606. [DOI] [PubMed] [Google Scholar]
- [310].Yu LM, Kazazian K, Shoichet MS, J. Biomed. Mater. Res, Part A 2007, 82, 243. [DOI] [PubMed] [Google Scholar]
- [311].Li H, Wijekoon A, Leipzig ND, PLoS One 2012, 7, e48824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Li H, Wijekoon A, Leipzig ND, Ann. Biomed. Eng. 2014, 42, 1456. [DOI] [PubMed] [Google Scholar]
- [313].Holmes TC, de Lacalle S, Su X, Liu G, Rich A, Zhang S, Proc. Natl. Acad. Sci. U. S. A. 2000, 97, 6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Zhang ZX, Zheng QX, Wu YC, Hao DJ, Biotechnol. Bioprocess Eng. 2010, 15, 545. [Google Scholar]
- [315].Ortinau S, Schmich J, Block S, Liedmann A, Jonas L, Weiss DG, Helm CA, Rolfs A, Frech MJ, Biomed. Eng. Online 2010, 9, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [316].Cheng TY, Chen MH, Chang WH, Huang MY, Wang TW, Biomaterials 2013, 34, 2005. [DOI] [PubMed] [Google Scholar]
- [317].Sun Y, Li W, Wu X, Zhang N, Zhang Y, Ouyang S, Song X, Fang X, Seeram R, Xue W, He L, Wu W, ACS Appl. Mater. Interfaces 2016, 8, 2348. [DOI] [PubMed] [Google Scholar]
- [318].Caprini A, Silva D, Zanoni I, Cunha C, Volonte C, Vescovi A, Gelain F, New Biotechnol. 2013, 30, 552. [DOI] [PubMed] [Google Scholar]
- [319].Pugliese R, Fontana F, Marchini A, Gelain F, Acta Biomater. 2018, 66, 258. [DOI] [PubMed] [Google Scholar]
- [320].Marchini A, Raspa A, Pugliese R, El Malek MA, Pastori V, Lecchi M, Vescovi AL, Gelain F, Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [321].Hospodiuk M, Dey M, Sosnoski D, Ozbolat IT, Biotechnol. Adv. 2017, 35, 217. [DOI] [PubMed] [Google Scholar]
- [322].Knowlton S, Anand S, Shah T, Tasoglu S, Trends Neurosci. 2018, 41, 31. [DOI] [PubMed] [Google Scholar]
- [323].Xu T, Gregory CA, Molnar P, Cui X, Jalota S, Bhaduri SB, Boland T, Biomaterials 2006, 27, 3580. [DOI] [PubMed] [Google Scholar]
- [324].Lee W, Pinckney J, Lee V, Lee JH, Fischer K, Polio S, Park JK, Yoo SS, Neuroreport 2009, 20, 798. [DOI] [PubMed] [Google Scholar]
- [325].Hsieh FY, Lin HH, Hsu SH, Biomaterials 2015, 71, 48. [DOI] [PubMed] [Google Scholar]
- [326].Zhou X, Cui H, Nowicki M, Miao S, Lee SJ, Masood F, Harris BT, Zhang LG, ACS Appl. Mater. Interfaces 2018, 10, 8993. [DOI] [PubMed] [Google Scholar]
- [327].Lozano R, Stevens L, Thompson BC, Gilmore KJ, Gorkin R 3rd, Stewart EM, in het Panhuis M, Romero-Ortega M, Wallace GG, Biomaterials 2015, 67, 264. [DOI] [PubMed] [Google Scholar]
- [328].Gu Q, Tomaskovic-Crook E, Lozano R, Chen Y, Kapsa RM, Zhou Q, Wallace GG, Crook JM, Adv. Healthcare Mater. 2016, 5, 1429. [DOI] [PubMed] [Google Scholar]
- [329].Gu Q, Tomaskovic-Crook E, Wallace GG, Crook JM, Adv. Healthcare Mater. 2017, 6, 1700175. [DOI] [PubMed] [Google Scholar]
- [330].Cheng K, Lai Y, Kisaalita WS, Biomaterials 2008, 29, 2802. [DOI] [PubMed] [Google Scholar]
- [331].Lai Y, Asthana A, Cheng K, Kisaalita WS, PLoS One 2011, 6, e26821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [332].Lai Y, Cheng K, Kisaalita W, PLoS One 2012, 7, e45074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [333].Li N, Zhang Q, Gao S, Song Q, Huang R, Wang L, Liu L, Dai J, Tang M, Cheng G, Sci. Rep. 2013, 3, 1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [334].Jakobsson A, Ottosson M, Zalis MC, O’Carroll D, Johansson UE, Johansson F, Nanomedicine : nanotechnology, biology, and medicine 2017, 13, 1563. [DOI] [PubMed] [Google Scholar]
- [335].Harley BA, Leung JH, Silva EC, Gibson LJ, Acta Biomater. 2007, 3, 463. [DOI] [PubMed] [Google Scholar]
- [336].Haugh MG, Murphy CM, O’Brien FJ, Tissue Eng., Part C 2010, 16, 887. [DOI] [PubMed] [Google Scholar]
- [337].Wang TW, Spector M, Acta Biomater. 2009, 5, 2371. [DOI] [PubMed] [Google Scholar]
- [338].Elias PZ, Spector M, J. Neurosci. Methods 2012, 209, 199. [DOI] [PubMed] [Google Scholar]
- [339].Hosseinkhani H, Hiraoka Y, Li CH, Chen YR, Yu DS, Hong PD, Ou KL, ACS Chem. Neurosci. 2013, 4, 1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Jurga M, Dainiak MB, Sarnowska A, Jablonska A, Tripathi A, Plieva FM, Savina IN, Strojek L, Jungvid H, Kumar A, Lukomska B, Domanska-Janik K, Forraz N, McGuckin CP, Biomaterials 2011, 32, 3423. [DOI] [PubMed] [Google Scholar]
- [341].Tang-Schomer MD, White JD, Tien LW, Schmitt LI, Valentin TM, Graziano DJ, Hopkins AM, Omenetto FG, Haydon PG, Kaplan DL, Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 13811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [342].Liaudanskaya V, Jgamadze D, Berk AN, Bischoff DJ, Gu BJ, Hawks-Mayer H, Whalen MJ, Chen HI, Kaplan DL, Biomaterials 2019, 192, 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [343].Zhang D, Pekkanen-Mattila M, Shahsavani M, Falk A, Teixeira AI, Herland A, Biomaterials 2014, 35, 1420. [DOI] [PubMed] [Google Scholar]
- [344].Labour MN, Vigier S, Lerner D, Marcilhac A, Belamie E, Acta Biomater. 2016, 37, 38. [DOI] [PubMed] [Google Scholar]
- [345].Papadimitriou C, Celikkaya H, Cosacak MI, Mashkaryan V, Bray L, Bhattarai P, Brandt K, Hollak H, Chen X, He S, Antos CL, Lin W, Thomas AK, Dahl A, Kurth T, Friedrichs J, Zhang Y, Freudenberg U, Werner C, Kizil C, Dev. Cell 2018, 46, 85. [DOI] [PubMed] [Google Scholar]
- [346].Cantley WL, Du C, Lomoio S, Depalma T, Peirent E, Kleinknecht D, Hunter M, Tang-Schomer MD, Tesco G, Kaplan DL, ACS Biomater. Sci. Eng. 2018, 4, 4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [347].Watson PMD, Kavanagh E, Allenby G, Vassey M, SLAS Discovery 2017, 22, 583. [DOI] [PubMed] [Google Scholar]
- [348].Xiao W, Sohrabi A, Seidlits SK, Future Sci. OA 2017, 3, Fso189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [349].Ananthanarayanan B, Kim Y, Kumar S, Biomaterials 2011, 32, 7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [350].Wang C, Tong X, Yang F, Mol. Pharmaceutics 2014, 11, 2115. [DOI] [PubMed] [Google Scholar]
- [351].Florczyk SJ, Wang K, Jana S, Wood DL, Sytsma SK, Sham J, Kievit FM, Zhang M, Biomaterials 2013, 34, 10143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [352].Sood D, Tang-Schomer M, Pouli D, Mizzoni C, Raia N, Tai A, Arkun K, Wu J, Black LD 3rd, Scheffler B, Georgakoudi I, Steindler DA, Kaplan DL, Nat. Commun. 2019, 10, 4529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [353].Knowlton S, Onal S, Yu CH, Zhao JJ, Tasoglu S, Trends Biotechnol. 2015, 33, 504. [DOI] [PubMed] [Google Scholar]
- [354].Dai X, Ma C, Lan Q, Xu T, Biofabrication 2016, 8, 045005. [DOI] [PubMed] [Google Scholar]
- [355].Pedron S, Becka E, Harley BA, Adv. Mater. 2015, 27, 1567. [DOI] [PubMed] [Google Scholar]
- [356].Heinrich MA, Bansal R, Lammers T, Zhang YS, Michel Schiffelers R, Prakash J, Adv. Mater. 2019, 31, e1806590. [DOI] [PubMed] [Google Scholar]
- [357].Caragher S, Chalmers AJ, Gomez-Roman N, Cancers 2019, 11, 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [358].Gomez-Roman N, Stevenson K, Gilmour L, Hamilton G, Chalmers AJ, Neuro-oncology 2017, 19, 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [359].Bar-Kochba E, Scimone MT, Estrada JB, Franck C, Sci. Rep. 2016, 6, 30550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [360].Schwartz MP, Hou Z, Propson NE, Zhang J, Engstrom CJ, Santos Costa V, Jiang P, Nguyen BK, Bolin JM, Daly W, Wang Y, Stewart R, Page CD, Murphy WL, Thomson JA, Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [361].Ranga A, Girgin M, Meinhardt A, Eberle D, Caiazzo M, Tanaka EM, Lutolf MP, Proc. Natl. Acad. Sci. U. S. A. 2016, 113, E6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [362].Kratochvil MJ, Seymour AJ, Li TL, Paşca SP, Kuo CJ, Heilshorn SC, Nat. Rev. Mater. 2019, 4, 606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [363].Cederquist GY, Asciolla JJ, Tchieu J, Walsh RM, Cornacchia D, Resh MD, Studer L, Nat. Biotechnol. 2019, 37, 436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [364].Knight GT, Lundin BF, Iyer N, Ashton LM, Sethares WA, Willett RM, Ashton RS, eLife 2018, 7, e37549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [365].Lam J, Truong NF, Segura T, Acta Biomater. 2014, 10, 1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [366].Boni R, Ali A, Shavandi A, Clarkson AN, J. Biomed. Sci. 2018, 25, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [367].Ilkhanizadeh S, Teixeira AI, Hermanson O, Biomaterials 2007, 28, 3936. [DOI] [PubMed] [Google Scholar]
- [368].Zhao G, Qing H, Huang G, Genin GM, Lu TJ, Luo Z, Xu F, Zhang X, NPG Asia Mater. 2018, 10, 982. [Google Scholar]
- [369].Zhao YH, Niu CM, Shi JQ, Wang YY, Yang YM, Wang HB, Neural Regener. Res. 2018, 13, 1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [370].Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, Sutcliffe M, Boulanger J, Tripodi M, Derivery E, Paulsen O, Lakatos A, Lancaster MA, Nat. Neurosci. 2019, 22, 669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [371].Ashammakhi N, Ahadian S, Xu C, Montazerian H, Ko H, Nasiri R, Barros N, Khademhosseini A, Mater. Today Bio 2019, 1, 100008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [372].Ma W, Tavakoli T, Chen S, Maric D, Liu JL, O’Shaughnessy TJ, Barker JL, Tissue Eng., Part A 2008, 14, 1673. [DOI] [PubMed] [Google Scholar]
- [373].Raimondi MT, Albani D, Giordano C, Trends Mol. Med. 2019, 25, 737. [DOI] [PubMed] [Google Scholar]
- [374].Kato-Negishi M, Morimoto Y, Onoe H, Takeuchi S, Adv. Healthcare Mater. 2013, 2, 1564. [DOI] [PubMed] [Google Scholar]
- [375].Hasan MF, Berdichevsky Y, Micromachines 2016, 7, 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [376].Ndyabawe K, Kisaalita WS, Drug discovery today 2019, 24, 1725. [DOI] [PubMed] [Google Scholar]
- [377].De Sandre-Giovannoli A, Bernard R, Cau P, Navarro C, Amiel J, Boccaccio I, Lyonnet S, Stewart CL, Munnich A, Le Merrer M, Lévy N, Science 2003, 300, 2055. [DOI] [PubMed] [Google Scholar]
- [378].Eriksson M, Brown WT, Gordon LB, Glynn MW, Singer J, Scott L, Erdos MR, Robbins CM, Moses TY, Berglund P, Dutra A, Pak E, Durkin S, Csoka AB, Boehnke M, Glover TW, Collins FS, Nature 2003, 423, 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [379].Miller JD, Ganat YM, Kishinevsky S, Bowman RL, Liu B, Tu EY, Mandal PK, Vera E, Shim JW, Kriks S, Taldone T, Fusaki N, Tomishima MJ, Krainc D, Milner TA, Rossi DJ, Studer L, Cell Stem Cell 2013, 13, 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [380].Mertens J, Paquola ACM, Ku M, Hatch E, Bohnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy JR, Goncalves JT, Toda T, Kim Y, Winkler J, Yao J, Hetzer MW, Gage FH, Cell Stem Cell 2015, 17, 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [381].White RR, Vijg J, Mol. Cell 2016, 63, 729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [382].Raj DD, Jaarsma D, Holtman IR, Olah M, Ferreira FM, Schaafsma W, Brouwer N, Meijer MM, de Waard MC, van der Pluijm I, Brandt R, Kreft KL, Laman JD, de Haan G, Biber KP, Hoeijmakers JH, Eggen BJ, Boddeke HW, Neurobiol. Aging 2014, 35, 2147. [DOI] [PubMed] [Google Scholar]
- [383].de Waard MC, van der Pluijm I, Zuiderveen Borgesius N, Comley LH, Haasdijk ED, Rijksen Y, Ridwan Y, Zondag G, Hoeijmakers JH, Elgersma Y, Gillingwater TH, Jaarsma D, Acta Neuropathol. 2010, 120, 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [384].Borgesius NZ, de Waard MC, van der Pluijm I, Omrani A, Zondag GC, van der Horst GT, Melton DW, Hoeijmakers JH, Jaarsma D, Elgersma Y, J. Neurosci. 2011, 31, 12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [385].Vegh MJ, de Waard MC, van der Pluijm I, Ridwan Y, Sassen MJ, van Nierop P, van der Schors RC, Li KW, Hoeijmakers JH, Smit AB, van Kesteren RE, J. Proteome Res. 2012, 11, 1855. [DOI] [PubMed] [Google Scholar]
- [386].de Graaf EL, Vermeij WP, de Waard MC, Rijksen Y, van der Pluijm I, Hoogenraad CC, Hoeijmakers JH, Altelaar AF, Heck AJ, Mol. Cell. Proteomics 2013, 12, 1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [387].Vera E, Bosco N, Studer L, Cell Rep. 2016, 17, 1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [388].Lai Y, Kisaalita WS, J. Lab. Autom. 2012, 17, 284. [DOI] [PubMed] [Google Scholar]
- [389].Domansky K, Leslie DC, McKinney J, Fraser JP, Sliz JD, Hamkins-Indik T, Hamilton GA, Bahinski A, Ingber DE, Lab Chip 2013, 13, 3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [390].Bjornsson CS, Lin G, Al-Kofahi Y, Narayanaswamy A, Smith KL, Shain W, Roysam B, Neurosci J. Methods 2008, 170, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [391].Jahnke HG, Braesigk A, Mack TG, Ponick S, Striggow F, Robitzki AA, Biosens. Bioelectron. 2012, 32, 250. [DOI] [PubMed] [Google Scholar]
- [392].Dana H, Marom A, Paluch S, Dvorkin R, Brosh I, Shoham S, Nat. Commun. 2014, 5, 3997. [DOI] [PMC free article] [PubMed] [Google Scholar]


