Abstract
During genome replication, telomerase adds repeats to the ends of chromosomes to balance the loss of telomeric DNA. The regulation of telomerase activity is of medical relevance, as it has been implicated in human diseases such as cancer, as well as in aging. Until recently, structural information on this enzyme that would facilitate its clinical manipulation had been lacking due to telomerase very low abundance in cells. Recent cryo-EM structures of both the human and Tetrahymena thermophila telomerases have provided a picture of both the shared catalytic core of telomerase and its interaction with species-specific factors that play different roles in telomerase RNP assembly and function. We discuss also progress towards an understanding of telomerase RNP biogenesis and telomere recruitment from recent studies.
Keywords: telomerase, RNP, telomere, cryo-EM
Introduction
Telomerase is a ribonucleoprotein (RNP) complex that maintains telomeres by synthesizing terminal telomeric repeats in compensation for incomplete genome replication [1]. For repeat synthesis, telomerase relies on two components: a specialized telomerase reverse transcriptase (TERT) and a template provided by telomerase RNA (TER or hTR in human) [2]. Telomerase activity is highly regulated through human development. Early embryos and some adult stem cells have active telomerase, but telomerase activity is undetectable in all but a few somatic cells [3–5]. During oncogenesis, telomerase is reactivated, allowing cancer cells to gain proliferation immortality [6]. Human diseases such as dyskeratosis congenita, aplastic anemia, and pulmonary fibrosis can arise from mutations that compromise telomerase function [7].
Besides TERT and TER, additional factors required for telomerase function in vivo are also associated with telomerase holoenzymes [8]. While TERT is highly conserved, TER differs significantly in size, predicted secondary structure and biogenesis pathways across different eukaryotic species [9,10]. Consequently, there is a remarkable diversity of holoenzyme composition and architecture (Figure 1). Structural studies of telomerase have proved extremely challenging due to its very low abundance. Telomerase holoenzyme from the ciliate Tetrahymena thermophila was the first to be fully characterized biochemically and structurally [11–14]. On the other hand, human telomerase composition was unclear due to contradictory conclusions [15–18]. In 2013, negative-stain electron microscopy (EM) structures of both T. thermophila and human telomerase at 25–30 Å resolution were reported, revealing their overall 3D architectures and the remarkable differences between them [12,14,17]. Taking advantage of recent developments in cryo-EM, structures of human and T. thermophila telomerase holoenzymes have been determined at higher resolution [13,19,20]. The structure of a complete fungal telomerase has yet to be determined, but recently additional progress has been made in defining both subunit composition and subunit structures [9,10,21–25]. In this review, we first discuss the cryo-EM structures of the human and T. thermophila telomerase (Figure 2) [19,20]. These structures not only provide detailed architectural insight into the common design principles of the catalytic core of telomerase holoenzymes, but also reveal the physical coordination of additional species-specific factors and their roles in telomerase RNP assembly. Subsequently, we highlight recent progress in our understanding of telomerase RNP biogenesis and telomere recruitment in T. thermophila, human, Saccharomyces cerevisiae and Schizosaccharomyces pombe.
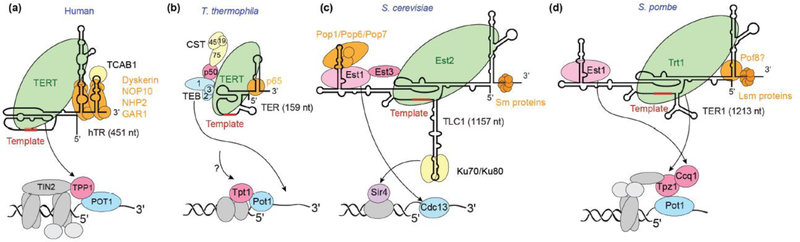
Figure 1.
Telomerase holoenzymes and telomerase recruitment to telomeres in (a) human, (b) T. thermophila, (c) S. cerevisiae, and (d) S. pombe (illustrations not drawn to scale). Telomerase holoenzyme and telomere protein complex schematics are shown, together with predicted secondary structures of TERs, and their lengths in nucleotides (nt) are indicated in parenthesis. TERTs are colored in light green and other holoenzyme proteins are placed and colored based on their predicted or known interaction networks. Below the telomerase holoenzyme schematics are schematics of telomere proteins (critical ones highlighted in colors) that are important for telomerase recruitment. Black arrows indicate interactions that recruit and activate telomerase at telomeres. In (b), it is possible that T. thermophila TERT interacts with Tpt1 by analogy to human and S. pombe systems, but this interaction may be replaced by a p50-TERT interaction.
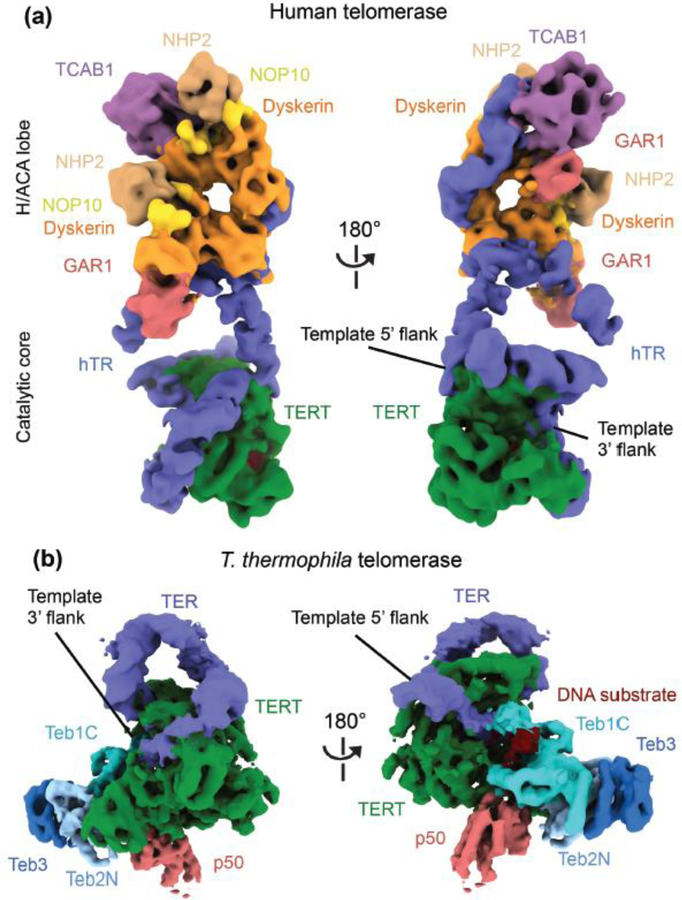
Figure 2.
Cryo-EM structures of (a) human and (b) T. thermophila telomerase holoenzymes in two views [19,20]. Subunits are colored as labeled. A telomeric DNA substrate is bound to template in both structures, but the template and template-paired DNA are buried in the views shown; only the single-stranded 5’ region of unpaired DNA in the T. thermophila structure has visualized density rendered in these illustrations. In (b), the depicted 4.8 Å T. thermophila telomerase structure was obtained from focused refinement of a structural core containing only TERT, TER, p50 and some of the TEB complex; p65 and the p75-p45-p19 heterotrimer complex are absent in this map (see Figure 1a for the complete holoenzyme). Only single domains of TEB subunits were resolved (Teb1C is Teb1 C-terminal domain, Teb2N is Teb2 N-terminal domain).
Cryo-EM structures of the human and T. thermophila telomerase holoenzymes
Transient overexpression of human TERT and hTR in HEK 293T cells is commonly used in telomerase studies because of telomerase low natural abundance and inefficient reconstitution from purified subunits. Single molecule fluorescence and negative stain EM studies showed that RNPs produced by this method are heterogeneous in terms of oligomeric states and catalytic activity [17,18]. Guided by activity assays and negative-stain EM, we developed a purification protocol that enriched for the active RNP population and allowed structural determination of human telomerase by cryo-EM (Figure 1a and 2a) [19]. Although the purification was performed via hTR and a tag on TERT, other protein factors required for biogenesis and localization were also present in the purified sample. Single-particle EM analysis showed that human telomerase has a bilobal architecture, similar to that observed in the previous 30 Å negative stain reconstruction [17], with the two lobes adopting a wide range of relative positions with respect to each other. Through extensive localized refinement/classification, cryo-EM maps at 7–8 Å resolution were obtained for each of the two lobes individually, allowing unambiguous fitting of known crystal and NMR structures or homology models of components, and idealized RNA helical segments. The resulting structure revealed an assembly of ten protein subunits around an hTR scaffold, with TERT located in one lobe, bound to a DNA substrate, and a single TCAB1 and two of each mature H/ACA protein (dyskerin, GAR1, NHP2 and NOP10; described below) in the other lobe (Figure 2a). We refer to the two lobes as the catalytic core and H/ACA lobe, respectively. Our interpretation of the cryo-EM density is different from that of the previous low-resolution negative-stain reconstruction, where each lobe was proposed to contain a copy of TERT and hTR [17]. A monomeric TERT and hTR subunit composition in human holoenzyme parallels that in holoenzymes from single-cell eukaryotes [12,26].
The catalytic core of human telomerase is formed by TERT and just the two activity-essential domains of hTR: the template/pseudoknot (t/PK) and phylogenetically defined conserved regions 4/5 (CR4/5) (Figure 1a, 3a–c) [27], in agreement with previous findings that catalytically active RNP can be assembled in both rabbit reticulocyte lysate and cells from just TERT and hTRmin, which contains only t/PK and CR4/5 connected by a short linker [28,29]. There are no observed RNA or protein interactions between the H/ACA RNP lobe and the catalytic core, consistent with the bypass of the H/ACA-domain requirement for active telomerase assembly using a different 3’ end-processing pathway [29]. However, a recent study suggested a direct role for TCAB1-hTR interaction in folding CR4/5 [30]. Given that TCAB1 and CR4/5 are distant in our structure (Figure 2a), the subtle TCAB1 influence on hTR CR4/5 could reflect an indirect TCAB1 role, for example in CR4/5 ψ modification [31,32] and/or RNP stability overall.
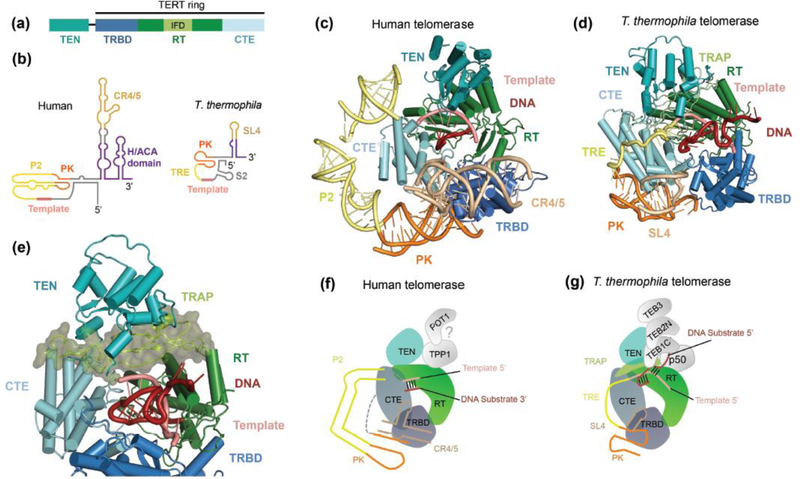
Figure 3.
Structural analysis of the catalytic core of the human and T. thermophila telomerase structures [19,20]. (a) Domain architecture of TERT. The IFD (light green) is embedded in the RT domain (dark green). (b) Secondary structure schematics of human and T. thermophila TERs. RNA domains described in the text are labeled; in addition, T. thermophila stem 2 (S2) is indicated, which is the template 5’ flanking region labeled in Figure 2b. The domain colors shown in (a) and (b) are used for the subsequent panels, which look down into the active site. (c and d) Catalytic cores of human and T. thermophila telomerase, respectively. (e) Close-up view of TRAP in the IFD, highlighted in space-filling representation. (f and g) Schematic representation of the architectures of the catalytic core in human and T. thermophila telomerase, respectively. The positioning of human TPP1 and POT1 is only hypothetical. The interactions observed in T. thermophila telomerase holoenzyme between TERT, p50 and TEB may or may not be paralleled by TERT, TPP1 and POT1 in human telomerase-telomere complexes.
The TERT telomerase RNA binding domain (TRBD), reverse transcriptase (RT) domain, and C-terminal extension (CTE) form a so-called TERT ring (Figure 3a) with a central cavity that accommodates the RNA template-DNA substrate duplex (Figure 3c and d), as first revealed by a Tribolium castaneum TERT structure [33]. The telomerase essential N-terminal (TEN) domain packs against the TERT ring above the active site cavity (Figure 3c and d), as first evident in the negative stain EM structure of telomerase holoenzyme from T. thermophila [12]. In the catalytic core, RNA tertiary structure forms a rigid layer outside the TERT TRBD and CTE domains, with folding of individual RNA elements consistent with prior hTR structural studies [34,35].
Vertebrate TERs have a double-hairpin H/ACA motif (Figure 3b) that is critical for nascent RNA processing to mature hTR and for mature hTR biological stability and RNP accumulation [36,37]. This H/ACA motif is shared with a large family of H/ACA small nucleolar RNAs (snoRNAs) and small Cajal body RNAs (scaRNAs). The H/ACA motif assembles with a set of four evolutionarily conserved proteins, dyskerin, GAR1, NHP2 and NOP10, on each of the hairpins, forming an H/ACA RNP [38]. The human telomerase structure not only confirmed the presence of a full H/ACA RNP in human telomerase (Figure 1a and 2a) [19], but also yielded the first structural insight into an intact eukaryotic H/ACA RNP. The heterotetramers on the two H/ACA hairpins are held together, not only by interactions with hTR, but also by newly revealed interactions between the two dyskerin molecules. Telomerase deficiency caused by H/ACA protein missense mutations underlie several forms of the bone marrow failure syndrome dyskeratosis congenita [7,39]. It was unclear from the previous single-hairpin H/ACA RNP structures [40,41] how these mutations affect the integrity of the two-hairpin RNP, and how they could affect hTR accumulation more severely than other snoRNAs or scaRNAs. Intriguingly, a large number of disease mutations in dyskerin are clustered at the dyskerin-dyskerin interface [19]. Telomerase depends on the 3’ hairpin to assemble proteins on the 5’ hairpin, and this asymmetry would make the cross-hairpin dyskerin interactions particularly critical in the hTR H/ACA RNP.
Additional insight into the telomerase catalytic core came from T. thermophila telomerase holoenzyme cryo-EM structure and its comparison to human telomerase (Figure 2b and 3d) [13]. As revealed by previous proteomic and biochemical studies and the 9.4 Å cryo-EM reconstruction of the complex, T. thermophila telomerase comprises a catalytic core of TERT, TER and the La-family protein p65, and two distinct Replication Protein A (RPA)-like complexes, p75-p45-p19 (analogous to CTC1-STN1-TEN1, CST) and Teb1-Teb2-Teb3 (TEB), which are tethered to the catalytic core via the p50 protein (Figure 1b) [11–14]. Teb2 and Teb3 are the only holoenzyme subunits that are not telomerase-specific, as they are also subunits of RPA [42]. Recently, Jiang et al., captured the catalytic core with a DNA substrate using a locked-nucleic-acid containing oligonucleotide to stabilize the DNA-RNA duplex [20]. Using focused classification and refinement, a 4.8 Å reconstruction was obtained for the most stable part of the complex, which included TERT, TER, domains from the TEB complex and p50 (Figure 2b). The improved resolution allowed tentative modeling of the insertion in fingers domain (IFD) of the RT domain, which is absent in the evolutionarily streamlined T. castaneum TERT. This region, named TRAP, forms interactions with the TEN domain above the active site (Figure 3d and e), suggesting an importance of this interface for the TEN domain contribution to processive repeat synthesis [8].
Despite the significant differences in the RNA structures for the ciliate and human holoenzymes (Figure 3b), there are clear similarities in the catalytic core. As initially observed for the streamlined T. castaneum TERT [33], the TERT ring encloses the RNA/DNA duplex (Figure 3c and d). The PK and CR4/5 of hTR bind at the interface between the TRBD and CTE domains of TERT, similar to the PK and stem 4 loop (SL4) of T. thermophila TER (Figure 3c, d, f and g). Despite expanded complexity and rigidity of the P2 stem connecting the template and PK in hTR, relative to the single-stranded (ss) template recognition element (TRE) connecting template to PK in T. thermophila TER, both straddle the CTE (Figure 3c and d). In the T. thermophila holoenzyme, due to the obstacles of the TRAP and TEN domain, the DNA substrate 5’ end and template 3’ end emerge from the active site cavity in opposite directions. The RNA winds over the CTE, while the DNA turns towards the TEB complex and also forms an additional 3 bp with residues 5’ of the template (Figure 3e and g), as anticipated by previous cross-linking data [43]. The p50-TEB complex in T. thermophila contributes critically to telomerase recruitment and activation [14,42]. Teb1 mediates telomeric repeat ssDNA binding via three tandem oligonucleotide/oligosaccharide-binding (OB) folds. As predicted by previous biochemical studies [11], the C-terminal OB-fold of Teb1 (Teb1C) interacts with the ssDNA exiting the TERT active site (Figure 2b and 3g). TEB is ciliate-specific. However, by analogy, it is possible that ssDNA exiting the template duplex of human telomerase holoenzyme could turn from the CTE to cross over the RT domain towards one possible speculative positioning of TPP1 and POT1 (Figure 3f), discussed below. NMR studies of the TEN domain of telomerase from a thermophilic yeast suggest a possible duplex splitting role that warrants future investigation [44].
Telomerase assembly
Establishment of the complex networks of interactions between TERT, TER and other holoenzyme subunits observed in the cryo-EM structures involves step-wise pathways of RNP subunit assembly and folding. Although yeast and metazoan TERs are transcripts of RNA Pol II, ciliate TERs are RNA Pol III transcripts that require La-family proteins such as p65 in T. thermophila for stability and folding (Figure 1a) [45–47]. Fungal TERs require 3’ association of the heptameric Sm-protein complex for their processing and accumulation (Figure 1c), a pathway shared with spliceosomal small nuclear RNAs [48,49]. In S. pombe, the Sm complex is later replaced by an Sm-like (Lsm) complex [50]. Curiously, Lsm loading is stimulated by Pof8, a La-family RNA-binding protein and recently discovered subunit of S. pombe telomerase holoenzyme (Figure 1d) [22–24]. Also, budding yeast S. cerevisiae telomerase RNA TLC1 was found to associate with the trimeric Pop1/Pop6/Pop7 protein complex via a domain similar to the P3 domain of the RNAs of eukaryotic Ribonuclease P and Ribonuclease for Mitochondrial RNA Processing (RNase P/MRP) [21]. This module of TLC1 functions in maintaining stable co-assembly of two holoenzyme subunits, Est1 and Est2/TERT (Figure 1c).
In the catalytic core of human and T. thermophila telomerase holoenzymes, the t/PK makes a large arc across several domains of TERT (Figure 3). One remaining open question is the role of alternative conformations of TER en route to this t/PK fold in active telomerase RNP. In the absence of TERT, T. thermophila TER does not have the PK fold [51]. Instead, the template and PK sequence have the alternative fold of a long stem-loop [52]. The alternative conformation could serve as an intermediate in RNP assembly that restrains PK formation until template has threaded across the active site surface of TERT ring. An intermediate hTR structure en route to the active hTR fold is also possible, although mutagenesis results suggest that any secondary structure rearrangement would be less extensive or critical than is the case for T. thermophila TER [53].
Telomerase recruitment to telomeres
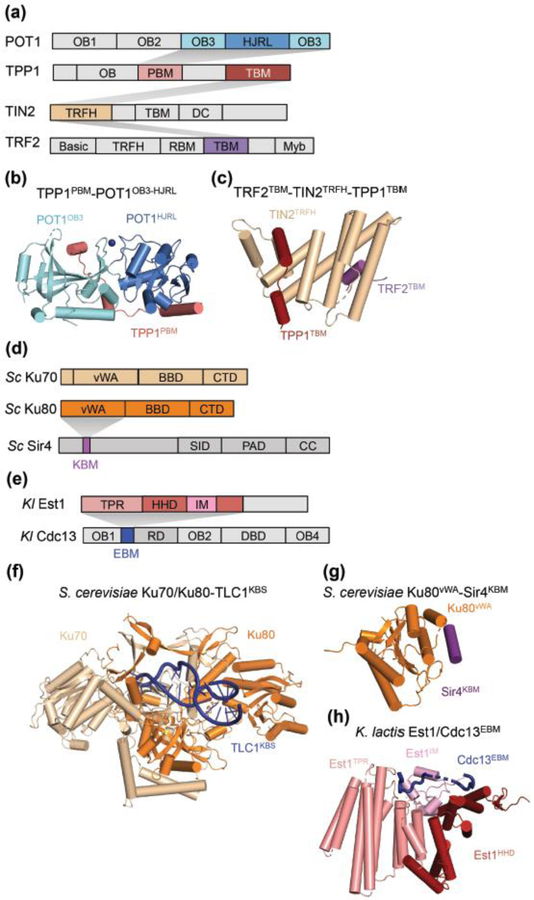
Similar to the diversity observed in telomerase holoenzyme composition and structure, telomeres are divergent in length, sequence and bound protein factors across eukaryotic species. As a consequence, mechanisms by which telomerase is recruited to telomeres are also species-specific (Figure 1) [54]. In humans, telomerase is recruited to telomeres by the shelterin complex, via the N-terminal OB-fold domain of TPP1 [55]. The molecular specificity of TPP1-POT1 interactions was defined recently from structures of the C-terminal portion of POT1, consisting of an OB-fold domain and a Holliday-junction resolvase-like (HJRL) domain inserted within the OB-fold, in complex with the POT1-binding motif (PBM) of TPP1 (Figure 4a and b) [56,57]. A structure of the telomeric repeat factor homology (TRFH) domain of TIN2 in complex with the distinct TIN2-binding motifs (TBMs) of TPP1 and TRF2 was also determined (Figure 4c) [58]. However, a structural understanding of how TPP1 and POT1 regulate telomerase recruitment, activation and inhibition at telomeres remains elusive.
Figure 4.
Structures of protein domains and sub-complexes with important roles in telomerase recruitment to telomeres. Structures recently determined are shown in colors. The shaded wedges indicate interactions. (a) Domain architectures of human POT1, TPP1, TIN2 and TRF2. OB indicates OB-folds. POT1 has three OB-fold domains and a Holliday junction resolvase-like (HJRL) domain embedded within the third OB-fold. TPP1 has an OB-fold domain, a POT1-binding motif (PBM), and a TIN2-binding motif (TBM). TIN2 has a TRFH domain, a TRFH-binding motif (TBM) and a dyskeratosis congenita (DC) disease mutation hotspot region. TRF2 has a basic domain, a TRFH domain, a RAP1-binding motif (RBM), a TIN2-binding motif (TBM) and a Myb DNA-binding domain. (b) Structure of the third OB-fold (OB3) and HJRL domains of POT1 in complex with the PBM of TPP1 (PDB 5H65) [56,57]. (c) Structure of the TRFH domain of TIN2 in complex with the TBMs of TPP1 and TRF2 (PDB 5XYF) [58]. (d) Domain architecture of S. cerevisiae Ku70, Ku80 and Sir4. Each of Ku70 and Ku80 contains a von Willebrand factor type A domain (vWA), a β-barrel domain (BBD) and a C-terminal α-helical domain (CTD). Sir4 has a Ku-binding motif (KBM), a Sir2-interacting domain (SID), a partitioning and anchoring domain (PAD), and a coiled-coil (CC) domain. (e) Domain architecture of K. lactis Est1 and Cdc13. Est1 has a tetratricopeptide repeat domain (TPR), a helical hairpin domain (HHD), and an insertion motif (IM). Cdc13 has three OB-fold domains, OB1, OB2 and OB4, an Est1-binding motif (EBM), a recruitment domain (RD), and a DNA-binding domain (DBD). (f) Structure of S. cerevisiae Ku70/Ku80 and the Ku binding site of TLC1 (PDB 5Y58) [25]. (g) Structure of S. cerevisiae Ku80 vWA domain with Sir4 KBM (PDB 5Y59) [25]. (h) Structure of K. lactis Est1 in complex with EBM of Cdc13 (PDB 5Y5A) [25].
Recent structural work has yielded considerable insight into telomerase recruitment in budding yeast [25]. In S. cerevisiae, telomerase recruitment is mediated by two pathways. In the first, Ku70/80 heterodimer, assembled with TLC1 as part of the budding yeast telomerase holoenzyme, targets telomerase to the telomeric double-stranded region via Ku interaction with Sir4, a subunit of the silent information regulator complex [59]. Recent crystal structures of Ku bound to the Ku-binding site (KBS) of TLC1 and of the Ku80 von Willebrand factor type A (vWA) domain with the Sir4 Ku-binding motif (KBM), together with associated mutational analyses, support the significance of Ku interactions to bridge TLC1 and Sir4 for telomerase recruitment (Figure 4d, f and g) [25]. The second pathway involves the telomeric repeat ssDNA binding protein Cdc13, which interacts with Est1, a subunit of the holoenzyme, and recruits telomerase to the telomeric 3’-overhang [54]. Chen et al. identified an Est1-binding motif (EBM) within the recruitment domain located between the first and second OB-folds of Cdc13, and determined the crystal structure of a large part of K. lactis Est1 and the EBM of Cdc13 (Figure 4e and h) [25]. Residues at the interface of Est1 and Cdc13 are highly conserved between K. lactis and S. cerevisiae. Disruption of the interface in S. cerevisiae predicted from K. lactis homology reduces telomerase association with telomeres in S-phase [25]. It remains to be understood how the interplay of the two recruitment pathways determines the activity of telomerase at telomeres. It is also of interest to gain additional biochemical and structural understanding of the dynamic interactions that recruit and activate S. pombe telomerase function at telomeres [60].
Conclusions
The recent cryo-EM structures of the T. thermophila and human telomerase holoenzymes have greatly advanced our structural understanding of how their components form such intricate networks of protein-protein and protein-RNA interactions to yield the functional RNPs. They provide a framework for further biochemical, functional and structural work. Particularly, improvement in the resolution of these EM structures will be necessary for understanding the biochemical principles of telomerase activity and for future drug design targeting telomerase. Crystallography and functional studies have also provided invaluable insight into telomerase assembly, regulation and telomere recruitment.
Acknowledgements
This work was funded by N.I.H. grant GM054198 to K.C. T.H.D.N. is a Fellow of the University of California, Berkeley Miller Institute for Basic Research in Science. E.N. is a Howard Hughes Medical Investigator.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declarations of interest: none
References
- 1.Blackburn EH, Greider CW, Szostak JW: Telomeres and telomerase: the path from maize, Tetrahymena and yeast to human cancer and aging. Nat Med 2006, 12:1133–1138. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH, Collins K: Telomerase: an RNP enzyme synthesizes DNA. Cold Spring Harb Perspect Biol 2011, 3:205–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aubert G: Telomere dynamics and aging. Prog Mol Biol Transl Sci 2014, 125:89–111. [DOI] [PubMed] [Google Scholar]
- 4.Hiyama E, Hiyama K: Telomere and telomerase in stem cells. Br J Cancer 2007, 96:1020–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collins K, Mitchell JR: Telomerase in the human organism. Oncogene 2002, 21:564. [DOI] [PubMed] [Google Scholar]
- 6.Shay JW, Wright WE: Role of telomeres and telomerase in cancer. Semin Cancer Biol 2011, 21:349–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanley SE, Armanios M: The short and long telomere syndromes: paired paradigms for molecular medicine. Curr Opin Genet Dev 2015, 33:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu RA, Upton HE, Vogan JM, Collins K: Telomerase Mechanism of Telomere Synthesis. Annu Rev Biochem 2017, 86:439–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Podlevsky JD, Chen JJ: Evolutionary perspectives of telomerase RNA structure and function. RNA Biol 2016:1–13. [DOI] [PMC free article] [PubMed]
- 10.Vasianovich Y, Wellinger RJ: Life and death of yeast telomerase RNA. J Mol Biol 2017. [DOI] [PubMed]
- 11.Min B, Collins K: An RPA-related sequence-specific DNA-binding subunit of telomerase holoenzyme is required for elongation processivity and telomere maintenance. Mol Cell 2009, 36:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang J, Miracco EJ, Hong K, Eckert B, Chan H, Cash DD, Min B, Zhou ZH, Collins K, Feigon J: The architecture of Tetrahymena telomerase holoenzyme. Nature 2013, 496:187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang J, Chan H, Cash DD, Miracco EJ, Ogorzalek Loo RR, Upton HE, Cascio D, O’Brien Johnson R, Collins K, Loo JA, et al. : Structure of Tetrahymena telomerase reveals previously unknown subunits, functions, and interactions. Science 2015, 350:aab4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong K, Upton H, Miracco EJ, Jiang J, Zhou ZH, Feigon J, Collins K: Tetrahymena Telomerase Holoenzyme Assembly, Activation, and Inhibition by Domains of the p50 Central Hub. Mol Cell Biol 2013, 33:3962–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen SB, Graham ME, Lovrecz GO, Bache N, Robinson PJ, Reddel RR: Protein composition of catalytically active human telomerase from immortal cells. Science 2007, 315:1850–1853. [DOI] [PubMed] [Google Scholar]
- 16.Egan ED, Collins K: Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol Cell Biol 2010, 30:2775–2786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sauerwald A, Sandin S, Cristofari G, Scheres SH, Lingner J, Rhodes D: Structure of active dimeric human telomerase. Nat Struct Mol Biol 2013, 20:454–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu RA, Dagdas YS, Yilmaz ST, Yildiz A, Collins K: Single-molecule imaging of telomerase reverse transcriptase in human telomerase holoenzyme and minimal RNP complexes. eLife 2015, 4:10.7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.**.Nguyen THD, Tam J, Wu RA, Greber BJ, Toso D, Nogales E, Collins K: Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 2018, 557:190–195.This paper reports the first cryo-EM reconstruction of human telomerase holoenzyme bound to a DNA substrate at subnanometer resolution, allowing all subunits to be fitted. The 11-sububnit complex has two lobes linked by two buldged stems that cross from one lobe, containing TERT and the two activity essential-domains of hTR, to the other lobe, containing a H/ACA RNP from a large family of RNPs including snoRNPs and scaRNPs. This study settles a longstanding debate regarding vertebrate telomerase subunit composition.
- 20.**.Jiang J, Wang Y, Sušac L, Chan H, Basu R, Zhou ZH, Feigon J: Structure of Telomerase with Telomeric DNA. Cell 2018, 173:1179–1190.This paper reports the cryo-EM reconstruction of T. thermophila telomerase holoenzyme bound to a locked-nucleic-acid oligonucleotide. The resolution of a conformationally stable subset of the holoenzyme subunits containing TERT, TER, some domains of the p50 and TEB subunits was improved to 4.8 Å compared to the previous 9.4 Å reconstruction of the full holoenzyme in Ref. [13], allowing the DNA substrate and the IFD region to be modeled. The potentially ciliate-specific path of DNA 5’ of the template duplex and the positioning of the TRAP IFD segment inserted into the RT domain have important implications for telomerase recruitment and telomere elongation in T. thermophila.
- 21.*.Lemieux B, Laterreur N, Perederina A, Noel JF, Dubois ML, Krasilnikov AS, Wellinger RJ: Active Yeast Telomerase Shares Subunits with Ribonucleoproteins RNase P and RNase MRP. Cell 2016, 165:1171–1181.This paper describes the identification and characterisation of Pop1, Pop6 and Pop7 as part of S. cerevisiae telomerase holoenzyme. These subunits are shared with RNase P and RNase MRP.
- 22.*.Páez-Moscoso DJ, Pan L, Sigauke RF, Schroeder MR, Tang W, Baumann P: Pof8 is a La-related protein and a constitutive component of telomerase in fission yeast. Nature Commun 2018, 9:587.describe the identification of Pof8, a La-family protein, as a component of S. pombe telomerase holoenzyme. Pof8 was shown to promote Lsm loading during telomerase assembly.
- 23.*.Mennie AK, Moser BA, Nakamura TM: LARP7-like protein Pof8 regulates telomerase assembly and poly(A)+TERRA expression in fission yeast. Nature Commun 2018, 9:586.describe the identification of Pof8, a La-family protein, as a component of S. pombe telomerase holoenzyme. Pof8 was shown to promote Lsm loading during telomerase assembly.
- 24.*.Collopy LC, Ware TL, Goncalves T, í Kongsstovu S, Yang Q, Amelina H, Pinder C, Alenazi A, Moiseeva V, Pearson SR, et al. : LARP7 family proteins have conserved function in telomerase assembly. Nature Commun 2018, 9:557.describe the identification of Pof8, a La-family protein, as a component of S. pombe telomerase holoenzyme. Pof8 was shown to promote Lsm loading during telomerase assembly.
- 25.**.Chen H, Xue J, Churikov D, Hass EP, Shi S, Lemon LD, Luciano P, Bertuch AA, Zappulla DC, Géli V, et al. : Structural Insights into Yeast Telomerase Recruitment to Telomeres. Cell 2018, 172:331–343.This paper reports the structures of S. cerevisiae Ku70/Ku80-TLC1KBS, Ku80vWA- Sir4KBM and K. lactis Est1-Cdc13EBM. These structures detail the molecular specificity of protein-protein and protein-RNA interactions crucial for telomerase recruitment to telomeres in S. cerevisiae.
- 26.Bajon E, Laterreur N, Wellinger RJ: A single templating RNA in yeast telomerase. Cell Rep 2015, 12:441–448. [DOI] [PubMed] [Google Scholar]
- 27.Chen JL, Blasco MA, Greider CW: Secondary structure of vertebrate telomerase RNA. Cell 2000, 100:503–514. [DOI] [PubMed] [Google Scholar]
- 28.Wu RA, Collins K: Human telomerase specialization for repeat synthesis by unique handling of primer-template duplex. EMBO J 2014, 33:921–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.*.Vogan JM, Zhang X, Youmans DT, Regalado SG, Johnson JZ, Hockemeyer D, Collins K: Minimized human telomerase maintains telomeres and resolves endogenous roles of H/ACA proteins, TCAB1, and Cajal bodies. eLife 2016, 5:10.7554.This paper develops a minimal cellular hTR, which lacks the binding site for the H/ACA proteins and Cajal body localization factor TCAB1. This minimal hTR can assemble with TERT to form biologically active enzyme that can maintain telomere in vivo. The findings give important insight into the requirements for telomerase biogenesis and trafficking in vertebrate cells.
- 30.Chen L, Roake CM, Freund A, Batista PJ, Tian S, Yin YA, Gajera CR, Lin S, Lee B, Pech MF, et al. : An Activity Switch in Human Telomerase Based on RNA Conformation and Shaped by TCAB1. Cell 2018, 174:218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim N, Theimer CA, Mitchell JR, Collins K, and Feigon J, Effect of pseudouridylation on the structure and activity of the catalytically essential P6.1 hairpin in human telomerase RNA. Nucleic Acids Res, 2010. [DOI] [PMC free article] [PubMed]
- 32.Zemora G, Handl S, Waldsich C: Human telomerase reverse transcriptase binds to a pre-organized hTR in vivo exposing its template. Nucleic Acids Res 2016, 44:413–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell M, Gillis A, Futahashi M, Fujiwara H, Skordalakes E: Structural basis for telomerase catalytic subunit TERT binding to RNA template and telomeric DNA. Nat Struct Mol Biol 2010, 17:513–518. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y, Yesselman JD, Zhang Q, Kang M, Feigon J: Structural conservation in the template/pseudoknot domain of vertebrate telomerase RNA from teleost fish to human. Proc Natl Acad Sci U S A 2016, 35:E5125–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parks JW, Stone MD: Single-Molecule Studies of Telomeres and Telomerase. RNA 2017, 46:357–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell JR, Cheng J, Collins K: A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3’ end. Mol Cell Biol 1999, 19:567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitchell JR, Collins K: Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase in vivo and in vitro. Mol Cell 2000, 6:361–371. [DOI] [PubMed] [Google Scholar]
- 38.Massenet S, Bertrand E, Verheggen C: Assembly and trafficking of box C/D and H/ACA snoRNPs. RNA Biol 2017, 14:680–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wegman-Ostrosky T, Savage SA: The genomics of inherited bone marrow failure: from mechanism to the clinic. Br J Haematol 2017, 177:526–542. [DOI] [PubMed] [Google Scholar]
- 40.Li L, Ye K: Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 2006, 443:302–307. [DOI] [PubMed] [Google Scholar]
- 41.Li S, Duan J, Li D, Yang B, Dong M, Ye K: Reconstitution and structural analysis of the yeast box H/ACA RNA-guided pseudouridine synthase. Genes Dev 2011, 25:2409–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Upton HE, Chan H, Feigon J, Collins K: Shared Subunits of Tetrahymena Telomerase Holoenzyme and Replication Protein A Have Different Functions in Different Cellular Complexes. J Biol Chem 2017, 292:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goldin S, Kertesz Rosenfeld K, Manor H: Tracing the path of DNA substrates in active Tetrahymena telomerase holoenzyme complexes: mapping of DNA contact sites in the RNA subunit. Nucleic Acids Res 2012, 40:7430–7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrova OA, Mantsyzov AB, Rodina EV, Efimov SV, Hackenberg C, Hakanpää J, Klochkov VV, Lebedev AA, Chugunova AA, Malyavko AN, et al. : Structure and function of the N-terminal domain of the yeast telomerase reverse transcriptase. Nucleic Acids Res 2018, 46:1525–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stone MS, Mihalusova M, O’Connor CM, Prathapam R, Collins K, Zhuang X: Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature 2007, 446:458–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh M, Wang Z, Koo BK, Patel A, Cascio D, Collins K, Feigon J: Structural basis for telomerase RNA recognition and RNP assembly by the holoenzyme La family protein p65. Mol Cell 2012. [DOI] [PMC free article] [PubMed]
- 47.Witkin KL, Collins K: Holoenzyme proteins required for the physiological assembly and activity of telomerase. Genes Dev 2004, 18:1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seto AG, Zaug AJ, Sobel SG, Wolin SL, Cech TR: Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 1999, 401:177–180. [DOI] [PubMed] [Google Scholar]
- 49.Tang W, Kannan R, Blanchette M, Baumann P: Telomerase RNA biogenesis involves sequential binding by Sm and Lsm complexes. Nature 2012, 484:260–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Box JA, Bunch JT, Tang W, Baumann P: Spliceosomal cleavage generates the 3’ end of telomerase RNA. Nature 2008, 456:910–914. [DOI] [PubMed] [Google Scholar]
- 51.Sperger JM, Cech TR: A stem-loop of Tetrahymena telomerase RNA distant from the template potentiates RNA folding and telomerase activity. Biochemistry 2001, 40:7005–7016. [DOI] [PubMed] [Google Scholar]
- 52.Cash DD, Feigon J: Structure and folding of the Tetrahymena telomerase RNA pseudoknot. Nucleic Acids Res 2017, 45:482–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Deshpande AP, Collins K: Mechanisms of template handling and pseudoknot folding in human telomerase and their manipulation to expand the sequence repertoire of processive repeat synthesis. Nucleic Acids Res 2018, 46:7886–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nandakumar J, Cech TR: Finding the end: recruitment of telomerase to telomeres. Nat Rev Mol Cell Biol 2013, 14:69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hockemeyer D, Collins K: Control of human telomerase action at telomeres. Nat Struct Mol Biol 2015, 22:848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.*.Chen C, Gu P, Wu J, Chen X, Niu S, Sun H, Wu L, Li N, Peng J, Shi S, et al. : Structural insights into POT1-TPP1 interaction and POT1 C-terminal mutations in human cancer. Nat Commun 2017, 8:14929.report the structure of POT1 C-terminal OB-fold and HJRL domains in complex with TPP1PBM, revealing the molecular specificity of POT1-TPP1 interactions. Additional biochemical characterization suggest that some cancer-associated mutations in POT1 interfere with TPP1-POT1 interactions.
- 57.*.Rice C, Shastrula PK, Kossenkov AV, Hills R, Baird DM, Showe LC, Doukov T, Janicki S, Skordalakes E: Structural and functional analysis of the human POT1-TPP1 telomeric complex. Nat Commun 2017, 8:14928.report the structure of POT1 C-terminal OB-fold and HJRL domains in complex with TPP1PBM, revealing the molecular specificity of POT1-TPP1 interactions. Additional biochemical characterization suggest that some cancer-associated mutations in POT1 interfere with TPP1-POT1 interactions.
- 58.Hu C, Rai R, Huang C, Broton C, Long J, Xu Y, Xue J, Lei M, Chang S, Chen Y: Structural and functional analyses of the mammalian TIN2-TPP1-TRF2 telomeric complex. Cell Res 2017, 27:1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hass EP, Zappulla DC: The Ku subunit of telomerase binds Sir4 to recruit telomerase to lengthen telomeres in S. cerevisiae. eLife 2015, 4:e07750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Armstrong CA, Tomita K: Fundamental mechanisms of telomerase action in yeasts and mammals: understanding telomeres and telomerase in cancer cells. Open Biol 2017, 7:160338. [DOI] [PMC free article] [PubMed] [Google Scholar]