Supplemental Digital Content is available in the text.
Keywords: decision-making, machine learning, reperfusion, stroke, tissue survival, triage
Abstract
Background and Purpose:
The ISLES challenge (Ischemic Stroke Lesion Segmentation) enables globally diverse teams to compete to develop advanced tools for stroke lesion analysis with machine learning. Detection of irreversibly damaged tissue on computed tomography perfusion (CTP) is often necessary to determine eligibility for late-time-window thrombectomy. Therefore, the aim of ISLES-2018 was to segment infarcted tissue on CTP based on diffusion-weighted imaging as a reference standard.
Methods:
The data, from 4 centers, consisted of 103 cases of acute anterior circulation large artery occlusion stroke who underwent diffusion-weighted imaging rapidly after CTP. Diffusion-weighted imaging lesion segmentation was performed manually and acted as a reference standard. The data were separated into 63 cases for training and 40 for testing, upon which quality metrics (dice score coefficient, Hausdorff distance, absolute lesion volume difference, etc) were computed to rank methods based on their overall performance.
Results:
Twenty-four different teams participated in the challenge. Median time to CTP was 185 minutes (interquartile range, 180–238), the time between CTP and magnetic resonance imaging was 36 minutes (interquartile range, 25–79), and the median infarct lesion size was 15.2 mL (interquartile range, 5.7–45). The best performance for Dice score coefficient and absolute volume difference were 0.51 and 10.1 mL, respectively, from different teams. Based on the ranking criteria, the top team’s algorithm demonstrated for average Dice score coefficient and average absolute volume difference 0.51 and 10.2 mL, respectively, outperforming the conventional threshold-based method (dice score coefficient, 0.3; volume difference, 15.3). Diverse algorithms were used, almost all based on deep learning, with top-ranked approaches making use of the raw perfusion data as well as methods to synthetically generate complementary information to boost prediction performance.
Conclusions:
Machine learning methods may predict infarcted tissue from CTP with improved accuracy compared with threshold-based methods used in clinical routine. This dataset will remain public and can be used to test improvement in algorithms over time.
The DEFUSE 3 (Endovascular Therapy Following Imaging Evaluation for Ischemic Stroke 3) and DAWN (Clinical Mismatch in the Triage of Wake Up and Late Presenting Strokes Undergoing Neurointervention With Trevo) trials demonstrated that endovascular treatment is highly efficacious in patients seen 6 to 24 hours after onset.1,2 Both trials selected patients predominantly using computed tomography (CT) perfusion (CTP), and as a consequence, CTP-based selection of patients in the late time window is now in the American Heart Association guidelines.3 These events have spawned a surge in the adoption of CTP in centers worldwide. The dynamic CTP images acquired by the scanner need postprocessing to obtain estimates of the volumes of the infarct core and hypoperfused regions. These volumes are then used to determine suitability for treatment. Although current threshold-based CTP algorithms4–6 are capable of identifying patients with a high response to treatment,7 correspondence of the CTP-derived measurements to reference standard diffusion-weighted imaging (DWI) lesions is still suboptimal.4 Given the inherent complexity of acute ischemic stroke lesion development, data-driven machine learning methods could be used to improve the estimate of core infarcted tissue.
The ISLES challenge (Ischemic Stroke Lesion Segmentation) was created in 2015 to encourage researchers around the world to develop advanced tools for stroke lesion analysis.8 ISLES publicly provides standardized, high-quality datasets to overcome the limitations of varying dataset sizes and heterogeneity in postprocessing, making it possible to compare novel approaches in a fair way.9,10 An increasing number of teams have participated in this challenge, which is annually organized in conjunction with the international conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). Over time, there has been intense development in the algorithms used, starting from classical machine learning tools and now focusing almost exclusively on deep learning techniques.9
Based on the development of clinical trials and community feedback, the aim of the ISLES 2018 was to predict the infarction core based on CTP imaging from acute ischemic stroke patients, using DWI-based manual segmentation from magnetic resonance imaging (MRI) acquired shortly after. This cohort was initially reported on by Cereda et al4 using threshold-based method. This report summarizes the logistics of the challenge, its results in comparison with a commercially available threshold-based software for CTP postprocessing, and the methods used by teams along with the results of the top-performing teams.
Methods
We point readers toward the publicly available data set, which since the challenge in 2018 has already been used in other studies,11,12 to encourage more rapid innovation. The data can be requested at https://www.smir.ch/ISLES/Start2018.
Patients and Image Acquisition
The patients were part of 2 prospective ischemic stroke trials13,14 diagnosed with acute large artery occlusive ischemic stroke without signs of hemorrhage ≤8 hours after last seen normal. Ethics approval was obtained from the local institutional review board at each center. MRI was acquired within 3 hours of CTP. DWI was coregistered to the CTP acquisition by aligning both to the Montreal Neurological Institute atlas. The cases were previously reported on by Cereda et al4 but exclusively using a threshold-based method and without the use of training/validation splitting. Unlike the Cereda study, tissue regions were not excluded if there was evidence of partial reperfusion (normally perfused DWI lesions with time to maximum ≤4 s on the CTP study), with the goal of making the challenge generalizable to real-world conditions.
The goal of the challenge was to accurately segment infarct core from CTP, which was defined by manual segmentation of DWI on subsequent MRI. No treatment occurred between the CTP and DWI. The segmentations, which acted as the reference standard for the challenge, were manually delineated by a single investigator, a stroke neurologist with >10 years of experience, and then subjected to group review until acceptance of the delineated lesion by the other group members. All investigators were blinded to baseline perfusion and all other imaging.
In total, datasets from 103 patients presenting with acute large artery occlusion anterior circulation ischemic stroke from 3 US centers and 1 Australian center were used in the challenge (Table 1). The CTP images were acquired from all 4 major manufacturers (GE, Philips, Siemens, and Toshiba). More details are available in the study by Cereda et al.4
Table 1.
Demographics of the Patient Population

Training and Testing Data Provided
The data were split into 2 groups—training group and testing group—randomized by vendor strata to ensure a balance between vendors in the two sets. To test the robustness of the algorithms, cases with no lesions were included in the testing set. To include a wide range of possible lesions, 40 patients were included in the test set for the challenge. In this group, the reference standard lesion segmentations were withheld from the teams, and the performance of the algorithm was determined by the organizing committee using predetermined criteria (see below). The remaining 63 cases comprised the training set, for which the competitors received the CTP images, DWI images, and DWI-based segmentations, which allowed them to train their models.
Teams were given the following types of training data: (1) motion-corrected dynamic CTP source data, which had been resampled to a standardized 1-s temporal resolution; (2) postprocessed perfusion maps, that is, cerebral blood flow (CBF), cerebral blood volume, mean transit time, and time to maximum, calculated by conventional thresholding method (RAPID; iSchemaview, Menlo Park, CA). No threshold-based lesion segmentations were provided; (3) DWI lesion segmentation in binary form and (4) the DWI images themselves. Of note, noncontrast enhanced CT scans were not provided to the teams. For the test set, only the CTP raw data and the postprocessed maps were provided (ie, no segmentations or DWI images). All images were provided at 256×256 in-plane resolution and native slice thickness with one exception: Toshiba Aquilion One 320 slice data (0.5-mm native resolution) was downsampled to 10-mm thickness by volume averaging sets of 20 slices. CTP covered from 2.4 to 16 cm in the axial direction depending on hardware and prediction of the lesion for each case was performed only within this image volume.
Challenge
Organization of the data and assessment of the algorithms were the responsibility of an international team of experts in artificial intelligence, neurology, and neuroradiology from Belgium, Germany, Switzerland, the United Kingdom, and the United States, who did not participate in the challenge.
Information about the challenge, its background, and aims were published online on the official website of the challenge (http://www.isles-challenge.org) that included a link to the training set, which was used to validate and optimize the method used by each participant. Shortly before the 2018 Medical Image Computing and Computer Assisted Intervention conference, the raw CTP data from the test sets were released. The teams were asked to run their algorithms and upload their segmentation results along with the submission of a short abstract describing their algorithmic approach. Finally, the results from each participant were compared with the reference standard manual infarct segmentations, and the teams were ranked using predetermined evaluation metrics.
Evaluation Metrics
The following metrics were used for evaluation: the Dice score coefficient (DSC), Hausdorff distance (HD), average and absolute lesion volume difference (VD), precision, recall, and average symmetrical surface distance. DSC is a measure of overlap between the reference standard on DWI and the predicted lesion from CTP, hence it tests the resemblance between the two lesions, while HD calculates the largest distance between the two contours representing the prediction lesion and the reference standard on DWI. For more details, the reader is pointed to Winzeck et al9 and Maier et al.8 The same metrics were assessed using the suggested parameter (relative CBF, <0.38)4 from the threshold-based method, using the relative CBF core as the predicted infarct core segmentation.
A case-wise approach was used to calculate ranks for each team, following the rationale of Maier et al8 as patient cases can have different degrees of complexity in predicting stroke outcome. DSC was the most important as it combines precision and sensitivity. First, DSC, HD, and average symmetrical surface distance were calculated for each case, for each team, with high DSC and low HD resulting in a high rank for an individual case. The mean of these ranks per case provided a case-specific rank. Calculating the average of all case-specific ranks through all cases for each team resulted in the team’s final overall rank.
Statistical Analysis
For each team and for the threshold-based method, the mean and SD of DSC, mean absolute VD, precision, and recall were calculated. Furthermore, the HD and average symmetrical surface distance from each case were compared to define the best case from each team. Wilcoxon signed-rank test (for nonuniformly distributed data) was used to compare results among teams. Furthermore, based on DSC and absolute VD, the best- and worst-performing test cases from the top 5 teams were defined and analyzed according to lesion volume and lesion location.
Results
Challenge Characteristics
Demographics of the patients and scanners included in the study can be found in Table 1. Median time from last seen normal to CTP was 185 minutes (interquartile range [IQR], 180–238). Time between completion of CT and the start of MRI ranged from 15 to 181 minutes (median, 36 minutes). Lesion volume ranged from 0 to 309 mL (average, 32 mL). In 6 patients, there was no diffusion lesion present (ie, zero core volume).
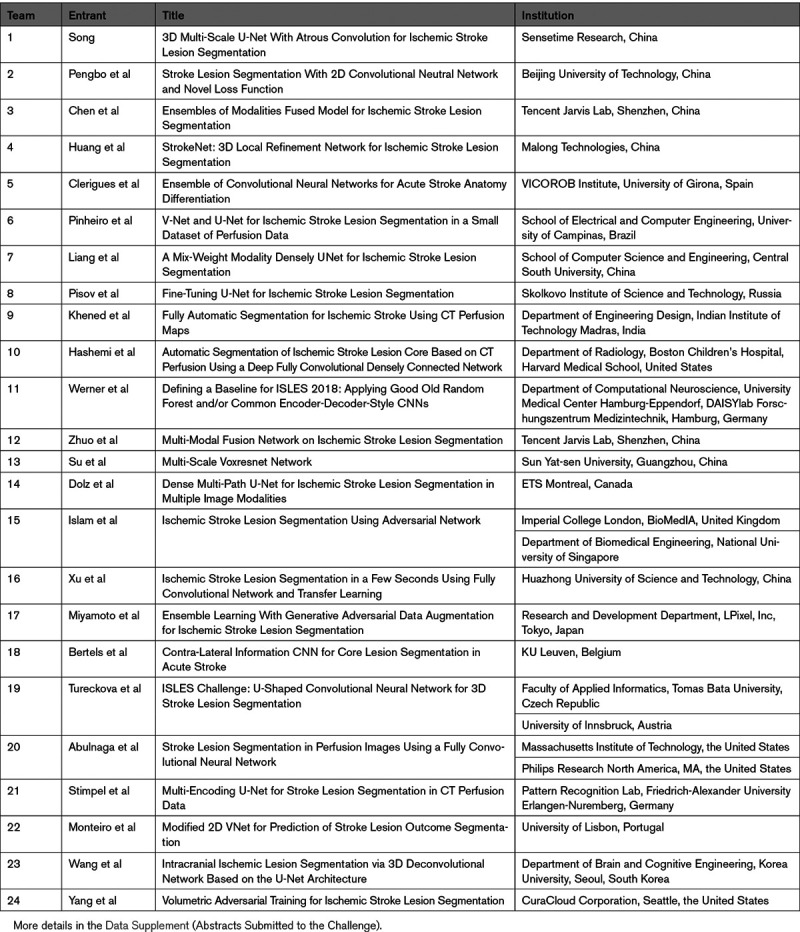
Teams from 15 different countries, as well as multinational collaboration teams, were registered. In total, 24 teams affiliated to research institutes, university hospitals, and industry participated and submitted results on all 40 test cases (Table 2).
Table 2.
Investigator, Title of the Submitted Abstracts, and Affiliation of the Participating Teams According to the Overall Ranking
Best-Performing Teams for Overall Performance
Based on the mean ranking of their performance on DSC, HD, and average symmetrical surface distance, the winner was Song from East China Normal University (team No. 1). He used a deep learning generative adversarial network algorithm in which the CTP data (raw and postprocessed maps) were used to create a pseudo-DWI image based on a U-net architecture. This pseudo-DWI image was compared with the true DWI image using a discriminator that attempted to determine whether the image presented was the real or pseudo-DWI. The features that were useful for making the distinction were fed back into the generator to improve its ability to predict the pseudo-DWI lesion. Once trained, the pseudo-DWI image was segmented using a separate deep learning algorithm that had been trained on a large number of self-segmented DWI scans.
Team No. 2 (Pengbo et al; Beijing University of Technology) used a convolutional neural network utilizing a 2-dimensional U-net with residual connection as the model backbone. To balance the gradients of the positive and negative areas in the training phase and highlight the stroke lesions, they proposed a novel loss function that contains a weight cross-entropy loss and generalized Dice score loss. Based on experiments during the training phase, this hybrid loss was found to be more stable.
Team No. 3 (Chen et al; Youtu Laboratory, Tencent) used a 2.5-dimensional framework, based on an ensemble of networks, including U-nets, to extract and fuse information from different modalities.
For more details about methods used by each team, please see the Data Supplement or refer to the proceeding of the challenge.15
Best-Performing Teams for Specific Metrics
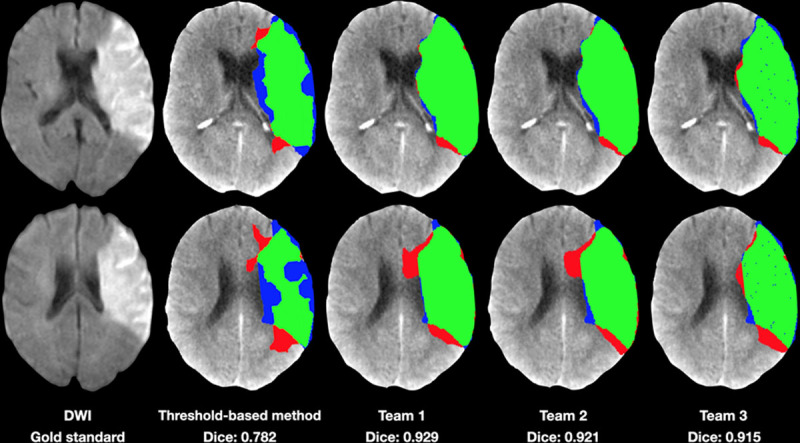
The top-ranking team (team No. 1; Song) achieved an average DSC of 0.51±0.31 (mean±SD). Several teams achieved similar performance (Table 3). This result is superior to the DSC achieved with a traditional threshold-based CBF method (0.34±0.29; P<0.001). The top-ranking team for average absolute lesion VD was team No. 2, Pengbo et al (10.0±10.5 mL). Their results on the absolute volume metric compared favorably to the threshold-based CBF method (15.3±16.4 mL; P=0.002). Figure 1 is a case example showing predictions for infarct core for each of the top 3 teams along with the conventional threshold-based method. The results of all teams compared with the results obtained with a threshold-based CBF method are shown in Table 3 and Figures 2 and 3.
Table 3.
Results of Each Team Obtained at the Time of Challenge Submission and the Results Obtained From a Threshold-Based Method, Using Relative Cerebral Blood Flow <0.38
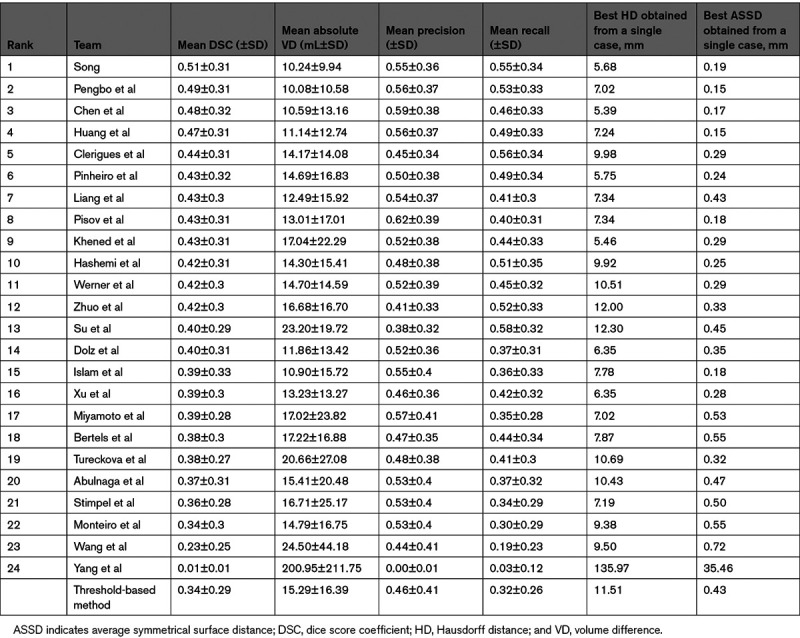
Figure 1.
Case example illustrating a patient with a large left middle cerebral artery infarction as seen on the dice score coefficient (DWI; far left). The estimated infarction core by threshold-based method and by the three top teams is highlighted. Green, true positive; blue, false negative; and red, false positive. The results of team 3 show multiple little blue dots (low probability), possibly due to artifacts resulting from a lack of regularization of the network or postprocessing.
Figure 2.
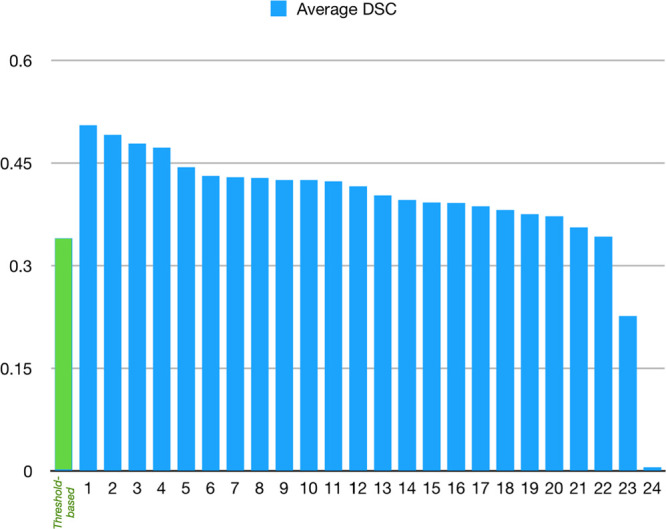
Chart showing the average of dice score coefficient (DSC) for each team compared with the results obtained from the threshold-based method.
Figure 3.
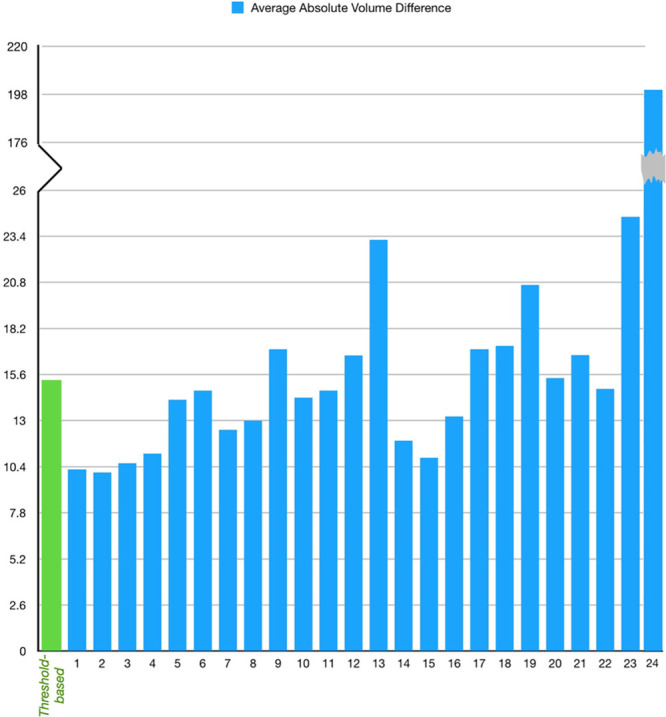
The average absolute volume difference between reference standard and automatically predicted infarction volume from the 24 teams compared with the threshold-based method.
Best- and Worst-Performing Cases
From the top 5 teams, 8 cases were identified as the best-performing cases, and 9 cases as the worst-performing cases, based on DSC. Twenty cases were identified as the best cases and 13 cases were identified as the worst cases based on absolute VD. Due to the overlap between teams, the number of best and worst cases was varying in each category.
Dice Score Coefficient
The median DWI volume for the best-performing cases was 72.8 mL (IQR, 42.45–103.1), and the median dice was 0.86 (IQR, 0.81–0.91); all cases were M1 lesions. The median DWI volume for the worst-performing cases was 6.7 mL (IQR, 2.8–9), and the median dice was 0.28 (IQR, 0.2–0.37; Figure I in the Data Supplement). In these cases, 66.7% were M1 lesions and 33.3% were M2.
Absolute VD
The median DWI volume from the best-performing cases was 7.2 mL (IQR, 3–20.7), and the median absolute VD was 2.6 mL (IQR, 0.64–5.06). In these cases, 70% were M1 lesions, 15% were M2, and 15% showed no occlusion. The median DWI volume from the worst-performing cases was 41.3 mL (IQR, 6.7–76.4), and the median absolute VD was 26.41 mL (IQR, 9.36–36.87; Figure II in the Data Supplement). In these cases, 92.3% were M1 lesions, 7.7% were M2, and 15.4% were P1.
Discussion
In this article, we describe the structure and rationale of the concluded ISLES 2018 stroke segmentation challenge. Many diverse teams throughout the world, in industry and academia, participated. The top teams demonstrated significantly better performance than a CBF-based threshold method (relative CBF, <0.38) that has previously been shown to best match DWI volume.4 The top teams almost exclusively incorporated deep learning methods using convolution neural networks, with a wide variety of network architectures, input data, and training methods.
Using machine learning algorithms to detect and segment abnormal imaging findings, including ischemic lesions, is an active research field that has been growing in recent years.16–18 However, building a satisfactory algorithm is an active process, requiring continuous improvement and testing. For this reason, having a well-characterized data set upon which to test algorithms is critical, since it enables the identification of improved performance. Furthermore, having a standardized data set enables researchers to directly test their algorithms and compare them to the results of other researchers, thus accelerating progress on the most promising methods.
For this reason, the ISLES challenge was initiated in 2015 to provide a platform with processed data that allows a continuous validation of different algorithms in a fair manner.10 The images provided are already preprocessed and annotated by experts in the field, so researchers can apply their method directly to the given data, generating results that are not affected by preprocessing steps or processing software. The datasets from the 2018 challenge and the previous years are available on the challenge website (http://www.isles-challenge.org) to allow further evaluation and testing of new algorithms. During the last several years of the challenge, there has been obvious progress in the algorithms used, starting from classical machine learning tools to advanced deep learning techniques. Lessons learned from previous ISLES challenges were summarized in the previously published articles (Maier et al8 and Winzeck et al9).
CT is increasingly being used for acute stroke triage, given its lower cost, widespread availability, and only few exclusion criteria. Core infarct size is an important predictor of outcome in acute stroke and represents the minimal lesion that can be expected after thrombectomy. Its relationship to at-risk or penumbral tissue has been used as entry criteria for recent thrombectomy trials in the late time window (6–24 hours).1 However, one of the disadvantages of CT, in contrast to MRI, is the challenge of accurately defining infarction core.19 Different criteria have been proposed over the years to estimate core from CTP data, with a threshold of relative CBF <0.38 of normal tissue being the suggested metric in the software used in this study.4 In contrast, DWI is generally recognized as a better estimate of irreversibly damaged tissue and serves as a marker of infarct core for MRI studies. DWI-based signal hyperintensity was used in this study as a reference standard for infarct core, which was to be predicted from CTP data acquired at a median time difference of 36 minutes. Recent reports of trained artificial intelligence algorithms show that they can perform well on narrow sets of input data but fail when exposed to real-world conditions.20 For this reason, the cases included in this challenge were chosen to be widely representative, from multiple institutions and scanned on all 4 major CT manufacturers. We additionally included cases in which there was potential reperfusion of infarcted tissue—a known challenge for CTP prediction. Of note, these regions were excluded from the earlier study of this cohort using threshold-based method. Thus, the structure of the challenge will reward algorithms that can generalize well to different case characteristics, which is critical for the clinical use of artificial intelligence methods.
Some of the teams showed innovation in their algorithm designs. The top team combined the provided raw perfusion data to the processed perfusion maps in an integrated network to boost their model and improve the prediction of the DWI lesion, as it has also been shown in the study by Pinto et al for the stroke prediction from MRI data.21 Despite the majority of approaches using neural networks of similar architectures, the performance variations indicate the difficulty of designing a successful machine learning algorithm for stroke lesion segmentation.
Performance differences among teams were observed (Table 3), alluding to potential overfitting and lack of regularization, stemming from the low sample size of the training dataset. Second, we observed that solutions having strong heuristics for postprocessing routines (eg, hard-coded elimination of a number of isolated objects) were also outperformed by solutions where regularization was performed implicitly via standard deep learning solutions, such as data augmentation and dropout.
Possible causes for poor algorithm performance include reperfusion of dead tissue, imperfect registration between the DWI and CTP which can have a large impact on the overlap between very small lesions and make it impossible to achieve a high DSC for such cases.
Teams varied in performance in each metric, for example, DSC was higher in team 1, but team 2 showed better VD, as DSC measures the relative overlap between the predicted lesion and the reference standard, whereas the VD is not influenced by the amount of overlap. Hence these metrics do not measure the same feature, so they do not correlate perfectly. In fact, there is a trade-off between these two metrics, that is, to obtain an optimal DSC, it is often better to allow for oversegmentation.22
The case-based analysis of the best- and worst-performing cases from the top 5 teams shows that the bigger the lesion, the better the DSC but the larger the absolute VD. There was no apparent correlation between performance and the anatomic or territorial location of the lesion.
Previous works aimed to use machine learning for stroke lesion segmentation, mostly focusing on MRI studies, giving the advantage of higher tissue contrast and DWI as a sensitive parameter for infarction core. In this study, we show the performance of deep learning techniques on CTP with MRI as a reference standard. We also show the shift toward more advanced deep learning techniques such as generative adversarial network. Also, the value of using raw perfusion data (not only the postprocessed perfusion maps) to reach a more accurate prediction is demonstrated.
The results of this challenge show the advantages of applying deep learning in stroke patients. The algorithms from top teams compared favorably with the state-of-the-art CTP postprocessing tool used in clinical practice, showing more accurate prediction of infarction core, which allows for better decisions regarding use of invasive therapy. Therefore, our results support the necessity of shifting the conventional stroke workflow toward the integration of modern computational algorithms in decision-making, which will improve stroke care and patient management.
There were several limitations to our study. Due to the strict criteria of including patients with CTP and DWI within 3 hours of each other, the dataset is relatively small. This points out the need for different centers to collaborate and share data to improve the ability of researchers to improve their algorithms. Also, the reference standard was defined as diffusion restriction on MRI, which was always performed after CTP. This may mean that the predictions do not exactly match the infarct core at the time of the CTP but rather may reflect some degree of lesion growth. Of course, it was not technically possible to acquire both modalities at the exact same time, and the retrospective nature of the dataset precluded randomization of CTP and MRI. Another potential limitation is that of spontaneous partial reperfusion, which would alter the CTP lesion severity and possibly introduce some degree of DWI lesion reversal that would render the CTP-to-DWI relationship much less predictable.23,24 Another limitation is the coverage on the z axis. The ISLES 2018 datasets were acquired in the period 2004 to 2012, and some scans were, therefore, acquired on older systems. While there have not been any major breakthroughs in improved contrast-to-noise of CTP, scanners generally have increased in terms of brain coverage along the patient z axis of CTP over the last 20 years. Seventy-six of 103 datasets were under 8-cm coverage along the patient z axis, whereas with modern scanners with wider detector arrays, at least 8 cm is usually imaged. One could speculate that larger spatial coverage could provide more context for the infarct prediction and potentially improve it. The limited spatial coverage will also result in lower absolute volumetric error than what would have been seen in whole-brain studies due to the smaller image volume acquired. So absolute volumetric error is biased by low coverage in some cases and not likely to indicate performance in a dataset with larger spatial coverage, but it still serves to compare between participating groups. Finally, the noncontrast CT data were not provided to the teams despite being typically available in the acute stroke setting. The presence of noncontrast CT hypodensity contains useful information about infarct core and might have helped the teams with their predictions. However, some of this information is present in the baseline (ie, prebolus arrival) CTP raw images, and some of the top-performing teams took advantage of this. Also, we did not include CT angiography data, given the wide variety of CT angiography methods in the cases. Consideration will be given to including coregistered noncontrast CT and CT angiography in future challenges.
Conclusions
This report summarizes the results of the ISLES 2018 challenge to predict core infarct lesions using CTP in acute large artery occlusive stroke. The top-ranked teams used various deep learning convolutional neural network approaches, including some state-of-the-art methods such as generative adversarial networks. The performance of the top groups was significantly better than threshold-dependent, rules-based models. The ISLES 2018 challenge data remain publicly available for researchers to improve methods for automatically segmenting infarct core on CTP.
Sources of Funding
Dr Robben is supported by Flanders Innovation and Entrepreneurship.
Disclosures
Dr Christensen has an equity interest from iSchemaView. Dr Lansberg is a consultant for Biogen, Genentech, NuvOx Pharma, and Nektar Therapeutics. Dr Parsons reports research collaborations with Siemens, Toshiba/Canon, and Apollo Medical Imaging. Dr Robben is an employee of icometrix; he received a grant from Flanders Innovation and Entrepreneurship; in addition, Dr Robben has a pending patent application to AIF selection. Dr Zaharchuk has received research funding from the National Institutes of Health, GE Healthcare, and Bayer Healthcare and equipment support from Nvidia; he is a board member and holds stock in Subtle Medical. The other authors report no conflicts.
Supplemental Materials
List of Contributors
Abstracts Submitted to the Challenge
Online Figures I and II
Supplementary Material
Nonstandard Abbreviations and Acronyms
- CBF
- cerebral blood flow
- CT
- computed tomography
- CTP
- computed tomography perfusion
- DSC
- dice score coefficient
- DWI
- diffusion-weighted imaging
- HD
- Hausdorff distance
- IQR
- interquartile range
- ISLES
- Ischemic Stroke Lesion Segmentation
- MRI
- magnetic resonance imaging
- VD
- volume difference
A. Hakim and S. Christensen contributed equally.
M. Reyes and G. Zaharchuk contributed equally.
The Data Supplement is available with this article at https://www.ahajournals.org/doi/suppl/10.1161/STROKEAHA.120.030696.
For Sources of Funding and Disclosures, see page 2336.
Contributor Information
Arsany Hakim, Email: arsany.hakim@insel.ch.
Søren Christensen, Email: sorenc@stanford.edu.
Stefan Winzeck, Email: sw742@cam.ac.uk.
Maarten G. Lansberg, Email: lansberg@stanford.edu.
Mark W. Parsons, Email: mark.parsons@unsw.edu.au.
Christian Lucas, Email: christian.lucas@hzg.de.
David Robben, Email: david.robben@kuleuven.be.
Roland Wiest, Email: roland.wiest@insel.ch.
Mauricio Reyes, Email: mauricio.reyes@med.unibe.ch.
References
- 1.Nogueira RG, Jadhav AP, Haussen DC, Bonafe A, Budzik RF, Bhuva P, Yavagal DR, Ribo M, Cognard C, Hanel RA, et al. ; DAWN Trial Investigators. Thrombectomy 6 to 24 hours after stroke with a mismatch between deficit and infarct. N Engl J Med. 2018;378:11–21. doi: 10.1056/NEJMoa1706442 [DOI] [PubMed] [Google Scholar]
- 2.Albers GW, Marks MP, Kemp S, Christensen S, Tsai JP, Ortega-Gutierrez S, McTaggart RA, Torbey MT, Kim-Tenser M, Leslie-Mazwi T, et al. ; DEFUSE 3 Investigators. Thrombectomy for stroke at 6 to 16 hours with selection by perfusion imaging. N Engl J Med. 2018;378:708–718. doi: 10.1056/NEJMoa1713973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Powers WJ, Rabinstein AA, Ackerson T, Adeoye OM, Bambakidis NC, Becker K, Biller J, Brown M, Demaerschalk BM, Hoh B, et al. ; American Heart Association Stroke Council. 2018 guidelines for the early management of patients with acute ischemic stroke: a guideline for healthcare professionals from the American Heart Association/American Stroke Association. Stroke. 2018;49:e46–e110. doi: 10.1161/STR.0000000000000158 [DOI] [PubMed] [Google Scholar]
- 4.Cereda CW, Christensen S, Campbell BCV, Mishra NK, Mlynash M, Levi C, Straka M, Wintermark M, Bammer R, Albers GW, et al. A benchmarking tool to evaluate computer tomography perfusion infarct core predictions against a DWI standard. J Cereb Blood Flow Metab. 2016;36:1780–1789. doi: 10.1177/0271678X15610586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mokin M, Levy EI, Saver JL, Siddiqui AH, Goyal M, Bonafé A, Cognard C, Jahan R, Albers GW; SWIFT PRIME Investigators. Predictive value of RAPID assessed perfusion thresholds on final infarct volume in SWIFT PRIME (Solitaire With the Intention for Thrombectomy as Primary Endovascular Treatment). Stroke. 2017;48:932–938. doi: 10.1161/STROKEAHA.116.015472 [DOI] [PubMed] [Google Scholar]
- 6.Austein F, Riedel C, Kerby T, Meyne J, Binder A, Lindner T, Huhndorf M, Wodarg F, Jansen O. Comparison of perfusion CT software to predict the final infarct volume after thrombectomy. Stroke. 2016;47:2311–2317. doi: 10.1161/STROKEAHA.116.013147 [DOI] [PubMed] [Google Scholar]
- 7.Menjot De Champfleur N, Saver JL, Goyal M, Jahan R, Diener HC, Bonafe A, Levy E, Pereira V, Cognard C, Yavagal D, et al. Efficacy of stent-retriever thrombectomy in magnetic resonance imaging versus computed tomographic perfusion-selected patients in SWIFT PRIME trial (Solitaire FR With the Intention for Thrombectomy as Primary Endovascular Treatment for Acute Ischemic Stroke). Stroke. 2017;48:1560–1566. [DOI] [PubMed] [Google Scholar]
- 8.Maier O, Menze BH, von der Gablentz J, Hani L, Heinrich MP, Liebrand M, Winzeck S, Basit A, Bentley P, Chen L, et al. ISLES 2015 - A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Med Image Anal. 2017;35:250–269. doi: 10.1016/j.media.2016.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winzeck S, Hakim A, McKinley R, Pinto JAADSR, Alves V, Silva C, Pisov M, Krivov E, Belyaev M, Monteiro M, et al. ISLES 2016 and 2017-benchmarking ischemic stroke lesion outcome prediction based on multispectral MRI. Front Neurol. 2018;9:679. doi: 10.3389/fneur.2018.00679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pinto A, Mckinley R, Alves V, Wiest R, Silva CA, Reyes M. Stroke lesion outcome prediction based on MRI imaging combined with clinical information. Front Neurol. 2018;9:1060. doi: 10.3389/fneur.2018.01060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kervadec H, Bouchtiba J, Desrosiers C, Granger É, Dolz J, Ben Ayed I. Boundary loss for highly unbalanced segmentation. Med Image Anal. 2021;67:101851. doi: 10.1016/j.media.2020.101851 [DOI] [PubMed] [Google Scholar]
- 12.Chen Y, Chen J, Wei D, Li Y, Zheng Y. OctopusNet: a deep learning segmentation network for multi-modal medical images. Multiscale Multimodal Medical Imaging. 2019. Springer [Google Scholar]
- 13.Lansberg MG, Straka M, Kemp S, Mlynash M, Wechsler LR, Jovin TG, Wilder MJ, Lutsep HL, Czartoski TJ, Bernstein RA, et al. ; DEFUSE 2 Study Investigators. MRI profile and response to endovascular reperfusion after stroke (DEFUSE 2): a prospective cohort study. Lancet Neurol. 2012;11:860–867. doi: 10.1016/S1474-4422(12)70203-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin L, Bivard A, Levi CR, Parsons MW. Comparison of computed tomographic and magnetic resonance perfusion measurements in acute ischemic stroke: back-to-back quantitative analysis. Stroke. 2014;45:1727–1732. doi: 10.1161/STROKEAHA.114.005419 [DOI] [PubMed] [Google Scholar]
- 15.Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T. Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Part I. 2019. Springer [Google Scholar]
- 16.Kamal H, Lopez V, Sheth SA. Machine learning in acute ischemic stroke neuroimaging. Front Neurol. 2018;9:945. doi: 10.3389/fneur.2018.00945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E. Convolutional neural networks for radiologic images: a radiologist’s guide. Radiology. 2019;290:590–606. doi: 10.1148/radiol.2018180547 [DOI] [PubMed] [Google Scholar]
- 18.Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep learning in neuroradiology. AJNR Am J Neuroradiol. 2018;39:1776–1784. doi: 10.3174/ajnr.A5543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Copen WA, Morais LT, Wu O, Schwamm LH, Schaefer PW, González RG, Yoo AJ. In acute stroke, can CT perfusion-derived cerebral blood volume maps substitute for diffusion-weighted imaging in identifying the ischemic core?. PLoS One. 2015;10:e0133566. doi: 10.1371/journal.pone.0133566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee EJ, Kim YH, Kim N, Kang DW. Deep into the brain: artificial intelligence in stroke imaging. J Stroke. 2017;19:277–285. doi: 10.5853/jos.2017.02054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pinto A, Pereira S, Meier R, Alves V, Wiest R, Silva CA, Reyes M. Enhancing clinical MRI perfusion maps with data-driven maps of complementary nature for lesion outcome prediction. In: Medical Image Computing and Computer Assisted Intervention – MICCAI 2018. 2018. Springer [Google Scholar]
- 22.Bertels J, Robben D, Vandermeulen D, Suetens P. Optimization with soft dice can lead to a volumetric bias. In: BrainLes 2019: Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries Part I. 2020. Springer [Google Scholar]
- 23.Campbell BC, Purushotham A, Christensen S, Desmond PM, Nagakane Y, Parsons MW, Lansberg MG, Mlynash M, Straka M, De Silva DA, et al. ; EPITHET–DEFUSE Investigators. The infarct core is well represented by the acute diffusion lesion: sustained reversal is infrequent. J Cereb Blood Flow Metab. 2012;32:50–56. doi: 10.1038/jcbfm.2011.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soize S, Tisserand M, Charron S, Turc G, Ben Hassen W, Labeyrie MA, Legrand L, Mas JL, Pierot L, Meder JF, et al. How sustained is 24-hour diffusion-weighted imaging lesion reversal? Serial magnetic resonance imaging in a patient cohort thrombolyzed within 4.5 hours of stroke onset. Stroke. 2015;46:704–710. doi: 10.1161/STROKEAHA.114.008322 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


