Abstract
Objectives
To assess the efforts deployed by different nations and territories in sequencing SARS-CoV-2 isolates, thus enabling detection of variants, known and novel, of concern.
Methods
The sources of over one million full genome sequences of SARS-CoV-2 virus available in the COVID-19 virus Mutation Tracker (CovMT) were analyzed to determine the number of variants in the RBD region of the genome determining infectivity detected in the various nations and territories.
Results
The number of detected variants increased as the square root of sequencing effort by nations. Eight nations have contributed 79% of all SARS-CoV-2 isolates that have been sequenced, with two-thirds of all unique variants, adding to 1118 RBD variants, reported by five nations. The median number of sequenced isolates required to detect, on average, one novel RBD variant is 24.05, which is a threshold achieved by 70 nations.
Conclusions
Many developing nations have not contributed any sequences due to lack of capacity. This poses a risk of dangerous virus variants in these under-sampled regions spreading globally before being detected. A collaborative program to sequence SARS-CoV-2 isolates, and other pathogens of concern, is needed to monitor, track, and control the pandemic.
Keywords: SARS-CoV-2, Variants, Mutations, Sequencing, Capacity, Detection
Graphical abstract
The rapid diversification of SARS-CoV-2 variants – with the total number (i.e., unique sequences across the entire genome) reaching 539,933 as of 11 April 2021 (COVID-19 virus Mutation Tracker (CovMT), https://www.cbrc.kaust.edu.sa/covmt, Alam et al. 2021) based on genomic data from a world-leading SARS-CoV-2 repository (gisaid.org) – is raising concern, as some of these variants, particularly the E484K mutation, may overcome the immune defenses produced by previously infected or vaccinated people (Tada et al. 2021).
Novel SARS-CoV-2 variants are initially diluted in a population of infected people, thereby leading to low detection power, enabling the more infective variants to reach high levels and possibly percolate across national borders before being detected. Detecting new SARS-CoV-2 variants requires whole genome sequencing, with the total number of good-quality reported genome sequences of isolates (with >90% base coverage) reaching 1,010,872 by 11 April 2021. Detecting variants is particularly important in the RBD region, as this is believed to determine SARS-CoV-2 infectivity (Greaney et al. 2021).
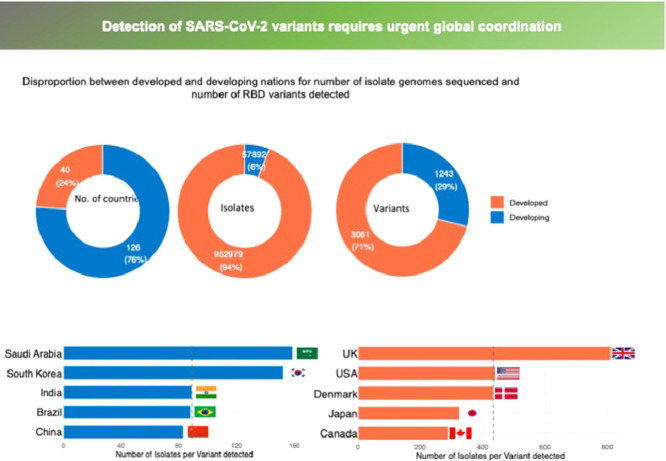
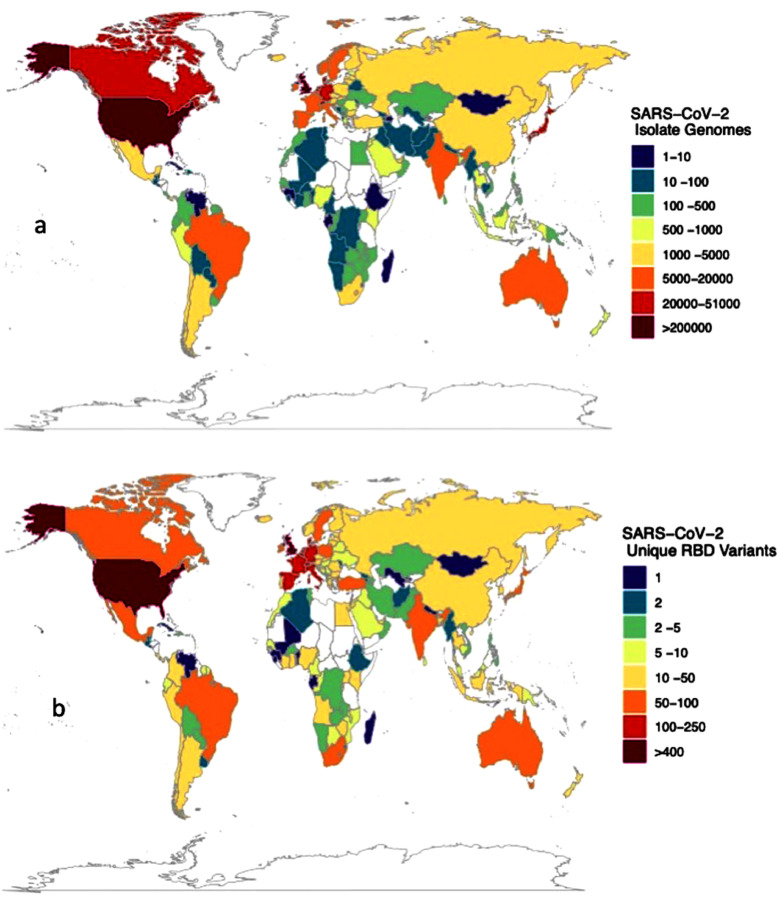
Most of the reported RBD variants were first detected in the UK, USA, Denmark, Germany, Mexico, Switzerland, and South Africa, accounting for 64% of the 1118 RBD variants reported to date, including the so-called UK (N501Y, B.1.1.7), South African (K417N+E484K+N501Y, B.1.351) and Brazilian (E484K+N501Y, P.1 and E484K, P.2) variants of concern (Sabino et al. 2021). One of the most concerning RBD mutations is E484K, also reported to be acquired by the UK variant, which has been involved in evasion of antibodies, rendering existing vaccines less effective (Greaney et al. 2021, Sabino et al. 2021). However, 87 nations, including most developing nations and territories particularly in Africa and Island states, have not yet reported any RBD variants (Figure 1 a) because no isolate sampled in those nations and territories has been sequenced. Sequencing effort is highly skewed (Furuse 2021), with the top eight nations contributing to this effort (UK, USA, Denmark, Japan, Australia, Canada, Switzerland, and the Netherlands, in order of contribution), with more than 10,000 reported isolate genomes sequenced from each and having reported 82% of all genome sequences globally (Figure 1b).
Figure 1.
(a) The number of isolate genomes sequenced and (b) the number of RBD variants detected across nations, as reported by 11 April, 2021. 11 April.
Data obtained from world variants table available at COVID-19 virus Mutation Tracker (CovMT) (1), where RBD variants are operationally defined as each set of SARS-CoV-2 genomes that generate the exact same amino acid sequence in the RBD region of the spike protein.
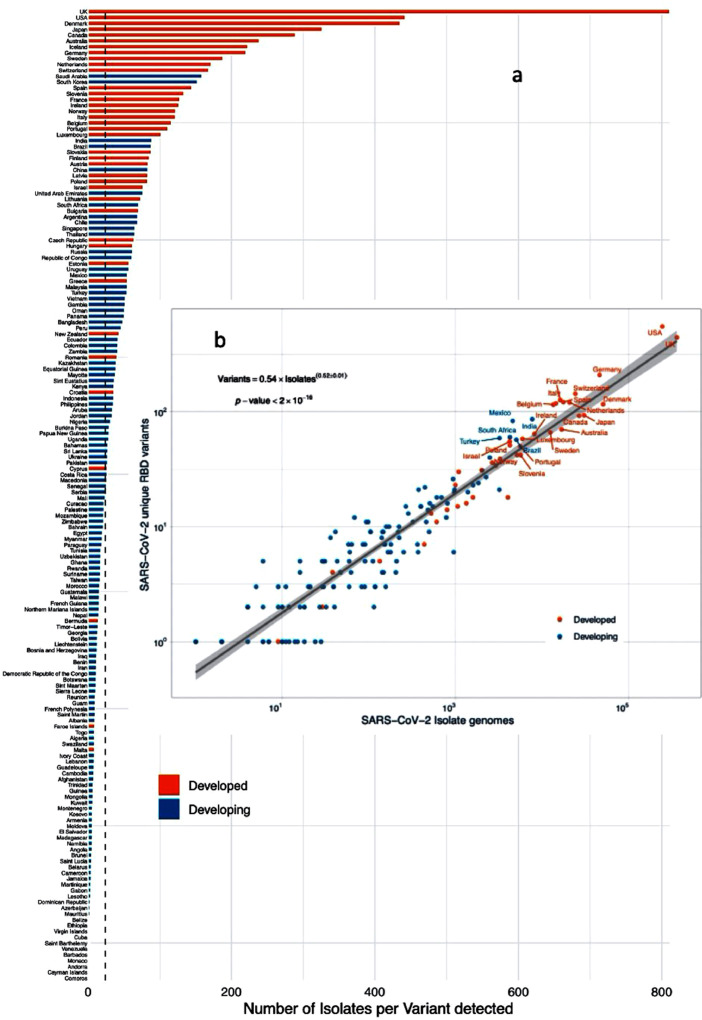
The number of RBD variants detected in isolates sequenced in any one nation increases with the square root of sequencing effort, with > 10,000 sequenced isolates required to considerably reduce the rate of discovery with further sequencing effort (Figure 2 ). The median number of sequenced isolates required to detect, on average, one novel RBD variant is 24.05, which is a threshold achieved by 70 nations (Figure 2). Hence, many nations with large numbers of infected cases, particularly developing nations but also some developed nations (Figure 2), have fallen short in the sequencing effort required to detect new RBD variants that may be present in populations.
Figure 2.
(a) The number of isolate genomes sequenced per RBD variants detected in developed and developing nations. The vertical dotted line shows the median number of isolates sequenced to detect the new RBD variant (24.05). (b) The relationship between the number of RBD variants detected and the number of isolate genomes sequenced in developed and developing nations. The solid line shows the fitted power law (equation in the insert) and the corresponding regression equation.
SARS-CoV-2 sequence data are mostly derived from clinical diagnostic samples, particularly from infected individuals with high viral loads (VL), that provide enough RNA for the sequencing of nearly complete genomes (Chiara et al. 2021). Handling the samples for RNA extraction requires using biosafety level (BSL) 2 laboratories, and extracted RNA should be stored at –80 °C to avoid degradation. Sequencing uses various next-generation sequencing strategies, mostly Illumina sequencing, with specific library protocols (Chiara et al. 2021). The sequences that are retrieved need to then be assembled to construct a high-quality full viral genome. Chiara et al. (2021) provided a useful overview of the different procedures involved. The resulting viral genomes should then be made available by depositing them in the GISAID EpiCov portal (Shu and McCauley 2017), which is the most widely used repository of SARS-CoV-2 genomic data. However, while relatively standard, these technologies and the infrastructure and skill sets required may not be available everywhere, particularly in developing nations.
Next-generation whole-genome sequencing of SARS-CoV-2 virus isolates is essential for tracing the spread and transmission chains of outbreaks, as well as for monitoring evolution and diversification (Chiara el al. 2021). Hence, the COVID-19 pandemic has led to unprecedented levels of full-genome sequencing in efforts to detect new variants and deploy defense strategies, such as mobility limitation and quarantine, as exemplified efforts across nations to limit the spread of the variants referred as UK (B.1.1.7), South African (501Y.V2), and Brazilian (P1) variants, some of which are more infective and/or may escape immune defenses (Kuzmina et al. 2021, Li et al. 2021).
Delivering the full power of whole-genome sequencing to detect, assess, and manage risks associated with the evolution of new SARS-CoV-2 variants requires a global effort, particularly in areas experiencing high numbers of infections, as these are the areas where new variants are more likely to be originating. Wide inequality in the capacity for advanced genomic sequencing is a manifestation of the growing gap in R&D capacity in health science between developing and developed nations. In a pandemic situation, the risk of failing to detect potentially dangerous variants until they have spread and become prominent is shared by all nations. Massive national efforts to sequence SARS-CoV-2 variants isolated from populations may be ineffective if dangerous variants are not detected where they originate, allowing them to spread. Hence, a mechanism is required for nations with demonstrated high capacity, such as the top 10 nations in sequencing efforts (Figure 1), to assist with sequencing of samples collected in developing nations lacking the capacity. The World Health Organization recently provided a guideline to improve sequencing efforts (https://apps.who.int/iris/rest/bitstreams/1326052/retrieve) and suggested that sequencing samples be sent to an established international sequencing laboratory in a third country where sequencing capacity maybe lacking (https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/).
Creating a global, coherent, and collaborative program for pathogen sequencing is not just required to respond to the COVID-19 pandemic. A permanent mechanism is required to maintain an effective monitoring and prevention system in the future. In a world with unprecedented connectivity, global collaboration and generosity in sharing genomic sequencing capacity and data is not just an act of generosity – it is also an act of self-interest for every nation.
Acknowledgments
Funding
This work was supported by King Abdullah University of Science and Technology through funding provided to TG.
Conflicts of Interests
The authors declare no conflict of interests.
Ethical Approval
This research did not require ethical approval.
References
- Alam I., Radovanovic A., Incitti R., Kamau A.A., Alarwai M., Azahar E.I., et al. CovMT: an interactive SARS-CoV-2 mutation tracker, with a focus on critical variants. The Lancet Infectious Diseases. 2021 doi: 10.1016/S1473-3099(21)00078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiara M., D'Erchia C.M., Gissi A.M., Manzari C., Parisi C., Resta A., et al. Next-generation sequencing of SARS-CoV-2 genomes: challenges, applications and opportunities. Briefings in Bioinformatics. 2021;22:616–630. doi: 10.1093/bib/bbaa297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse Y. Genomic sequencing effort for SARS-CoV-2 by country during the pandemic. Int J Infect Dis. 2021;103:305–307. doi: 10.1016/j.ijid.2020.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., et al. Complete mapping of mutations to the SARSCoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57. doi: 10.1016/j.chom.2020.11.007. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmina A., Khalaila Y., Voloshin O., Keren-Naus A., Bohehm L., Raviv Y., et al. SARS CoV-2 spike variants exhibit differential infectivity and neutralization resistance to convalescent or post-vaccination sera. Cell Host Microbe. 2021;29:522–528. doi: 10.1016/j.chom.2021.03.008. e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., et al. SARS-CoV-2 501Y. V2 variants lack higher infectivity but do have immune escape. Cell. 2021 doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino E.C., Buss L.F., Carvalho M.P., et al. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397:452–455. doi: 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data—from vision to reality. Eurosurveillance. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada T., Dcosta B.M., Samanovic-Golden M., et al. 2021. Neutralization of viruses with European, South African, and United States SARS-CoV-2 variant spike proteins by convalescent sera and BNT162b2 mRNA vaccine-elicited antibodies. bioRxiv. [Google Scholar]