Abstract
The calcium-sensing receptor (CaSR) is a G protein-coupled receptor that plays a fundamental role in extracellular calcium (Ca2+e) homeostasis by regulating parathyroid hormone release and urinary calcium excretion. The CaSR has been described to activate all four G protein subfamilies (Gαq/11, Gαi/o, Gα12/13, Gαs), and mutations in the receptor that cause hyper/hypocalcaemia, have been described to bias receptor signalling. However, many of these studies are based on measurements of second messengers or gene transcription that occurs many steps downstream of receptor activation and can represent convergence points of several signalling pathways. Therefore, to assess CaSR-mediated G protein activation directly, we took advantage of a recently described NanoBiT G protein dissociation assay system. Our studies, performed in HEK293 cells stably expressing CaSR, demonstrate that Ca2+e stimulation activates all Gαq/11 family and several Gαi/o family proteins, although Gαz was not activated. CaSR stimulated dissociation of Gα12/13 and Gαs from Gβ-subunits, but this occurred at a slower rate than that of other Gα-subunits. Investigation of cDNA expression of G proteins in three tissues abundantly expressing CaSR, the parathyroids, kidneys and pancreas, showed Gα11, Gαz, Gαi1 and Gα13 genes were highly expressed in parathyroid tissue, indicating CaSR most likely activates Gα11 and Gαi1 in parathyroids. In kidney and pancreas, the majority of G proteins were similarly expressed, suggesting CaSR may activate multiple G proteins in these cells. Thus, these studies validate a single assay system that can be used to robustly assess CaSR variants and biased signalling and could be utilised in the development of new pharmacological compounds targeting CaSR.
Keywords: calcium homeostasis, calcium signalling, G protein-coupled receptor, parathyroid and bone, signal transduction
Introduction
The calcium-sensing receptor (CaSR) is a class C homo-dimeric G protein-coupled receptor (GPCR) that plays a fundamental role in extracellular calcium (Ca2+e) homeostasis by regulating parathyroid hormone (PTH) release and urinary calcium excretion. Consistent with its critical role in maintaining serum calcium concentrations, the CaSR is highly expressed on the surface of parathyroid and kidney cells (Riccardi & Brown 2010). The CaSR is also widely expressed on other tissues, including significant expression on pancreatic islet α- and β-cells (Regard et al. 2008), and has been described to mediate a diverse range of functions, including inflammation, bronchoconstriction, gastrointestinal hormone secretion and glucose metabolism (Hofer et al. 2000, Rossol et al. 2012, Yarova et al. 2015, Zietek & Daniel 2015).
The importance of CaSR in the regulation of Ca2+e is highlighted in patients with germline mutations of the receptor that present with disorders of calcium homeostasis. Inactivating mutations cause familial hypocalciuric hypercalcaemia type-1 (FHH1), characterised by lifelong elevated serum calcium, moderate-to-high PTH concentrations, and low renal calcium excretion, and rarely cause neonatal severe hyperparathyroidism, which can be fatal if untreated (Pollak et al. 1993). Activating CaSR mutations cause autosomal dominant hypocalcaemia type-1 (ADH1), characterised by mild-to-moderate hypocalcaemia, with inappropriately low-to-normal serum PTH (Pearce et al. 1996). In addition, inactivating mutations in the G protein-α11 (Gα11), by which the CaSR signals and the adaptor protein-2 σ-subunit (AP2σ), which regulates endocytosis, cause FHH2 and FHH3, respectively; whereas activating Gα11 mutations cause ADH2 (Mannstadt et al. 2013, Nesbit et al. 2013a,b).
On stimulation by elevations in Ca2+e, the CaSR activates diverse signalling pathways. In most cell types, CaSR is understood to signal via Gαq/11 to activate phospholipase C (PLC)-mediated increases in intracellular calcium and mitogen-activated protein kinase (MAPK), and Gαi/o to inhibit adenylate cyclase and reduce cAMP (Kifor et al. 1997, Brown & MacLeod 2001, Hofer & Brown 2003). Additionally, CaSR has been described to activate cytoskeletal remodelling, attributed to Gα12/13 pathways in some cell types (e.g. MDCK cell-lines) (Huang et al. 2004) and Gαq/11 in others (Pi et al. 2002). Finally, in breast cancer and AtT20 pituitary tumour cells, CaSR activates elevations in cAMP by switching of CaSR coupling from Gαq/11 and Gαi/o to Gαs (Mamillapalli et al. 2008, Mamillapalli & Wysolmerski 2010).
The specific coupling of CaSR to individual G protein pathways was investigated in early studies by radio-ligand measurements of phospholipases; while subsequent studies have measured a range of second messengers in cells depleted of Gα proteins by chemical (e.g. YM-254890 or pertussis toxin that inhibit Gq/11 and Gi/o, respectively) or genetic manipulation (by siRNA knockdown or CRISPR-Cas9) (Kifor et al. 1997, Pi et al. 2002, Gorvin et al. 2018d). Although these studies have successfully identified many components of the CaSR signalling pathway, these methodologies are associated with several disadvantages, including variable sensitivity and complexity of assays; poor targeting of individual G proteins by chemical inhibitors and siRNAs, and the available CRISPR-generated cell-lines have deletions of whole G protein subfamilies rather than individual G proteins (Takasaki et al. 2004, Devost et al. 2017, Gundry et al. 2017). These limitations, combined with the distinct activation kinetics of second messengers, and recognition that multiple G proteins converge on the same signal proteins or genes (Goldsmith & Dhanasekaran 2007), impair the researcher’s ability to compare GPCR stimulation of different signalling pathways using many assay platforms.
The ability to assess GPCR activation of individual G proteins is increasingly important as it is recognised that receptors may couple preferentially to different signalling pathways in diverse tissues, and disease-causing mutations may confer alternative receptor conformations that couple differentially to intracellular signalling pathways (Wingler & Lefkowitz 2020). Such tissue- and mutation-specific bias has been described for the CaSR indicating that assessment of multiple signalling pathways may be required to assess pathogenicity of receptor variants identified in patient samples (Leach et al. 2012, Gorvin et al. 2018c). Moreover, allosteric modulators of the CaSR exhibit signalling bias, indicating it may be possible to develop additional pharmacological compounds that convey unique functional selectivity on individual G proteins to separate desirable from adverse (i.e. off-target) effects (Hauser et al. 2017). Therefore, a direct measure of G protein coupling to CaSR would be beneficial to aid in future drug design, in assigning decisions regarding the pathogenicity of receptor variants, and in identifying biased signalling by receptor mutants.
Recently, a NanoBiT G protein dissociation assay system that assesses activation of multiple Gα subunits was described (Inoue et al. 2019). This assay utilises a split nano-luciferase (NanoLuc), in which the large part of NanoLuc (LgBiT) is tagged to the Gα-subunits and the small part (SmBiT) is tagged to the Gβ-subunit. Thus G protein activation can be monitored by agonist-induced reductions in luciferase due to separation of the Gα- and Gβγ-subunits following receptor stimulation. The development of this assay afforded an opportunity to interrogate the promiscuity of CaSR coupling to G proteins without the problem of signal pathway cross-talk. These studies, combined with gene expression analyses, indicate that the physiological effects of CaSR are likely mediated by a combination of G protein activation and expression of individual Gα-proteins in different tissues. This simple assay system is easily scalable, measures in real-time and could be used for drug screening and assessment of pathogenicity and signalling bias of receptor variants by multiple CaSR signalling pathways.
Methods
Cell culture and transfection
Adherent HEK293 (AdHEK) cells were purchased from Agilent Technologies. HEK293 cells with deletion of the Gαq/11, Gαi/o, Gα12/13 or Gαs/l family of G proteins (G protein knockout cells) were described previously (Inoue et al. 2019). AdHEK cells and G protein knockout cells were maintained in DMEM-Glutamax media (Sigma) with 10% foetal bovine serum (FBS, Sigma) at 37°C, 5% CO2. The full-length CaSR cDNA with an N-terminal FLAG tag was PCR amplified (reagents from Promega) from total human kidney cDNA (Ambion) and cloned into a pcDNA3.1 expression vector (Life Technologies). The full-length somatostatin receptor-5 (SSTR5) was PCR amplified from SSTR5-Tango (a gift from Bryan Roth (Addgene plasmid # 66506; RRID:Addgene_66506) (Kroeze et al. 2015)) and cloned into pcDNA3.1. Expression constructs were sequence-verified by Source Bioscience (Nottingham, UK). NanoBiT G protein dissociation and the IP3 biosensor constructs were described in detail previously (Inoue et al. 2019). The pGloSensor-20F plasmid was purchased from Promega. Transfections were performed using Lipofectamine 2000 (LifeTechnologies), according to the manufacturer’s instructions. To generate AdHEK-CaSR, cells stably transfecting pcDNA3.1-FLAG-CaSR cells were plated in six-well plates, transfected with 1 µg DNA, then single clones selected using cell culture media containing 500 µg/mL geneticin (Gibco).
Western blot analysis
Western blot analysis was performed as previously described (Gorvin et al. 2018d). Endogenous calnexin was used as a loading control. Lysates were resuspended in Laemmli buffer, boiled and separated on 6% SDS-PAGE gels. Following transfer to polyvinylidene difluoride membrane (ThermoFisher), blots were blocked in 5% marvel/TBS-T, then probed with anti-CaSR (ADD, Abcam), anti-Gα11 (SantaCruz Biotechnology), anti-Gα12 (SantaCruz), and anti-calnexin (Millipore) antibodies. Blots were visualised using the Immuno-Star WesternC kit (BioRadK) on a BioRad Chemidoc XRS+ system. For cell surface expression of CaSR, plasma membrane fractions were extracted using the Plasma Membrane Protein Extraction kit (Abcam). Plasma membrane calcium adenosine triphosphatase (PMCA1) (Abcam) was used as a housekeeping protein for the plasma membrane fraction.
cAMP GloSensor assays
Cells were plated in six-well plates and transfected with 50 ng pGloSensor-20F plasmid. Twenty-four hours later cells were re-plated in 96-well plates in FluoroBrite DMEM media (ThermoScientific) containing 10% FBS. On the following day cells were incubated with 100 µL of equilibration media consisting of Hank’s buffered saline solution (HBSS, calcium and magnesium-free) containing 2% (v/v) dilution of the GloSensor cAMP Reagent stock solution according to manufacturer’s instructions. Cells were incubated for 2 h at 37°C. Basal luminescence was read on a PheraStar FSX (BMGLabtech) for 8 min, then agonist added with 10 µM forskolin (Sigma), and plates read for a further 30 min.
NanoBiT G protein dissociation and IP3 assays
Cells were plated in six-well plates and transfected 24 h later with: 250 ng LgBiT-Gα plasmid, 500 ng SmBiT-Gβ plasmid, 500 ng untagged Gγ2 plasmid. For studies shown in Supplementary Figs 1, 2 and 3 (see section on supplementary materials given at the end of this article), AdHEK were also transiently transfected with 250 ng of FLAG-CaSR, while studies of SSTR5 were transiently transfected with pcDNA3.1-SSTR5. For reactions with LgBiT-Gα11 cells were co-transfected with RIC8A, as previously described (Inoue et al. 2019). Following 24 h, cells were harvested in DMEM-fluorobrite (ThermoScientific) with 10% FCS and 4mM L-glutamine and seeded in 8 wells of a 96-well plate. NanoBiT assays were performed the following day using NanoGlo reagent (Promega). Media was changed to HBSS without antibiotics (containing 0.1 mM CaCl2) on the morning of the assay. Approximately 4 h later, each well was loaded with 40 µL substrate and baseline signal read on a Pherastar FSX plate reader (BMGLabtech, Aylesbury, UK) at 37°C for four cycles (equivalent to 8 min). Vehicle and agonists were prepared in Hank’s Balanced Saline Solution (HBSS, Sigma) at 10× concentration and added to wells once baselines were stable and responses recorded immediately following agonist addition for up to 25 min. All responses were normalised to that treated with vehicle at 0 min. For SSTR5 studies cells were exposed to either vehicle (DMSO) or 50 nM somatostatin (purchased from Sigma). The consequent fold-change values were fitted to a four-parameter sigmoidal concentration-response using GraphPad Prism. IP3 biosensor assays were performed similarly using 200 ng LgBiT-IP3R2-SmBiT plasmid. For IP3 measurements, basal luminescence was read on a PheraStar FSX (BMGLabtech) for 8 min, then agonist added, and plates read for a further 30 min.
Gene expression analysis
Analysis of gene expression in parathyroid, kidney and pancreatic tissues was performed using the Gene Expression Omnibus (GEO) website (https://www.ncbi.nlm.nih.gov/geo/) using the search term ‘parathyroid’, ‘kidney’, ‘pancreas’ and ‘human’, accession date August 2020. Two datasets, #GSE83421 (Balenga et al. 2017) and #GSE2193 (Shyamsundar et al. 2005), were identified and used in analyses using GEO2R for parathyroid tissue. These comprised data from 9 normal parathyroids and 25 adenoma tissues from patients with primary hyperparathyroidism. For kidney and pancreas expression the following datasets were used: GSM12660, GSM12661, GSM12663, GSM12659, GDS181, GDS1085, GDS1663 (Su et al. 2002, Shyamsundar et al. 2005). Gene expression values for a panel of housekeeping genes: actin, beta (ACTB); phosphoglycerate kinase 1 (PGK1); peptidylprolyl isomerase A (PPIA); beta-2-microglobulin (B2M); transferrin receptor (TFRC); glucuronidase, beta (GUSB); hypoxanthine phosphoribosyltransferase 1 (HPRT1); TATA box binding protein (TBP); and tubulin, beta (TUBB) were acquired and the mean value used for normalisation using Microsoft Excel 2013. Expression of each G protein subunit was expressed relative to the geometric mean of the housekeeping genes in each sample and plotted using GraphPad Prism 7. A cut-off of representation in at least four datasets was used for analysis of expression between genes. To compare expression within each G protein subfamily, the relative expression of each gene was expressed as a fold-change to a representative gene (GNA11, GNAI1, GNA12, GNAS). Statistical analyses were performed using ANOVA between two or more datasets and correction for multiple testing applied. A value of P < 0.05 was considered significant. Comparisons between the gene expression data and proteome datasets could not be made as there is insufficient proteomic data available on parathyroid tissues.
Results
CaSR can couple to all members of the Gq/11 subfamily
Previous studies have indicated that CaSR is able to couple promiscuously to multiple G proteins (Kifor et al. 1997, Brown & MacLeod 2001, Hofer & Brown 2003, Huang et al. 2004). However, these studies are largely based on measurements of second messengers or changes in gene transcription, which can only indicate the G protein subfamily and not necessarily the specific G protein activated. Furthermore, as many signalling pathways share downstream signal components, it may be difficult to differentiate the effects of different G proteins. We therefore used an unbiased approach to determine which specific G proteins are activated by CaSR using a recently described NanoBiT G protein dissociation assay system (Inoue et al. 2019). NanoBiT utilises a split nano-luciferase (NanoLuc) in which the large part of NanoLuc (LgBiT) is tagged to the Gα subunits and the small part (SmBiT) is tagged to the Gβ-subunit. Under basal conditions, the heterotrimeric Gαβγ protein is stable and the LgBiT and SmBiT are associated with yielding high levels of luciferase. On activation of the GPCR, Gα and Gβγ dissociate and luciferase levels reduce.
To investigate G protein activation by the CaSR, we first assessed Gα-Gβ3 dissociation with six different concentrations of Ca2+e in adherent HEK293 (AdHEK) transiently expressing the CaSR. Robust concentration-dependent increases in G protein dissociation, including within the physiological range of Ca2+e (~1–3 mM), were observed for several members of the Gαq/11 and Gαi/o families (Supplementary Fig. 1). However, G protein dissociation was only observed for some G proteins (Gαi2, Gαi3, Gα12) at concentrations >3 mM Ca2+e. Therefore, for subsequent studies, we used 5 mM Ca2+e, as this concentration was the closest to the physiological range that would allow us to assess dissociation of the largest number of G proteins. G protein dissociation in response to 5 mM Ca2+e was not observed in AdHEK cells lacking the CaSR (Supplementary Fig. 2). Overexpression of other untagged Gα proteins did not affect G protein coupling of NanoBiT constructs (Supplementary Fig. 3), nor did deletion of other Gα proteins (Supplementary Fig. 4).
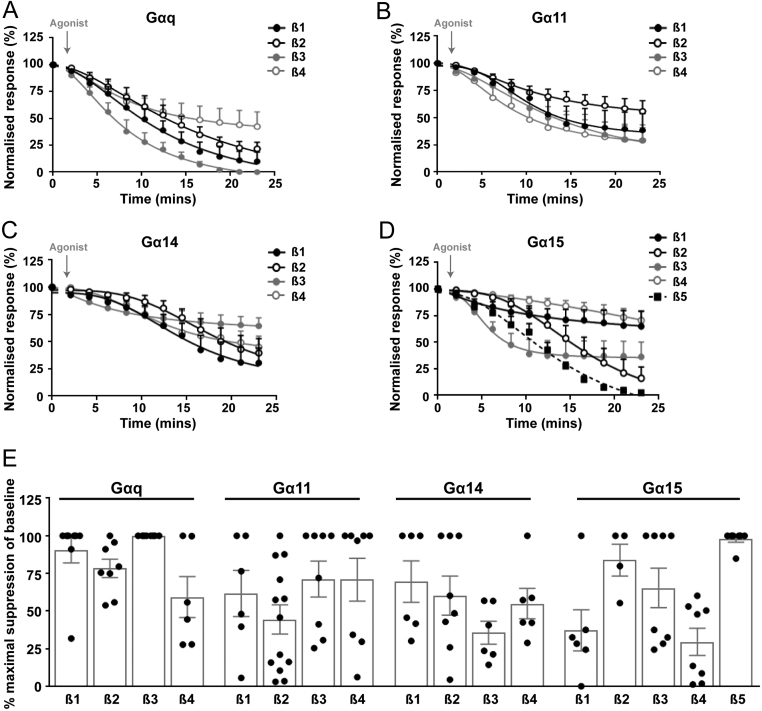
To assess G protein coupling and dissociation between 12 Gα and 5 Gβ subunits we generated a AdHEK cell-line stably expressing the CaSR with an N-terminal FLAG tag (Supplementary Fig. 5). These cells were demonstrated to overexpress CaSR and to activate IP3 accumulation and cAMP depletion on stimulation with increasing doses of Ca2+e (Supplementary Fig. 5). All subsequent assays were performed in one clone of this cell-line (clone A, named AdHEK-CaSR). Overexpression of NanoBiT constructs had no effect on CaSR total or cell surface expression, nor on the expression of other G proteins (Supplementary Fig. 6). Assays were performed with the 12 available Gα-LgBiT-subunits in combination with 5 Gβ-SmBiT-subunits. Cells were exposed to low concentrations of Ca2+e (0.1 mM) or an activating concentration of 5 mM Ca2+e. The Gαq/11 family was first investigated as GNA11 is known to be highly expressed in parathyroid tissue and mutations in this gene cause FHH2 and ADH2 (Mannstadt et al. 2013, Nesbit et al. 2013a). The four Gα proteins (q, 11, 14, 15) were assessed with all Gβ-subunits. Cells in which the relative luminescence units (RLU) were below 1000 were excluded as this value was not significantly greater than background luminescence (Supplementary Table 1). Analyses of the Gq/11 family demonstrated that all four α-subunits could be activated following exposure of cells to 5 mM Ca2+e (Fig. 1A, B, C, D and Supplementary Fig. 7). CaSR produced greater activation responses with Gαq and Gα11 than the other family members (Fig. 1A, B, C, D, E and Supplementary Table 2), and therefore it is more likely that these proteins are activated by the receptor when present in CaSR expressing tissues.
Figure 1.
CaSR activation of the Gq/11 family by NanoBiT G protein dissociation assays. NanoBiT dissociation assays of AdHEK-CaSR cells transiently transfected with: (A) LgBiT-Gαq, (B) LgBiT-Gα11, (C) LgBiT-Gα14, (D) LgBiT-Gα15 with SmBiT-Gβ subunits (Gβ1–5) and unlabelled Gγ2 following treatment with 5 mM Ca2+e. Pre-calcium baselines are not shown. The first baseline value is shown. (E) Percentage suppression of baseline from A, B, C and D. Statistical analyses comparing values of all Gα-Gβ dissociations are shown in Supplementary Table 1. For all panels only those Gα and Gβ pairs that yielded relative luminescence units (RLU) over a threshold of 1 × 103 (i.e. above background luminescence) are shown. All responses were normalised to those under basal conditions (0.1 mM Ca2+e). Dissociation curves for 5 mM Ca2+e are shown (curves with 0.1 mM Ca2+e are shown in Supplementary Fig. 7). Curves show mean±s.e.m. for n = 6–11 independent assays.
CaSR activates several members of the Gi/o subfamily but not Gαz
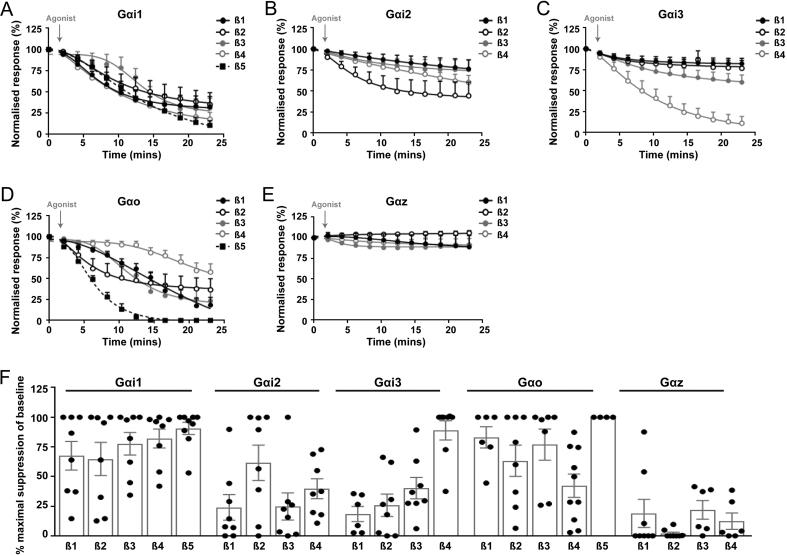
CaSR has been described to reduce cAMP in many studies, and signalling by these pathways has been demonstrated to play a role in PTH secretion in parathyroid cells (Brown et al. 1978). Five members of the Gi/o subfamily were investigated by NanoBiT dissociation assays. CaSR robustly activated Gαi1 when combined with any Gβ-subunit, and Gαo similarly activated all Gβ-subunits, although activation, when combined with β4, was slow (Fig. 2 and Supplementary Fig. 8). Gαi2 and Gαi3 were activated only when combined with one Gβ-subunit and Gαz was not activated at all (Fig. 2). This was not due to inactivity of the LgBiT-Gαz plasmid as other GPCRs could activate this G protein (Supplementary Fig. 9). Thus, activation of the Gi/o family in CaSR expressing cells is unlikely to occur by Gαz and activation of some family members (Gαi2 and Gαi3) will be dependent on β-subunit expression.
Figure 2.
CaSR activation of the Gi/o family by NanoBiT G protein dissociation assays. NanoBiT dissociation assays of AdHEK-CaSR cells transiently transfected with: (A) LgBiT-Gαi1, (B) LgBiT-Gαi2, (C) LgBiT-Gαi3, (D) LgBiT-Gαo and (E) LgBiT-Gαz with SmBiT-Gβ subunits (Gβ1–5) and unlabelled Gγ2 following treatment with 5mM Ca2+e. Pre-calcium baselines are not shown. The first baseline value is shown. (F) Percentage suppression of baseline from A, B, C, D and E. Statistical analyses comparing values of all Gα-Gβ dissociations are shown in Supplementary Table 1. For all panels only those Gα and Gβ pairs that yielded relative luminescence units (RLU) over a threshold of 1 × 103 (i.e. above background luminescence) are shown. All responses were normalised to those under basal conditions (0.1 mM Ca2+e). Dissociation curves for 5 mM Ca2+e are shown (curves with 0.1 mM Ca2+e are shown in Supplementary Figure 8). Curves show mean ± s.e.m. for n = 6–10 independent assays.
Other G protein families are activated slowly by the CaSR
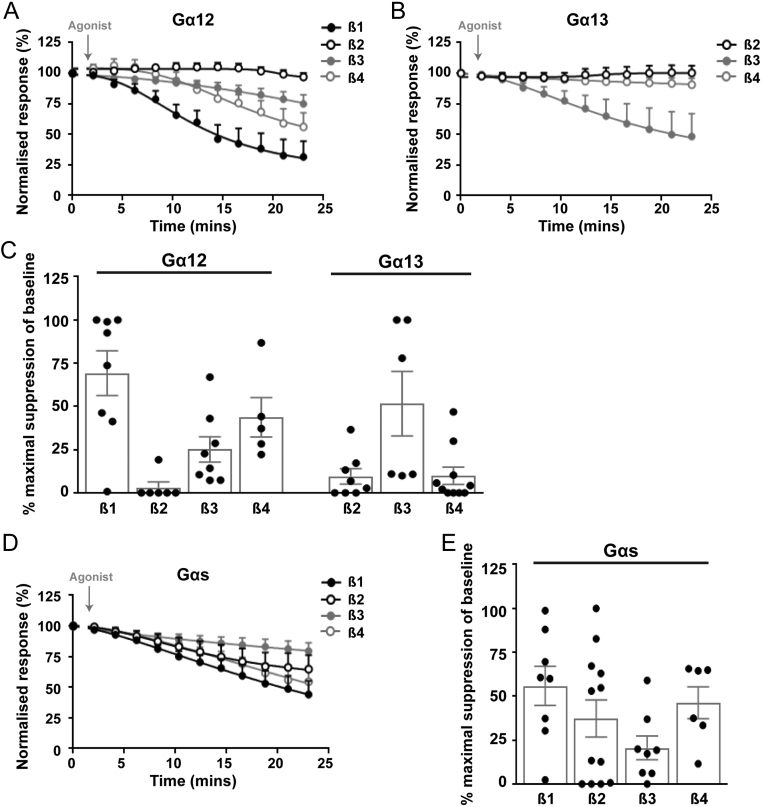
CaSR has been reported to activate G12/13 inducing cytoskeletal changes in some cells, while other studies were unable to show any signalling, indicating the pathway may be cell type specific (Pi et al. 2002, Huang et al. 2004, Davies et al. 2006). NanoBiT dissociation assays in AdHEK-CaSR cells showed Gα12 was activated by CaSR when combined with three β-subunits, β1, β2 and β3 (Fig. 3 and Supplementary Fig. 10). However, activation of Gα12 with Gβ2 and Gβ3 took more than 12 min, indicating it is unlikely rapid CaSR activation is induced by coupling between these subunits. Similarly, Gα13 could only be activated when combined with Gβ4. Thus, activation of Gα12/13 by CaSR may only occur when specific β-subunits are present in cells and may explain some of the discrepancies observed between published studies investigating CaSR signalling by this pathway.
Figure 3.
CaSR activation of the G12/13 family by NanoBiT G protein dissociation assays. NanoBiT dissociation assays of AdHEK-CaSR cells transiently transfected with: (A) LgBiT-Gα12 and (B) LgBiT-Gα13 with SmBiT-Gβ subunits (Gβ1–5) and unlabelled Gγ2 following treatment with 5 mM Ca2+e. Pre-calcium baselines are not shown. The first baseline value is shown. (C) Percentage suppression of baseline from A and B. (D) NanoBiT dissociation assays of AdHEK-CaSR cells transiently transfected with: LgBiT-Gαs, SmBiT-Gβ subunits (Gβ1–5) and unlabelled Gγ2 following treatment with 5 mM Ca2+e. Pre-calcium baselines are not shown. The first baseline value is shown. (E) Percentage suppression of baseline from D. Statistical analyses comparing values of all Gα-Gβ dissociations are shown in Supplementary Table 1. For all panels only those Gα and Gβ pairs that yielded relative luminescence units (RLU) over a threshold of 1 × 103 (i.e. above background luminescence) are shown. All responses were normalised to those under basal conditions (0.1 mM Ca2+e). Dissociation curves for 5 mM Ca2+e are shown (curves with 0.1 mM Ca2+e are shown in Supplementary Figs 10 and 11). Curves show mean ± s.e.m. for n = 6–12 independent assays.
Previous studies of CaSR in cancer cell-lines, including MCF-7 human breast cancer cells and mouse pituitary AtT20 cells, have shown that CaSR switches from preferentially coupling to Gq/11 and Gi/o pathways to exclusively signalling by a Gs pathway (Mamillapalli et al. 2008, Mamillapalli & Wysolmerski 2010). Combinations of Gαs with Gβ1–4 achieved luminescence values above the threshold, indicating efficient activation. However, agonist-induced responses were only significantly greater than basal responses from 12 min after stimulations in cells transfected with Gβ1 and after 15 min of stimulation in cells expressing Gβ2 and Gβ4 (Fig. 3 and Supplementary Fig. 11). Therefore, CaSR may activate Gαs and Gα12/13 more slowly than other Gα proteins, and this could explain why in vitro assays often fail to detect CaSR activation of these pathways.
Gα11, Gαi1 and Gαz are most abundantly expressed in human parathyroid tissue
Our NanoBiT dissociation studies have demonstrated which G proteins are capable of coupling to CaSR. However, CaSR coupling in native human tissue will be dependent on the expression of the individual G proteins in that tissue. For example, previous studies using qRT-PCR have indicated that the gene encoding Gα11 (GNA11) is expressed at higher levels than that of Gαq (GNAQ) (Nesbit et al. 2013a); while in bovine parathyroid, genes for Gα11, Gαs, Gαi2, Gα12 and Gαz were highly expressed (Varrault et al. 1995). We sought to verify these findings in a larger dataset and investigate G protein expression in the three tissues in which CaSR is most abundantly expressed: the parathyroid glands, the kidneys and the pancreas.
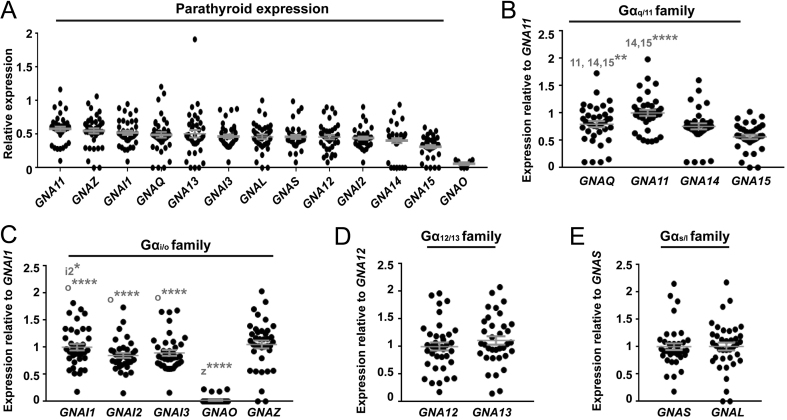
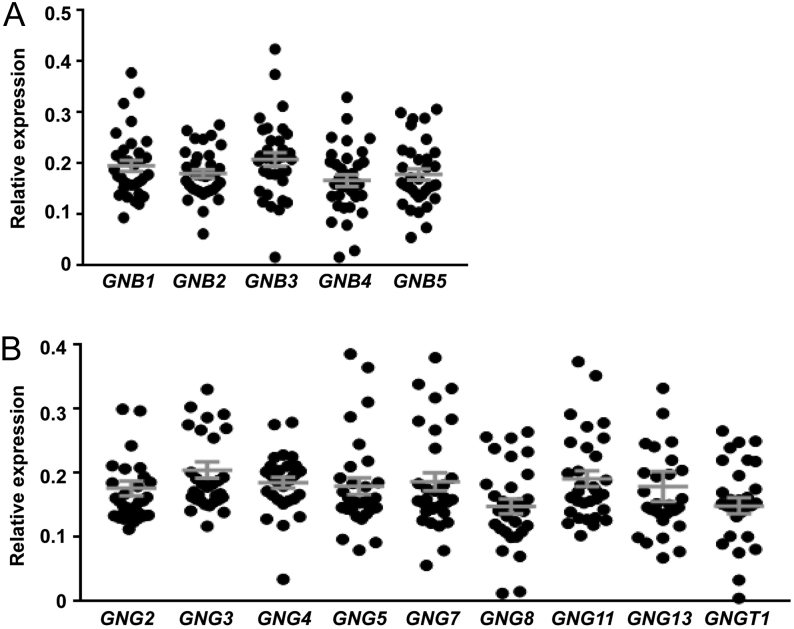
We first searched the Gene Expression Omnibus (GEO) repository to identify existing gene expression datasets from human parathyroid tissue. This identified: one dataset from expression profiling by array of normal parathyroid glands (n = 6) and adenoma tissue from patients with primary hyperparathyroidism (n = 25) (GEO accession #GSE83421) (Balenga et al. 2017); and one dataset from expression profiling of normal human tissues (n = 3 parathyroid tissues) (GEO accession #GSE2193) (Shyamsundar et al. 2005). The expression of G protein α, β or γ-subunits was normalised to a panel of housekeeping genes. As no differences were detected between expression levels of any of these genes in the normal vs adenoma tissues, all data were combined. This demonstrated 13 Gα-subunits, five Gβ-subunits and 9 Gγ-subunits were expressed in the majority of parathyroid tissues (Figs 4 and 5). The expression of all Gα subunits was compared in each sample by expression as a ratio of abundance. No Gβ- or Gγ-subunits were consistently overexpressed (Fig. 5). However, these studies confirmed that GNA11 has greater expression than GNAQ (Fig. 4A, B and Supplementary Table 3). Additionally, three other G protein subunits (GNAZ, GNAI1 and GNA13) had comparable expression to GNA11 (Fig. 4 and Supplementary Table 3). As the NanoBiT dissociation assays demonstrated no or very little coupling to GNAZ and GNA13 (Figs 2 and 3), it is unlikely that CaSR signals via these G proteins in human parathyroid glands; however, this remains to be verified in parathyroid cells. Thus, the higher expression of genes encoding Gα11 and Gαi1 may, in part, explain the propensity for CaSR to activate Ca2+i and reduce cAMP signalling pathways, and the inability of other Gαq/11 family members to fully compensate for mutant Gα11 in FHH2 and ADH2 patients.
Figure 4.
Expression of G protein α-subunits in human parathyroid tissue. (A) The mRNA expression of 13 Gα-subunits in human parathyroid tissue shown as a relative fold-change compared to the geometric mean of control genes. Statistical analyses comparing expression between subunits is shown in Supplementary Table 1. (B, C, D and E) To compare expression of genes within subfamilies, each gene was expressed as a fold-change compared to either: (B) GNA11; (C) GNAI1; (D) GNA12; and (E) GNAS. Statistical analyses for panels B, C, D, E and F were performed by one-way ANOVA with multiple comparisons and subunits with which comparisons are made are indicated in grey. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
Figure 5.
Expression of G protein β-subunits in human parathyroid tissue. The mRNA expression of (A) all five Gβ-subunits and (B) nine Gγ-subunits in human parathyroid tissue shown as a relative fold-change compared to the geometric mean of control genes. Statistical analyses were performed by one-way ANOVA and subunits with which comparisons are made are indicated in grey. **P < 0.01, *P < 0.05.
Gαs is the most abundantly expressed G protein in human kidney and pancreatic tissue
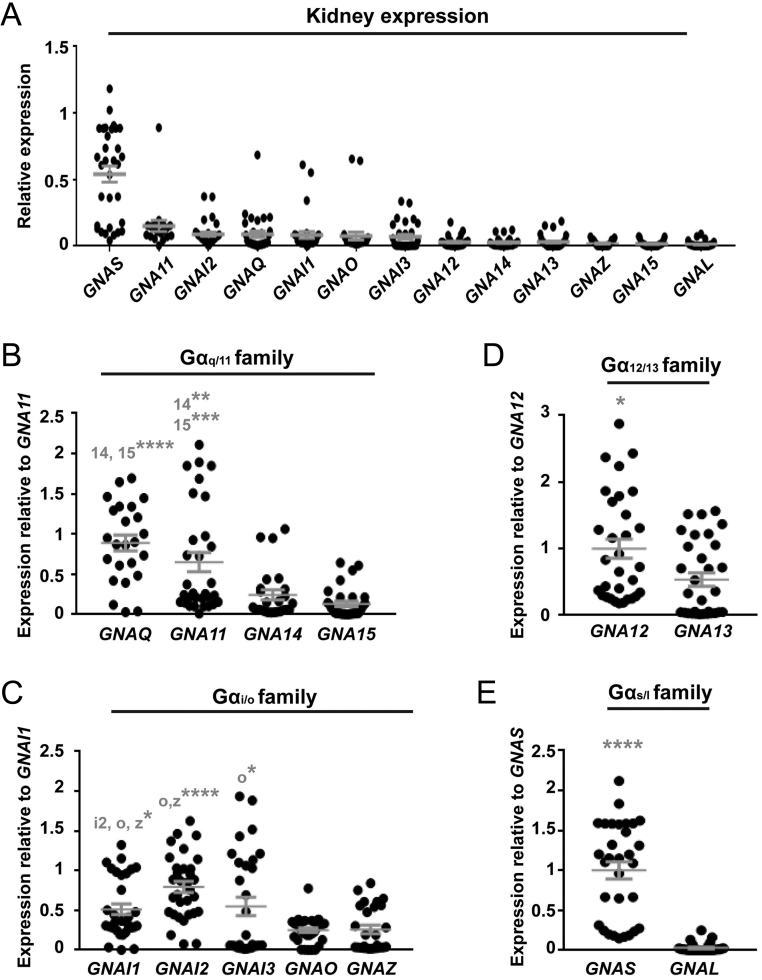
Similar analyses were conducted investigating G protein expression in human kidney tissue in the GEO repository. Only datasets investigating expression in non-diseased tissue were included in analyses. Whole kidney samples were used, as datasets for individual kidney cell types were not available. GNAS was the most abundantly expressed G protein in kidney tissue (Fig. 6 and Supplementary Table 4). When the data were reanalysed to focus on individual subfamily members, GNAQ and GNA11 were similarly expressed, and both were more abundant than GNA14 and GNA15 (Fig. 6B). The GNAL, GNAO and GNAZ genes were poorly expressed, and GNA12 was expressed at significantly greater levels than GNA13 (Fig. 6C and D). Thus, within the kidney, CaSR may be able to signal by Gαq or Gα11, and may explain why patients with FHH2 and ADH2 mutations have milder symptoms than patients with CaSR mutations in FHH1 or ADH1 (Nesbit et al. 2013a, Gorvin et al. 2018a). Additionally, CaSR may signal by several other G proteins that are similarly expressed in kidney tissue.
Figure 6.
Expression of G protein α-subunits in human kidney tissue. (A) The mRNA expression of 13 Gα-subunits in human kidney tissue shown as a relative fold-change compared to the geometric mean of control genes. Statistical analyses comparing expression between subunits is shown in Supplementary Table 2. (B, C, D and E) To compare expression of genes within subfamilies, each gene was expressed as a fold-change compared to either: (B) GNA11; (C) GNAI1; (D) GNA12; and (E) GNAS. Statistical analyses were performed by one-way ANOVA and subunits with which comparisons are made are indicated in grey. ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05.
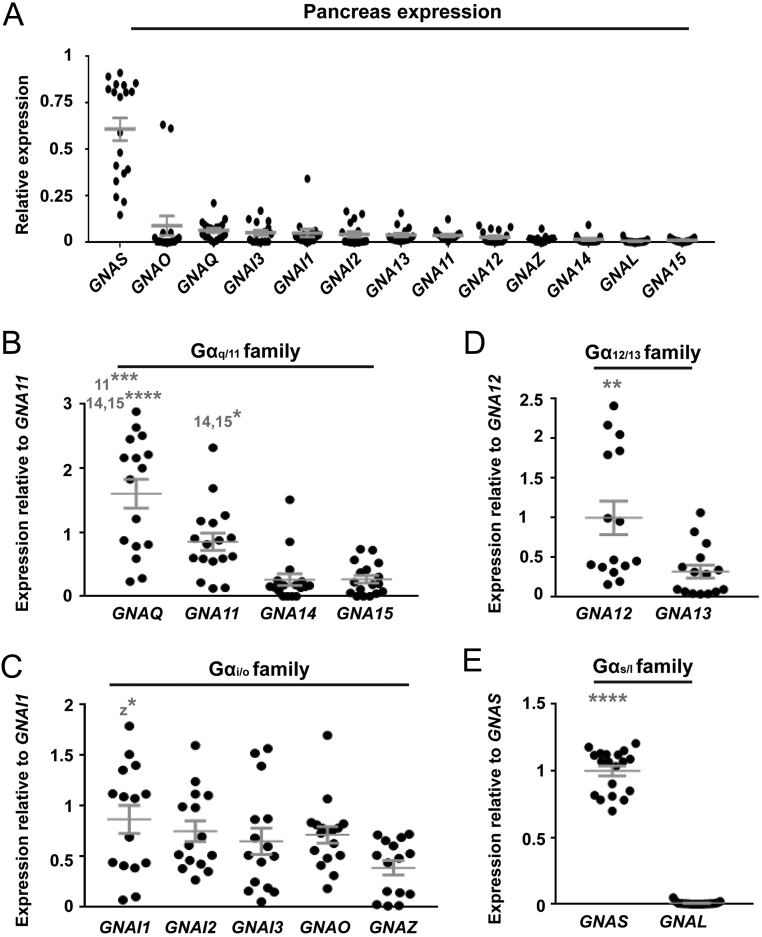
The CaSR is highly expressed in pancreatic tissue, and roles have been described in α- and β-cells (Squires et al. 2000, Regard et al. 2008). A number of gene expression datasets are available in the GEO repository, including from the whole pancreas and individual cell types. We chose to analyse whole pancreas samples rather than individual cell types as: CaSR is expressed in multiple cell types; these samples were collected from non-disease tissue; and there were enough samples to perform statistical analyses on. These analyses demonstrated that GNAS is the most highly expressed G protein in pancreatic tissue (Fig. 7), while members of the Gi/o and Gq/11 families were expressed at similar levels (Fig. 7 and Supplementary Table 5). Analyses within subfamilies demonstrated GNAQ and GNA11 to be more abundantly expressed than GNA14 and GNA15, with GNAQ most highly expressed (Fig. 7B). Few differences were observed between subfamily members in the Gi/o family, while Gα12 was more highly expressed than GNA13 (Fig. 7). Therefore, CaSR could couple to many G proteins within pancreatic tissue, and this may explain why no overt pancreatic phenotypes (e.g. diabetes, variations in blood glucose or insulin levels) have been reported in patients with GNA11 mutations.
Figure 7.
Expression of G protein α-subunits in human pancreatic tissue. (A) The mRNA expression of 13 Gα-subunits in human pancreatic tissue shown as a relative fold-change compared to the geometric mean of control genes. Statistical analyses comparing expression between subunits is shown in Supplementary Table 3. (B, C, D and E) To compare expression of genes within subfamilies, each gene was expressed as a fold-change compared to either: (B) GNA11; (C) GNAI1; (D) GNA12; and (E) GNAS. Statistical analyses were performed by one-way ANOVA and subunits with which comparisons are made are indicated in grey. ****P < 0.0001, ***P < 0.001, *P < 0.05.
Discussion
Our studies confirm that the CaSR can activate multiple G proteins, and this may account for the increasingly varied functions that have been described for the receptor in a wide range of tissues (Hofer et al. 2000, Rossol et al. 2012, Yarova et al. 2015, Zietek & Daniel 2015). These studies indicate that GPCR coupling in different tissues is most likely governed by the ability of the receptor to activate an individual G protein, as well as the expression of G proteins in different tissues. Thus, in parathyroid tissue, in which CaSR has its predominant role, GNA11 and GNAI1 are two of the most abundantly expressed G proteins, and have amongst the highest G protein activity, consistent with previous studies that indicate that parathyroid cells typically activate Gq/11 and Gi/o pathways (Ca2+i mobilisation and reductions in cAMP, respectively) (Kifor et al. 1997, Brown & MacLeod 2001, Hofer & Brown 2003). Other G proteins that are highly expressed in parathyroid tissue, such as Gαz, were not activated by CaSR in the NanoBiT dissociation assays, indicating it is unlikely to be important in calcium-mediated responses. These findings would need to be replicated in parathyroid cells to determine whether Gαz is activated by CaSR in this cell type.
These studies showed that CaSR activation of some members of the Gi/o and Gq/11 subfamilies is comparable, indicating that these pathways may have similar importance in CaSR-mediated parathyroid activities, as indicated by early studies of PTH secretion (Conigrave 2016). Despite this, many studies of CaSR variants assess only changes in Ca2+i, and thus disease-causing mutations in CaSR could be overlooked by these approaches. It is likely researchers have focussed on Ca2+i pathways rather than cAMP as fewer robust assays to assess cAMP were available until recent years. Newer assays, such as the cAMP Glosensor system, which allows cAMP to be assessed kinetically, have been increasingly used to assess cAMP signalling by GPCRs (Kumar et al. 2017, Lotta et al. 2019). However, these assays often still require forskolin treatment to elevate intracellular cAMP levels, and thus increase assay-to-assay variability. Therefore, the development of the NanoBiT dissociation assay will allow both Gq/11 and Gi/o pathways to be compared in cells expressing CaSR variant proteins without the need to pre-treat cells with forskolin.
Our studies showed that CaSR could couple to Gα12 and Gα13, as indicated by previous studies (Huang et al. 2004), but that rates of activation were slower than with other G protein families. Slow rates of nucleotide exchange and GTP hydrolysis for G12/13 have been described for other GPCRs, and it has been suggested that this may facilitate temporal control over cell processes that require prolonged signalling (Siehler 2009). These slower activation rates could explain why some laboratories have been unable to detect CaSR-mediated G12/13 signalling. Alternatively, G12/13 signalling may not have been detected in previous studies, as CaSR appears to activate only select combinations of G12/13 and Gβ-subunits (Fig. 3), despite coupling between Gα12/13 and Gβ1-4 (Supplementary Table 1). Post-translational modifications, including palmitoylation and myristoylation, can affect G-protein recruitment and activation (Bhattacharyya & Wedegaertner 2000), and this may have affected the ability of Gα12/13 to activate some Gβ subunits. Further studies investigating the effects of lipid modifications on CaSR signalling by G12/13 would be required to determine whether this could explain differences between activation within members of the same subfamilies.
Previous studies have suggested that patients with FHH2 due to Gα11 mutations may have a milder phenotype than patients with CaSR mutations, as other G proteins could compensate for Gα11 functions (Nesbit et al. 2013a, Gorvin et al. 2018a). Our studies demonstrate that CaSR can activate all members of the Gq/11 subfamily, and therefore, in principle, these proteins could compensate for impaired Gα11 functionality. However, consistent with previous studies, GNA11 was expressed at significantly greater levels than GNAQ in human parathyroid tissue, which may explain why Gαq cannot entirely compensate for Gα11, and patients with inactivating Gα11 mutations develop mild hypercalcaemia (Nesbit et al. 2013a). In contrast to parathyroid tissue, expression of GNA11 and GNAQ are not significantly different in kidney tissue, thus compensation by other subunits may be present in kidney cells, providing a reason why phenotypes may be milder than that observed in FHH1 and ADH1. Further investigation of CaSR coupling to mutant Gα11 in cells depleted of other Gq/11 subfamily members may be required to investigate Gα-protein compensation in greater detail.
In pancreatic tissue, expression of Gα-subunits of the Gi/o family were not significantly different to some members of the Gq/11 family. The NanoBiT dissociation assays demonstrated that CaSR robustly activates Gαi1 and Gαo, both of which are expressed at comparable levels in the pancreas. Thus, CaSR-mediated functions in pancreatic cells may arise by coupling to either G protein family. Activation of the Gi/o family that negatively modulates cAMP signalling and is important in reducing hormone secretion by other GPCRs (Braun 2014), could explain how CaSR activation impairs hormone secretion in pancreatic cells. However, previous studies in pancreatic β-cells failed to show a role for CaSR-mediated cAMP depletion in insulin secretion (Squires et al. 2000), and therefore further studies of CaSR coupling in pancreatic β-cells may be required.
Studies of GPCR signalling using a NanoBiT G protein dissociation assay system has several advantages over other methodologies to study GPCR activation. The ability to assess multiple G protein pathways in parallel reduces the need to perform different assays to assess individual signalling pathways, and avoids the need to chemically or genetically inhibit gene expression, allowing the researcher to directly compare responses by each G protein. This will revolutionise the ability to assess biased signalling by disease-causing CaSR mutations which has been described in several studies (Leach et al. 2012, Gorvin et al. 2018b,c) and will simplify comparisons between CaSR allosteric modulators. Furthermore, the simplicity of the methodology and availability of these plasmids will allow researchers in different laboratories to directly compare results, increasing the robustness and reproducibility of data yielded from these assays. The ability to collect kinetic data easily, which many assays that have been used previously to measure GPCR signalling cannot do (e.g. IP-one, LANCE cAMP, pERK AlphaScreen, luciferase reporter genes), will increase our understanding of GPCR temporal signalling. This was demonstrated for the CaSR as our NanoBiT assays showed activation of Gα12/13 and Gαs occurs more slowly than activation of other Gα-proteins. If this assay system is to be adopted by multiple laboratories, it will be important that a uniform methodology is used. For example, the time between agonist addition and measurement of luminescence should always be the same in all laboratories, to allow rates of dissociation to be compared.
A disadvantage of this study is that it was conducted in a single cell-line, adherent HEK293 overexpressing CaSR, which could be criticised as an artificial system. We chose this cell-line as HEK293: are easy to grow and transfect; are available to most research labs so findings can be replicated; are routinely used in CaSR research; and cultures of parathyroid and kidney cell-lines are not widely available. Moreover, the purpose of this study was to determine whether CaSR can couple to all G protein subunits and establish a simple assay system to assess CaSR variants and potentially screen pharmacological compounds to determine their ability to engender signalling bias. Future studies investigating endogenous G protein activation by CaSR in specific cell types may become possible with recently published BRET constructs (Maziarz et al. 2020). However, these assays still require transfection and, therefore, may be difficult in primary cells. Our study could also be criticised for only assessing one CaSR ligand. We assessed Ca2+e as it is the best characterised ligand in in vitro assays of CaSR signalling and is the major physiological agonist of the receptor. Other studies have described CaSR activation by other ligands, including aromatic amino acids and polyamines (Conigrave et al. 2000, 2004, Thomsen et al. 2012). However, many of these act as modulators of Ca2+e-mediated responses, or physiological functions are unknown. Future studies could utilise the NanoBiT dissociation assay system to compare the effects of these ligands by a single methodology. Finally, the NanoBiT assay we describe will be unable to detect G protein activation in which Gα-Gβγ dissociation does not occur. Studies have indicated that such activation in the absence of Gα-Gβγ dissociation may occur in some cell types (e.g. cone photoreceptors (Rosenzweig et al. 2007)). However, the number of GPCRs able to utilise such signalling is unknown, and it is possible that some studies that reported the absence of dissociation may not have measured signal kinetics, and thus may have missed slower dissociation rates that have been reported by some laboratories (Digby et al. 2008). The NanoBiT dissociation assay, will be able to detect these differences in dissociation rates as shown for Gα12/13 and Gαs.
In conclusion, we have demonstrated that CaSR differentially activates multiple Gα proteins. Physiological effects of CaSR are likely mediated by a combination of G protein activation and expression of individual Gα-proteins in different tissues. These studies highlight a simple, single assay system that can be used to robustly assess biased signalling and could be utilised in the development of new pharmacological compounds targeting the CaSR.
Supplementary Material
Declaration of interest
The authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Funding
This work was funded by: start-up funds to C M G from the Centre of Membrane Proteins and Receptors (COMPARE) and an Academy of Medical Sciences Springboard Award (Ref: SBF004|1034, to C M G), which is supported by the British Heart Foundation, Diabetes UK, the Global Challenges Research Fund, the Government Department of Business, Energy and Industrial Strategy and the Wellcome Trust. A I was funded by PRIME (19gm5910013) and Leading Advanced Projects for Medical Innovation (LEAP) (JP19gm0010004) from the Japan Agency for Medical Research and Development.
References
- Balenga N, Azimzadeh P, Hogue JA, Staats PN, Shi Y, Koh J, Dressman H, Olson Jr JA. 2017. Orphan adhesion GPCR GPR64/ADGRG2 is overexpressed in parathyroid tumors and attenuates calcium-sensing receptor-mediated signaling. Journal of Bone and Mineral Research 32 654–666. ( 10.1002/jbmr.3023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya R, Wedegaertner PB. 2000. Galpha 13 requires palmitoylation for plasma membrane localization, Rho-dependent signaling, and promotion of p115-RhoGEF membrane binding. Journal of Biological Chemistry 275 14992–14999. ( 10.1074/jbc.M000415200) [DOI] [PubMed] [Google Scholar]
- Braun M.2014. The somatostatin receptor in human pancreatic beta-cells. Vitamins and Hormones 95 165–193. ( 10.1016/B978-0-12-800174-5.00007-7) [DOI] [PubMed] [Google Scholar]
- Brown EM, MacLeod RJ. 2001. Extracellular calcium sensing and extracellular calcium signaling. Physiological Reviews 81 239–297. ( 10.1152/physrev.2001.81.1.239) [DOI] [PubMed] [Google Scholar]
- Brown EM, Gardner DG, Windeck RA, Aurbach GD. 1978. Relationship of intracellular 3′,5′-adenosine monophosphate accumulation to parathyroid hormone release from dispersed bovine parathyroid cells. Endocrinology 103 2323–2333. ( 10.1210/endo-103-6-2323) [DOI] [PubMed] [Google Scholar]
- Conigrave AD.2016. The calcium-sensing receptor and the parathyroid: past, present, future. Frontiers in Physiology 7 563. ( 10.3389/fphys.2016.00563) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conigrave AD, Quinn SJ, Brown EM. 2000. L-amino acid sensing by the extracellular Ca2+-sensing receptor. PNAS 97 4814–4819. ( 10.1073/pnas.97.9.4814) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conigrave AD, Mun HC, Delbridge L, Quinn SJ, Wilkinson M, Brown EM. 2004. L-amino acids regulate parathyroid hormone secretion. Journal of Biological Chemistry 279 38151–38159. ( 10.1074/jbc.M406373200) [DOI] [PubMed] [Google Scholar]
- Davies SL, Gibbons CE, Vizard T, Ward DT. 2006. Ca2+-sensing receptor induces Rho kinase-mediated actin stress fiber assembly and altered cell morphology, but not in response to aromatic amino acids. American Journal of Physiology: Cell Physiology 290 C1543–C. ( 10.1152/ajpcell.00482.2005) [DOI] [PubMed] [Google Scholar]
- Devost D, Sleno R, Petrin D, Zhang A, Shinjo Y, Okde R, Aoki J, Inoue A, Hebert TE. 2017. Conformational profiling of the AT1 angiotensin II receptor reflects biased agonism, G protein coupling, and cellular context. Journal of Biological Chemistry 292 5443–5456. ( 10.1074/jbc.M116.763854) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Digby GJ, Sethi PR, Lambert NA. 2008. Differential dissociation of G protein heterotrimers. Journal of Physiology 586 3325–3335. ( 10.1113/jphysiol.2008.153965) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsmith ZG, Dhanasekaran DN. 2007. G protein regulation of MAPK networks. Oncogene 26 3122–3142. ( 10.1038/sj.onc.1210407) [DOI] [PubMed] [Google Scholar]
- Gorvin CM, Hannan FM, Cranston T, Valta H, Makitie O, Schalin-Jantti C, Thakker RV. 2018a. Cinacalcet rectifies hypercalcemia in a patient with familial hypocalciuric hypercalcemia type 2 (FHH2) caused by a germline loss-of-function Galpha11 mutation. Journal of Bone and Mineral Research 33 32–41. ( 10.1002/jbmr.3241) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvin CM, Frost M, Malinauskas T, Cranston T, Boon H, Siebold C, Jones EY, Hannan FM, Thakker RV. 2018b. Calcium-sensing receptor residues with loss- and gain-of-function mutations are located in regions of conformational change and cause signalling bias. Human Molecular Genetics 27 3720–3733. ( 10.1093/hmg/ddy263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvin CM, Babinsky VN, Malinauskas T, Nissen PH, Schou AJ, Hanyaloglu AC, Siebold C, Jones EY, Hannan FM, Thakker RV. 2018c. A calcium-sensing receptor mutation causing hypocalcemia disrupts a transmembrane salt bridge to activate beta-arrestin-biased signaling. Science Signaling 11 eaan3714. ( 10.1126/scisignal.aan3714) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorvin CM, Rogers A, Hastoy B, Tarasov AI, Frost M, Sposini S, Inoue A, Whyte MP, Rorsman P, Hanyaloglu ACet al. 2018d. AP2sigma mutations impair calcium-sensing receptor trafficking and signaling, and show an endosomal pathway to spatially direct G-protein selectivity. Cell Reports 22 1054–1066. ( 10.1016/j.celrep.2017.12.089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundry J, Glenn R, Alagesan P, Rajagopal S. 2017. A practical guide to approaching biased agonism at G protein coupled receptors. Frontiers in Neuroscience 11 17. ( 10.3389/fnins.2017.00017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser AS, Attwood MM, Rask-Andersen M, Schioth HB, Gloriam DE. 2017. Trends in GPCR drug discovery: new agents, targets and indications. Nature Reviews: Drug Discovery 16 829–842. ( 10.1038/nrd.2017.178) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofer AM, Brown EM. 2003. Extracellular calcium sensing and signalling. Nature Reviews: Molecular Cell Biology 4 530–538. ( 10.1038/nrm1154) [DOI] [PubMed] [Google Scholar]
- Hofer AM, Curci S, Doble MA, Brown EM, Soybel DI. 2000. Intercellular communication mediated by the extracellular calcium-sensing receptor. Nature Cell Biology 2 392–398. ( 10.1038/35017020) [DOI] [PubMed] [Google Scholar]
- Huang C, Hujer KM, Wu Z, Miller RT. 2004. The Ca2+-sensing receptor couples to Galpha12/13 to activate phospholipase D in Madin-Darby canine kidney cells. American Journal of Physiology: Cell Physiology 286 C22–C. ( 10.1152/ajpcell.00229.2003) [DOI] [PubMed] [Google Scholar]
- Inoue A, Raimondi F, Kadji FMN, Singh G, Kishi T, Uwamizu A, Ono Y, Shinjo Y, Ishida S, Arang Net al. 2019. Illuminating G-protein-coupling selectivity of GPCRs. Cell 177 1933, .e25–1947.e25. ( 10.1016/j.cell.2019.04.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kifor O, Diaz R, Butters R, Brown EM. 1997. The Ca2+-sensing receptor (CaR) activates phospholipases C, A2, and D in bovine parathyroid and CaR-transfected, human embryonic kidney (HEK293) cells. Journal of Bone and Mineral Research 12 715–725. ( 10.1359/jbmr.1997.12.5.715) [DOI] [PubMed] [Google Scholar]
- Kroeze WK, Sassano MF, Huang XP, Lansu K, McCorvy JD, Giguere PM, Sciaky N, Roth BL. 2015. Presto-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nature Structural and Molecular Biology 22 362–369. ( 10.1038/nsmb.3014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar BA, Kumari P, Sona C, Yadav PN. 2017. GloSensor assay for discovery of GPCR-selective ligands. Methods in Cell Biology 142 27–50. ( 10.1016/bs.mcb.2017.07.012) [DOI] [PubMed] [Google Scholar]
- Leach K, Wen A, Davey AE, Sexton PM, Conigrave AD, Christopoulos A. 2012. Identification of molecular phenotypes and biased signaling induced by naturally occurring mutations of the human calcium-sensing receptor. Endocrinology 153 4304–4316. ( 10.1210/en.2012-1449) [DOI] [PubMed] [Google Scholar]
- Lotta LA, Mokrosinski J, Mendes de Oliveira E, Li C, Sharp SJ, Luan J, Brouwers B, Ayinampudi V, Bowker N, Kerrison Net al. 2019. Human gain-of-function MC4R variants show signaling bias and protect against obesity. Cell 177 597, .e9–607.e9. ( 10.1016/j.cell.2019.03.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamillapalli R, Wysolmerski J. 2010. The calcium-sensing receptor couples to Galpha(s) and regulates PTHrP and ACTH secretion in pituitary cells. Journal of Endocrinology 204 287–297. ( 10.1677/JOE-09-0183) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamillapalli R, VanHouten J, Zawalich W, Wysolmerski J. 2008. Switching of G-protein usage by the calcium-sensing receptor reverses its effect on parathyroid hormone-related protein secretion in normal versus malignant breast cells. Journal of Biological Chemistry 283 24435–24447. ( 10.1074/jbc.M801738200) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannstadt M, Harris M, Bravenboer B, Chitturi S, Dreijerink KM, Lambright DG, Lim ET, Daly MJ, Gabriel S, Juppner H. 2013. Germline mutations affecting Galpha11 in hypoparathyroidism. New England Journal of Medicine 368 2532–2534. ( 10.1056/NEJMc1300278) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maziarz M, Park JC, Leyme A, Marivin A, Garcia-Lopez A, Patel PP, Garcia-Marcos M. 2020. Revealing the activity of trimeric G-proteins in live cells with a versatile biosensor design. Cell 182 770, .e16–785.e16. ( 10.1016/j.cell.2020.06.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbit MA, Hannan FM, Howles SA, Babinsky VN, Head RA, Cranston T, Rust N, Hobbs MR, Heath 3rd H, Thakker RV. 2013a. Mutations affecting G-protein subunit alpha11 in hypercalcemia and hypocalcemia. New England Journal of Medicine 368 2476–2486. ( 10.1056/NEJMoa1300253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesbit MA, Hannan FM, Howles SA, Reed AA, Cranston T, Thakker CE, Gregory L, Rimmer AJ, Rust N, Graham Uet al. 2013b. Mutations in AP2S1 cause familial hypocalciuric hypercalcemia type 3. Nature Genetics 45 93–97. ( 10.1038/ng.2492) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce SH, Williamson C, Kifor O, Bai M, Coulthard MG, Davies M, Lewis-Barned N, McCredie D, Powell H, Kendall-Taylor Pet al. 1996. A familial syndrome of hypocalcemia with hypercalciuria due to mutations in the calcium-sensing receptor. New England Journal of Medicine 335 1115–1122. ( 10.1056/NEJM199610103351505) [DOI] [PubMed] [Google Scholar]
- Pi M, Spurney RF, Tu Q, Hinson T, Quarles LD. 2002. Calcium-sensing receptor activation of rho involves filamin and rho-guanine nucleotide exchange factor. Endocrinology 143 3830–3838. ( 10.1210/en.2002-220240) [DOI] [PubMed] [Google Scholar]
- Pollak MR, Brown EM, Chou YH, Hebert SC, Marx SJ, Steinmann B, Levi T, Seidman CE, Seidman JG. 1993. Mutations in the human Ca(2+)-sensing receptor gene cause familial hypocalciuric hypercalcemia and neonatal severe hyperparathyroidism. Cell 75 1297–1303. ( 10.1016/0092-8674(9390617-y) [DOI] [PubMed] [Google Scholar]
- Regard JB, Sato IT, Coughlin SR. 2008. Anatomical profiling of G protein-coupled receptor expression. Cell 135 561–571. ( 10.1016/j.cell.2008.08.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccardi D, Brown EM. 2010. Physiology and pathophysiology of the calcium-sensing receptor in the kidney. American Journal of Physiology: Renal Physiology 298 F485–F. ( 10.1152/ajprenal.00608.2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig DH, Nair KS, Wei J, Wang Q, Garwin G, Saari JC, Chen CK, Smrcka AV, Swaroop A, Lem Jet al. 2007. Subunit dissociation and diffusion determine the subcellular localization of rod and cone transducins. Journal of Neuroscience 27 5484–5494. ( 10.1523/JNEUROSCI.1421-07.2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossol M, Pierer M, Raulien N, Quandt D, Meusch U, Rothe K, Schubert K, Schoneberg T, Schaefer M, Krugel Uet al. 2012. Extracellular Ca2+ is a danger signal activating the NLRP3 inflammasome through G protein-coupled calcium sensing receptors. Nature Communications 3 1329. ( 10.1038/ncomms2339) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyamsundar R, Kim YH, Higgins JP, Montgomery K, Jorden M, Sethuraman A, van de Rijn M, Botstein D, Brown PO, Pollack JR. 2005. A DNA microarray survey of gene expression in normal human tissues. Genome Biology 6 R22. ( 10.1186/gb-2005-6-3-r22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siehler S.2009. Regulation of RhoGEF proteins by G12/13-coupled receptors. British Journal of Pharmacology 158 41–49. ( 10.1111/j.1476-5381.2009.00121.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squires PE, Harris TE, Persaud SJ, Curtis SB, Buchan AM, Jones PM. 2000. The extracellular calcium-sensing receptor on human beta-cells negatively modulates insulin secretion. Diabetes 49 409–417. ( 10.2337/diabetes.49.3.409) [DOI] [PubMed] [Google Scholar]
- Su AI, Cooke MP, Ching KA, Hakak Y, Walker JR, Wiltshire T, Orth AP, Vega RG, Sapinoso LM, Moqrich Aet al. 2002. Large-scale analysis of the human and mouse transcriptomes. PNAS 99 4465–4470. ( 10.1073/pnas.012025199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasaki J, Saito T, Taniguchi M, Kawasaki T, Moritani Y, Hayashi K, Kobori M. 2004. A novel Galphaq/11-selective inhibitor. Journal of Biological Chemistry 279 47438–47445. ( 10.1074/jbc.M408846200) [DOI] [PubMed] [Google Scholar]
- Thomsen AR, Hvidtfeldt M, Brauner-Osborne H. 2012. Biased agonism of the calcium-sensing receptor. Cell Calcium 51 107–116. ( 10.1016/j.ceca.2011.11.009) [DOI] [PubMed] [Google Scholar]
- Varrault A, Pena MS, Goldsmith PK, Mithal A, Brown EM, Spiegel AM. 1995. Expression of G protein alpha-subunits in bovine parathyroid. Endocrinology 136 4390–4396. ( 10.1210/endo.136.10.7664659) [DOI] [PubMed] [Google Scholar]
- Wingler LM, Lefkowitz RJ. 2020. Conformational basis of G protein-coupled receptor signaling versatility. Trends in Cell Biology 30 736–747. ( 10.1016/j.tcb.2020.06.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarova PL, Stewart AL, Sathish V, Britt Jr RD, Thompson MA, AP PL, Freeman LM, Aravamudan B, Kita H, Brennan SCet al. 2015. Calcium-sensing receptor antagonists abrogate airway hyperresponsiveness and inflammation in allergic asthma. Science Translational Medicine 7 284ra260.( 10.1126/scitranslmed.aaa0282) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zietek T, Daniel H. 2015. Intestinal nutrient sensing and blood glucose control. Current Opinion in Clinical Nutrition and Metabolic Care 18 381–388. ( 10.1097/MCO.0000000000000187) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a