Abstract
PURPOSE
Cancer classification is foundational for patient care and oncology research. Systems such as International Classification of Diseases for Oncology (ICD-O), Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT), and National Cancer Institute Thesaurus (NCIt) provide large sets of cancer classification terminologies but they lack a dynamic modernized cancer classification platform that addresses the fast-evolving needs in clinical reporting of genomic sequencing results and associated oncology research.
METHODS
To meet these needs, we have developed OncoTree, an open-source cancer classification system. It is maintained by a cross-institutional committee of oncologists, pathologists, scientists, and engineers, accessible via an open-source Web user interface and an application programming interface.
RESULTS
OncoTree currently includes 868 tumor types across 32 organ sites. OncoTree has been adopted as the tumor classification system for American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE), a large genomic and clinical data-sharing consortium, and for clinical molecular testing efforts at Memorial Sloan Kettering Cancer Center and Dana-Farber Cancer Institute. It is also used by precision oncology tools such as OncoKB and cBioPortal for Cancer Genomics.
CONCLUSION
OncoTree is a dynamic and flexible community-driven cancer classification platform encompassing rare and common cancers that provides clinically relevant and appropriately granular cancer classification for clinical decision support systems and oncology research.
INTRODUCTION
Structured cancer classification systems are an integral part of cancer care for diagnosis, treatment, and cancer research. Standardized cancer ontologies are essential for the homogenized and refined diagnosis of the disease and play a crucial role in understanding and traversing the evolution of tumors and cancer incidences and epidemiology.1 There are formal classification systems such as the Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT)2 and the International Classification of Diseases for Oncology (ICD-O)3 as well as terminology systems such as National Cancer Institute Thesaurus (NCIt),4 Unified Medical Language System (UMLS),5 and Medical Subject Headings (MeSH).6 The field of cancer care is evolving rapidly, and classification systems need to evolve in step to accommodate new research data and to support current evidence-based clinical decisions. As the decision-making processes in oncology are becoming more complex and data-driven, clinical decision support systems are increasingly used by clinicians to improve patient care and optimize clinical workflows. Cancer classification is one of the fundamental provisions of clinical decision support systems. However, classification systems such as ICD-O and SNOMED-CT were not designed specifically to support the computational needs of such decision support systems or to include specific, common genomic alterations of cancer specimens.7 Moreover, cancer classification systems such as ICD-O and SNOMED-CT are slowly iterative, taking years to adopt newer entities, while struggling to incorporate newly defined tumor entities, especially rare tumors.
CONTEXT
Key Objective
OncoTree is an open-source cancer classification system that was developed to meet the fast-evolving needs in clinical reporting of genomic sequencing results and associated oncology research.
Knowledge Generated
The open-source cancer classification system currently includes 868 tumor types across 32 organ sites encompassing rare and common cancers, providing relevant and appropriately granular cancer classification for clinical decision support systems and oncology research. It is maintained by a cross-institutional committee of oncologists, pathologists, scientists, and engineers, accessible via an open-source Web user interface and an application programming interface.
Relevance
OncoTree has been adopted as the tumor classification system for American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) and for clinical molecular testing efforts at Memorial Sloan Kettering (MSK) and Dana-Farber Cancer Institute (DFCI). It is also used by precision oncology tools such as OncoKB, cBioPortal for Cancer Genomics, and MatchMiner. OncoTree is not intended to replace any existing systems but works in synchrony with them by providing tools and mappings between different ontologies.
To address the limitations of current cancer classification systems, we developed OncoTree, a hierarchical cancer classification platform that was specifically designed to agilely address the fast-evolving computational needs of a clinical decision support tool and to facilitate cancer research. OncoTree is open-source, open-access, and community-driven—reviewed by a multi-institutional committee of physicians, pathologists, and scientists with oncology expertise. It was initially developed at Memorial Sloan Kettering (MSK) and later adopted by American Association for Cancer Research (AACR) Project Genomics Evidence Neoplasia Information Exchange (GENIE) as its main cancer classification system. It is now maintained by a multi-institutional community of disease experts, scientists, and engineers. OncoTree is publicly available at ref. 8 (Fig 1), which includes a user interface for viewing and searching tumor types as well as an application programming interface (API) for programmatic data access. OncoTree is intended to work in conjunction with other tumor classification systems, including ICD-O, NCIt, and UMLS. Direct mappings between OncoTree and NCIt/UMLS are provided.
FIG 1.
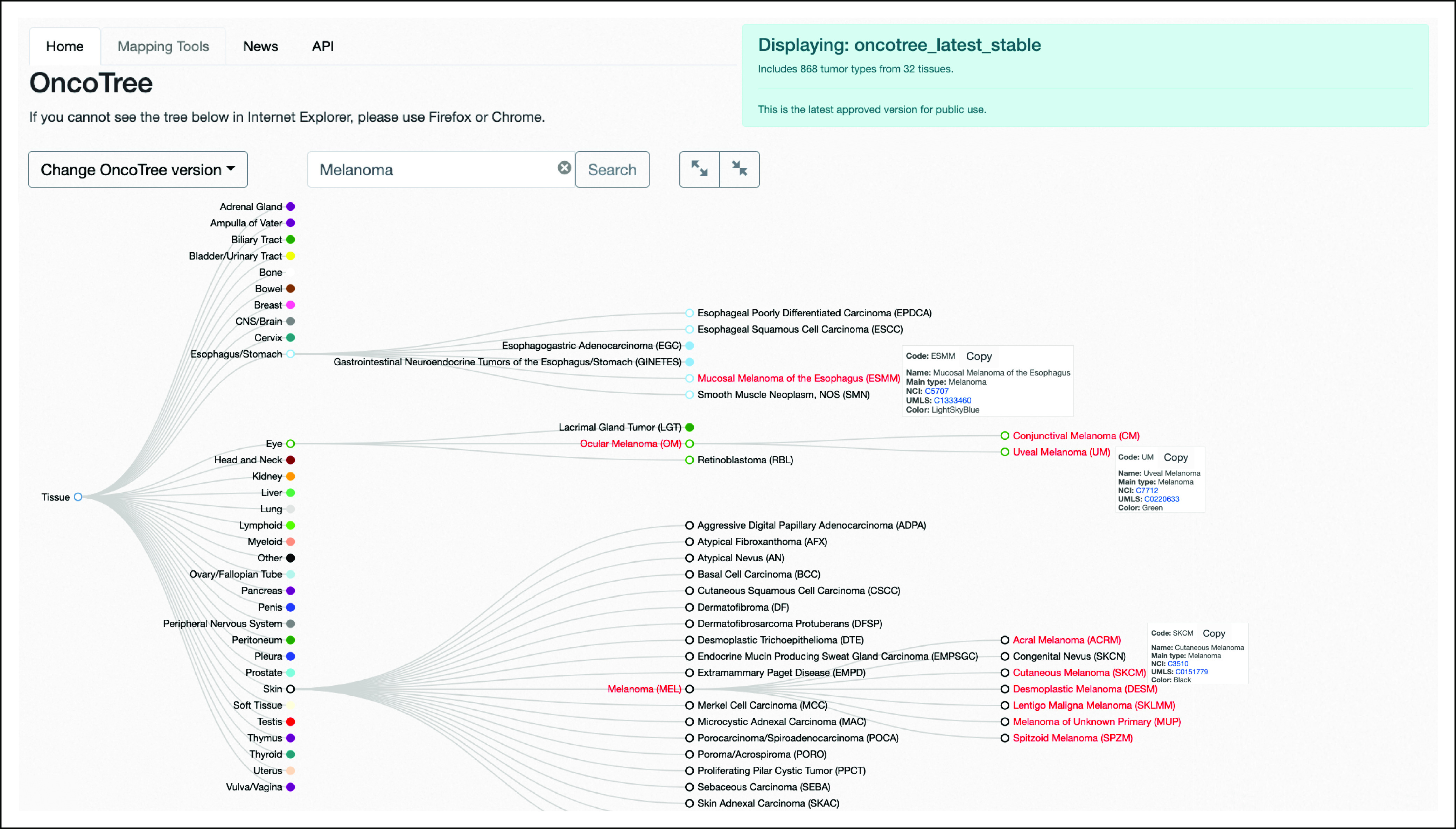
OncoTree homepage and tree structure. All cancer types are represented by a node on the tree. All subclassifications are connected to parent nodes through branches. The location of the cancer is based on the cell of origin and histologic architecture. Although all melanomas fall under the main type Melanoma, the location of each cancer on the tree is based on the organ of tumor origin. In this case, Mucosal Melanoma of the Esophagus is located under the Esophagus/Stomach node; Uveal Melanoma is located under Ocular Melanoma, which itself is nested under the Eye node; and Cutaneous Melanoma is located under Melanoma, which itself is nested under the Skin node. This structure of the tree allows connecting nodes across branches on the basis of histology.
OncoTree Structure: Hierarchically Organized and Clinically Relevant Cancer Classification
According to the ICD-O and NCI SEER cancer classification, cancers can be classified by histology in tandem with the location in the body where the cancer first developed.9 By design, the tree structure of OncoTree supports both these classifications: its root node represents a tissue and the branches and leaves include different histological and genomic subclassifications. The primary level (level 1) of the tree comprises 32 tissue sites.
Each node in the tree has an OncoTree code, which is its unique identifier, and a main cancer type, which is a category under which the tumor type can be grouped. The main type is independent of tissue and is of more general clinical specificity for cancer classification. Under the main type, nodes of higher conceptual granularity can be characterized. As an example, a node can be found in OncoTree representing Ocular Melanoma with OncoTree code OM and Melanoma as its assigned main type. OM is directly under the level 1 tissue Eye. Ocular Melanoma’s main type of Melanoma is determined by its histology and not the location of origin (Fig 1).
In addition to both cell of origin and histologic architecture, the OncoTree team have begun to curate genotype-specific cancer subtypes for hematological tumors on the basis of the 2016 WHO classification system,10,11 with further modifications made by OncoTree committee members. This high level of granularity is especially important in a point-of-care setting as genetic abnormalities may be associated with patient diagnosis, prognosis, and treatment options. For example, imatinib is FDA-approved for the treatment of patients with chronic myelogenous leukemia (CML) harboring the BCR-ABL1 fusion,12 and OncoTree includes the node Chronic Myeloid Leukemia, BCR-ABL1+, which is a subtype of CML. This level of detail in classification helps clinical decision support tools such as OncoKB to accurately map clinical implications (diagnostic, prognostic, and therapeutic) on the basis of the genotypic information of patients.
OncoTree Website: An Intuitive User Interface to Query, Explore, and View the Curated Data
OncoTree is available through a public Web resource.8 The current stable version (2020-10-01) of OncoTree comprises 868 tumor types branching from 31 tissue nodes and one additional node (called Other), which is used to categorize tumor types that cannot be attributed to a single tissue, including cancer of unknown primary and mixed tumors.
The OncoTree website also provides access to previous versions of OncoTree to enable version control and migration of OncoTree codes, which is essential for downstream applications. To help users upgrade or downgrade to a different version of OncoTree, the OncoTree Mapping tool in the form of a Python script was developed to facilitate the mapping of OncoTree codes to align older version entries with new versions. The Mapping Tool tab allows the user to download a script along with detailed instructions and tutorials to map between OncoTree versions.
Mapping of OncoTree codes to other ontologies including NCIt, UMLS, ICD-O, and HemOnc is also made available through the OncoTree GitHub page along with a documented script to automate the mappings.
OncoTree Web API: Supporting Programmatic Access to the Curated Data
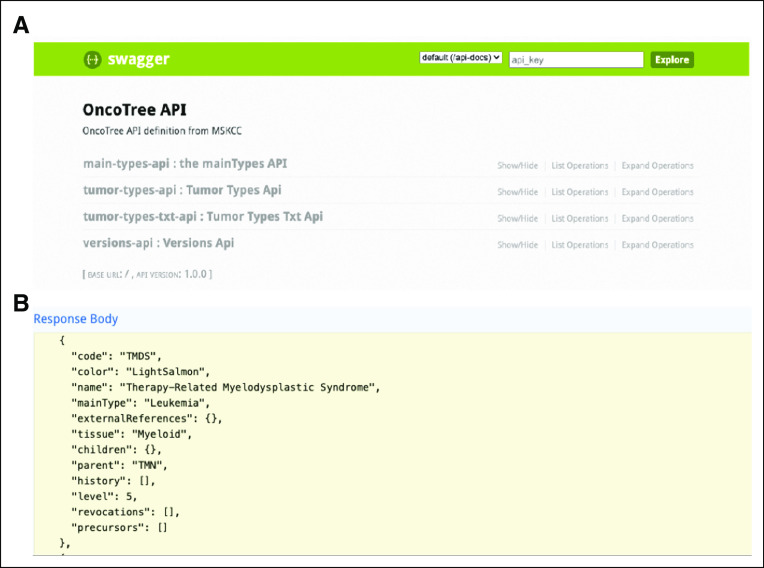
The OncoTree website includes a Web service API to facilitate programmatic access to OncoTree data (Fig 2A). The API was designed and developed on the basis of the OpenAPI specification.13 Interactive API documentation is automatically generated by the Swagger UI tool.14 API clients can be generated by the Swagger Codegen tool,15 which supports various programming languages, including Python, Java, JavaScript, R, and Perl.
FIG 2.
The OncoTree API. (A) Interactive documentation of the OncoTree API. (B) The API returns all curated attributes, which include cancer type full name, a unique OncoTree code of the tumor type, the tissue type, and the OncoTree code for the parent tumor type and any mapping to external sources. Historical information if applicable are also returned. API, application programming interface.
The primary purpose of the OncoTree API is to query and list the curated tumor types in OncoTree. For example, a list of all tumor types for the most current release of OncoTree is available through ref. 16. Querying the API returns all curated attributes of each Tumor Type including the tumor type full name, a unique OncoTree code of the tumor type, the tissue type, and the OncoTree code for the parent Tumor Type in the OncoTree. Historical information showing relationships to earlier versions of the OncoTree are available through attributes history, precursors, and revocations (these assist the mapping tool, which will translate OncoTree codes between different OncoTree releases; Fig 2B).
A tree representation with embedded subcancer types can also be retrieved through ref. 17, and a search function for querying by attribute value is available through ref. 18.
The main type of each OncoTree tumor type is available via ref. 19.
By default, all the above API services will provide responses to queries on the basis of the most current release of OncoTree. Additional releases are also available for retrieval by using the optional parameter version to specify the desired OncoTree version to use such as in ref. 20. A list of all available releases is available through ref. 21. OncoTree is stored and managed in an agile data governance system called TopBraid in a hierarchical ontology format.
OncoTree Governance and Releases: Expert-Curated and Community-Driven Process
OncoTree development is supervised by a multi-institute and multi-disciplinary committee consisting of solid tumor pathologists and hematopathologists, oncologists, and laboratory and translational scientists. Proposed changes to OncoTree are reviewed and approved or rejected by its governing body (Fig 3). Since OncoTree is an evolving system, many changes occur as a result of direct observations made by committee members in the clinic or because of updates to external classification systems such as ICD-O and NCIt. Additionally, a public mailing list on Google Groups is available in which anyone can suggest changes to OncoTree. These additions and modifications are collated along with metadata (Table 1), and the optimal location within the OncoTree is curated from suggested feedback. New nodes are added to the database, and checks are in place to avoid any duplicates and to validate that the addition is complete and properly formatted to circumvent potential system errors. Suggestions regarding the addition of new nodes or the placement of nodes within the tree are made available through the development version (Table 2) of OncoTree and sent to the OncoTree Committee. The tumor type, its metadata, location on the tree, and the evidence that it represents a distinct clinical and/or biological entity are reviewed by the members of the OncoTree committee and if accepted are pushed immediately to the candidate release for use in internal pathology reports. New nodes or changes to the location of a node are then included in the latest stable version of the OncoTree, which is released on the first day of each month. Typically, the review process takes a week or less, allowing for the rapid incorporation of newly identified tumor subtypes into clinical reports. Comprehensive rereview of the structure of an entire tissue type may take about a month. Committee members and the broader community of OncoTree users can also report errors, which are reviewed and resolved by the OncoTree team under the supervision of the committee.
FIG 3.
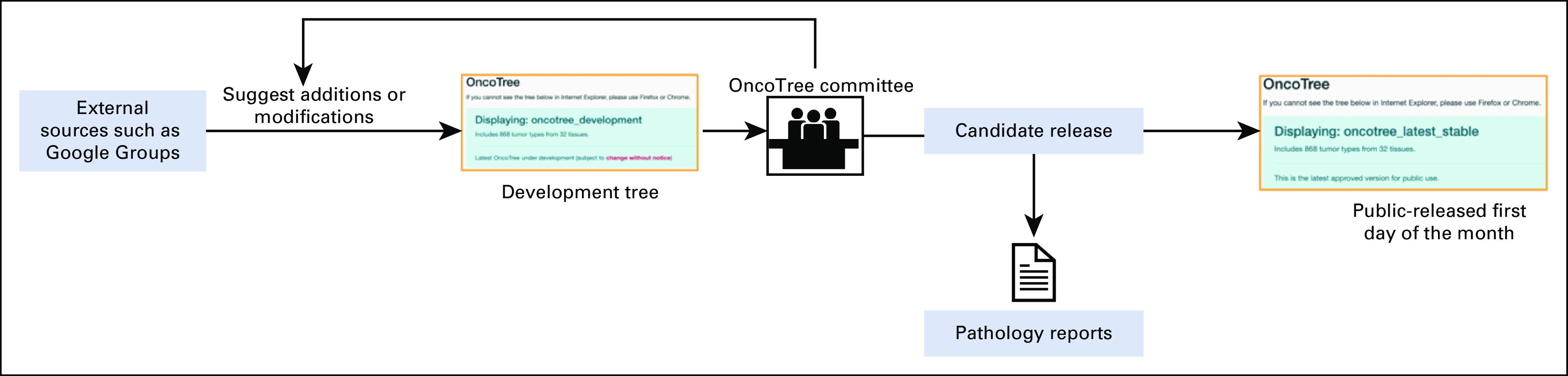
Overview of the curation process in OncoTree. OncoTree relies on a variety of sources for new additions and modifications. All new suggestions and updates are reviewed by the OncoTree committee and if approved are applied to the next OncoTree release for downstream applications.
TABLE 1.
Metadata for Each Cancer Node Is Described by Five Specific Properties
TABLE 2.
Multiple Versions of the OncoTree System Are Maintained: Latest Stable, Candidate, and Development, in Addition to All Previous Versions of OncoTree
OncoTree Applications
OncoTree was originally created to support cancer subtype classification of samples analyzed by The Cancer Genome Atlas (TCGA), the results of which were made available to the broader scientific community via cBioPortal for Cancer Genomics,22 a platform for exploring, visualizing, and analyzing multidimensional cancer genomics data.23 All tumor samples in cBioPortal, which now includes > 80,000 samples in the public version, are annotated with OncoTree classifications, thus enabling standardization across all cancer studies and allowing coherent queries (Fig 4).
FIG 4.
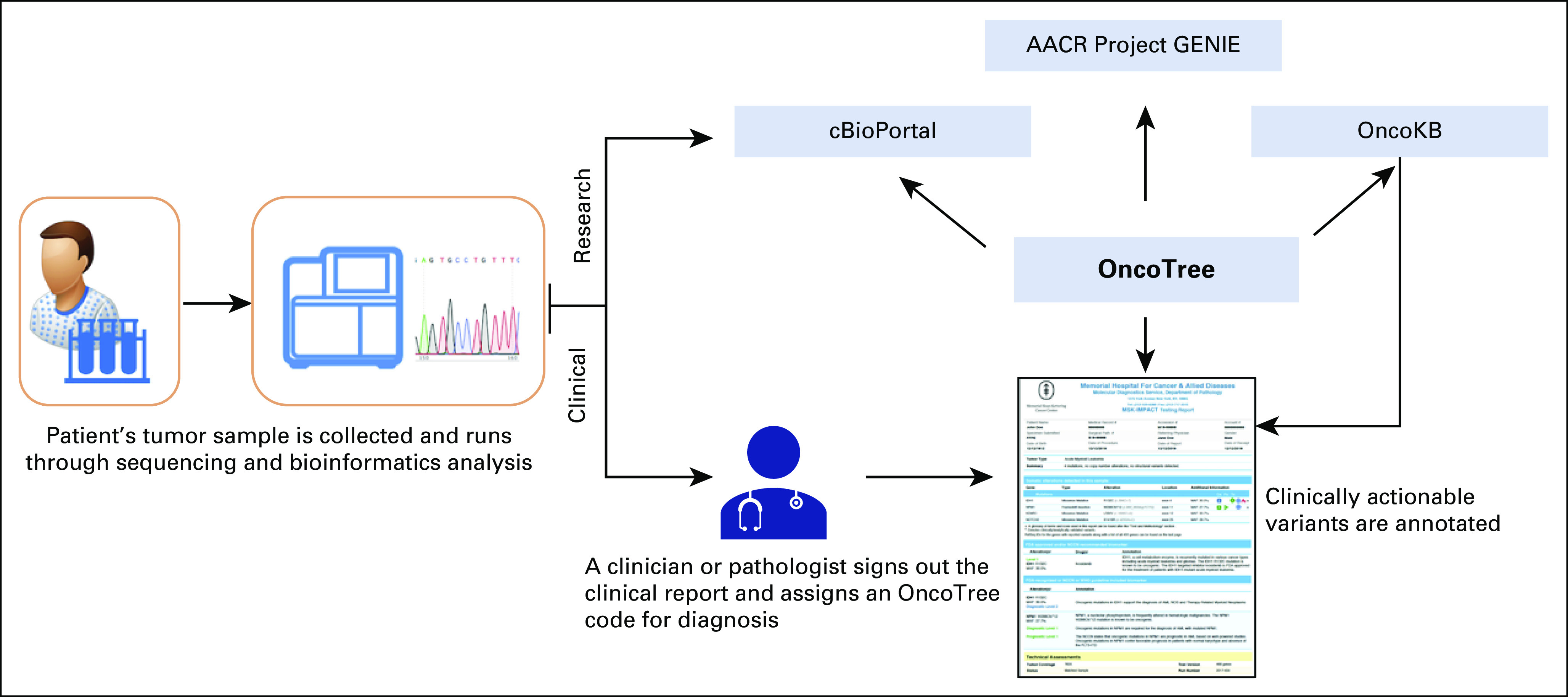
Overview of the different tools and resources that use OncoTree as a cancer classification vocabulary. From the clinical aspect, OncoTree codes are assigned in each genomic patient report at MSK by a clinician. In research, projects such as AACR Project GENIE and cBioPortal use OncoTree to assign codes to all tumor samples in their cohorts. OncoKB also uses OncoTree diagnosis codes to curate the knowledge base and to annotate clinically actionable variants in the genomic patient reports at MSK. AACR, American Association for Cancer Research; GENIE, Genomics Evidence Neoplasia Information Exchange; MSK, Memorial Sloan Kettering.
OncoTree was subsequently adopted by AACR Project GENIE, an international consortium created to facilitate the integration and sharing of integrated clinical genomic data sets.24 As of July 2020, 96,000 tumor and blood samples from 18 institutes have been annotated with OncoTree classifications. As AACR Project GENIE is a multi-institutional project, standardizing vocabularies for genomic and clinical data is essential for cross-institutional data integration. Across all institutes, demographic data are normalized using the North American Association of Central Cancer Registries (NAACR) vocabulary,25 and the cancer diagnosis of each individual sample profiled is mapped to an OncoTree code. The highly collaborative nature of AACR Project GENIE helped to fill gaps and to refine the structure of OncoTree. As a result of the inclusion of rarer tumor types or rare subtypes of common cancers not included in the TCGA data set, OncoTree became a more robust classification system. To facilitate the incorporation of AACR Project GENIE data into the National Cancer Institute’s Genomic Data Commons (GDC), OncoTree codes have been mapped to ontologies used by the GDC.26
OncoTree is also used by OncoKB,27 a precision oncology knowledge base containing information about the biological effects and clinical implications of genomic alterations in cancer.28 OncoKB uses the OncoTree classifications in conjunction with genomic information (gene and alterations) to annotate therapeutic, prognostic, and diagnostic data for various cancer types.
OncoTree is also used as a cancer classification system for the MSK-IMPACT initiative, a large-scale, prospective clinical sequencing effort at MSK in which samples from clinical sequencing platforms are assigned OncoTree codes on the basis of their clinical and genomic reports. Almost 60,000 MSK-IMPACT samples have now been annotated with OncoTree codes, and the data are shared with the broader research community via AACR Project GENIE.29 The hierarchical structure of OncoTree provides a key advantage here as it allows molecular pathologists to assign a cancer subtype with varying levels of precision on the basis of the available supportive clinical data.
OncoTree has also been integrated into MatchMiner, an open-source computational platform for matching prospectively generated patient-specific genomic profiles from clinical tumor sequencing to precision medicine clinical trials in which eligibility is increasingly both tumor subtype–restricted and genomic subtype–restricted.30
The goal of OncoTree is to streamline clinical reporting of tumor- and plasma-based genomic profiling results and to facilitate collaborative clinical and translational research. The recognition that tumors arising within the same organ can have different genomic drivers and that these genomic drivers can be prognostic or predictive biomarkers of drug response has led to reconsideration of how tumors are subclassified and the incorporation of genomic features into tumor subtype classification schema. The accelerating shift to precision oncology–based treatment paradigms has also created the need for a cancer subtype classification system that can be quickly modified to incorporate newly recognized molecularly distinct tumor subtypes.
Although there is overlap between OncoTree and other classification systems, OncoTree provides unique benefits in that it is easy to navigate; has an API for straightforward downloads, data extraction, and integration with clinical reporting systems; and facilitates mappings between different ontologies. OncoTree can also be rapidly modified as new tumor subtypes are identified within the context of ongoing clinical and translational research studies. An agile computational ontology system such as OncoTree can thus play an important role in precision oncology by providing physicians with an accurate and up-to-date tumor classification reference that captures common, rare, and newly evolving tumor types encountered in a clinical setting and that can be linked to cutting-edge clinical trials of novel biological therapies targeting specific molecularly defined tumor subtypes.
Additionally, academic molecular pathologists and private companies offering tumor and cell-free NGS-based clinical diagnostics do not always have the full clinical history of a patient including immunohistochemical stains or other clinical features used by other cancer subtype classification systems. The hierarchical structure of OncoTree facilitates the workflow of these clinical molecular diagnostic laboratories by allowing for tumor subtype classification at different levels of precision on the basis of the often limited clinical information available in real time to molecular pathologists. OncoTree is also used by OncoKB, a precision oncology knowledge base that supports clinical decision making by providing insight into the level of evidence supporting the use of a molecular alteration as a predictive biomarker of drug response. Although some biomarkers are tumor type–agnostic, such as tumor mutation burden and microsatellite instability, most molecular biomarkers contained in OncoKB are tumor type–specific, and OncoTree is a critical component of this decision support tool.31–33
OncoTree remains under active development with changes and additions to OncoTree being suggested, discussed, and approved by a multi-institutional and multidisciplinary committee of experts on a continuous basis. Moreover, there are now efforts to extend existing OncoTree code mappings to NCIt, UMLS, HemOnc, and ICD-10 and to create new mapping tools to additional systems including MeSH and SNOMED-CT. All existing mappings are available on the OncoTree GitHub page34 along with a mapping script that provides the users to map certain ontologies to and from OncoTree codes. Importantly, OncoTree was not developed with the goal of replacing other cancer classification systems, such as ICD-O, which officially recognizes new tumor entities in due time. Rather, OncoTree serves as a complementary system to harmonize terminologies and concepts with other systems such as ICD-O and provides an interoperable and transparent hierarchical classification platform to share cancer data that incorporates our most current understanding of how genomics and other biological or clinical features influence cancer subtype classification.
In conclusion, our understanding of the biological differences within individual cancers arising from the same organ site has increased exponentially in recent years. We have designed OncoTree to be flexible so that it can serve a diversity of cancer community knowledge endeavors and data initiatives as well as to help meet the rapidly evolving needs of both ontology research and clinical decision support for precision oncology.
SUPPORT
Supported by Marie-Josée and Henry R. Kravis Center for Molecular Oncology, the Robertson Foundation and Prostate Cancer Foundation (to N.S.), and a National Cancer Institute Cancer Center Core Grant (P30-CA008748).
AUTHOR CONTRIBUTIONS
Conception and design: Ritika Kundra, Hongxin Zhang, Robert Sheridan, Sahussapont Joseph Sirintrapun, Avery Wang, Angelica Ochoa, Manda Wilson, Benjamin Gross, Yichao Sun, Ramyasree Madupuri, Maria Arcila, Anas Younes, Andrew Intlekofer, Yelena Y. Janjigian, Tara A. Soumerai, Ingo K. Mellinghoff, Ethan Cerami, Angela Waanders, Jianjiong Gao, David Solit, and Nikolaus Schultz
Financial support: David Solit, Nikolaus Schultz
Administrative support: Benjamin Gross, Dalicia Reales, David Hyman, Tara A. Soumerai, Stuart Gardos, David Solit
Provision of study materials or patients: Sahussapont Joseph Sirintrapun, Hikmat A. Al-Ahmadie, Katherine Janeway, David Hyman, Anas Younes, James J. Harding, Ghassan K. Abou-Alfa, Ingo K. Mellinghoff, Abraham A. Hakimi, Matthew Fury, David Solit
Collection and assembly of data: Ritika Kundra, Hongxin Zhang, Sahussapont Joseph Sirintrapun, Avery Wang, Angelica Ochoa, Manda Wilson, Baby A. Satravada, Dalicia Reales, Efsevia Vakiani, Hikmat A. Al-Ahmadie, Ahmet Dogan, Maria Arcila, Cristina Viaplana, Katherine Janeway, Matthew Ducar, Lynette Sholl, Snjezana Dogan, Iker Huerga Sanchez, Aijaz Syed, Anoop Balakrishnan Rema, Moriah Nissan, Gopakumar V. Iyer, Rajmohan Murali, Nancy Bouvier, Robert A. Soslow, Andrew Intlekofer, James J. Harding, Meighan M. Gallagher, Ingo K. Mellinghoff, Matthew Fury, Jason T. Huse, Aditya Bagrodia, Stuart Gardos, Pichai Raman, Priyanka Shivdasani, David Solit, and Nikolaus Schultz
Data analysis and interpretation: Ritika Kundra, Hongxin Zhang, Sahussapont Joseph Sirintrapun, Angelica Ochoa, Hikmat A. Al-Ahmadie, Ahmet Dogan, Maria Arcila, Ahmet Zehir, Steven Maron, Michael F. Berger, Matthew Ducar, Lynette Sholl, Philippe Bedard, Lea F. Surrey, Aijaz Syed, Debyani Chakravarty, Sarah Suehnholz, Rajmohan Murali, David Hyman, Anas Younes, Andrew Intlekofer, James J. Harding, Richard D. Carvajal, Paul J. Sabbatini, Ghassan K. Abou-Alfa, Luc Morris, Yelena Y. Janjigian, Ingo K. Mellinghoff, Abraham A. Hakimi, Meera Hameed, Stacy Thomas, Tali Mazor, Priti Kumari, Suzanne MacFarland, Scott Newman, David Solit, and Nikolaus Schultz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO’s conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/cci/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Robert Sheridan
Stock and Other Ownership Interests: Agios, Alnylam, Alphabet, Amazon Inc, Amgen, Biogen, Bluebird Bio, Bristol-Myers Squibb, Clementia Pharmaceuticals, General Electric Company, Johnson & Johnson, Merck, Northwest Biotherapeutics, Pfizer, 3M, Celene Corp, DowDuPont, Portola Pharmaceuticals, Fortive Corp
Patents, Royalties, Other Intellectual Property: A collaboration with the laboratory of Thomas Tuschl at The Rockefeller University between 2004 and 2012 led to the publication of several papers on the discovery of novel microRNA and other small interfering molecules in mammals and other species. A patent was awarded related to this work. The MSK collaborators (including myself) do not hold inventor rights in the intellectual property, but The Rockefeller University designated a portion of any received royalties to be paid to me in recognition of the facilitation of this research and these discoveries. I have received occasional payments from The Rockefeller University, but no payments have been received in the last two years. I do not know whether future payments might be made to me related to this work.
Avery Wang
Stock and Other Ownership Interests: Gilead Sciences
Angelica Ochoa
Stock and Other Ownership Interests: Infinity Pharmaceuticals, AstraZeneca/MedImmune
Hikmat A. Al-Ahmadie
Consulting or Advisory Role: Bristol-Myers Squibb, EMD Serono, AstraZeneca/MedImmune, Janssen Biotech
Ahmet Dogan
Consulting or Advisory Role: Seattle Genetics, Roche, Takeda, EUSA Pharma, Abbvie
Research Funding: Roche/Genentech
Maria Arcila
Honoraria: Invivoscribe, Biocartis
Consulting or Advisory Role: AstraZeneca
Travel, Accommodations, Expenses: AstraZeneca, Invivoscribe, Raindance Technologies
Ahmet Zehir
Honoraria: Illumina
Steven Maron
Stock and Other Ownership Interests: Calithera Biosciences
Honoraria: RMEI Medical Education, MedEd, Clinical Care Options
Consulting or Advisory Role: Health Advances, Natera, Basilea
Research Funding: Roche/Genentech, Guardant Health
Travel, Accommodations, Expenses: Merck, Bayer
Michael F. Berger
Research Funding: Grail
Patents, Royalties, Other Intellectual Property: Provisional patent pending for Systems and Methods for Detecting Cancer via cfDNA Screening
Katherine Janeway
Honoraria: Foundation Medicine
Consulting or Advisory Role: Bayer, Ipsen
Travel, Accommodations, Expenses: Bayer
Lynette Sholl
Honoraria: AstraZeneca
Consulting or Advisory Role: Loxo, EMD Serono
Research Funding: Roche/Genentech
Philippe Bedard
Research Funding: Bristol-Myers Squibb, Sanofi, AstraZeneca, Genentech/Roche, Servier, GlaxoSmithKline, Novartis, SignalChem, PTC Therapeutics, Nektar, Merck, Seattle Genetics, Mersana, Immunomedics, Lilly
Iker Huerga Sanchez
Employment: Memorial Sloan-Kettering Cancer Center
Stock and Other Ownership Interests: Tempus
Consulting or Advisory Role: AstraZeneca
Anoop Balakrishnan Rema
Stock and Other Ownership Interests: Array BioPharma, AnaptysBio, Loxo
Gopakumar V. Iyer
Consulting or Advisory Role: Bayer, Janssen, Mirati Therapeutics
Research Funding: Mirati Therapeutics, Novartis, Debiopharm Group, Bayer
Robert A. Soslow
Speakers' Bureau: Ebix/Oakstone
Patents, Royalties, Other Intellectual Property: Methods for diagnosis and treatment of endometrial cancer. Assignee: 2015—The Regents of the University of California August 18, 2015, Application: Compositions and methods for the diagnosis and treatment—2016 of ovarian cancers that are associated with reduced smarca4 gene expression or protein function, Royalties from published books: Cambridge University Press and Springer Publishing
David Hyman
Employment: Lilly, Loxo Oncology
Stock and Other Ownership Interests: Fount Therapeutics
Consulting or Advisory Role: Chugai Pharma, Boehringer Ingelheim, AstraZeneca, Pfizer, Bayer, Genentech, Fount Therapeutics, Lilly
Research Funding: AstraZeneca, Puma Biotechnology, Loxo, Bayer
Travel, Accommodations, Expenses: Genentech, Chugai Pharma
Anas Younes
Employment: AstraZeneca
Stock and Other Ownership Interests: AstraZeneca
Honoraria: Merck, Roche, Takeda, Janssen, Abbvie, Curis, Epizyme
Consulting or Advisory Role: Bio-Path Holdings, Inc, Xynomic Pharma, Epizyme, Roche, Celgene, HCM
Research Funding: Janssen, Curis, Roche, Genentech, Merck, Bristol-Myers Squibb, Syndax
Other Relationship: AstraZeneca
James J. Harding
Consulting or Advisory Role: Bristol-Myers Squibb, CytomX Therapeutics, Lilly, Eisai, Imvax, Merck, Exelixis, Zymeworks, Adaptimmune
Research Funding: Bristol-Myers Squibb, Pfizer, Lilly, Novartis, Incyte, Calithera Biosciences, Polaris, Yiviva, Debiopharm Group, Zymeworks, Boehringer Ingelheim
Richard D. Carvajal
Consulting or Advisory Role: Merck, Aura Biosciences, Bristol-Myers Squibb, Castle Biosciences, Immunocore, I-Mab, PureTech, Genzyme, Sorrento Therapeutics, Chimeron Bio, Rgenix, InxMed, Pierre Fabre, TriSalus Life Sciences
Research Funding: Amgen, Astellas Pharma, AstraZeneca, Bayer, Bellicum Pharmaceuticals, Bristol-Myers Squibb, Corvus Pharmaceuticals, Lilly, Immunocore, Incyte, Macrogenics, Merck, Mirati Therapeutics, Novartis, Pfizer, Plexxikon, Roche/Genentech, Array BioPharma, IDEAYA Biosciences, Regeneron
Paul J. Sabbatini
Honoraria: UpToDate
Consulting or Advisory Role: UpToDate
Research Funding: Bristol-Myers Squibb, Ludwig Institute for Cancer Research
Ghassan K. Abou-Alfa
Consulting or Advisory Role: Celgene, Silenseed, Sillajen, Gilead Sciences, Agios, Bayer, Eisai, Ipsen, Merck Serono, AstraZeneca, CytomX Therapeutics, BeiGene, Genoscience Pharma, LAM Therapeutics, Lilly, Loxo, Minapharma, QED Therapeutics, RedHill Biopharma, SOBI, twoXAR, Yiviva, Flatiron Health, Roche/Genentech, Autem Medical, Berry Genomics, Gilead Sciences, Incyte, TheraBionic, Vector Health
Research Funding: Bayer, Exelixis, CASI Pharmaceuticals, AstraZeneca, Bristol-Myers Squibb, Incyte, Agios, Polaris, Puma Biotechnology, QED Therapeutics
Travel, Accommodations, Expenses: Polaris
Luc Morris
Consulting or Advisory Role: Rakuten Aspyrian
Research Funding: AstraZeneca, Bristol-Myers Squibb
Patents, Royalties, Other Intellectual Property: Employer pending patent application, listed as inventor
Yelena Y. Janjigian
Stock and Other Ownership Interests: Rgenix
Consulting or Advisory Role: Pfizer, Merck, Bristol-Myers Squibb, Merck Serono, Daiichi Sankyo, Ono Pharmaceutical, Michael J. Hennessy Associates, Jounce Therapeutics, Rgenix, Bayer, Imugene, AstraZeneca, Lilly, Paradigm, Zymeworks, Seattle Genetics, Physicans' Education Resource, Merck Sharpe and Dohme Corp, Peerview
Research Funding: Boehringer Ingelheim, Bayer, Lilly, Amgen, Roche, Genentech, Rgenix, Bristol-Myers Squibb, Merck
Meighan M. Gallagher
Employment: ExecuPharm, Inc
Tara A. Soumerai
Consulting or Advisory Role: Verastem
Research Funding: Genentech/Roche, BeiGene, TG Therapeutics
Ingo K. Mellinghoff
Honoraria: Roche
Consulting or Advisory Role: Agios, Puma Biotechnology, Debiopharm Group, Black Diamond Therapeutics, Voyager Therapeutics
Research Funding: General Electric, Amgen, Lilly
Travel, Accommodations, Expenses: Voyager Therapeutics, AstraZeneca, Roche, Puma Biotechnology, Agios
Matthew Fury
Employment: Regeneron
Stock and Other Ownership Interests: Regeneron
Research Funding: Regeneron
Patents, Royalties, Other Intellectual Property: Regeneron
Jason T. Huse
Research Funding: Taiho Pharmaceutical
Travel, Accommodations, Expenses: Taiho Pharmaceutical
Pichai Raman
Employment: GlaxoSmithKline
Consulting or Advisory Role: Scholar Rock
Scott Newman
Employment: Sema4
David Solit
Stock and Other Ownership Interests: Loxo, Scorpion Therapeutics, Inc
Consulting or Advisory Role: Pfizer, Loxo, Illumina, Vividion Therapeutics, Lilly, QED Therapeutics, BridgeBio Pharma, Scorpion Therapeutics, Inc
No other potential conflicts of interest were reported.
REFERENCES
- 1.Salto-Tellez M, Cree IA.Cancer taxonomy: Pathology beyond pathology Eur J Cancer 11557–602019 [DOI] [PubMed] [Google Scholar]
- 2.International Health Terminology Standards Development Organisation . SNOMED-CT. http://www.ihtsdo.org/snomed-ct/ [Google Scholar]
- 3.International Classification of Diseases. https://www.who.int/classifications/icd/en/ [Google Scholar]
- 4.National Cancer Institute Thesaurus. https://ncithesaurus.nci.nih.gov/ [Google Scholar]
- 5.Bodenreider O.The Unified Medical Language System (UMLS): Integrating biomedical terminology Nucleic Acids Res 32D267–D2702004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.MeSH (Medical Subject Headings) https://www.ncbi.nlm.nih.gov/mesh [Google Scholar]
- 7.Cree IA, Indave BI. Commentary: Cancer research quality and tumour classification. Tumor Biol. 2020;42 doi: 10.1177/1010428320907544. [DOI] [PubMed] [Google Scholar]
- 8.OncoTree http://oncotree.info/
- 9.National Cancer Institute's Surveillance, Epidemiology and End Results (SEER) Program. https://training.seer.cancer.gov/disease/categories/classification.html [Google Scholar]
- 10.Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms Blood 1272375–23902016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia Blood 1272391–24052016 [DOI] [PubMed] [Google Scholar]
- 12.US Food and Drug Administration https://www.accessdata.fda.gov/drugsatfda_docs/label/2008/021588s024lbl.pdf
- 13.OpenAPI https://openapi.org
- 14.Swagger UI https://swagger.io/tools/swagger-ui/
- 15.Swagger Codegen https://swagger.io/tools/swagger-codegen/
- 16.OncoTree http://oncotree.info/api/tumorTypes
- 17.OncoTree http://oncotree.info/api/tumorTypes/tree
- 18.OncoTree http://oncotree.info/api/tumorTypes/search
- 19.OncoTree http://oncotree.info/api/mainTypes
- 20.OncoTree http://oncotree.info/api/tumorTypes?version=oncotree_2019_12_01
- 21.OncoTree http://oncotree.info/api/versions
- 22.cBioPortal . cBioPortal for Cancer Genomics. https://www.cbioportal.org/ [Google Scholar]
- 23.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.AACR Project GENIE Consortium AACR Project GENIE: Powering precision medicine through an international consortium Cancer Discov 7818–8312017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.North American Association of Central Cancer Registries (NAACR) https://www.naaccr.org/
- 26.Thomas S, Lichtenberg T, Dang K, et al. Linked Entity Attribute Pair (LEAP): A harmonization framework for data pooling JCO Clin Cancer Inform 4691–6992020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.OncoKB . Precision Oncology Knowledge Base. https://www.oncokb.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A precision oncology knowledge base JCO Precis Oncol10.1200/PO.17.00011 [epub ahead of print on May 16, 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients Nat Med 23703–7132017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindsay J, Del Vecchio Fitz C, Zwiesler Z, et al. MatchMiner: An open source computational platform for real-time matching of cancer patients to precision medicine clinical trials using genomic and clinical criteria. bioRxiv. 2017 [Google Scholar]
- 31.Hyman D, Piha-Paul S, Won H, et al. HER kinase inhibition in patients with HER2- and HER3-mutant cancers Nature 554189–1942018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency N Engl J Med 3722509–25202015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Snyder A, Makarov V, Merghoub T, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma N Engl J Med 3712189–21992014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.GitHub, Inc https://github.com/cBioPortal/oncotree