Tseng et al. show the Bro1 V domain stimulates Vps4 to promote ESCRT-III–driven intralumenal vesicle formation. This mode of stimulation is required for coordinated cargo incorporation during ESCRT-mediated multivesicular body biogenesis.
Abstract
Endosomal sorting complexes required for transport (ESCRT-0, -I, -II, -III) execute cargo sorting and intralumenal vesicle (ILV) formation during conversion of endosomes to multivesicular bodies (MVBs). The AAA-ATPase Vps4 regulates the ESCRT-III polymer to facilitate membrane remodeling and ILV scission during MVB biogenesis. Here, we show that the conserved V domain of ESCRT-associated protein Bro1 (the yeast homologue of mammalian proteins ALIX and HD-PTP) directly stimulates Vps4. This activity is required for MVB cargo sorting. Furthermore, the Bro1 V domain alone supports Vps4/ESCRT–driven ILV formation in vivo without efficient MVB cargo sorting. These results reveal a novel activity of the V domains of Bro1 homologues in licensing ESCRT-III–dependent ILV formation and suggest a role in coordinating cargo sorting with membrane remodeling during MVB sorting. Moreover, ubiquitin binding enhances V domain stimulation of Vps4 to promote ILV formation via the Bro1–Vps4–ESCRT-III axis, uncovering a novel role for ubiquitin during MVB biogenesis in addition to facilitating cargo recognition.
Introduction
The endosomal sorting complexes required for transport (ESCRTs) have been implicated in a number of cellular membrane remodeling processes, including intralumenal vesicle (ILV) generation during the conversion of endosomes into multivesicular bodies (MVBs; relevant to both lysosomal degradation and exosome biogenesis), abscission during cytokinesis, viral budding, nuclear pore surveillance, autophagy, and membrane repair (Baietti et al., 2012; Lee et al., 2007a; Webster et al., 2014; Skowyra et al., 2018; Takahashi et al., 2018; reviewed by Hanson and Cashikar, 2012; Vietri et al., 2020; Votteler and Sundquist, 2013). These studies have highlighted a conserved role for ESCRT-III and associated factors in membrane deformation. ESCRT-III–dependent membrane remodeling is coordinated with upstream events, such as cargo recognition during MVB sorting (reviewed by Piper and Lehner, 2011; Williams and Urbé, 2007). Ubiquitylated cargos destined for lysosomal destruction are actively recognized by a host of ubiquitin (Ub)–binding domains within ESCRT-0, -I, and -II, as well as in the Bro1 domain family proteins, and sequestration of these ubiquitylated cargos forms microdomains at which ESCRT-II activates ESCRT-III polymerization to drive ILV formation (reviewed by Schmidt and Teis, 2012; Williams and Urbé, 2007). Depletion of Ub from the site of MVB sorting precludes cargo sorting as well as ILV formation itself, indicating a level of coordination between cargo recognition and ESCRT-III–mediated ILV generation (MacDonald et al., 2012a; Stringer and Piper, 2011). ESCRT-III assembly, remodeling, and disassembly drive ILV formation in vivo, although how this cycle is regulated to ensure cargo transfer into ILVs remains unresolved.
ESCRT-III subunits are monomeric in the cytosol and undergo an ordered polymerization on membranes into filamentous spirals responsible for membrane remodeling (reviewed by Schmidt and Teis, 2012). ESCRT-III function is intimately connected to the AAA-ATPase Vps4, which supports both dynamic exchange of subunits during ESCRT-III polymerization and ESCRT-III disassembly at the completion of the reaction (Adell and Teis, 2011; Adell et al., 2017; Babst et al., 1997; Babst et al., 1998; Davies et al., 2010; Mierzwa et al., 2017; Pfitzner et al., 2020). A cohort of regulators, including ESCRT-III itself, the Vps4 cofactor Vta1/LIP5, and Bro1, serve to coordinate the activities of ESCRT-III and Vps4 to optimize ESCRT function during MVB sorting (Azmi et al., 2006; Azmi et al., 2008; Merrill and Hanson, 2010; Shim et al., 2008; Wemmer et al., 2011). These studies have highlighted the importance of Vps4 regulation during MVB sorting but have not resolved how these factors act in concert to enable MVB biogenesis.
The Bro1 domain family members, including the aforementioned yeast Bro1 as well as mammalian ALIX/PDCD6IP and HD-PTP/PTPN23, make multiple contributions to ESCRT-mediated events through a conserved three-domain architecture: the N-terminal Bro1 domain (BOD) binds the ESCRT-III subunit CHMP4/Snf7; the V domain, a V-shaped structure formed by two helix bundle arms, interacts with both Ub and YPXnL motifs found within Gag, Syntenin, and Rfu1; and the C-terminal proline-rich region (PRR) facilitates associations with ESCRT-I and other factors (e.g., Ub isopeptidase Doa4; Baietti et al., 2012; Buysse et al., 2020; Carlton et al., 2008; Johnson et al., 2017; Kimura et al., 2014; Lee et al., 2016; Luhtala and Odorizzi, 2004; McCullough et al., 2008; Nikko and André, 2007; Richter et al., 2013; Wemmer et al., 2011; Zhai et al., 2011). Bro1 domain family members contribute to (1) Ub-dependent and Ub-independent cargo recognition in concert with or in parallel to the early ESCRTs and (2) regulating ESCRT-III dynamics by facilitating CHMP4/Snf7 activation and inhibiting Vps4 disassembly of ESCRT-III. These diverse contributions suggest Bro1 domain family members may serve roles coordinating cargo entry into budding ILVs during MVB sorting.
Here, we find that Bro1 V domain supports ESCRT-dependent MVB biogenesis in vivo by facilitating ILV formation. The V domain interacts with Vps4’s MIT domain and stimulates Vps4 ATPase activity, and this stimulation is further enhanced by the V domain binding to Ub. This mode of Vps4 stimulation is critical for Bro1 function, since V domain mutations that preserve Vps4 and Ub binding but specifically perturb the ability to stimulate Vps4 ATPase activity showed defects in MVB cargo sorting and ILV formation in vivo. These results indicate that ESCRT-driven ILV formation is separable from cargo sorting and suggest that Bro1 domain family members coordinate ubiquitylated cargo sorting into ILVs while promoting ILV formation via the Vps4–ESCRT-III axis.
Results
Bro1 V domain promotes ESCRT-dependent ILV formation
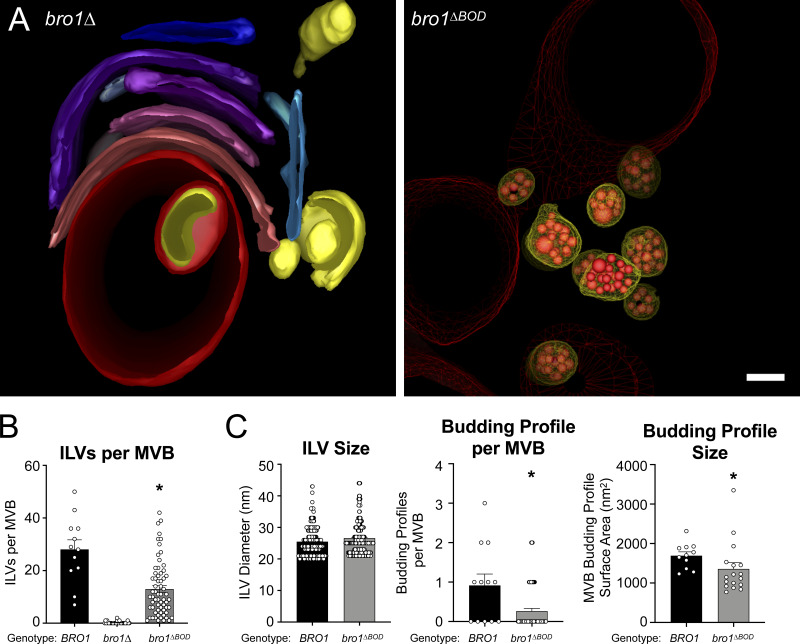
The Bro1 BOD interacts with the ESCRT-III subunit Snf7 (Kim et al., 2005) to promote ESCRT-III polymerization through both facilitating Snf7 activation (Tang et al., 2016) and inhibiting Vps4-mediated disassembly of ESCRT-III (Wemmer et al., 2011). Disruption of Bro1–Snf7 association abrogates MVB cargo sorting, but some degree of ILV formation is retained (Wemmer et al., 2011). To better understand this unexpected phenomenon, we examined the phenotype of cells expressing Bro1 lacking its N-terminal BOD (bro1ΔBOD, aa 370–844) as the sole allele of BRO1. Cells expressing Bro1ΔBOD displayed spherical endosomal structures containing ILVs (Fig. 1 A and Video 1). In contrast, cells lacking Bro1 altogether (bro1Δ) were deficient in ILVs and displayed multilamellar endosomal stacks characteristic of “class E vps” endosomal compartments (Luhtala and Odorizzi, 2004; Fig. 1 A and Video 2; a rare bro1Δ cell with ILVs is shown in Fig. S1 A). Compared with WT cells, bro1ΔBOD cells showed reductions in ILVs per MVB, budding profile (BP) frequency and BP surface area (Fig. 1, B and C; and Video 3), however the sizes of ILVs were equivalent to those in WT cells (Fig. 1 C). While aspects of the bro1ΔBOD MVBs differed from those in WT cells, the striking result was their presence in bro1ΔBOD cells in contrast to their absence in bro1Δ cells (Fig. 1 B). This result indicates that Bro1 BOD is not required to support ILV formation, consistent with the previous analysis of Snf7 defective for Bro1 binding (Wemmer et al., 2011).
Figure 1.
Bro1ΔBOD supports ILV formation. (A) Three-dimensional models reconstructed from 200-nm thick-section electron tomograms of bro1Δ (GOY65) and bro1ΔBOD (bro1Δ::TEF1p-bro1ΔBOD; CTY2) cells. The bro1Δ cells have class E compartments, which are flattened stacks of endosomal membranes that generally lack internal vesicles; these stacks are shown in different colors to differentiate individual membranes. For bro1ΔBOD, the limiting membrane of MVBs are labeled yellow, the ILVs are highlighted in red, and the vacuole limiting membrane is labeled as red mesh. Scale bar = 100 nm. (B) WT (SEY6210), bro1Δ (GOY65), and bro1ΔBOD (CTY2) were analyzed by ET and quantified for number of ILV per MVB. Asterisk indicates statistically significant differences compared with WT and bro1Δ (P < 0.0001). (C) WT (SEY6210) and bro1ΔBOD (CTY2) were analyzed by ET and quantified to assess ILV size (diameter), individual BP size (surface area), and the frequency of incomplete ILV budding events (BPs per MVB); 12 MVBs from WT cells (SEY6210) containing 337 ILVs and 11 budding intermediates (BPs), 64 MVBs from bro1ΔBOD cells (CTY2) containing 831 ILVs and 17 budding intermediates, and 32 class E endosomal compartments from bro1Δ cells (GOY65) were quantified. Data are represented as mean ± SEM. Asterisks indicate statistically significant differences compared with WT (BP per MVB, P = 0.0016; BP size, P = 0.0048).
Video 1.
Tomogram of MVB in a bro1ΔBOD cell. MVBs in a bro1ΔBOD cell exhibit WT-like MVB morphology. This video shows nine MVBs that are adjacent to the vacuole (red mesh), while the limiting membrane of the MVB is colored yellow and the ILVs are in red. Intermediate BPs are highlighted in green. Scale bar = 100 nm. Frame rate, 50 frames/s.
Video 2.
Tomogram of MVB in a bro1Δ cell.bro1Δ cell has class E compartments, which are flattened stacks of endosomal membranes that generally lack internal vesicles. Cisternal class E compartment stacks are shown in different colors to differentiate individual membranes. The vacuolar limiting membrane is labeled red. Scale bar = 100 nm. Frame rate, 50 frames/s.
Figure S1.
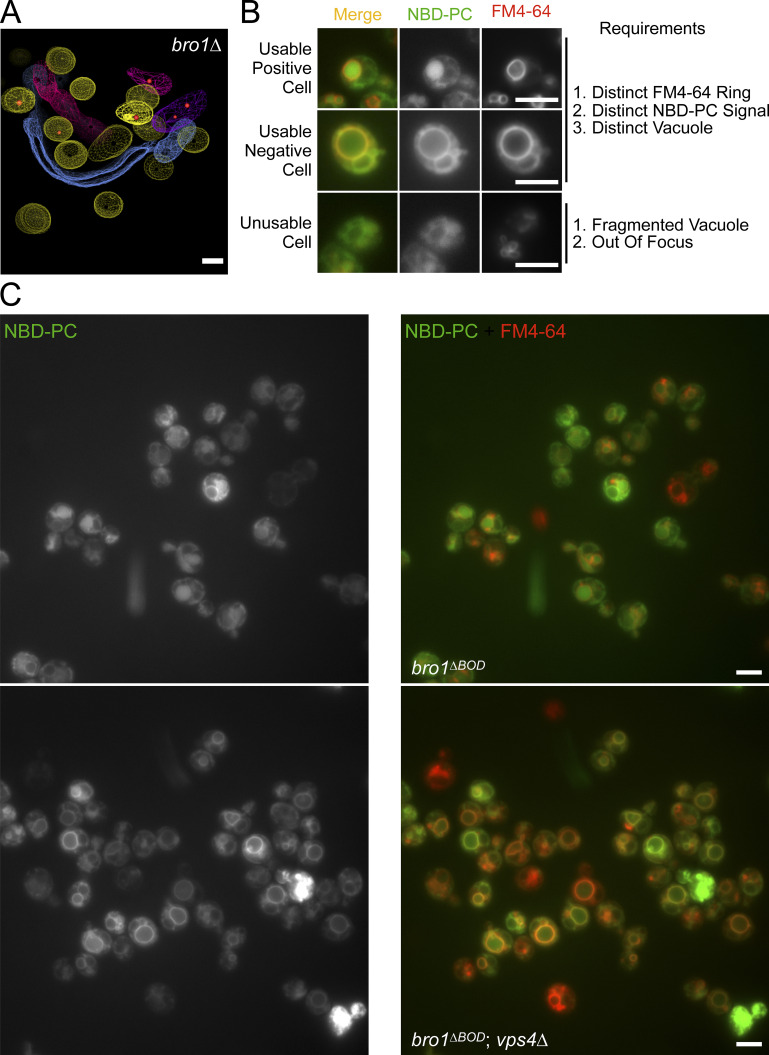
Representative images of cells stained with NBD-PC and FM4-64. This figure complements Fig. 3. (A) Three-dimensional models reconstructed from 200-nm thick-section electron tomograms of bro1Δ (GOY65) cells. This image depicts rare MVBs that contains an ILV. Normal-like endosomes are highlighted by yellow limiting membrane, while other colors depict flattened or tubular endosomes devoid of vesicles. The ILVs are highlighted in red. Scale bar = 100 nm. (B) Selection criteria for scoring positive NBD-PC cells. A usable cell is defined by having (1) readily identifiable FM4-64 labeling of vacuole membrane, (2) readily identifiable NBD-PC signal, and (3) defined vacuoles. An unusable cell is defined by having (1) a fragmented vacuole and (2) out of focus. A positive cell is defined by a diffuse NBD-PC signal within the lumen while lacking a distinct ring on the limiting membrane of the vacuole (defined by FM4-64), while a negative cell is defined by the colocalization of NBD-PC and FM4-64 on the limiting membrane of the vacuole. Scale bars = 5 µm. (C) Representative micrographs of bro1ΔBOD (bro1Δ::TEF1p-bro1ΔBOD; CTY2) and bro1ΔBOD vps4Δ (CTY5) cells stained with NBD-PC to assess lipid MVB sorting and FM4-64 to label vacuoles. Scale bars = 5 µm.
Video 3.
Tomogram of MVB in a WT cell. MVBs in WT yeast are roughly spherical membrane structures that contain smaller membrane-bound vesicles. This video shows two WT MVBs that are adjacent to the vacuole (red mesh). The limiting endosomal membrane is shown in yellow, and ILVs are in red. Scale bar = 100 nm. Frame rate, 50 frames/s.
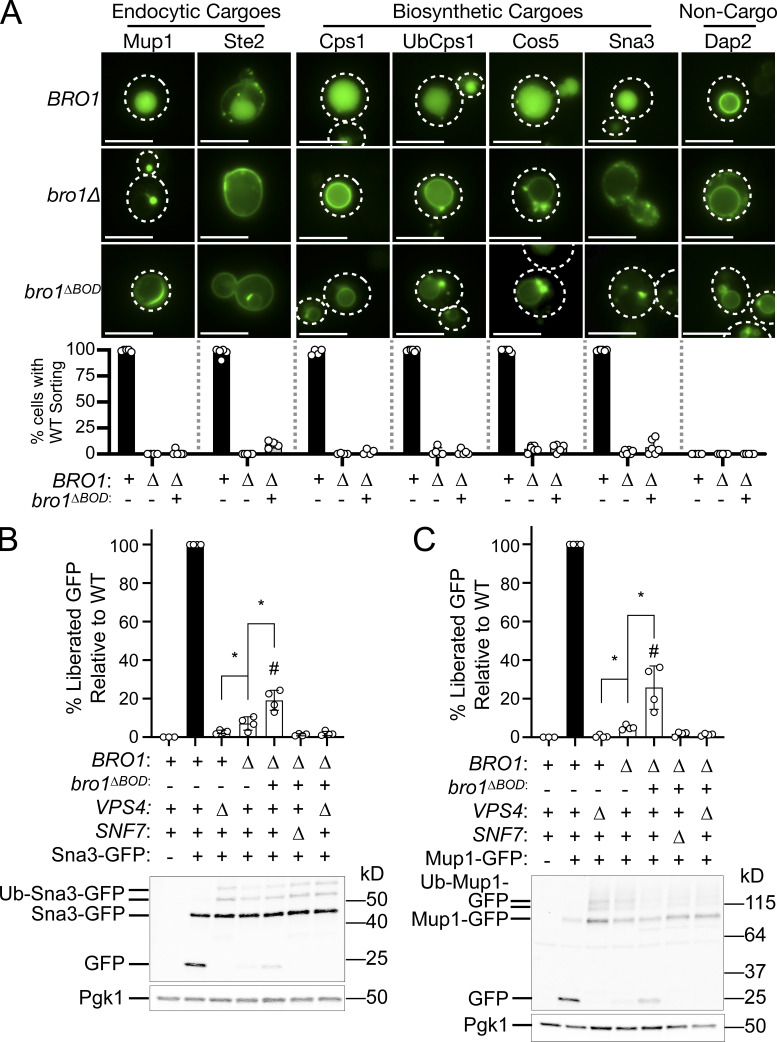
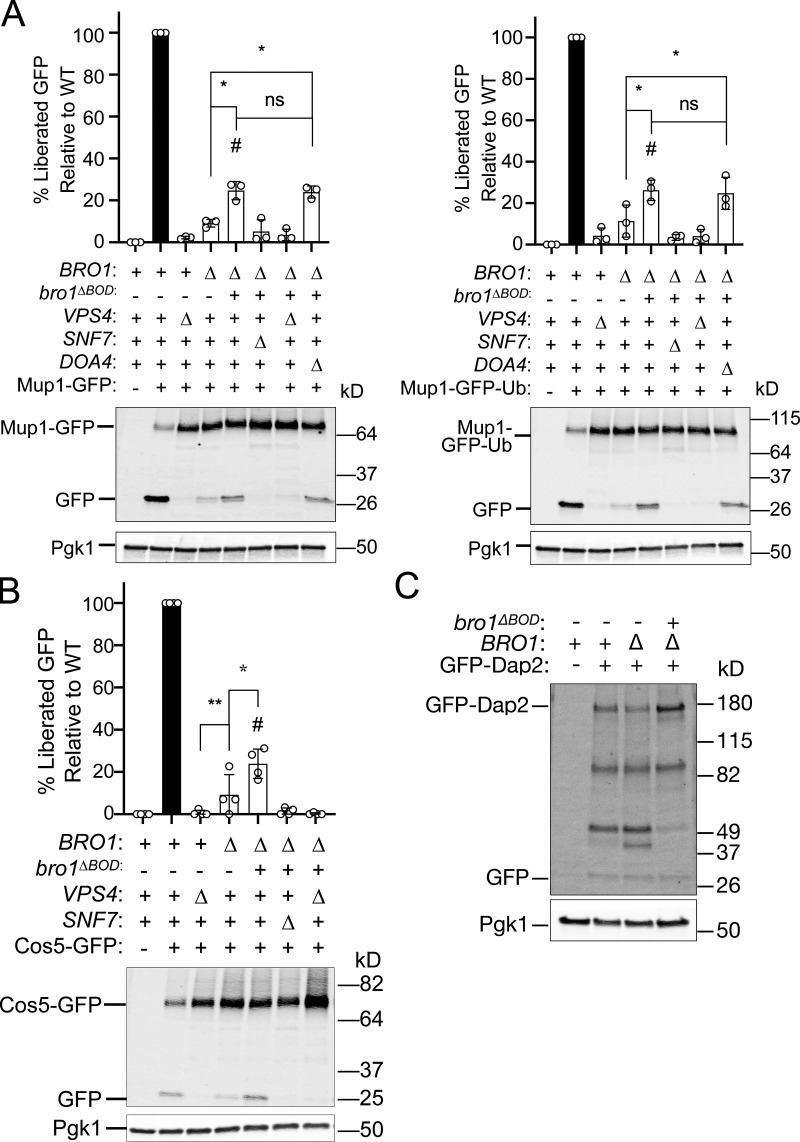
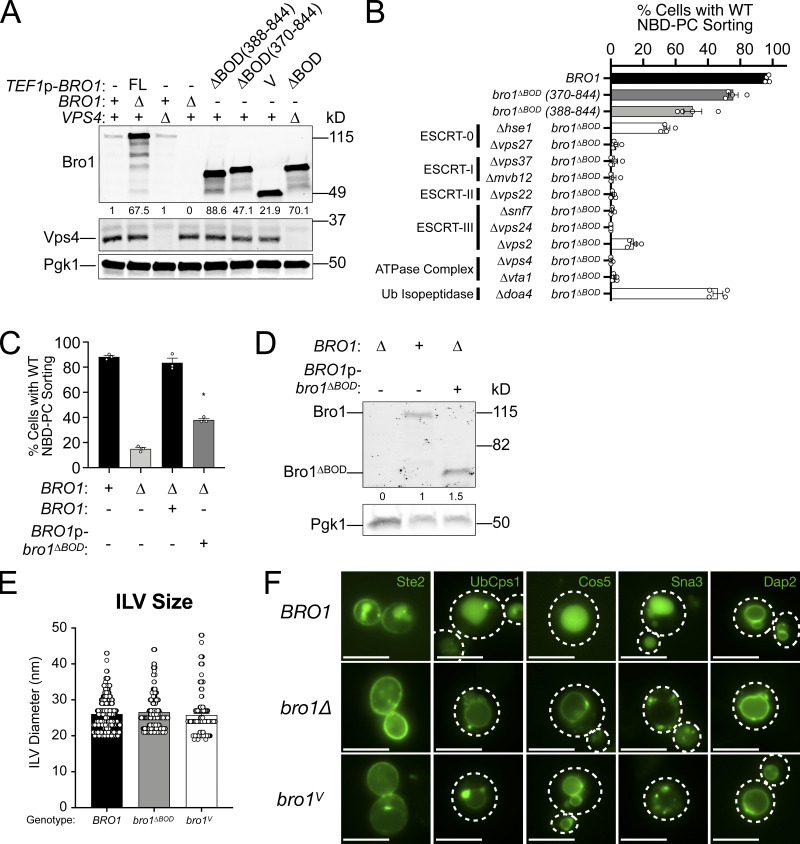
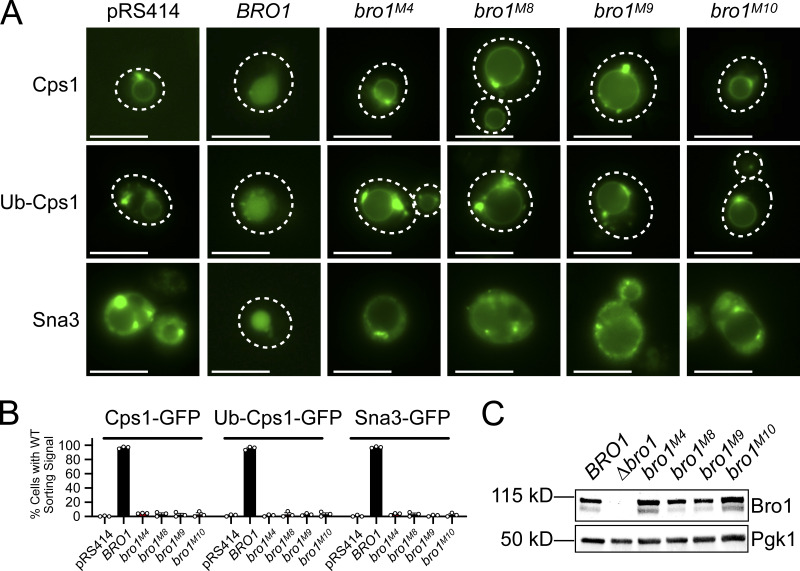
To determine whether Bro1ΔBOD could mediate MVB cargo sorting in addition to ILV formation, we examined the localization of MVB cargos delivered from endocytic (Ste2 and Mup1) and biosynthetic pathways (Cps1, Ub-Cps1, Cos5, and Sna3) as well as a vacuolar limiting membrane protein (DPAP-B/Dap2) typically excluded from MVB sorting. Though bro1ΔBOD cells made MVBs, MVB cargo proteins failed to efficiently sort into MVBs and instead were localized to the limiting membrane of the vacuole and perivacuolar endosomal compartments (Fig. 2 A). To complement this microscopy analysis, immunoblot analyses of four GFP-tagged MVB cargos (Sna3, Cos5, Mup1, and Mup1-Ub) were performed (Fig. 2, B and C; and Fig. S2, A and B). Delivery to the vacuolar lumen results in liberation of an ∼24-kD fragment of GFP, permitting assessment of MVB sorting through quantifying GFP liberation. While both bro1Δ and bro1ΔBOD cells exhibited reduced GFP liberation compared with WT for all MVB cargos examined, bro1ΔBOD cells permitted more MVB sorting than bro1Δ cells. GFP liberation was dependent on Vps4 and Snf7, validating the analysis. These sorting defects in bro1ΔBOD cells did not appear to be related to perturbed cargo ubiquitylation, since (1) immunoblotting showed substantial Ub modification (Fig. 2, B and C), (2) fusion of Ub to MVB cargo did not impact sorting (Ub-Cps1 in Fig. 2 A; Mup1-GFP-Ub in Fig. S2 A), and (3) loss of Ub isopeptidase Doa4 did not compromise GFP liberation from Mup1-GFP or Mup1-GFP-Ub in bro1ΔBOD cells (Fig. S2 A). The increase in MVB sorting in bro1ΔBOD cells compared with bro1Δ cells does not appear to be related to nonspecific entry of endosomal cargos in ILVs, as GFP liberation from Dap2 is not increased in bro1ΔBOD cells (Fig. S2 C) and GFP-Dap2 sorting is not apparent in the microscopy analysis (Fig. 2 A). The small increase in MVB cargo GFP liberation in bro1ΔBOD cells compared with bro1Δ cells is consistent with the increased presence of MVBs in bro1ΔBOD cells; however, the more striking result is the reduced MVB sorting in bro1ΔBOD and bro1Δ cells compared with WT evident in both immunoblot and microscopy analyses. Together, these results indicate that Bro1ΔBOD cannot support efficient MVB cargo sorting despite its ability to facilitate MVB biogenesis.
Figure 2.
Bro1ΔBOD does not support efficient MVB cargo sorting. (A) WT (SEY6210), bro1Δ (GOY65), or bro1ΔBOD (bro1Δ::TEF1p-bro1ΔBOD; CTY2) cells were transformed with the indicated GFP-tagged cargo plasmid to assess MVB sorting using live-cell fluorescence microscopy. Percentage of cells with WT sorting signal was quantified and calculated from at least 183 cells from four independent experiments performed on four different days from three different transformations. White dashed lines indicate cell boundaries. Scale bars = 5 µm. (B and C) Representative immunoblots showing processing of Sna3-GFP (B) or Mup1-GFP (C), including liberated GFP and ubiquitylated species in WT (SEY6210), vps4Δ (MBY3), bro1Δ (GOY65), bro1ΔBOD (bro1Δ::TEF1p- bro1ΔBOD ; CTY2), bro1ΔBOD snf7Δ (CTY12), or bro1ΔBOD vps4Δ (CTY5) cells. Pgk1 served as loading control. Data were quantified from four independent experiments performed on four separate days from two transformations. Data are represented as mean ± SD. Asterisks indicate a statistically significant difference (P < 0.05; B: BRO1∆ vs. VPS4∆ P = 0.04, BRO1∆ vs. bro1∆BOD P = 0.008; C: BRO1∆ vs. VPS4∆ P = 0.0006, BRO1∆ vs. bro1∆BOD P = 0.0255), and number signs indicate a statistically significant difference compared with WT (P < 0.0001).
Figure S2.
Bro1ΔBOD does not support efficient MVB cargo sorting of Mup1, Mup1-Ub, and Cos5. This figure complements Fig. 2. (A) Representative immunoblots showing processing of Mup1-GFP or Mup1-GFP-Ub, including liberated GFP in WT (SEY6210), vps4Δ (MBY3), bro1Δ (GOY65), bro1ΔBOD (bro1Δ::TEF1p- bro1ΔBOD ; CTY2), bro1ΔBOD snf7Δ (CTY12), bro1ΔBOD vps4Δ (CTY5), or bro1ΔBOD doa4Δ (CTY13) cells. Pgk1 serves as loading control. (B) Representative immunoblots processing of Cos5-GFP, including liberated GFP in WT (SEY6210), vps4Δ (MBY3), bro1Δ (GOY65), bro1ΔBOD (bro1Δ::TEF1p- bro1ΔBOD ; CTY2), bro1ΔBOD snf7Δ (CTY12), or bro1ΔBOD vps4Δ (CTY5) cells. Pgk1 serves as loading control. Data were quantified from three independent experiments performed on three separate days from two transformations. Data are represented as mean ± SD, Asterisks indicate a statistically significant difference (P < 0.05), and number signs indicate statistically significant difference compared with WT (P < 0.0001). (C) Lysates generated from bro1Δ (GOY65), WT (SEY6210), and TEF1p-bro1ΔBOD (CTY2) cells expressing GFP-Dap2 were analyzed by immunoblotting using antibodies against GFP and Pgk1 (loading control).
To investigate whether Bro1ΔBOD-mediated ILV formation was dependent upon other ESCRTs, we analyzed MVB sorting of NBD-phosphatidylcholine (NBD-PC), a fluorescent lipid that partitions into ILVs in an ESCRT-dependent manner (Bilodeau et al., 2002; Hanson et al., 2002; Shields et al., 2009; Fig. 3, A–C; and Fig. S1). 98% of WT cells displayed lumenal NBD-PC sorting, and overexpression of full-length Bro1 from the TEF1 promoter had a slight dominant negative effect (85%). Deletion of VPS4 or BRO1 eliminated or dramatically reduced the percentage of cells with lumenal NBD-PC signal, consistent with ESCRT dependence (Figs. 3 C and S3 A). Expression of Bro1ΔBOD enhanced NBD-PC MVB sorting (77% of bro1ΔBOD cells displayed lumenal NBD-PC signal), and this sorting was dependent upon ESCRT-0, ESCRT-I, ESCRT-II, ESCRT-III, and Vps4 (Figs. 3 C and S3 B). These data reveal that Bro1ΔBOD uses the canonical ESCRT-dependent pathway for ILV formation. Bro1ΔBOD expressed from the BRO1 promoter also enhanced NBD-PC MVB sorting compared with bro1Δ cells (Fig. S3, C and D), albeit to a lesser extent than Bro1ΔBOD expressed from the TEF1 promoter. These results indicate that overexpression is not required for Bro1ΔBOD-mediated ILV formation as well as a dose–response relationship for this activity.
Figure 3.
Bro1ΔBOD sorting of NBD-PC into the vacuolar lumen is dependent on the Vps4–ESCRT machinery. (A) Domain cartoon of Bro1 (aa 1–844) with interacting factors annotated. (B) Sample micrographs of WT (SEY6210), bro1Δ (GOY65), and bro1ΔBOD (bro1Δ::TEF1p- bro1ΔBOD; CTY2) cells costained with NBD-PC and FM4-64, revealing endpoints of the observed phenotypes. Scale bars = 5 µm. (C) NBD-PC– and FM4-64–stained WT (SEY6210), TEF1p-BRO1 (CTY1), vps4Δ (MBY3), bro1Δ (GOY65), TEF1p- bro1ΔBOD (CTY2), TEF1p-bro1V (CTY4), bro1ΔBOD vps4Δ (CTY5), bro1ΔBOD vps27Δ (CTY29), bro1ΔBOD vps37Δ (CTY21), bro1ΔBOD vps22Δ (CTY24), bro1ΔBOD snf7Δ (CTY12), bro1ΔBOD vps4Δ(CTY5), and bro1ΔBOD doa4Δ(CTY13) cells were analyzed by live-cell fluorescence microscopy and quantified for the frequency of cells able to support NBD-PC trafficking to the vacuolar lumen. Data are represented as mean ± SEM. Asterisks indicate statistically significant differences (P < 0.006), and number signs indicate statistically significant difference compared with bro1Δ (P < 0.0001). Percentage of cells with WT sorting signal was quantified and calculated from at least 94 cells from four independent experiments performed on four different days. (D and E) bro1V (CTY4) were analyzed by ET. 3D reconstructions of the tomogram are shown in D, and ILVs per MVB are quantified in E. 20 MVBs from WT (SEY6210), 32 MVBs from bro1Δ (GOY65), 64 MVBs from bro1ΔBOD (CTY2) and 72 MVBs from bro1V (CTY4) were quantified. Data are represented as mean ± SEM. The limiting membrane of normal-like MVBs are labeled yellow, while the limiting membrane of tubular/aberrant MVBs are shown in different colors. ILVs are highlighted in red. A minimum of 13 MVBs from at least 10 cells were quantified. Scale bar = 100 nm. Asterisks indicate statistically significant differences comapred to bro1 (P < 0.0001). (F) WT (SEY6210), bro1Δ (GOY65), or bro1V (bro1Δ::TEF1p-bro1V; CTY4) cells were transformed with the GFP-CPS plasmid to assess MVB sorting using live-cell fluorescence microscopy. The percentage of cells with WT sorting signal was quantified and calculated from at least 156 cells from three independent experiments performed on three different days from two different transformations. White dashed lines indicate cell boundaries. Scale bars = 5 µm.
Figure S3.
bro1ΔBOD-supported ILV formation requires the Vps4–ESCRT machinery. This figure complements Fig. 3. (A) Lysates generated from WT (SEY6210), vps4Δ (MBY3), bro1Δ (GOY65), TEF1p-BRO1 (CTY1), bro1ΔBOD (370–844; bro1Δ::TEF1p-bro1ΔBOD; CTY2), bro1ΔBODΔUBD (388–844; bro1Δ::TEF1p-bro1ΔBODΔUBD; CTY3), TEF1p-bro1V (CTY4), and bro1ΔBOD vps4Δ (CTY5) cells were analyzed by immunoblotting using antibodies against Bro1, Vps4, and Pgk1. Numbers below the Bro1 blot indicate expression levels normalized to BRO1 expression. (B) WT (SEY6210), bro1ΔBOD (370–844; CTY2), bro1ΔBODΔUBD (388–844; CTY3), bro1ΔBOD hse1Δ (CTY30), bro1ΔBOD vps27Δ (CTY29), bro1ΔBOD vps37Δ (CTY21), bro1ΔBOD mvb12Δ (CTY22), bro1ΔBOD vps22Δ (CTY24), bro1ΔBOD snf7Δ (CTY12), bro1ΔBOD vps24Δ (CTY18), bro1ΔBOD vps2Δ (CTY26), bro1ΔBOD vta1Δ (CTY27), bro1ΔBOD vps4Δ (CTY5), and bro1ΔBOD doa4Δ (CTY13) cells were analyzed by live-cell fluorescence microscopy and quantified for the frequency of cells able to support NBD-PC trafficking to the vacuolar lumen. Data are represented as mean ± SEM. (C) WT (SEY6210), GOY65, and GOY65 cells transformed with BRO1 and BRO1p-bro1ΔBOD were analyzed by live-cell fluorescence microscopy and quantified for the frequency of cells able to support NBD-PC trafficking to the vacuolar lumen. Error bars indicate SEM. Asterisk indicates statistically significant differences compared with WT (P = 0.0002). (D) Lysates generated from bro1Δ (GOY65) transformed with empty vector, BRO1, and BRO1p-bro1ΔBOD were analyzed by immunoblotting using antibodies against Bro1 and Pgk1. Numbers below the Bro1 blot indicate expression levels normalized to BRO1 expression. (E) WT (SEY6210), bro1ΔBOD (CTY2), and bro1V (CTY4) were analyzed by ET and quantified to assess ILV diameter. 337 ILVs from WT (SEY6210), 831 ILVs from bro1ΔBOD (CTY2), and 225 ILVs from bro1V (CTY4) were quantified. Data are represented as mean ± SEM. Data are represented as mean ± SEM. (F) WT (SEY6210), bro1Δ (GOY65), or bro1V (bro1Δ::TEF1p-bro1V; CTY4) cells were transformed with the indicated GFP-tagged cargo plasmid to assess MVB sorting using live-cell fluorescence microscopy. White dashed lines indicate cell boundaries. Scale bars = 5 µm.
Bro1ΔBOD contains both the V domain as well as a C-terminal PRR that mediates interaction with the Ub isopeptidase Doa4 (Luhtala and Odorizzi, 2004). We found that Bro1ΔBOD-mediated NBD-PC sorting was not dependent on Doa4 and that the Bro1 V domain alone (bro1V) supports NBD-PC sorting into the vacuole lumen (Fig. 3 C), demonstrating that PRR and its associations are not required for this activity. This finding was supported by EM tomography analysis wherein bro1V was able to support ILV formation (Fig. 3 D and Video 4), albeit at levels less than WT or bro1ΔBOD cells (Fig. 3 E). However, the sizes of the ILVs in bro1V cells were indistinguishable from those in WT or bro1ΔBOD cells (Fig. S3 E). While bro1V cells exhibited ILV formation, Bro1 V domain was unable to support MVB cargo sorting (Figs. 3 F and S3 F), as observed with bro1ΔBOD cells. In total, these results implicate the V domain in promoting Bro1–Vps4–ESCRT-III–driven ILV formation.
Video 4.
Tomogram of MVB in a bro1V cell. This tomogram depicts 14 MVBs in a bro1V cell. The majority of these MVBs exhibit normal-like MVB morphology (yellow), with relatively large BPs (green). One tubular MVB is visible (cyan). ILVs are in red. Scale bar = 100 nm. Frame rate, 50 frames/s.
Bro1 V domain stimulates Vps4 in vitro
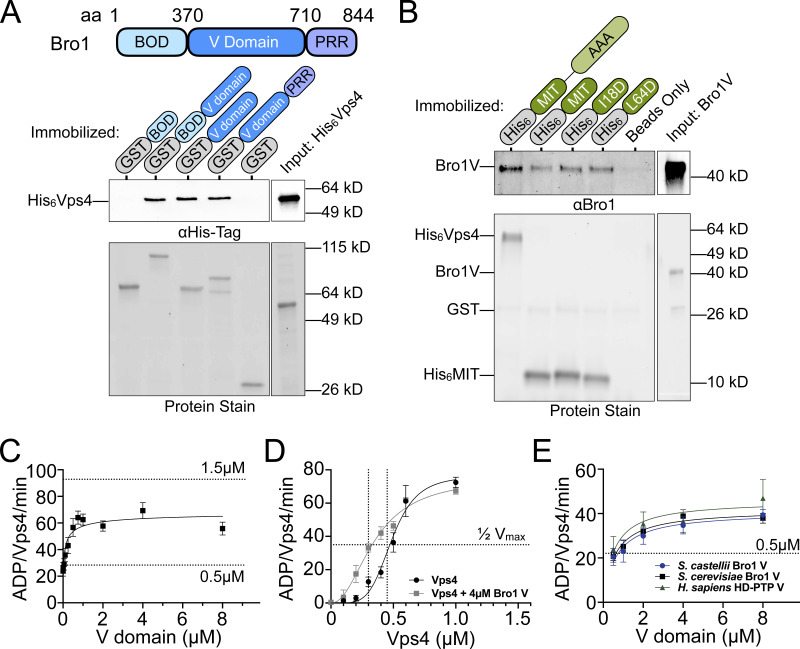
The ability of the Bro1 V domain to support Vps4–ESCRT-III–driven ILV formation along with the reported interaction between Bro1 and Vps4 (Vajjhala et al., 2007) suggested that the V domain might activate Vps4. Binding studies using a series of recombinant GST-tagged Bro1 protein fragments and His6-tagged Vps4 protein fragments showed that the Bro1 V domain binds the N-terminal Vps4 MIT domain (Fig. 4, A and B). Two distinct surfaces of the MIT domain mediate interactions with MIM1 and MIM2 elements in ESCRT-III subunits (Kieffer et al., 2008; Obita et al., 2007; Stuchell-Brereton et al., 2007); however, mutations (I18D, MIM2; L64D, MIM1) disrupting these modes of association did not impair MIT–V domain association. These results suggest that the Bro1 V domain binds the Vps4 MIT domain in a manner distinct from MIM1 and MIM2.
Figure 4.
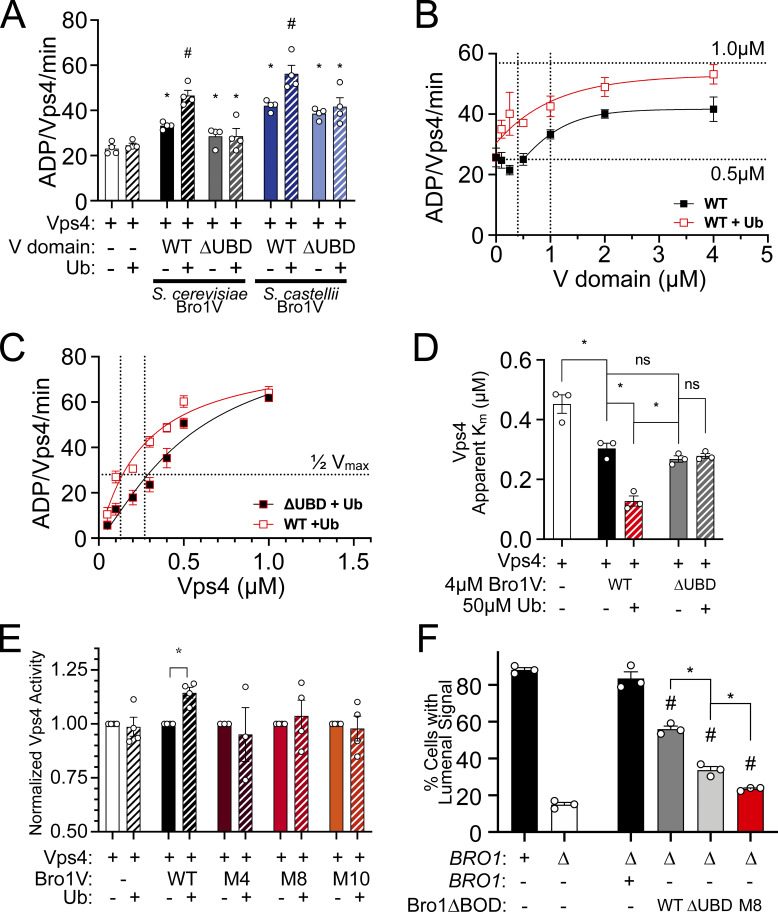
V domain stimulates Vps4 ATPase activity in vitro. (A) GST-Bro1 fragments and GST-bound beads were incubated with His6-Vps4. Bound material was visualized by both EZBiolab Instant-Band protein stain and immunoblotting with penta-His antibody. (B) His6-Vps4, His6-MIT (WT, I18D or L64D), and Ni-NTA beads were incubated with Bro1V. Bound material was visualized by both EZBiolab Instant-Band protein stain and immunoblotting with anti-Bro1 antiserum. (C) Vps4-specific activity with titration of Bro1V (10 nM to 8 µM). Vps4 (0.5 µM) ATPase assays were conducted using the indicated conditions and resolved by thin-layer chromatography for quantitation and calculation of hydrolysis rates. Dashed lines indicate Vps4-specific activity for 0.5 µM or 1.5 µM Vps4 alone, as indicated. Bro1V alone did not exhibit measurable ATP hydrolysis. (D) Vps4 titrations were performed with or without 4 µM Bro1V. Vps4-specific activity is presented. The vertical dotted line indicates the Vps4 apparent Km ±Bro1V. (E) V domains of S. cerevisiae Bro1, S. castellii Bro1, and H. sapiens HD-PTP (0.5–4 µM) were titrated against 0.5 µM S. cerevisiae Vps4. Specific activity of Vps4 is expressed as ADP generated per Vps4 molecule per minute. Data are represented as mean ± SEM.
The possibility that the Bro1 V domain modulates Vps4 activity was assessed using ATPase assays with purified recombinant proteins (Azmi et al., 2006; Babst et al., 1997). Vps4 exhibits concentration-dependent increases in specific activity due to Vps4 oligomer formation, with maximal activity observed at 1.5 µM Vps4 (Azmi et al., 2006; Babst et al., 1998; Davies et al., 2010). Titration of the Bro1 V domain (Bro1V) into ATPase assays with a submaximal concentration of Vps4 (0.5 µM) showed a concentration-dependent stimulation of activity, with a maximum of 2.6-fold over basal activity (Fig. 4 C).
Stimulation of Vps4 did not surpass the inherent maximal Vps4-specific activity, suggesting Bro1V stimulates Vps4 oligomerization without further enhancing activity of the Vps4 oligomer. To examine this directly, Vps4 titration was performed in the presence of 4 µM Bro1V. Bro1V addition reduced the Vps4 apparent Michaelis constant (Km) but did not enhance maximal specific activity (Fig. 4 D), supporting enhanced oligomerization as the mechanism by which Bro1V stimulates Vps4. Furthermore, V domains from Saccharomyces castellii Bro1 (residues 370–709) and human HD-PTP (residues 364–695) stimulated Saccharomyces cerevisiae Vps4 ATPase activity similarly in a dose-dependent manner (Fig. 4 E). Together, these results reveal that the Bro1 V domain enhances Vps4 ATPase activity directly and suggest this stimulatory activity is conserved among homologues.
Generation of Bro1 V domain mutants defective for Vps4 stimulation
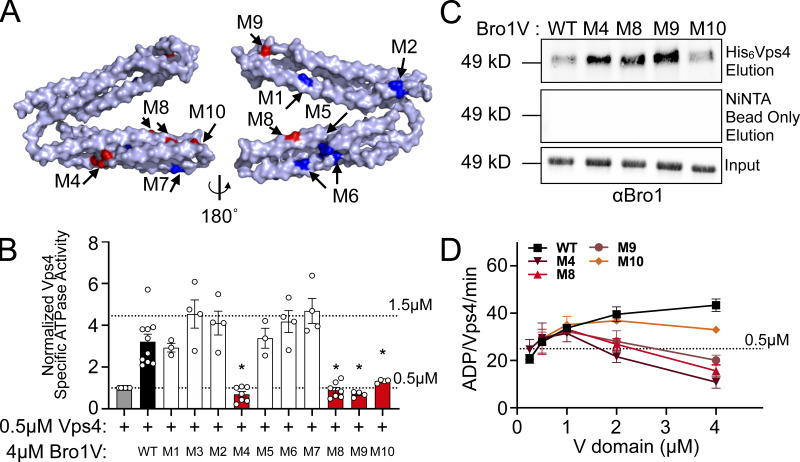
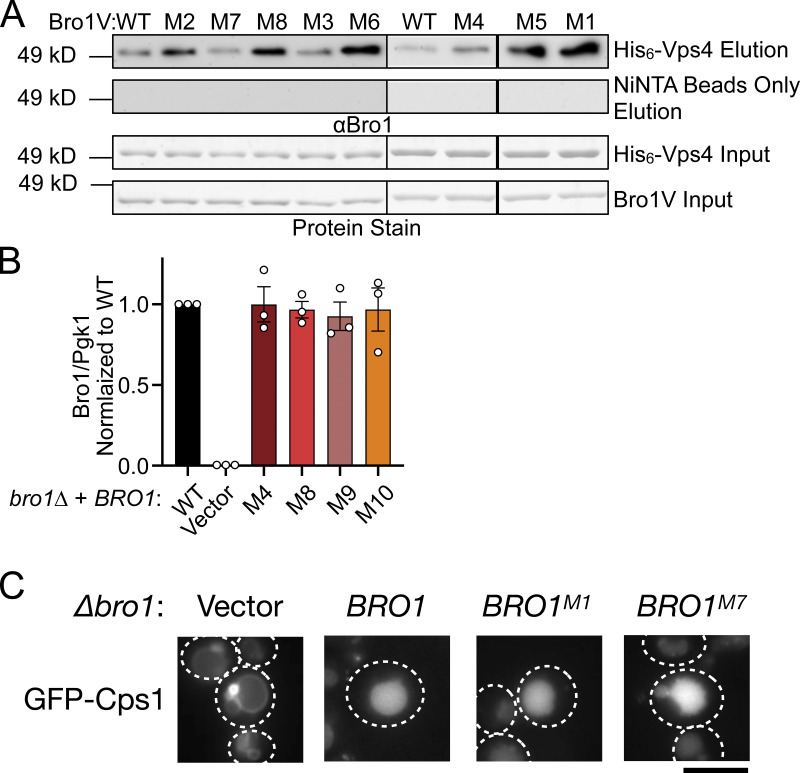
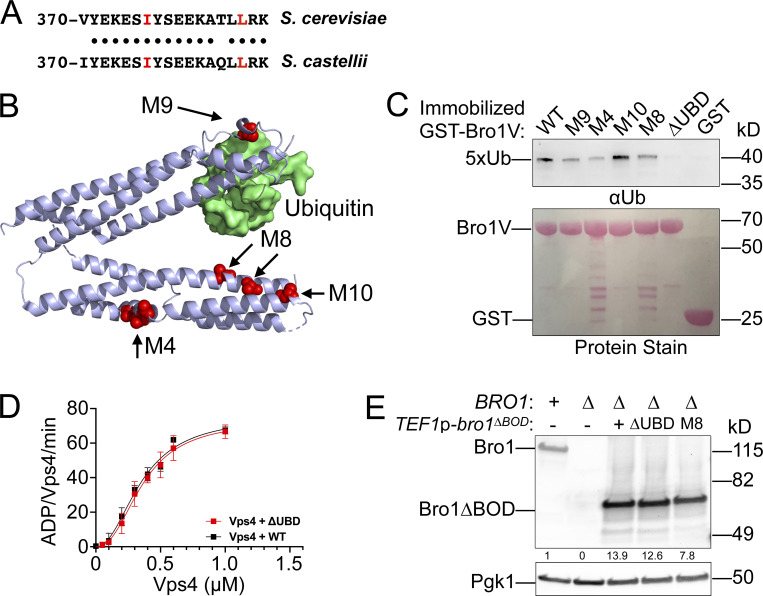
Mutagenesis of the V domain was undertaken with the goals of dissecting the mode of stimulation (i.e., whether binding was sufficient or additional biochemical processes were at play) as well as determining its biological significance. Residues conserved between yeast Bro1 and HD-PTP were targeted, with the goal of generating Bro1 mutants specifically altered in in their ability to stimulate Vps4 activity while retaining their ability to bind Vps4 (Figs. 5 and S4 A). Vps4 stimulation was measured using ATPase reactions containing WT or mutant forms of Bro1V. Six of the Bro1V mutants tested stimulated Vps4 activity like WT Bro1V, whereas four mutants—Bro1VM4: V505A, H508A, I512A; Bro1VM8: E686A, L691D; Bro1VM9: T587D; and Bro1VM10: K481D—did not stimulate Vps4 ATPase activity (Fig. 5 B) but retained their ability to bind Vps4 (Figs. 5 C and S4 A). The ability of these mutants to associate with Vps4 (in some cases better than WT) led us to perform titration experiments to assess the impact on Vps4 ATPase activity. Bro1V mutants were unable to stimulate Vps4 at higher concentrations (>1 µM), yet these mutants could stimulate Vps4 ATPase activity at lower concentrations (≤1 µM; Fig. 5 D). This profile contrasted with the behavior of WT Bro1V that continued to stimulate Vps4 ATPase activity even at the highest concentrations tested. Together, these analyses suggest that Bro1V stimulation of Vps4 relies on determinants beyond mere binding and provided tools to determine whether Bro1V-stimulated Vps4 activity was important in vivo for MVB sorting and biogenesis.
Figure 5.
Bro1 V domain mutations disrupt Vps4 stimulation in vitro without disrupting binding. (A) Model of the V domain based on the S. castellii Bro1 V domain crystal structure (Protein Data Bank accession no. 4JIO, chain A) with amino acid substitution mutations impacting V domain stimulation of Vps4 indicated in red. Conserved residues that when mutated did not impact Vps4 stimulation in vitro are indicated in blue. See Table S4 for individual mutations. (B) Stimulation of Vps4 ATPase activity (0.5 µM) by 4 µM Bro1V(370–709) and Bro1V mutants represented as normalized Vps4 ATPase activity of at least three experiments done in duplicate. Error bars indicate SD, and asterisks indicate a statistically significant difference compared with WT (P < 0.05). (C) Immobilized His6-Vps4 or Ni-NTA beads alone were incubated with Bro1V, Bro1VM4, Bro1VM8, Bro1VM9, and Bro1VM10. Bound material was visualized by immunoblotting with anti-Bro1 antiserum. (D) Vps4 (0.5 µM) ATPase activities with titration of Bro1V, Bro1VM4, Bro1VM8, Bro1VM9, and Bro1VM10 (0.25–5 µM). Vps4-specific activity is expressed as ADP generated per Vps4 molecule per minute. Data are represented as mean ± SEM.
Figure S4.
Bro1V mutants bind Vps4. This figure complements Figs. 5 and 6. (A) Immobilized His6-Vps4 or Ni-NTA beads alone were incubated with Bro1V WT or Bro1V mutants, and bound material was analyzed by immunoblotting with anti-Bro1 antiserum. M, mutant. (B) Mutant protein expression levels normalized to WT. Bro1 expression levels were normalized to Pgk1 and subsequently normalized to WT/Pgk1 ratios. Quantified from three independent experiments performed on three different days from two sets of transformations. Data are represented as mean ± SEM. Immunoblots were probed against Bro1 and PGK1 using lysates of GOY65 transformed with empty plasmid (pRS414) or BRO1, bro1M4, bro1M8, bro1M9, or bro1M10 plasmids. Statistical analyses did not reveal differences between WT and mutant forms of Bro1. (C) bro1Δ (GOY65) cells were transformed with GFP-Cps1 and vector, BRO1, BRO1M1, or BRO1M7 plasmids to assess the impact of these mutations on GFP-Cps1 sorting. White dashed lines indicate cell boundaries. Scale bar = 5 µm.
Altered regulation of Vps4 activity by Bro1V perturbs ILV formation in vivo
MVB sorting was examined in bro1Δ cells expressing full-length alleles of BRO1 containing mutations in the V domain that altered Vps4 stimulation (Fig. 6 A). Three distinct model MVB cargos were used for this analysis: GFP-Cps1 for its dependence upon Ub modification for proper sorting, the chimera Ub-GFP-Cps1 for its lack of dependence upon exogenous Ub modification, and Sna3-GFP as a cargo that has been shown to access the MVB pathway both directly and via association with other ubiquitylated cargos (Katzmann et al., 2001; Katzmann et al., 2004; MacDonald et al., 2012b; Odorizzi et al., 1998; Oestreich et al., 2007; Reggiori and Pelham, 2001). Although all Bro1 mutants were expressed at levels similar to WT Bro1, none of the mutant strains (bro1M4, bro1M8, bro1M9, or bro1M10) mediated MVB sorting of any cargo (Figs. 6 and S4 B). In contrast, BRO1M1 and BRO1M7 displayed WT sorting of GFP-Cps1 (Fig. S4 C), correlating with Vps4 stimulation equivalent to WT (Fig. 5 B). The correlation between MVB cargo sorting in vivo and Vps4 stimulation observed with these mutants in vitro support the conclusion that Bro1 V domain stimulation of Vps4 is required for ESCRT-driven MVB cargo sorting in vivo.
Figure 6.
Bro1 V domain mutations disrupting Vps4 stimulation in vitro disrupt MVB sorting in vivo. (A)bro1Δ (GOY65) cells were transformed with empty plasmid (pRS414) or plasmids with the BRO1 promoter and BRO1, bro1M4, bro1M8, bro1M9, or bro1M10. Localizations of model MVB cargo GFP-Cps1, Ub-GFP-Cps1, or Sna3-GFP were determined using live-cell fluorescence microscopy to assess MVB sorting in these mutant contexts. White dashed lines indicate cell boundaries. Scale bars = 5 µm. (B) The percentage of cells with WT sorting signal was quantified and calculated from at least 100 cells from three independent experiments performed on three different days from three different transformations. (C) Representative immunoblots showing mutant protein expression levels, probing against Bro1 and Pgk1 as a loading control, using lysates of GOY65 transformed with empty plasmid (pRS414) or plasmids with the BRO1 promoter and BRO1, bro1M4, bro1M8, bro1M9, or bro1M10.
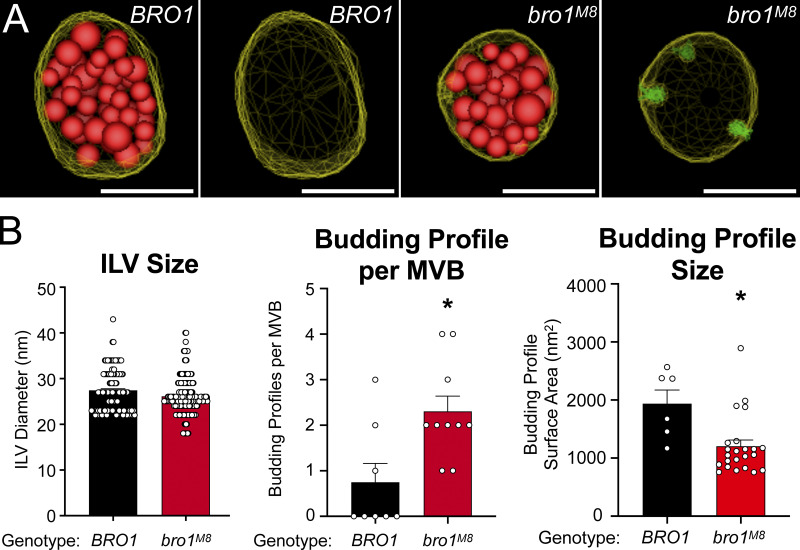
The impact of V domain stimulation on ILV formation was addressed in cells expressing bro1M8 (Fig. 7 A). ILVs in mutant bro1M8 cells were the same size as those in WT cells. However, bro1M8 cells had roughly twice as many ILV budding intermediates per MVB compared with WT cells, as well as reduction in the surface area of the BPs (Fig. 7 B and Video 5). These observations suggest that proper stimulation of Vps4 by Bro1 V domain contributes to ILV formation at the level of bud expansion.
Figure 7.
bro1M8 perturbs ILV formation in vivo. (A) Three-dimensional models reconstructed from 200-nm thick-section electron tomograms of bro1Δ (GOY65) with BRO1 or bro1M8 plasmids. The limiting membrane of the MVB is labeled yellow, the ILVs are highlighted in red, and the budding intermediates are colored in green. Scale bars = 100 nm. (B) Quantification of electron tomograms of GOY65 with either BRO1 or bro1M8 plotting ILV size, BP size, and BPs per MVB; 8 MVBs from bro1Δ (GOY65) with BRO1 plasmid containing 6 BPs and 141 ILVs and 10 MVBs from bro1Δ with bro1M8 plasmids containing 23 BPs and 204 ILVs were quantified. Data are represented as mean ± SEM. Asterisks indicate statistically significant differences compared with WT (P < 0.006).
Video 5.
Tomogram of MVB in a bro1Δ cell expressing Bro1M8. This tomogram depicts one MVBs in a bro1Δ cell expressing Bro1M8. This MVB contains several budding intermediates (green), while its limiting membrane is shown in yellow and ILVs are labeled red. Scale bar = 100 nm. Frame rate, 50 frames/s.
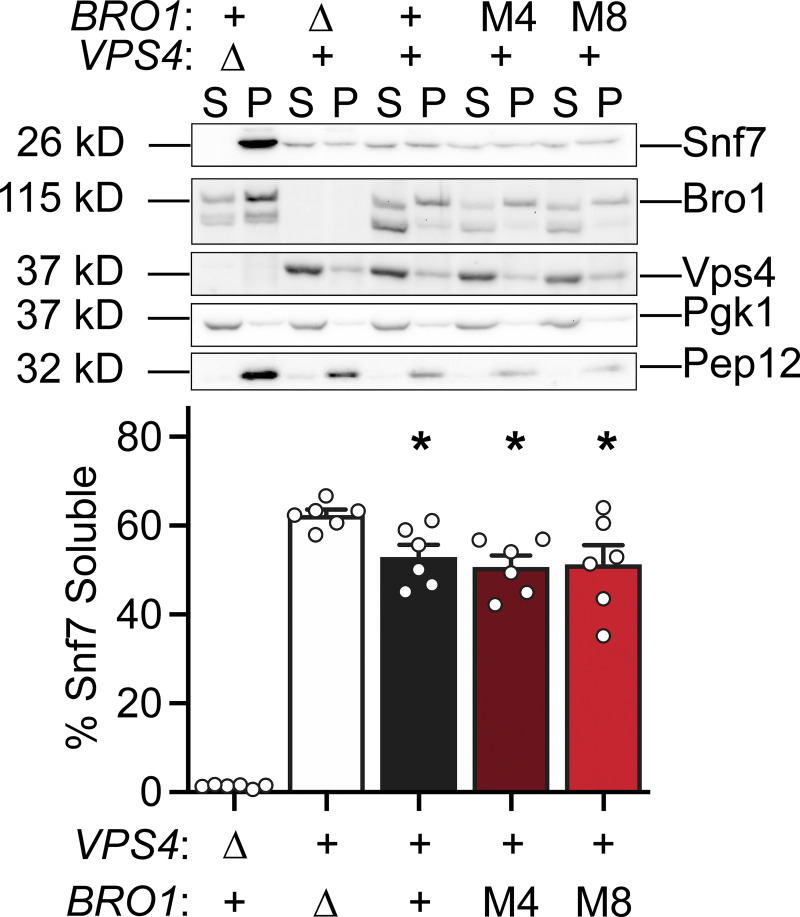
Previous studies have documented the accumulation of ESCRT-III on endosomal membranes when Vps4 activation is defective (Babst et al., 2002; Wemmer et al., 2011). Whether Bro1 stimulation of Vps4 affected the steady-state membrane association of ESCRT-III was addressed using subcellular fractionation of bro1M4 and bro1M8 cells. This analysis revealed indistinguishable levels of the ESCRT-III subunit Snf7 on membranes in WT, bro1M4, and bro1M8 cells (Fig. 8). In contrast, bro1Δ cells showed more soluble Snf7, while vps4Δ cells displayed near-complete Snf7 membrane association. These findings suggest that ESCRT-III polymerization and disassembly/recycling were normal in bro1M4 and bro1M8 cells, implying that Bro1 V domain–mediated Vps4 stimulation exerts its function at a biochemical step distinct from Vps4-mediated ESCRT-III disassembly.
Figure 8.
bro1M4 and bro1M8 display WT steady-state membrane-associated ESCRT-III in vivo. Subcellular fractionation was performed in bro1Δ (GOY65) cells transformed with an empty vector or BRO1, bro1M4, or bro1M8 plasmids. vps4Δ (MBY3) cells were used as a control highlighting the distribution upon complete loss of Vps4 function. Representative immunoblots indicating fractionation of Snf7, Bro1, and Vps4 are shown. Pep12 and Pgk1 were used as membrane and soluble markers, respectively. Quantification represents six experiments; data are represented as mean ± SEM. Asterisks indicate a statistically significant difference compared with bro1Δ (P < 0.05).
Ub binding enhances Bro1 V domain stimulation of Vps4 activity in vitro
Bro1 binds Ub via a conserved motif at the N-terminal portion of the V domain (Pashkova et al., 2013; Fig. S5, A and B). Therefore, the impact of Ub on V domain stimulation of Vps4 was addressed (Fig. 9 A). While Ub does not directly stimulate Vps4 in vitro, addition of Ub increased Bro1V stimulation of Vps4 activity using Bro1V from both S. cerevisiae and the related yeast S. castellii, for which the crystal structure is known (Pashkova et al., 2013). In contrast, addition of Ub to mutant Bro1VΔUBD (I377R) in which Ub binding was ablated (Pashkova et al., 2013; Fig. S5 C) had no effect on Vps4 stimulation. These results indicate a conserved mode of Ub-dependent regulation of the V domain’s impact on Vps4.
Figure S5.
Bro1V mutants bind Ub. This figure complements Fig. 9. (A) Sequence alignment of V domain aa 370–388 from S. castellii and S. cerevisiae. Conserved amino acids are indicated by black circles, and isoleucine 377 and leucine 386, critical for Ub binding, are highlighted in red. (B) Bro1 V domain mutations M4, M8, and M10 (red) that disrupt V domain stimulation of Vps4 ATPase activity are spatially separated from its Ub-binding site using S. castellii Bro1V crystal structure (Protein Data Bank accession no. 4JIO, chain A). (C) Immobilized GST-fused Bro1V, Bro1VM4, Bro1VM8, Bro1VM9, Bro1VM10, Bro1VΔUBD (I377R) and GST alone were incubated with V5 epitope-tagged linear penta-Ub. Bound material was visualized by both Ponceau S protein stain and immunoblotting for the V5 epitope. (D) Vps4 titrations were performed with 4 µM Bro1V WT or Bro1VΔUBD (L386R). Vps4-specific activity is presented as mean ± SEM. (E) Lysates generated from bro1Δ (GOY65) transformed with empty vector, BRO1, TEF1p-bro1ΔBOD, TEF1p-bro1ΔBOD,ΔUBD (ΔUBD:I377R,L386R) and TEF1p-bro1ΔBOD,M8 plasmids were analyzed by immunoblotting using antibodies against Bro1 and Pgk1. Numbers below the Bro1 blot indicate expression levels normalized to BRO1 expression.
Figure 9.
Ub potentiates V domain stimulation of Vps4 ATPase activity. (A) Vps4 (0.5 µM) ATPase specific activity in the presence of 1 µM S. cerevisiae Bro1V, S. cerevisiae Bro1VΔUBD (I377R), S. castellii Bro1V (370–708), and S. castellii Bro1VΔUBD (I377R) ± 50 µM mono-Ub. Data are represented as mean ± SEM. Asterisks indicate a statistically significant difference compared with Vps4 alone (P < 0.05), and number signs indicate a statistically significant difference compared with Vps4 + Bro1V − Ub (P < 0.05). (B) Bro1V titration performed in the presence of 0.5 µM Vps4 with or without 50 µM Ub. The vertical dotted line indicates the Bro1V concentration generating half-maximal stimulation within each context. (C) Vps4 titration (0.05–1.0 µM) in the presence of 4 µM Bro1V WT or Ub-binding mutant (L386R) and 50 µM Ub. Vps4-specific activity (ADP generated per Vps4 molecule per minute) is presented. Data are represented as mean ± SEM. Vertical dotted lines indicate the Vps4 apparent Km in each context. (D) The Vps4 apparent Km of Vps4 alone, Vps4 + 4 µM Bro1V, Vps4 + 4 µM Bro1V + 50 µM Ub, Vps4 + 4 µM Bro1VΔUBD (L386R), and Vps4 + 4 µM Bro1VΔUBD + 50 µM Ub was determined from Vps4 titration experiments in Figs. 4 D, 9 C, and S4 C. Asterisks indicate a statistically significant difference (P < 0.02). (E) Vps4 ATPase activity (0.5 µM) in the presence of 4 µM S. cerevisiae Bro1V, Bro1VM4, Bro1VM8, or Bro1VM10 without or with 50 µM Ub. Vps4-specific activity with Ub addition is normalized to activity without Ub. Data are represented as mean ± SEM. Asterisk indicates a statistically significant difference with or without Ub addition (P = 0.0012). (F) NBD-PC– and FM4-64–stained WT (SEY6210), bro1Δ (GOY65), and GOY65 reexpressing Bro1 or overexpressing Bro1ΔBOD, Bro1ΔBOD,ΔUBD (I377R, L386R), and Bro1ΔBOD,M8 were analyzed by live-cell fluorescence microscopy and quantified for the frequency of cells with NBD-PC in the vacuolar lumen. Data are represented as mean ± SEM. Asterisks indicate a statistically significant difference (P < 0.01), and number signs indicate a statistically significant difference compared with bro1Δ (P < 0.005).
How Ub impacts the stimulation of Vps4 by Bro1V was assessed using titration experiments in the presence or absence of Ub (Fig. 9 B). Ub increased the potency of Bro1V, wherein the Bro1V concentration yielding half-maximal stimulation of Vps4 was reduced from 1.0 µM to 0.4 µM. We also examined the impact of Ub-bound Bro1V on Vps4 oligomer assembly and/or activity of the assembled Vps4 oligomer. Whereas both WT Bro1V and Bro1VΔUBD (L386R) similarly reduced the apparent Km of Vps4, addition of Ub to WT Bro1V further reduced the Vps4 apparent Km (Fig. 9, C and D; and Fig. S5 D); however, a similar reduction was not observed with Bro1VΔUBD. These results suggest that Ub binding alters V domain conformation to enhance its association with Vps4 and promote Vps4 oligomerization without impacting activity of the assembled Vps4 oligomer.
Lastly, we explored whether the Ub enhancement of V domain Vps4 stimulation was dependent on the mutants identified through mutagenesis studies. Bro1V mutants impacting Vps4 stimulation are not confined to a single surface nor do they disrupt association with Vps4, suggesting that these mutations alter the conformation or dynamics of the V domain to impact Vps4 stimulation. While mutants M4, M8, and M10 are not located on the V domain arm that binds Ub (Fig. S5 B), binding studies were performed to examine whether these mutants indirectly impact Ub binding. Whereas Bro1VΔUBD (I377R) failed to bind Ub, Bro1VM4, Bro1VM8, and Bro1VM10 retained Ub-binding activity (Fig. S5 C), indicating these mutants were suitable for examining Ub-modulated Vps4 stimulation. However, Ub did not impact Bro1VM4, Bro1VM8, or Bro1VM10 regulation of Vps4 in contrast to Ub enhancement of WT Bro1V stimulation (Fig. 9 E). Together, these results demonstrate that the mechanism by which the V domain stimulates Vps4 transduces the additional stimulation observed when the V domain binds Ub.
Ub promotes ILV formation via the Bro1–Vps4–ESCRT-III axis
Bro1 has been proposed to work early in the MVB sorting process as a receptor for Ub cargo (Pashkova et al., 2013). Because this function is upstream of the Vps4–ESCRT-III ILV formation axis, it was not clear whether Ub binding to Bro1 V domain might play a role in downstream events as well. Ub contribution to ILV formation was assessed using Bro1ΔBOD and NBD-PC sorting, since Bro1ΔBOD drives ILV formation disconnected from efficient cargo sorting. Cells expressing Bro1ΔBOD defective for Ub binding (ΔUBD: I377R, L386R) showed reduced levels of NBD-PC sorting in comparison with WT, supporting the interpretation that Ub plays a role in ILV formation itself (Fig. 9 F); similar results were observed with truncation of the Ub-binding residues (aa 388–844; Fig. S2 B). In addition, mutant Bro1ΔBOD,M8, which contains a V domain unable to properly stimulate Vps4, showed a further reduction in NBD-PC sorting comparable to sorting observed in bro1Δ cells. These degrees of NBD-PC sorting with Bro1ΔBOD,ΔUBD and Bro1ΔBOD,M8 correlate with Bro1VΔUBD exhibiting basal, but not Ub-enhanced, stimulation of Vps4 in vitro (Fig. 9 A). These results indicate that (1) V domain stimulation of Vps4 contributes to Bro1ΔBOD ILV formation, and (2) Bro1 Ub binding enhances ILV formation via the Bro1–Vps4–ESCRT-III axis.
Discussion
Here, we reveal a new function of Bro1 in MVB biogenesis that is mediated by its V domain binding to and stimulating Vps4. This stimulation is further enhanced by Ub binding, suggesting that Bro1 interaction with Ub-cargo enhances Vps4 activity during ILV formation. Moreover, this mode of Vps4 regulation is important for the coordination of cargo sorting into ILVs. These data suggest Bro1 plays a role in “licensing” ESCRT-III and Vps4 to drive membrane remodeling in concert with cargo transfer into the ILV. The idea that Bro1 couples cargo sorting with ILV formation is underscored by the observations here and elsewhere that disconnecting Bro1 from some of its interactions separates the process of ILV formation with the process of efficiently sorting cargos into those ILVs. We propose that the central Bro1 activity of the V domain stimulating Vps4/ESCRT-III–driven ILV formation is coordinated with BOD and PRR activities to properly time ILV budding to enable normal MVB biogenesis.
Bro1 is one of several proteins that regulate Vps4 activity. ESCRT-III subunits and Vta1 also stimulate Vps4 activity (Azmi et al., 2006; Azmi et al., 2008; Merrill and Hanson, 2010; Shim et al., 2008). We found that the Bro1 V domain binds to the Vps4 MIT domain and increases Vps4 specific activity by facilitating Vps4 oligomerization. The Vps4 MIT domain also binds ESCRT-III subunits along two distinct surfaces that mediate association with MIM1 and MIM2 motifs found in Vps2 or Vps24 and Vps20 or Snf7, respectively (Kieffer et al., 2008; Obita et al., 2007; Stuchell-Brereton et al., 2007). Our data indicate that Bro1 V domain binds by a distinct mode, but it remains to be determined if V domain binding diminishes or enhances the ability of Vps4 to bind to MIM1- or MIM2-containing ESCRT-III subunits. Vps4 recruitment to the site of MVB sorting occurs in two phases: an early, minimal recruitment of Vps4 associated with stable ESCRT-III membrane association followed by a later recruitment of additional Vps4 implicated in ILV formation and eventual ESCRT-III disassembly (Adell et al., 2017). Our in vitro activity analyses indicate that Bro1 facilitates Vps4 oligomerization, yet exactly how V domain regulation of Vps4 activity impacts the dynamics and function of ESCRT-III remains to be determined. Overall, the level of polymerized membrane-associated ESCRT-III remains unperturbed in cells where Bro1 is unable to properly stimulate Vps4 ATPase activity. Moreover, the size of the ILVs themselves, an indicator of the fidelity of ESCRT-III–mediated scission, is unaltered as well. Together, these data suggest this role of Bro1 regulating Vps4 is transient, precise, and distinct from Vta1–ESCRT-III stimulation of Vps4 implicated in ILV scission and ESCRT-III disassembly. Bro1 mutant with altered Vps4 regulation exhibited reduced BP size, suggesting Bro1 stimulation of Vps4 contributes to ILV bud expansion. Bud expansion controlled by Vps4 should be coordinated with entry of cargo as well as deubiquitination of cargo just before its entry into ILVs, and the biochemical activities now described for Bro1 position it well for such a role. While future studies are required to expand our understanding of Bro1’s role in the Vps4–ESCRT-III ILV formation axis, this study unequivocally demonstrates Bro1 stimulation of Vps4 makes unique contributions to MVB biogenesis.
Analyses in vitro and in vivo indicate that V domain mutations (M4, M8, M9, and M10) retain partial Vps4 stimulation activity. These mutants retain interaction with Vps4 and Ub and stimulation of Vps4 ATPase activity at lower concentrations; however, these mutants are defective for MVB cargo sorting in vivo and stimulation of Vps4 ATPase activity at higher concentrations in vitro. We conclude Bro1V stimulation of Vps4 observed in vitro under high concentrations is the activity relevant for Bro1 function in MVB sorting in vivo. These mutations map to both arms of the V domain, suggesting they impart subtle conformational changes in the V domain that maintain association with Vps4 but disrupt stimulation. Further structural studies are needed to elucidate this mechanism.
Our results indicate that Vps4 responds to Ub through Bro1. Previous studies demonstrated a role for Ub in MVB sorting at the level of cargo recognition (Bilodeau et al., 2002; Katzmann et al., 2001; Reggiori and Pelham, 2001) as well as the dependence of ILV formation on the presence of ubiquitylated cargo (MacDonald et al., 2012a; Stringer and Piper, 2011). However, upstream effects on cargo recognition and early ESCRT membrane recruitment precluded examination of the role of Ub in ILV formation itself. The present studies indicate that Ub binding also contributes to ILV formation, as disruption of Ub binding through point mutation (Fig. 9 F) or deletion (Fig. S2 B) reduced Bro1ΔBOD NBD-PC MVB sorting, consistent with Ub-enhanced Bro1V stimulation of Vps4 in vitro. The source of Ub in vivo remains to be determined, but likely candidates would be Ub cargo, Ub-Cos proteins or tetraspanins, or ubiquitylated ESCRT machinery (Hicke et al., 2005; MacDonald et al., 2015; Piper and Katzmann, 2007). Another possibility is free Ub generated by Bro1 recruiting Ub isopeptidase Doa4; however, neither bro1ΔBOD MVB cargo sorting nor NBD-PC sorting was compromised by loss of Doa4, arguing against this possibility (Figs. 3 C and S2 A). Irrespective of the source of Ub, our results implicate a role for Bro1 Ub binding in modulating Vps4 activity during ILV formation in addition to Ub’s previously appreciated role as a cargo sorting determinant.
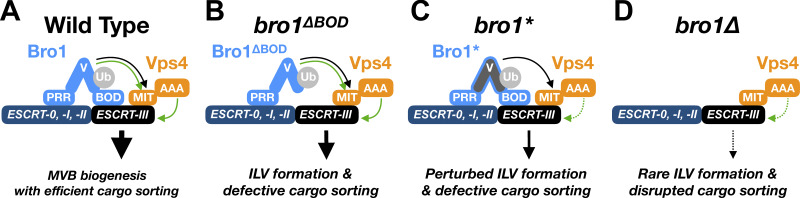
We suggest that Bro1 serves to “license” ILV formation (Fig. 10), typically coordinated with cargo entry, to enable efficient MVB sorting. This licensing concept is supported by our observation that highly expressed Bro1ΔBOD promoted more ILV formation, as assessed by NBD-PC sorting, than Bro1ΔBOD expressed at lower levels. Human HD-PTP V domain also stimulates Vps4 activity, indicating evolutionary conservation of this newly discovered biochemical activity. Bro1 domain family members have important yet unclear roles in myriad ESCRT-mediated processes. ALIX binds viral structural proteins, such as Gag, to facilitate retroviral budding as well as other factors (e.g., Syntenin and CEP55) to facilitate ESCRT-driven exosome biogenesis and cytokinesis (Baietti et al., 2012; Carlton and Martin-Serrano, 2007; Fisher et al., 2007; Larios et al., 2020; Lee et al., 2007b). Increased ALIX activity through overexpression or activating truncation enhances the release of human immunodeficiency virus particles as well as small extracellular vesicles (Fisher et al., 2007; Larios et al., 2020), consistent with ALIX serving as a licensing factor for these ESCRT-mediated events as well. Mutations in HD-PTP are linked to a congenital neurodevelopment disorder characterized by seizures and spasticity (Bend et al., 2020; Sowada et al., 2017). Moreover, HD-PTP homozygous mouse knockouts are embryonic lethal, and HD-PTP heterozygosity is linked to increased tumorigenesis (Gingras et al., 2009; Manteghi et al., 2016). In yeast, loss of Bro1 disrupts MVB sorting and ILV formation, leading to disrupted endosomal morphology (i.e., class E vps phenotype). Thus, understanding the biochemical mechanism that Bro1 uses to control Vps4 has wide-ranging ramifications and is likely to describe a shared function for other Bro1 domain family proteins.
Figure 10.
Model of Bro1 “licensing” ESCRT-III membrane remodeling.(A) Bro1 interacts with ESCRTs via interactions between PRR and early ESCRTs, as well as Bro1 domain (BOD) and ESCRT-III. Depicted are two activities of the Bro1 V domain, binding to the Vps4 MIT domain in a manner distinct from ESCRT-III MIM1 and MIM2 elements (curved black arrow), and stimulation of Vps4 ATPase activity (curved green arrow); Ub binding to the V domain potentiates Vps4 stimulation and promotes ILV formation (curved green arrow). These activities contribute to MVB biogenesis and efficient cargo sorting (straight black arrow). (B) When the BOD is deleted, Bro1ΔBOD (via the V domain) improperly activates Vps4 and facilitates ILV formation without efficient cargo sorting. (C) Bro1 without the ability to stimulate the Vps4 (Bro1*, e.g., mutants M4, M8, M9, M10) disrupts MVB cargo sorting and perturbs ILV formation. (D) The absence of BRO1 (bro1Δ) abrogates V domain stimulation of Vps4 as well as Bro1 interactions with early ESCRTs and ESCRT-III; thus, bro1Δ exhibits severe defects in both cargo sorting and ILV formation.
Methods
Plasmids and strains
For a complete list of plasmids used in this study, see Table S1. All restriction enzymes were obtained from New England Biolabs. All PCR reactions were performed using Platinum SuperFi II PCR Master Mix (Thermo Fisher Scientific). S. cerevisiae Vps4 MIT domain was amplified from SEY6210 genomic DNA and subsequently cloned into NdeI and SalI sites of pET28a to generate pET28-MIT. Similarly, MIT domain was amplified from pVPS4(I18D) and pVSP4(L64D) and subsequently cloned into pET28a to generate pET28b-MIT I18D and pET28b-MIT L64D. The sequence encoding the V domain of Bro1 (Bro1V; aa 370–709) was amplified from pGO216 (Wemmer et al., 2011) and cloned into the BamH1 and SalI sites of pGST-parallel1 (Sheffield et al., 1999) to generate pSD1. BRO1 was amplified from pGO187 (Odorizzi et al., 2003) and cloned into the SpeI and SalI site of pRS414 (Simons et al., 1987) to generate pSD12. Mutagenesis of Bro1V or BRO1 encoding constructs (pSD2-9, pSD13-14, pCT2, pCT4-8, pCT10, pCT13-16, pCT18, pCT20-24, pCT26, and pCT29-33 was performed using the GeneTailor Site-Direct Mutagenesis System (Life Technologies). The veracity of plasmids was confirmed by sequencing. The sequence encoding Bro1ΔPRR (aa 1–709) was amplified from pGO216 and cloned into BamH1 and SalI sites of pET28b (Novagen) and pGST-parallel1 for use generating and purifying the anti-Bro1 sera, respectively (see “Antibody generation”).
Two silent mutations (D133D and D153D) were introduced into the HIS3 cassette of pRS413 to remove undesired BglII sites by site-directed mutagenesis; the resulting plasmid was named pCT34. The sequence encoding the HIS3 cassette was amplified from pCT34 by PCR to introduce BamHI and PacI sites and cloned into the BglII and PacI sites of pFA6a-HisMX6; the resulting plasmid was named pCT35. The sequence encoding Bro1V (aa 370–709) was amplified from SEY6210 genomic DNA and cloned into NcoI and EcoRI sites of pCT35; the resulting plasmid was named pCT36. Subsequently, 500 bp BRO1 3′ UTR was amplified from SEY6210 genomic DNA and cloned into the EcoRI and SacI sites of pCT35; the resulting plasmid was named pCT37. BRO1 promoter was amplified from genomic DNA of SEY6210 and cloned into the SalI and EcoRI sites of pRS414; the resulting plasmid was named pCT38. pCT38 generated by deleting BOD (aa 1–369) in pSD12 using GeneTailor Site-Direct Mutagenesis System. Subsequently, ΔUBD mutations (I377R and L386R) were introduced by site-directed mutagenesis; the resulting plasmid was named pCT39. The fragment containing M8 mutation was cloned into the ClaI and NdeI sites of pCT38 from pSD14; the resulting plasmid was named pCT40. TEF1p-Bro1ΔBOD (aa 370–844) was amplified from CTY2 and cloned into the SalI and BamHI sites in pRS414; the resulting plasmid was named pCT41. Mutations (ΔUBD and M8) were amplified from pCT39 and pCT40 and cloned into NcoI–BamHI–digested pCT41 by NEBuilder HiFi DNA Assembly (New England Biolabs). DPAP-B-GFP was cloned from pGO89 (Odorizzi et al., 1998) into the NotI and SalI sites of pRS425; the resulting plasmid was DPAP-B-GFP. For a list of primers used to generate pCT34-43, please see Table S2.
For a complete list of strains used in this study, see Table S3. CTY1, CTY2, and CTY3 were generated by transforming PCR product from pCT34 into SEY6210.1, while CTY4 was generated by transforming PCR product from pCT36; for a list of primers used, please see Table S2. Transformants were plated on histidine drop-out synthetic plates and screened by immunoblotting. The altered bro1 allele was amplified and sequenced. CTY5, CTY11-13, CTY518, CTY21-22, CTY24, CTY27, CTY29-30, and JPY403 were generated using standard yeast genetics.
Protein expression and purification
Protein expression was performed in the BL21-DE3 bacterial strain using 0.5 mM IPTG at 16°C for 18–22 h. GST-Bro1V fusion protein and mutants thereof were purified using Glutathione Sepharose 4B (GE Healthcare), treated with AcTEV protease (Thermo Fisher Scientific) overnight at room temperature to remove the GST tag, incubated with ATP to dissociate chaperones, and subjected to size-exclusion chromatography (Superdex200 HiLoad 16/60) in 25 mM Hepes and 150 mM KCl. Purified proteins were tested for purity using SDS-PAGE and Coomassie staining and were analyzed to exclude contaminating ATPase activity. All Bro1V mutants examined in this study exhibited equivalent expression levels in bacteria, and mutants expressed in yeast were at levels similar to WT (Fig. 6 B). GST-Vps4 was expressed and purified as previously described for use in the ATPase assays (Babst et al., 1997). His6-Vps4 was purified as previously described (Davies et al., 2014), except the His6 fusion tag was not cleaved in order to use His6-Vps4 in the in vitro binding assays. His-Bro1ΔPRR was purified from BL21-DE3 using Ni2+-affinity chromatography (5 ml HiTrap Chelating FF column; GE Healthcare). GST-Bro1V(370–709) was purified using Glutathione Sepharose 4B (GE Healthcare) and eluted with reduced Glutathione (Thermo Fisher Scientific). V domains of S. castellii Bro1 and human HD-PTP were produced in Escherichia coli BL21 (DE3) were purified by TALON-Co2+ and size-exclusion chromatography and cleaved from their 6xHis tag with AcTEV protease (Pashkova et al., 2013). Mono-Ub was expressed in E. coli BL21 (DE3) from pPL5293. The protein was purified from bacterial cell lysate by precipitation with 5% perchloric acid and carboxymethyl cellulose cation exchange chromatography in 0.05 M ammonium acetate buffer, pH 4.5, followed by elution with 0.1 M ammonium acetate buffer, pH 5.5 (Sundd et al., 2002).
Antibody generation
Anti-Snf7 antiserum was generated against GST-Snf7. Antiserum was generated in a New Zealand rabbit (Covance). Bleeds were evaluated for detection of Snf7 (SEY6210 and snf7Δ). The Snf7 antibody was used at 1:5,000. Anti-Bro1 antiserum was generated against His6-Bro1ΔPRR. Antiserum was generated in a New Zealand rabbit (Covance). Antibodies were purified using GST-Bro1ΔPRR immobilized on an AminoLink column (Thermo Fisher Scientific) and verified for sensitivity and specificity using yeast extracts (SEY6210 and bro1Δ) and recombinant Bro1V. The purified Bro1 antisera was used at 1:1,000.
In vitro binding assays
Ni-NTA agarose beads (Qiagen) with or without His6-Vps4 (10 µg) were incubated for 1 h at 4°C in NiA buffer (25 mM NaH2PO4 and 300 mM NaCl, pH 7.5), washed with NiA buffer, and equilibrated with ATPase buffer (see “ATPase assays”). Binding reactions were performed in Handee Spin Columns (Pierce) with 6 µg Bro1V or mutants in ATPase buffer plus 0.02% Tween20 incubated for 1 h at 30°C. Reactions were washed four times with NiA buffer with 10 mM imidazole (Sigma-Aldrich), and the bound material was eluted with NiA buffer with 200 mM imidazole. Instant-Bands protein stain (EZBiolab) was used to visualize input and eluted material on a Typhoon FLA 7000 (GE Healthcare). Samples were resolved via SDS-PAGE and detected using immunoblotting against Bro1. Bro1V input and His6-Vps4 were detected via SDS-PAGE followed by Coomassie staining.
Glutathione Sepharose 4B GST-tagged protein purification resin (Cytiva Life Sciences) with or without lysate from E. coli BL21 (DE3) expressing GST-tagged Bro1 fragments were incubated for 1 hr at 4°C in PBS, washed with PBS and equilibrated with ATPase buffer. Binding reactions were performed in Handee Spin Columns with His6-Vps4 in ATPase buffer plus 0.02% Tween20 incubated for 1 h at 30°C. Reactions were washed six times with PBS plus 0.02% Tween20, and the bound material was eluted with GST elution buffer (10 mM reduced glutathione and 50 mM Tris-HCl, pH 8.0). Instant-Bands protein stain was used to visualize input and eluted material on a Typhoon FLA 7000. Samples were resolved via SDS-PAGE, and His6-Vps4 was detected using penta-His antibody (Qiagen) per the manufacturer’s instruction.
Three or more independent experiments were performed, with each experiment performed in replicates within the experiment, and representatives are shown. Signal was visualized with goat anti-rabbit IgG 680LT (product number [P/N]: 925–68021; Li-Cor Biosciences) or goat anti-mouse IgG 800CW (P/N: 925–32210; Li-Cor Biosciences) and detected using the Odyssey Infrared Imager with ImageStudio software (Li-Cor Biosciences). Signal was analyzed using ImageQuantTL (GE Healthcare) and Prism 9 (GraphPad).
ATPase assays
Measurement of Vps4 ATPase activity was performed in ATPase buffer (20 mM Hepes, 100 mM KOAc, and 5 mM MgOAc, pH 7.5) as previously described (Azmi et al., 2008; Babst et al., 1998; Davies et al., 2010; Davies et al., 2014; Norgan et al., 2013; Tan et al., 2015). All reactions were incubated at 30°C for 30 min before initiation by ATP addition (4 mM final concentration). Images were captured using a Typhoon FLA 7000 (GE Healthcare). Vps4 titration data represent ATPase activities from a minimum of three independent experiments, with each experiment performed in duplicate. Data represent ATPase activities from a minimum of three independent experiments, with each experiment performed in replicates within the experiment. Data were graphed, and statistical significance was assessed by t tests using Prism 9 (GraphPad). The Vps4 concentration used was 0.5 µM, because this concentration of Vps4 exhibits submaximal specific activity. ATPase assays in the presence of Ub were performed with the minimal concentration (50 µM) yielding maximal enhancement.
Fluorescence microscopy
For live-cell imaging, cells expressing GFP-tagged cargos were grown in minimal media at 30°C to mid-log phase (0.5 OD600) for live-cell fluorescence microscopy. Mup1-GFP induction was accomplished by adding 50 µM copper sulfate/chloride, as previously described (MacDonald et al., 2015). Colabeling with FM4-64 and C6-NBD-PC (1-palmitoyl-2-(6-((7-nitro-2-1,3-benzoxadiazol-4-yl)amino)hexanoyl)-sn-glycero-3-phosphocholine; Avanti Polar Lipids) was performed by growing cells in YPD or Synthetic Media containing 1 µM C6-NBD-PC and 250 nM FM4-64 for 30 min at 30°C. Cells were then harvested at mid-log phase and imaged in YPD or Synthetic Media. Images were captured at room temperature (25°C) using an Olympus IX70-S1F2 fluorescence microscope equipped with an Olympus UPIanApo 100× numerical aperture 1.35 oil objective with the complementing immersion oil (n = 1.516; Applied Precision), Standard DeltaVision filters, FITC and rhodamine, and a Photometrics CoolSNAP HQ charge-coupled device monochrome camera (Teledyne Photometrics). Image was acquired using Delta Vision softWoRx (version 3.5.1; Applied Precision) and subsequently processed by Fiji (version: 2.1.0/1.53c, National Institutes of Health; Schindelin et al., 2012). Captured images were exported under the standard DeltaVision file format and converted into 16-bit TIFF images using Bio-Formats Importer (available within Fiji). The contrast and brightness of images were subsequently adjusted within Fiji as well. Images were captured on three different days using two different sets of transformations. Verification of Bro1 and mutant Bro1 expression in these cells was confirmed by immunoblotting. Cells with WT cargo sorting signal (as indicated by predominant lumenal fluorescence and an absence of appreciable limiting membrane accumulation; see Fig. S1 B for an NBD-PC example) were scored manually. Data represent quantification from a minimum of three independent labeling experiments, with each experiment quantifying at least 100 cells. Statistical significance was assessed by t tests using Prism 9 (GraphPad).
Cargo GFP liberation analysis
Cells expressing GFP-tagged cargos grown in minimal media at 30°C to mid-log phase (0.5 OD600) were harvested and treated with 0.2 M sodium hydroxide for 5 min. Mup1-GFP and Mup1-GFP-Ub induction was accomplished by adding 50 µM copper sulfate/chloride, as previously described (MacDonald et al., 2015). Subsequently, cells were resuspended in urea buffer (40 mM Tris-HCl, pH 6.8, 8 M urea, 15% SDS wt/vol, 0.1 mM EDTA, 1% β-mercaptoethanol vol/vol, and 0.01% bromophenol blue wt/vol) and lysed with glass beads for 5 min (Katzmann et al., 1999). Sample were resolved via SDS-PAGE and immunoblotted for GFP (1:1,000; Roche) and Pgk1 (1:10,000; Life Technologies). Signal was visualized with goat anti-mouse IgG 800CW (P/N: 925–32210; Li-Cor Biosciences) and detected using the Odyssey Infrared Imager with ImageStudio software (Li-Cor Biosciences). Signal was analyzed using ImageQuantTL (GE Healthcare) and Prism 9 (GraphPad).
Subcellular fractionations
Subcellular fractionations were performed by osmotic lysis as previously described (Dimaano et al., 2008), except the cell extracts were centrifuged at 13,000 ×g for 10 min at 4°C to separate the soluble and pellet fractions (Tan et al., 2015). Samples (0.1 or 0.2 OD600 equivalents) were resolved via SDS-PAGE and immunoblotted for Snf7 (this study; 1:5,000), Vps4 (Babst et al., 1997; 1:1,000), and Bro1 (this study, 1:1,000). Pgk1 (1:10,000; Life Technologies) and Pep12 (1:1,000; Life Technologies). Immunoblots were developed using HRP-conjugated Goat anti-Rabbit (1:30,000; Life Technologies) or Goat anti-Mouse (1:1,000; Life Technologies), SuperSignal West Pico and SuperSignal West Femto substrates (Thermo Fisher Scientific), and the UVP Autochemi System. Signal was quantified using ImageQuant software (GE Healthcare). Signal linearity was confirmed via image histograms. Data represent three or more independent experiments (representative immunoblots are shown) and are graphed as mean with SEM. Statistical significance was assessed by t tests using Prism 9 (GraphPad).
Statistical analysis
Statistical significance was assessed by parametric unpaired two-tailed Student’s t tests using Prism 9 (GraphPad).
Molecular modeling
Molecular graphics and analyses of the S. castellii Bro1 V domain (Protein Data Bank accession no. 4JIO, chain A) were performed with the UCSF Chimera package and PyMOL (Pettersen et al., 2004; The PyMOL Molecular Graphics System). Chimera was developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by National Institute of General Medical Sciences [NIGMS] grant P41-GM103311).
Dual-axis ET
Yeast cells were high-pressure frozen and freeze substituted as previously described (Buysse et al., 2020; Johnson et al., 2017). Liquid cultures were harvested during log phase and filtered with 4.5-μm Millipore paper, collected into 0.5-mm aluminum hats, high-pressure frozen with a Wohlwend high-pressure freezer, and transferred to freeze-substituted media kept at liquid-nitrogen temperature until cryofixation. Cells were then freeze substituted in an automated freeze-substitution machine (Leica) at −90°C in cryomedia made from 0.1% uranyl acetate and 0.25% glutaraldehyde in anhydrous acetone, washed in pure acetone, and embedded at −60°C in Lowicryl HM20 (Polysciences). Samples were polymerized at −60°C and warmed slowly over 4 d. Plastic blocks were trimmed, and sections were cut in 80-nm thin sections and 250-nm thick sections with a Leica UC6 ultramicrotome and placed on rhodium-plated copper slot grids (Electron Microscopy Sciences) for thin-section transmission EM (TEM) and thick-section electron tomography (ET). TEM of hundreds of cells per strain is used to quality control freezing, embedding, and staining for tomography (Richter et al., 2013). Tomographic samples are en bloc stained with 0.1% uranyl acetate and 0.25% glutaraldehyde only, with no additional post-staining as before (Giddings, 2003). 100 cells were surveyed before at least 10 representative cells were selected to gather the tomography.
Thick sections were labeled with 15-nm fiducial gold on both sides and mapped on a Phillips CM10 (TEM) at 80 kV and tilt imaged with a Tecnai 30 (300 kV; FEI) with dual–tilt series images collected from +60° to −60° with 1° increments using a Gatan US4000 4k × 4k charge-coupled device camera. Tilt series were shot at 19,000× magnification with a 0.6534-nm working pixel (binning 2) and repeated at a 90° rotation for dual-axis tomography. Tomograms and models were constructed using IMOD software package (Kremer et al., 1996).
MVB membrane models from dual-axis electron tomograms are manually assigned from the inner leaflet every 4 nm and calculated using Imodmesh (3DMOD). We designated BPs by their negative curvature, since the majority of endosome limiting membrane curvature is positive or spherical in shape. BP models are drawn from the 0° rim at the outer leaflet, measured, and sorted by surface area using only BPs that have more than 750 nm2 or approximately half of the mean ILV surface (Wemmer et al., 2011). ILVs are spherical and measured using sphere-fitting models from the vesicle’s outer leaflet (the inner leaflet of the MVB limiting membrane) and ILV diameters are measured using these sphere models. Videos were made using IMOD and QuickTime (Apple). Data were analyzed and graphed using Prism 9 (GraphPad).
Online supplemental material
Fig. S1 shows endosomes reconstructed from an EM tomogram of bro1Δ yeast, an NBD-PC scoring guide, and representative images of bro1ΔBOD and bro1ΔBOD vps4Δ yeast labeled with NBD-PC and FM4-64. Fig. S2 shows that free GFPs liberated from bro1ΔBOD and bro1ΔBOD Δdoa4 expressing Mup1-GFP or Mup1-GFP-Ub are equivalent; GFP liberation from Cos5-GFP and GFP-Dap2 are also presented. Fig. S3 expands characterization of Bro1ΔBOD NBD-PC sorting ESCRT dependence; bro1V does not support MVB cargo sorting. Fig. S4 shows that Bro1V mutations do not disrupt its interaction between His6-Vps4 as well as levels of protein expression when introduced into BRO1; mutants M1 and M7 support GFP-Cps1 sorting when expressed in bro1Δ cells. Fig. S5 shows Bro1V mutants retain their ability to bind Ub. Video 1 shows the EM tomogram of bro1ΔBOD and computer reconstruction used in Fig. 1 A. Video 2 shows the EM tomogram of bro1Δ and computer reconstruction used in Fig. 1 A. Video 3 shows the EM tomogram of WT yeast and computer reconstruction used in Fig. 7 A. Video 4 shows the EM tomogram of bro1V and computer reconstruction used in Fig. 3 D. Video 5 shows the EM tomogram of bro1M8 and computer reconstruction used in Fig. 7 A. Table S1 lists plasmids used in this study. Table S2 lists primers used in this study. Table S3 lists yeast strains used in this study. Table S4 lists mutations analyzed in this study.
Supplementary Material
lists plasmids used in this study.
lists primers used in this study.
lists yeast strains used in this study.
lists mutations analyzed in this study.
Acknowledgments
We acknowledge Drs. Gina Razidlo and Jason Tan (Mayo Clinic, Rochester, MN) for their inquiries and thoughtful discussions.
S. Dean and C.-C. Tseng contributed to these studies as members of the Biochemistry and Molecular Biology Graduate Program and received financial support from the Mayo Clinic Graduate School of Biomedical Sciences. J. Staffenhagen contributed to these studies as a member of the Mayo Clinic Summer Undergraduate Research Program and received financial support from the Mayo Clinic Graduate School of Biomedical Sciences. This research was supported, in whole or in part, by National Institutes of Health grants R01 GM116826 (D.J. Katzmann), R01 GM065505 (G. Odorizzi), and R01 GM58202 (R.C. Piper) and a Fraternal Order of Eagles postdoctoral fellowship (B.A. Davies). S. Dean was supported by the National Science Foundation graduate research fellowship under grant DGE-NSF-01-02. C.-C. Tseng was supported by a Mayo Clinic Sydney Luckman Family predoctoral fellowship. J. Staffenhagen was supported by the Mayo Clinic summer undergraduate research fellowship. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Any opinion, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation.
The authors declare no competing financial interests.
Author contributions: B.A. Davies, I.F. Azmi, and D.J. Katzmann conceived of the study. C.-C. Tseng, S. Dean, B.A. Davies, and D.J. Katzmann designed and conducted the experiments along with J.A. Payne, J. Staffenhagen, N. Pashkova, R.C. Piper, M. West, and G. Odorizzi. D.J. Katzmann, R.C. Piper, C.-C. Tseng, and B.A. Davies wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
References
- Adell, M.A., and Teis D.. 2011. Assembly and disassembly of the ESCRT-III membrane scission complex. FEBS Lett. 585:3191–3196. 10.1016/j.febslet.2011.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adell, M.A.Y., Migliano S.M., Upadhyayula S., Bykov Y.S., Sprenger S., Pakdel M., Vogel G.F., Jih G., Skillern W., Behrouzi R., et al. 2017. Recruitment dynamics of ESCRT-III and Vps4 to endosomes and implications for reverse membrane budding. eLife. 6:e31652. 10.7554/eLife.31652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi, I., Davies B., Dimaano C., Payne J., Eckert D., Babst M., and Katzmann D.J.. 2006. Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J. Cell Biol. 172:705–717. 10.1083/jcb.200508166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azmi, I.F., Davies B.A., Xiao J., Babst M., Xu Z., and Katzmann D.J.. 2008. ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev. Cell. 14:50–61. 10.1016/j.devcel.2007.10.021 [DOI] [PubMed] [Google Scholar]
- Babst, M., Sato T.K., Banta L.M., and Emr S.D.. 1997. Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J. 16:1820–1831. 10.1093/emboj/16.8.1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Wendland B., Estepa E.J., and Emr S.D.. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993. 10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst, M., Katzmann D.J., Estepa-Sabal E.J., Meerloo T., and Emr S.D.. 2002. Escrt-III: an endosome-associated heterooligomeric protein complex required for mvb sorting. Dev. Cell. 3:271–282. 10.1016/S1534-5807(02)00220-4 [DOI] [PubMed] [Google Scholar]
- Baietti, M.F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., Ivarsson Y., Depoortere F., Coomans C., Vermeiren E., et al. 2012. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat. Cell Biol. 14:677–685. 10.1038/ncb2502 [DOI] [PubMed] [Google Scholar]
- Bend, R., Cohen L., Carter M.T., Lyons M.J., Niyazov D., Mikati M.A., Rojas S.K., Person R.E., Si Y., Wentzensen I.M., et al. Regeneron Genetics Center . 2020. Phenotype and mutation expansion of the PTPN23 associated disorder characterized by neurodevelopmental delay and structural brain abnormalities. Eur. J. Hum. Genet. 28:76–87. 10.1038/s41431-019-0487-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilodeau, P.S., Urbanowski J.L., Winistorfer S.C., and Piper R.C.. 2002. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 4:534–539. 10.1038/ncb815 [DOI] [PubMed] [Google Scholar]
- Buysse, D., Pfitzner A.-K., West M., Roux A., and Odorizzi G.. 2020. The ubiquitin hydrolase Doa4 directly binds Snf7 to inhibit recruitment of ESCRT-III remodeling factors in S. cerevisiae. J. Cell Sci. 133:jcs241455. 10.1242/jcs.241455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton, J.G., and Martin-Serrano J.. 2007. Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science. 316:1908–1912. 10.1126/science.1143422 [DOI] [PubMed] [Google Scholar]
- Carlton, J.G., Agromayor M., and Martin-Serrano J.. 2008. Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc. Natl. Acad. Sci. USA. 105:10541–10546. 10.1073/pnas.0802008105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, B.A., Azmi I.F., Payne J., Shestakova A., Horazdovsky B.F., Babst M., and Katzmann D.J.. 2010. Coordination of substrate binding and ATP hydrolysis in Vps4-mediated ESCRT-III disassembly. Mol. Biol. Cell. 21:3396–3408. 10.1091/mbc.e10-06-0512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, B.A., Norgan A.P., Payne J.A., Schulz M.E., Nichols M.D., Tan J.A., Xu Z., and Katzmann D.J.. 2014. Vps4 stimulatory element of the cofactor Vta1 contacts the ATPase Vps4 α7 and α9 to stimulate ATP hydrolysis. J. Biol. Chem. 289:28707–28718. 10.1074/jbc.M114.580696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimaano, C., Jones C.B., Hanono A., Curtiss M., and Babst M.. 2008. Ist1 regulates Vps4 localization and assembly. Mol. Biol. Cell. 19:465–474. 10.1091/mbc.e07-08-0747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, R.D., Chung H.-Y., Zhai Q., Robinson H., Sundquist W.I., and Hill C.P.. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell. 128:841–852. 10.1016/j.cell.2007.01.035 [DOI] [PubMed] [Google Scholar]
- Giddings, T.H.2003. Freeze-substitution protocols for improved visualization of membranes in high-pressure frozen samples. J. Microsc. 212:53–61. 10.1046/j.1365-2818.2003.01228.x [DOI] [PubMed] [Google Scholar]
- Gingras, M.C., Kharitidi D., Chénard V., Uetani N., Bouchard M., Tremblay M.L., and Pause A.. 2009. Expression analysis and essential role of the putative tyrosine phosphatase His-domain-containing protein tyrosine phosphatase (HD-PTP). Int. J. Dev. Biol. 53:1069–1074. 10.1387/ijdb.082820mg [DOI] [PubMed] [Google Scholar]
- Hanson, P.I., and Cashikar A.. 2012. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 28:337–362. 10.1146/annurev-cellbio-092910-154152 [DOI] [PubMed] [Google Scholar]
- Hanson, P.K., Grant A.M., and Nichols J.W.. 2002. NBD-labeled phosphatidylcholine enters the yeast vacuole via the pre-vacuolar compartment. J. Cell Sci. 115:2725–2733. 10.1242/jcs.115.13.2725 [DOI] [PubMed] [Google Scholar]
- Hicke, L., Schubert H.L., and Hill C.P.. 2005. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6:610–621. 10.1038/nrm1701 [DOI] [PubMed] [Google Scholar]
- Johnson, N., West M., and Odorizzi G.. 2017. Regulation of yeast ESCRT-III membrane scission activity by the Doa4 ubiquitin hydrolase. Mol. Biol. Cell. 28:661–672. 10.1091/mbc.e16-11-0761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., Epping E.A., and Moye-Rowley W.S.. 1999. Mutational disruption of plasma membrane trafficking of Saccharomyces cerevisiae Yor1p, a homologue of mammalian multidrug resistance protein. Mol. Cell. Biol. 19:2998–3009. 10.1128/MCB.19.4.2998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmann, D.J., Babst M., and Emr S.D.. 2001. Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell. 106:145–155. 10.1016/S0092-8674(01)00434-2 [DOI] [PubMed] [Google Scholar]
- Katzmann, D.J., Sarkar S., Chu T., Audhya A., and Emr S.D.. 2004. Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol. Biol. Cell. 15:468–480. 10.1091/mbc.e03-07-0473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer, C., Skalicky J.J., Morita E., De Domenico I., Ward D.M., Kaplan J., and Sundquist W.I.. 2008. Two distinct modes of ESCRT-III recognition are required for VPS4 functions in lysosomal protein targeting and HIV-1 budding. Dev. Cell. 15:62–73. 10.1016/j.devcel.2008.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Kim, J., Sitaraman S., Hierro A., Beach B.M., Odorizzi G., and Hurley J.H.. 2005. Structural basis for endosomal targeting by the Bro1 domain. Dev. Cell. 8:937–947. 10.1016/j.devcel.2005.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, Y., Kawawaki J., Kakiyama Y., Shimoda A., and Tanaka K.. 2014. The ESCRT-III adaptor protein Bro1 controls functions of regulator for free ubiquitin chains 1 (Rfu1) in ubiquitin homeostasis. J. Biol. Chem. 289:21760–21769. 10.1074/jbc.M114.550871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer, J.R., Mastronarde D.N., and McIntosh J.R.. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76. 10.1006/jsbi.1996.0013 [DOI] [PubMed] [Google Scholar]
- Larios, J., Mercier V., Roux A., and Gruenberg J.. 2020. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 219:e201904113. 10.1083/jcb.201904113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.-A., Beigneux A., Ahmad S.T., Young S.G., and Gao F.-B.. 2007a. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr. Biol. 17:1561–1567. 10.1016/j.cub.2007.07.029 [DOI] [PubMed] [Google Scholar]
- Lee, S., Joshi A., Nagashima K., Freed E.O., and Hurley J.H.. 2007b. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 14:194–199. 10.1038/nsmb1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J., Oh K.-J., Lee D., Kim B.Y., Choi J.S., Ku B., and Kim S.J.. 2016. Structural Study of the HD-PTP Bro1 Domain in a Complex with the Core Region of STAM2, a Subunit of ESCRT-0. PLoS One. 11:e0149113. 10.1371/journal.pone.0149113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie A. III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., and Pringle J.R.. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961. [DOI] [PubMed] [Google Scholar]
- Luhtala, N., and Odorizzi G.. 2004. Bro1 coordinates deubiquitination in the multivesicular body pathway by recruiting Doa4 to endosomes. J. Cell Biol. 166:717–729. 10.1083/jcb.200403139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, C., Buchkovich N.J., Stringer D.K., Emr S.D., and Piper R.C.. 2012a. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep. 13:331–338. 10.1038/embor.2012.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, C., Stringer D.K., and Piper R.C.. 2012b. Sna3 is an Rsp5 adaptor protein that relies on ubiquitination for its MVB sorting. Traffic. 13:586–598. 10.1111/j.1600-0854.2011.01326.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, C., Payne J.A., Aboian M., Smith W., Katzmann D.J., and Piper R.C.. 2015. A family of tetraspans organizes cargo for sorting into multivesicular bodies. Dev. Cell. 33:328–342. 10.1016/j.devcel.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manteghi, S., Gingras M.-C., Kharitidi D., Galarneau L., Marques M., Yan M., Cencic R., Robert F., Paquet M., Witcher M., et al. 2016. Haploinsufficiency of the ESCRT Component HD-PTP Predisposes to Cancer. Cell Rep. 15:1893–1900. 10.1016/j.celrep.2016.04.076 [DOI] [PubMed] [Google Scholar]
- McCullough, J., Fisher R.D., Whitby F.G., Sundquist W.I., and Hill C.P.. 2008. ALIX-CHMP4 interactions in the human ESCRT pathway. Proc. Natl. Acad. Sci. USA. 105:7687–7691. 10.1073/pnas.0801567105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill, S.A., and Hanson P.I.. 2010. Activation of human VPS4A by ESCRT-III proteins reveals ability of substrates to relieve enzyme autoinhibition. J. Biol. Chem. 285:35428–35438. 10.1074/jbc.M110.126318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa, B.E., Chiaruttini N., Redondo-Morata L., von Filseck J.M., König J., Larios J., Poser I., Müller-Reichert T., Scheuring S., Roux A., and Gerlich D.W.. 2017. Dynamic subunit turnover in ESCRT-III assemblies is regulated by Vps4 to mediate membrane remodelling during cytokinesis. Nat. Cell Biol. 19:787–798. 10.1038/ncb3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikko, E., and André B.. 2007. Split-ubiquitin two-hybrid assay to analyze protein-protein interactions at the endosome: application to Saccharomyces cerevisiae Bro1 interacting with ESCRT complexes, the Doa4 ubiquitin hydrolase, and the Rsp5 ubiquitin ligase. Eukaryot. Cell. 6:1266–1277. 10.1128/EC.00024-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norgan, A.P., Davies B.A., Azmi I.F., Schroeder A.S., Payne J.A., Lynch G.M., Xu Z., and Katzmann D.J.. 2013. Relief of autoinhibition enhances Vta1 activation of Vps4 via the Vps4 stimulatory element. J. Biol. Chem. 288:26147–26156. 10.1074/jbc.M113.494112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obita, T., Saksena S., Ghazi-Tabatabai S., Gill D.J., Perisic O., Emr S.D., and Williams R.L.. 2007. Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature. 449:735–739. 10.1038/nature06171 [DOI] [PubMed] [Google Scholar]
- Odorizzi, G., Babst M., and Emr S.D.. 1998. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 95:847–858. 10.1016/S0092-8674(00)81707-9 [DOI] [PubMed] [Google Scholar]
- Odorizzi, G., Katzmann D.J., Babst M., Audhya A., and Emr S.D.. 2003. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J. Cell Sci. 116:1893–1903. 10.1242/jcs.00395 [DOI] [PubMed] [Google Scholar]
- Oestreich, A.J., Davies B.A., Payne J.A., and Katzmann D.J.. 2007. Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway. Mol. Biol. Cell. 18:646–657. 10.1091/mbc.e06-07-0601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pashkova, N., Gakhar L., Winistorfer S.C., Sunshine A.B., Rich M., Dunham M.J., Yu L., and Piper R.C.. 2013. The yeast Alix homolog Bro1 functions as a ubiquitin receptor for protein sorting into multivesicular endosomes. Dev. Cell. 25:520–533. 10.1016/j.devcel.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen, E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., and Ferrin T.E.. 2004. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pfitzner, A.-K., Mercier V., Jiang X., Moser von Filseck J., Baum B., Šarić A., and Roux A.. 2020. An ESCRT-III Polymerization Sequence Drives Membrane Deformation and Fission. Cell. 182:1140–1155.e18. 10.1016/j.cell.2020.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, R.C., and Katzmann D.J.. 2007. Biogenesis and function of multivesicular bodies. Annu. Rev. Cell Dev. Biol. 23:519–547. 10.1146/annurev.cellbio.23.090506.123319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper, R.C., and Lehner P.J.. 2011. Endosomal transport via ubiquitination. Trends Cell Biol. 21:647–655. 10.1016/j.tcb.2011.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reggiori, F., and Pelham H.R.B.. 2001. Sorting of proteins into multivesicular bodies: ubiquitin-dependent and -independent targeting. EMBO J. 20:5176–5186. 10.1093/emboj/20.18.5176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, C.M., West M., and Odorizzi G.. 2013. Doa4 function in ILV budding is restricted through its interaction with the Vps20 subunit of ESCRT-III. J. Cell Sci. 126:1881–1890. 10.1242/jcs.122499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.S., Klionsky D.J., Banta L.M., and Emr S.D.. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8:4936–4948. 10.1128/MCB.8.11.4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., et al. 2012. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 9:676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt, O., and Teis D.. 2012. The ESCRT machinery. Curr. Biol. 22:R116–R120. 10.1016/j.cub.2012.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield, P., Garrard S., and Derewenda Z.. 1999. Overcoming expression and purification problems of RhoGDI using a family of “parallel” expression vectors. Protein Expr. Purif. 15:34–39. 10.1006/prep.1998.1003 [DOI] [PubMed] [Google Scholar]
- Shestakova, A., Hanono A., Drosner S., Curtiss M., Davies B.A., Katzmann D.J., and Babst M.. 2010. Assembly of the AAA ATPase Vps4 on ESCRT-III. Mol. Biol. Cell. 21:1059–1071. 10.1091/mbc.e09-07-0572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields, S.B., Oestreich A.J., Winistorfer S., Nguyen D., Payne J.A., Katzmann D.J., and Piper R.. 2009. ESCRT ubiquitin-binding domains function cooperatively during MVB cargo sorting. J. Cell Biol. 185:213–224. 10.1083/jcb.200811130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim, S., Merrill S.A., and Hanson P.I.. 2008. Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol. Biol. Cell. 19:2661–2672. 10.1091/mbc.e07-12-1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski, R.S., and Hieter P.. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27. 10.1093/genetics/122.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons, R.W., Houman F., and Kleckner N.. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 53:85–96. 10.1016/0378-1119(87)90095-3 [DOI] [PubMed] [Google Scholar]
- Skowyra, M.L., Schlesinger P.H., Naismith T.V., and Hanson P.I.. 2018. Triggered recruitment of ESCRT machinery promotes endolysosomal repair. Science. 360:eaar5078. 10.1126/science.aar5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowada, N., Hashem M.O., Yilmaz R., Hamad M., Kakar N., Thiele H., Arold S.T., Bode H., Alkuraya F.S., and Borck G.. 2017. Mutations of PTPN23 in developmental and epileptic encephalopathy. Hum. Genet. 136:1455–1461. 10.1007/s00439-017-1850-3 [DOI] [PubMed] [Google Scholar]
- Stringer, D.K., and Piper R.C.. 2011. A single ubiquitin is sufficient for cargo protein entry into MVBs in the absence of ESCRT ubiquitination. J. Cell Biol. 192:229–242. 10.1083/jcb.201008121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuchell-Brereton, M.D., Skalicky J.J., Kieffer C., Karren M.A., Ghaffarian S., and Sundquist W.I.. 2007. ESCRT-III recognition by VPS4 ATPases. Nature. 449:740–744. 10.1038/nature06172 [DOI] [PubMed] [Google Scholar]
- Sundd, M., Iverson N., Ibarra-Molero B., Sanchez-Ruiz J.M., and Robertson A.D.. 2002. Electrostatic interactions in ubiquitin: stabilization of carboxylates by lysine amino groups. Biochemistry. 41:7586–7596. 10.1021/bi025571d [DOI] [PubMed] [Google Scholar]
- Takahashi, Y., He H., Tang Z., Hattori T., Liu Y., Young M.M., Serfass J.M., Chen L., Gebru M., Chen C., et al. 2018. An autophagy assay reveals the ESCRT-III component CHMP2A as a regulator of phagophore closure. Nat. Commun. 9:2855. 10.1038/s41467-018-05254-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, J., Davies B.A., Payne J.A., Benson L.M., and Katzmann D.J.. 2015. Conformational Changes in the Endosomal Sorting Complex Required for the Transport III Subunit Ist1 Lead to Distinct Modes of ATPase Vps4 Regulation. J. Biol. Chem. 290:30053–30065. 10.1074/jbc.M115.665604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, S., Buchkovich N.J., Henne W.M., Banjade S., Kim Y.J., and Emr S.D.. 2016. ESCRT-III activation by parallel action of ESCRT-I/II and ESCRT-0/Bro1 during MVB biogenesis. eLife. 5:e15507. 10.7554/eLife.15507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System . Version 2.0. Schrödinger, LLC. https://pymol.org/2/ (Accessed May 29, 2018.)
- Vajjhala, P.R., Catchpoole E., Nguyen C.H., Kistler C., and Munn A.L.. 2007. Vps4 regulates a subset of protein interactions at the multivesicular endosome. FEBS J. 274:1894–1907. 10.1111/j.1742-4658.2007.05736.x [DOI] [PubMed] [Google Scholar]
- Vietri, M., Radulovic M., and Stenmark H.. 2020. The many functions of ESCRTs. Nat. Rev. Mol. Cell Biol. 21:25–42. 10.1038/s41580-019-0177-4 [DOI] [PubMed] [Google Scholar]
- Votteler, J., and Sundquist W.I.. 2013. Virus budding and the ESCRT pathway. Cell Host Microbe. 14:232–241. 10.1016/j.chom.2013.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, B.M., Colombi P., Jäger J., and Lusk C.P.. 2014. Surveillance of nuclear pore complex assembly by ESCRT-III/Vps4. Cell. 159:388–401. 10.1016/j.cell.2014.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer, M., Azmi I., West M., Davies B., Katzmann D., and Odorizzi G.. 2011. Bro1 binding to Snf7 regulates ESCRT-III membrane scission activity in yeast. J. Cell Biol. 192:295–306. 10.1083/jcb.201007018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R.L., and Urbé S.. 2007. The emerging shape of the ESCRT machinery. Nat. Rev. Mol. Cell Biol. 8:355–368. 10.1038/nrm2162 [DOI] [PubMed] [Google Scholar]
- Zhai, Q., Landesman M.B., Chung H.-Y., Dierkers A., Jeffries C.M., Trewhella J., Hill C.P., and Sundquist W.I.. 2011. Activation of the retroviral budding factor ALIX. J. Virol. 85:9222–9226. 10.1128/JVI.02653-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists plasmids used in this study.
lists primers used in this study.
lists yeast strains used in this study.
lists mutations analyzed in this study.