Bosch et al. discuss the role of lipid droplets as a front-line defense in the immune system.
Abstract
In the ongoing conflict between eukaryotic cells and pathogens, lipid droplets (LDs) emerge as a choke point in the battle for nutrients. While many pathogens seek the lipids stored in LDs to fuel an expensive lifestyle, innate immunity rewires lipid metabolism and weaponizes LDs to defend cells and animals. Viruses, bacteria, and parasites directly and remotely manipulate LDs to obtain substrates for metabolic energy, replication compartments, assembly platforms, membrane blocks, and tools for host colonization and/or evasion such as anti-inflammatory mediators, lipoviroparticles, and even exosomes. Host LDs counterattack such advances by synthesizing bioactive lipids and toxic nucleotides, organizing immune signaling platforms, and recruiting a plethora of antimicrobial proteins to provide a front-line defense against the invader. Here, we review the current state of this conflict. We will discuss why, when, and how LDs efficiently coordinate and precisely execute a plethora of immune defenses. In the age of antimicrobial resistance and viral pandemics, understanding innate immune strategies developed by eukaryotic cells to fight and defeat dangerous microorganisms may inform future anti-infective strategies.
Introduction
Most eukaryotic cells, from the simplest green algae to the tireless cardiomyocytes of the heart, survive environmental fluctuations using saved nutrients in the form of fat. Lipids are water-insoluble biomolecules that provide cells and organisms with unique resources (Muro et al., 2014). Three lipid classes will be emphasized throughout this review. First, phospholipids and cholesterol are critical membrane components that allow compartmentalization of organelles. Second, various lipids are essential structural components of second messengers (e.g., diacylglycerol and ceramides), inflammatory mediators (e.g., eicosanoids and resolvins), and molecules with key physiological roles (e.g., lipoproteins, bile salts, and ketones). Finally, fatty acids are highly reduced molecules that, when esterified in triacylglycerol (esters of one glycerol and three fatty acids), constitute the main energy reservoirs of eukaryotic cells (containing high energy per unit of storage mass).
The “cell’s pantry” storing these and other lipids are the atypical organelles called lipid droplets (LDs; Olzmann and Carvalho, 2019). Surrounded by a single monolayer of phospholipids, LDs organize as spherical assemblies to optimize the size of the valuable core of hydrophobic lipids within the cell’s aqueous environment (Fig. 1, A and B). The surface of LDs accommodates a plethora of resident proteins that exquisitely manage these resources. When needed, for example during fasting, LDs are activated by key energetic sensors to progressively deliver lipids to be locally transformed by LD proteins or crafted by other organelles into new membranes or signaling molecules. Also, during fasting, fatty acids are supplied by LDs to mitochondria through specific contact sites, enabling them to be oxidized by β oxidation. This, coupled to the citric acid cycle and oxidative phosphorylation, generates the metabolic energy invested by cells in obtaining new nutrients (Bosch et al., 2020a).
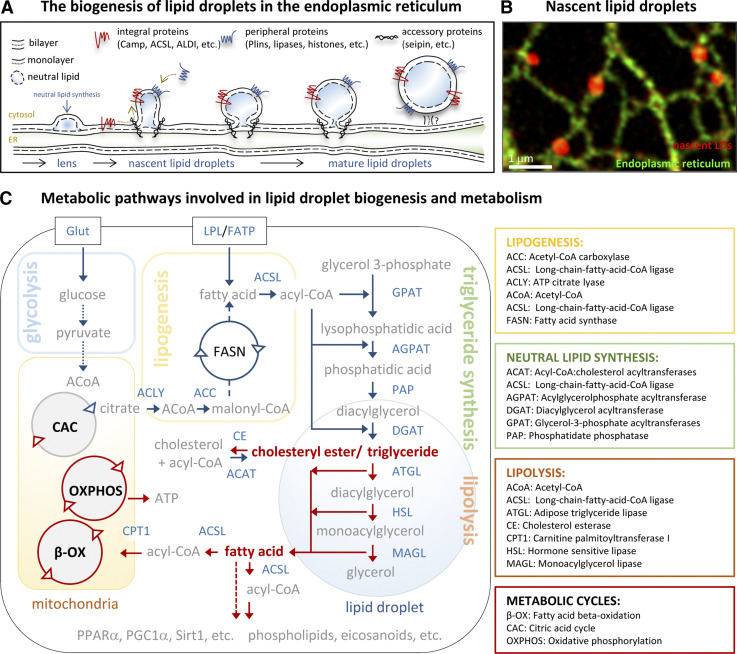
Figure 1.
Biogenesis and metabolism of LDs. (A and B) The biogenesis of LDs. (A) Schematic representation of the processes occurring in the ER membranes during LD biogenesis. After esterification, a neutral lipid lens separates in the ER bilayer. The lens laterally moves into nascent LDs, which progressively grow into mature LDs. Neutral lipid synthesis also occurs locally on LDs. LD proteins access the LD monolayer laterally from the ER membranes (red) or from the cytosol (blue) to regulate LD function. Accessory proteins, such as seipin, cooperate during the process. (B) The image illustrates LD formation (red) on ER membranes (green) in COS-7 cells that were treated for 7.5 min with oleic acid to induce LD formation. A specific marker for nascent LDs, a peptide formed by Aldi’s hydrophobic domain and caveolin-1 LD-targeting motif, was used to visualize LDs. Adapted with permission from the Journal of Cell Biology. (C) Main metabolic pathways involved in LDs biogenesis and metabolism. Fatty acids obtained from the extracellular environment or formed de novo by lipogenesis are esterified into triacylglycerols and stored within LDs in cells. Cholesterol is esterified into cholesteryl esters and accumulated within LDs. When these nutrients are needed, lipolysis is activated by the actions of LD lipases that produce fatty acids to be oxidized in mitochondria to generate ATP or produce molecules such as phospholipids or inflammatory mediators. Enzymes (detailed in boxes) are written with blue letters and intermediate molecules with gray letters. Blue arrows indicate fluxes during LD biogenesis and red arrows reactions occurring during LD catabolism.
Hence, it is not surprising that these rich organelles represent an attractive source of nutrients for microorganisms that subsist within cells. Pathogenic bacteria, viruses, and parasites guarantee their survival by chronically persisting or rapidly multiplying within the host. Lacking all (viruses) or key (bacteria) biosynthetic pathways, pathogens can mimic cellular organelles by using ingenious strategies to exploit LDs. Since Rudolf Virchow’s pioneering description in 1863 of fat-laden cells in biopsies of Mycobacterium leprae–infected patients (de Mattos et al., 2012; Virchow, 1867), the list of pathogens related to LDs has grown exponentially (Table S1, Table S2, and Table S3). Indeed, LDs accumulate in cells infected by some of the most medically significant pathogens, including infections caused by bacteria (e.g., tuberculosis), viruses (e.g., hepatitis), and parasites (e.g., malaria). The most widely accepted view is that host LDs are first induced and then hijacked by influential virulence factors secreted by pathogens to support a demanding lifestyle. However, successful innate immunity has been critical for survival, and eukaryotic cells have coevolved with invaders to develop a plethora of generic defense mechanisms. In this warlike scenario, the pathogen’s attraction for LDs emerges as a strategic location to organize a first line of cellular defense. In this review, we will revisit classical studies and highlight new findings from this point of view. We discuss evidence that LDs are not simply hijacked organelles but can be immune hubs regulating immunometabolism and immune defense. 10 recent and relevant publications to illustrate the multifaceted relationship of host lipids with pathogens, impossible to address here in all of its complexity, are recommended for further reading (Supplemental box).
LDs are nutrient storage organelles
LDs: Delivering flexibility for the eukaryotic lifestyle
The capacity to save nutrients in the face of environmental fluctuations represented a decisive advantage for eukaryotic cells. Triacylglycerol was adopted early as the preferred energy source, and cells developed LDs as the organelles to manage this resource. Very few prokaryotes (see under LDs and bacteria) can save nutrients in LDs, being totally conditioned for environmental fluctuations or being entirely dependent on the capacity of their hosts for nutrient acquisition. In contrast, the LD is an ancient organelle already present in the last eukaryotic common ancestor (Leyland et al., 2020; Lundquist et al., 2020). Triacylglycerol-enriched LDs are found in the simplest unicellular representatives of the three main eukaryotic clades, including green algae, diatoms, and yeast. LDs also accumulate molecules particularly relevant for some of those cell types such as pigments or vitamins (Davidi et al., 2015; Wake, 1974). Although the major structural LD proteins vary among eukaryotic clades, oleosins in plants and perilipins (Plins) in animals (Greenberg et al., 1991; Huang et al., 2013), ancestral mechanisms for triacylglycerol partitioning into LDs are envisaged by conservation of key regulatory proteins. For example, the ER-resident integral membrane protein seipin regulates the channeling of triacylglycerols into the LDs of plants, protists, fungi, and animals (Salo et al., 2019; Taurino et al., 2018).
The plasticity of LDs
Fundamental questions about LD biogenesis are still being addressed (Klemm and Ikonen, 2020). LD formation is triggered by lipids formed de novo (lipogenesis) and by lipids arriving to cells from the extracellular environment (Fig. 1 C). Fatty acids and cholesterol are esterified into neutral lipids by coordinated enzymatic cascades and cycles functioning on/around the ER (Pol et al., 2014). The accumulation of neutral lipids in ER bilayers promotes a gradual “demixing” of neutral lipids from membrane phospholipids and the formation of a neutral lipid “lens” between the leaflets of the bilayer (Renne et al., 2020; Fig. 1, A and B). As more lipid arrives and esterifies, the lens progressively grows into the cytosol to finally generate a phospholipid-surrounded spherical organelle with a hydrophobic core of neutral lipids. Structural proteins such as fat storage–inducing transmembrane 2, seipin, and Plins cooperate with the biosynthetic enzymes during this process (Olzmann and Carvalho, 2019). Once they are formed, LDs become regulatory metabolic hubs that ensure efficient delivery of lipids. Coordinated by energetic sensors, LD-resident proteins such as Plins (details in Box 1), acyl-CoA synthetases (ACSL), and lipases (ATGL, HSL, and monoacylglycerol lipase [MAGL]) regulate triacylglycerol hydrolysis and adjust the fatty acid and cholesterol supply to meet cellular demands (Fig. 1 B; Bosch et al., 2020a). In turn, Plins and lipases become signaling platforms by delivery fatty acids that activate key transcription factors and signaling molecules, such as peroxisome proliferator–activated receptor α (PPARα), the PPARγ coactivator 1-α (PGC1α), and the NAD-dependent lysine deacetylase sirtuin-1 (Sirt1), that are coordinated to increase the cellular mitochondrial content for coupling oxidative capacity to levels of substrates being generated (Najt et al., 2020).
Box 1. An overview of Plins
The discovery of Plins in the 1990s started to change the notion that LDs are simply cytosolic fat inclusions (Greenberg et al., 1991). Plins constitute a family of proteins that exclusively or majorly reside on LDs (Kimmel and Sztalryd, 2016; Sztalryd and Brasaemle, 2017). Plins (1–5) are encoded by different genes and show complementary tissue expression. Plins have the capacity to physically interact and modulate the activity of a plethora of LD proteins. Plin–protein interactions are finely regulated by phosphorylation, at several residues, by key energetic sensors such as protein kinase A (PKA) and 5′ adenosine monophosphate–activated protein kinase (AMPK).
The primary role of Plins is to coordinate inactivation/activation of LD lipases with the cellular energetic status (Bosch et al., 2020a). Under fed or insulin-stimulated conditions, Plins are nonphosphorylated, increasing the crowding on LDs and limiting the access and activity of the lipases ATGL and HSL, which are largely cytosolic. Plin1 in adipocytes (Subramanian et al., 2004); Plin2 and Plin3 in the majority of tissues (Yamaguchi et al., 2004); and Plin5 abundantly expressed in oxidative cells such as hepatocytes, type I myocytes, and cardiomyocytes (Wang et al., 2015) sequester CGI58, an essential coactivator of ATGL (Lass et al., 2006). During fasting, PKA becomes active to phosphorylate Plin1 (Tansey et al., 2003), Plin5 (Pollak et al., 2013), CGI58 (Sahu-Osen et al., 2015), ATGL (Pagnon et al., 2012), and HSL (Egan et al., 1992). After phosphorylation, Plin1 and Plin5 release CGI58, which activates ATGL. Further, phosphorylation of Plin1 and Plin5 facilitates the recruitment and activation of HSL (Sztalryd et al., 2003; Whytock et al., 2018). Consequently, stimulated lipolysis becomes active to produce fatty acids. Lipolysis is also regulated by chaperone-mediated autophagy. The AMPK-mediated phosphorylation of Plin2, which is not phosphorylated by PKA, activates its degradation by chaperone-mediated autophagy. Plin3 is degraded by similar mechanisms. Displacement of Plin2 and Plin3 from the LD surface reduces crowding and facilitates the recruitment of ATGL (Kaushik and Cuervo, 2015; Kaushik and Cuervo, 2016). In addition, Plins (especially Plin5) mediate formation of LD–mitochondrial contact sites (Boutant et al., 2017; Rogne et al., 2018; Varghese et al., 2019; Wang et al., 2011; Zhang et al., 2019). Finally, Plin5 can translocate to the nucleus to activate mitochondrial biogenesis via Sirt1 and PGC1α (Gallardo-Montejano et al., 2016; Najt et al., 2020).
As key LD components, Plins have been related to different aspects of immunity. Changes in Plin expression have been consistently reported in infected cells (see text). Plin levels are directly regulated by cytokines such as type II IFN and TNF, and it was suggested that Plin2 could organize complex clusters of defensive proteins on LDs (Bosch et al., 2020b). Indeed, Plin2 physically interacts with IFN-inducible proteins such as IGTP (IRGM3), a complex essential for cross-presentation of phagocytosed antigens by MHC class I in dendritic cells (Bougnères et al., 2009). The key role of Plins in the control of lipolysis is essential for proinflammatory molecule production. Adipocytes of the Plin1 knockout mice show a sustained inflammation (Sohn et al., 2018). Plins are also manipulated by pathogens to exploit LDs. The Chlamydia trachomatis LDA3 protein displaces Plin2 to promote an apparent translocation of host LDs into the bacterial inclusion (Cocchiaro et al., 2008). PV targets PV2BC, -2B and -2C proteins into LDs to displace Plin3, promoting LD (Zhang et al., 2019) clustering, dysregulated lipolysis, and production of fatty acids to generate phospholipids for the assembly of peplication compartments (Laufman et al., 2019). The HCV NS5A protein interacts with Plin3 to form and coordinate viral assembly platforms on LDs (Vogt et al., 2013).
LD formation exhibits remarkable plasticity, a trait needed to rapidly respond to unpredictable changes in nutrient availability. Arriving fatty acids are recruited into preexisting LDs within seconds after entering cells and, if needed, new LDs are formed in ∼10 min to accommodate more arriving lipids (Fig. 1 B; Kassan et al., 2013). The high efficiency of this process is achieved by forming platforms of functionally connected enzymes at sites of biogenesis (Pol et al., 2014).
Diatoms form and use LDs in nocturnal/diurnal cycles (Jallet et al., 2016) or when deprived of elements such as phosphorus, nitrogen, and silicon (Jaussaud et al., 2020). Yeast assemble LDs during the early stationary phase when nutrients such as inositol become limiting (Loewen et al., 2004). In metazoans, especially vertebrates, the formation of LDs is synchronized by endocrine cues to involve different cell types such as adipocytes, hepatocytes, myocytes, and cardiomyocytes (Bosch et al., 2020a). Fats and carbohydrates that are absorbed after feeding are partially converted into fatty acids, esterified in triacylglycerols, and stored in the LDs of adipocytes. During caloric restriction, adipocytes supply fatty acids to the liver, muscle, and heart (oxidative cells). These fatty acids are rapidly reesterified into the local LDs, which can then progressively supply lipids in nutrient-poor conditions (Bosch et al., 2020a).
While central in bioenergetics, unexpected LD functions beyond nutritional responses have been proposed in recent years (Welte and Gould, 2017). The remarkable plasticity of LD formation, distribution, and consumption could participate in the rapid response to a variety of stresses including the unfolded protein response (e.g., accumulating toxic proteins), oxidative damage (e.g., accumulating oxidized lipids), or when cells are challenged with pathogens (Henne et al., 2018). For example, hepatic LDs are essential for liver regeneration, a complex cellular process occurring in response to chemical, traumatic, and infectious injuries (Fernández et al., 2006). Indeed, accumulation of LDs is a phenotype commonly observed in cells and organs during systemic inflammation, polymicrobial sepsis, or endotoxic shock. Here, we discuss the role of LDs during microbial infections.
Accumulation of LDs in infected cells
Excellent reviews have been published recently to describe in detail formation of host LDs in cells infected with viruses, bacteria, and parasites (Libbing et al., 2019; Monson et al., 2021; Vallochi et al., 2018). A brief overview for selected microbes is provided next.
LDs and viruses
At the boundary between living and nonliving entities, viruses parasitize host lipids in all steps of their life cycle (Ketter and Randall, 2019). Despite having an extremely simple genome, viruses trigger massive alterations in host metabolism to establish a permissive environment for (i) genome amplification, (ii) virion assembly, and (iii) reorganizing the secretory pathway to egress. As key metabolic organelles, LDs are directly involved in generating these environments.
Positive-strand RNA viruses are by far the largest group infecting eukaryotic hosts, with many of them inducing and targeting LDs (Table S1). These viruses remodel intracellular membranes to generate viral replication compartments (Strating and van Kuppeveld, 2017). Enriched in cholesterol obtained from the ER and endosomes, replication compartments enable local concentration of RNA synthesis components and reduce their exposure to the innate immune sensors. The replication compartments formed by enteroviruses, such as the poliovirus (PV), are generated through membrane contact sites that recruit host LDs (Laufman et al., 2019). The viral proteins PV-2BC, -2B, and -2C are targeted to LDs to displace Plin3 and promote LD clustering. On LDs, the viral proteins PV-3A and -3AB interact with ATGL and HSL, which channel fatty acids into the phospholipids needed to form replication compartments. Indeed, inhibition of HSL abrogates replication compartment biogenesis and PV replication (Laufman et al., 2019).
Hepatitis C virus (HCV) also utilizes LDs to form replication compartments (Lee et al., 2019). In addition, LDs are assembly platforms for HCV. The HCV core interacts and activates diglyceride acyltransferase-1 (DGAT1) to increase triacylglycerol synthesis and LD formation (Vieyres and Pietschmann, 2019). DGAT1 inhibition impairs production of infectious virions (Herker et al., 2010). The dengue virus (DENV)–NS3 protein binds the small GTPase Rab18 on LDs and recruits fatty acid synthase (FASN, a key lipogenic enzyme; Heaton et al., 2010; Tang et al., 2014). HCV-NS5A interacts with DGAT1, Plin3, and Rab18, suggesting that viral components are bridged together by targeting abundant LD proteins (Camus et al., 2013; Salloum et al., 2013; Vogt et al., 2013). Because HCV-NS5A has RNA-binding properties, the protein may also transport viral RNAs from the replication compartments into LDs, facilitating interactions with the HCV core and virion encapsidation (Salloum et al., 2013). The affinity of RNAs for LDs was demonstrated many years ago (Dvorak et al., 2003), yet has remained poorly understood. The unique biophysical properties of the LD monolayer in combination with the underlying hydrophobic lipid core (Dhiman et al., 2020) could favor accretion and assembly of viral components.
The last step of HCV replication consists in the formation of lipoviroparticles occurring also in the proximity of LDs (Miyanari et al., 2007). Other viruses such as DENV and West Nile virus could follow similar mechanisms for producing infectious particles. By incorporating LD-triacylglycerol and interacting with very low-density lipoproteins, virions finally egress following the exocytic pathway (Faustino et al., 2014; Filipe and McLauchlan, 2015; Martins et al., 2019). Viral replication is a highly expensive process in terms of metabolic energy. Lipophagy provides fatty acids for the mitochondrial production of the ATP needed for DENV and HCV replication (Heaton and Randall, 2010). The DENV-NS4A exploits the acyltransferase activity of ancient ubiquitous protein 1 (AUP1) to trigger the autophagic degradation of LDs (Zhang et al., 2018). Intriguingly, in some cases, autophagy seems to be a host-driven defensive mechanism (Gassen et al., 2019), which is likely coordinated by host LDs (Schulze et al., 2017). In any case, infected cells accumulate antiviral proteins on LDs, reflecting an ongoing conflict.
LDs and bacteria
Only a limited number of bacterial species are known to be able to form LDs. The list includes some actinobacteria, such as the genera Rhodococcus or Mycobacterium, and cyanobacteria, including Anabaena (Zhang and Liu, 2017). Prokaryotes have developed a variety of nutrient acquisition strategies including, in a few cases, parasitizing eukaryotic cells. Obligate and facultative intracellular, Gram-positive and -negative, and even cytosolic bacteria induce and target LDs in host cells (Table S2). Host LDs are also affected by extracellular bacteria, including those that contribute to the gut microbiota (Tazi et al., 2018). Two representative examples of these are briefly summarized below.
C. trachomatis is a Gram-negative obligate intracellular human pathogen. Infection by C. trachomatis results in the accumulation of LDs in different cell types (Kumar et al., 2006; Saka et al., 2015). To manipulate LDs, C. trachomatis secretes into the cytosol proteins such as lipid metabolizing enzymes and LD-associated proteins (LDAs; Kumar et al., 2006; Soupene et al., 2015). On LDs, Ct-LDA3 displaces Plin2 and promotes, by unknown mechanisms, translocation of host LDs into the bacterial inclusion (Cocchiaro et al., 2008), with some host LD proteins completely translocating and other remaining on the phagolysosomal membrane (Soupene et al., 2012). Host LDs also provide cholesterol and fatty acids to produce membrane blocks (Peters and Byrne, 2015; Yao et al., 2015). Further, the fatty acids obtained from LDs are used for the synthesis of prostaglandin E2 (Fukuda et al., 2005), an immune suppressor induced by bacteria such as C. trachomatis, Chlamydia pneumoniae, and Coxiella burnetii to evade the immune response (Libbing et al., 2019). Suggesting that LDs support chlamydial replication, reducing LD formation by chemical inhibition of ACSL or acyl-CoA:cholesterol acyltransferase 1 (ACAT1) limits bacterial growth (Kumar et al., 2006; Peters and Byrne, 2015). However, the possibility that these drugs have off-target effects on key bacterial enzymes has been raised (Soupene and Kuypers, 2017).
Studies using cells genetically unable to form LDs demonstrate that LDs are not needed for C. trachomatis growth (Recuero-Checa et al., 2016). On the contrary, C. trachomatis–infected DGAT1/DGAT2 double knockout fibroblasts (unable to form LDs) showed more bacterial progeny at 48 h after infection (Sharma et al., 2018). Likewise, C. burnetii–infected ACAT1 knockout macrophages (unable to form LDs) also showed more bacterial progeny at 4 d after infection (Mulye et al., 2018). Perhaps supporting an antibacterial effect of LDs on chlamydial replication, Chlamydia muridarum inclusions interacting with the LDs of mouse polymorphonuclear leukocytes contain a higher number of aberrant reticulate bodies, which are reminiscent of the reticulate bodies produced in vitro when cells are treated with type II IFN or penicillin (Rank et al., 2011). The possibility that LDs directly function as antibiotic organelles is discussed later.
M. tuberculosis, or Koch’s bacilli, is a facultative intracellular bacterium. Macrophages infected with M. tuberculosis accumulate LDs (Peyron et al., 2008). Accumulation of host LDs is the result of PPAR dysregulation and correlates with enhanced bacterial growth (Kim et al., 2017). As observed for C. trachomatis, host LDs apparently migrate toward and are engulfed by the M. tuberculosis inclusion (Peyron et al., 2008). M. tuberculosis can persist latently for several years, forming granulomas in the lungs without manifesting clinical symptoms (Ehlers and Schaible, 2013). To support dormancy, M. tuberculosis synthesizes its own LDs. M. tuberculosis uses host triacylglycerols and bacterial neutral lipid synthesis enzymes (Daniel et al., 2011). In addition, M. tuberculosis expresses a complex cholesterol import system for persistence in murine lungs (Pandey and Sassetti, 2008). It was assumed that LD-laden macrophages support the maintenance and growth of persistent M. tuberculosis (Kim et al., 2010). However, as in the case of C. trachomatis, recent studies have challenged this view by their demonstration that host LDs are not needed for M. tuberculosis growth (Knight et al., 2018). On the contrary, host-driven LD formation, mediated by type II IFN, was essential to produce defensive eicosanoids (Knight et al., 2018). Further, in animal models of tuberculosis, the LDs of foamy macrophages mediate production of defensive cytokines (Jaisinghani et al., 2018). The hypothesis that LDs facilitate antimycobacterial responses forcing M. tuberculosis to shift into fat-saving survival mode has been proposed (Laval et al., 2021). The key role of LDs for production of inflammatory mediators and cytokines will be discussed in detail.
LDs and protozoan parasites
LDs also accumulate in cells infected by protozoan parasites (Table S3; Vallochi et al., 2018). As detailed for bacteria, mechanisms of docking and engulfment of LDs into the parasitophorous vacuole have been suggested for parasites such as Toxoplasma gondii and Leishmania major (Nolan et al., 2017; Rabhi et al., 2016). Intriguingly, host LD proteins such as Rab18 and Rab7 have been described within the parasitophorous vacuole (Nolan et al., 2017; Sant’Anna et al., 2009). In addition, like other eukaryotic cells, protozoans generate their own LDs, often in nutrient-rich conditions. LDs are commonly observed in the cytosol of animal and plant pathogens including Plasmodium falciparum (malaria), Trypanosoma cruzi (Chagas disease), Leishmania infantum chagasi (leishmaniasis), and Plasmodiophora brassicae (cabbage clubroot; Vallochi et al., 2018). Proteomic studies illustrate the complexity of protozoan LDs (Bi et al., 2016; Sant’Anna et al., 2009). T. gondii releases lipases into the cytosol to divert and use host LD resources (Charron and Sibley, 2002). T. gondii and P. falciparum express the necessary enzymes for neutral lipid synthesis (TgACAT1, TgACAT2, and PfDGAT; Lige et al., 2013; Vielemeyer et al., 2004). In addition to being a source of structural lipids, protozoan LD could accumulate toxic metabolites, such as heme-derived hemozoin in Plasmodium-infected cells, for detoxification or prevent the lipotoxicity caused by high concentrations of imported cholesterol (Vallochi et al., 2018). Lipid analogues with anti-proliferative properties are voraciously taken up by the parasites, resulting in parasite membrane defects, and ultimately, death (Coppens, 2006).
LDs and innate immunity
Mechanisms of innate immunity
All forms of life have some capacity for innate defense against environmental threats. In eukaryotic cells, front-line defense has been organized around rapid and generic mechanisms collectively defined as innate immunity (Paludan et al., 2021). Lower eukaryotic organisms, such as Caenorhabditis elegans and Drosophila, lack adaptive immunity and thus rely completely on innate immunity to fight against invaders (Salminen and Vale, 2020; Engelmann and Pujol, 2010). In higher organisms, innate immunity has developed complex endocrine and paracrine communication systems to synchronize host defense in a variety of professional and nonprofessional cell types.
To sense the presence of pathogens, eukaryotic cells have evolved receptors that detect essential microbial components, collectively referred to as pathogen-associated molecular patterns (PAMPs; Paludan et al., 2021; Fig. 2 A; and details in Box 2). Selected PAMPs include a variety of proteins, lipids, and nucleotides commonly found in pathogens. PAMPs are detected by pattern recognition receptors (PRRs), which are strategically positioned on the plasma membrane, on endocytic membranes, and in the cytosol of virtually all cell types. When activated by specific PAMP(s), each PRR recruits a set of accessory proteins and transduces danger signals into signaling molecules that relay responses through a series of intricate post-translational modifications (Fig. 2, B–D; Iwasaki and Medzhitov, 2015; Paludan et al., 2021). In the case of TLRs, activated signaling molecules in turn activate transcription factors, such as NF-κB, AP-1, IRF3, and IRF7, promoting expression of inflammatory cytokines, type I IFN, and a range of antimicrobial defense genes such as antimicrobial peptides (AMPs; Stocks et al., 2018).
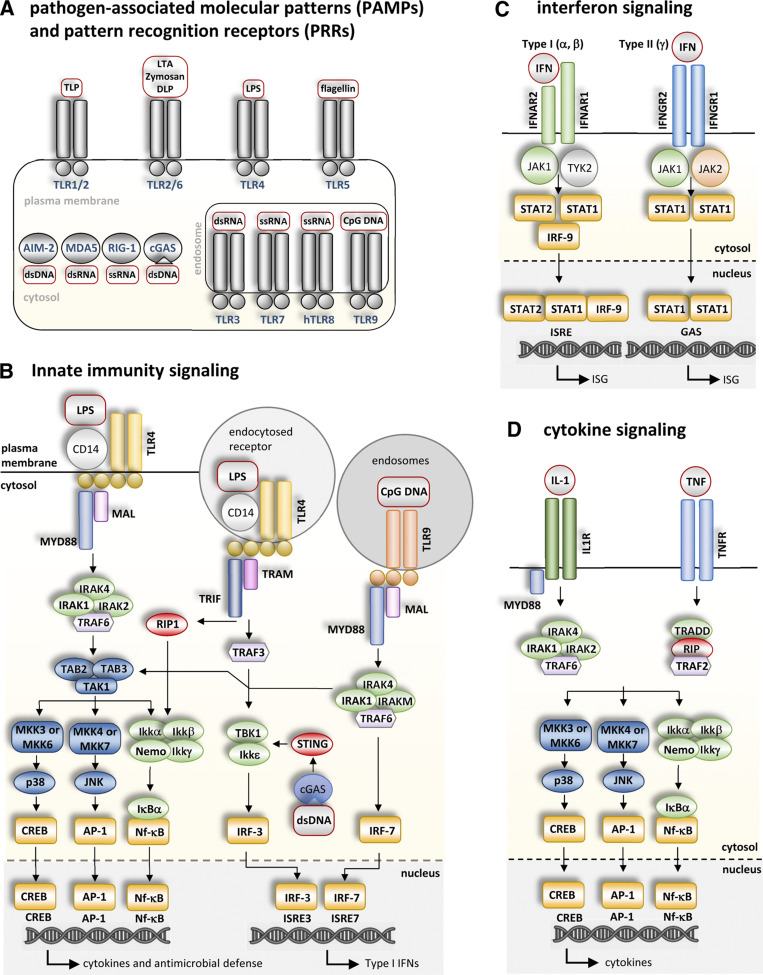
Figure 2.
Innate immunity signaling. (A) PAMPs and PRRs. Some examples of PAMPs (red squares) and specific PRRs (blue letters) that recognize them (see Box 2 for details). PRRs are positioned on the plasma membrane, endocytic membranes, and the cytosol. TLRs on the plasma membrane and endosomes, as well as nucleic acid-sensing cytosolic PRRs such as RIG-1, MDA5, cGAS, and AIM2 are some of the most widely studied PRRs. Selected PAMPs include a variety of proteins, lipids, and nucleotides frequently found in pathogens. PAMPs include molecules such as peptidoglycan (PGN), triacyl and diacyl lipopeptides (TLP and DLP), lipoteichoic acid (LTA), lipoarabinomannan (LAM), glycosylphosphatidylinositol (GPI)-anchored mucins (tGPI-mucin), LPS, single stranded (ss) and ds nucleotides (ss and dsRNA and DNA), and CpG-containing DNA (CpG DNA). (B) Innate immunity signaling. Schematic representation of PRR signaling during innate immune activation. Only specific PRRs directly mentioned in this review are shown (TLR4 on the plasma membrane and endocytic vesicles; TLR9 in endosomes; and cGAS in the cytoplasm). When activated by a specific PAMP(s), each PRR recruits a set of accessory proteins and transduces danger signals into signaling molecules that relay responses through a series of intricate post-translational modifications. Activated signaling molecules, in turn, activate transcription factors (yellow boxes) promoting expression of inflammatory cytokines, type I IFN, and a range of antimicrobial defense genes. Cytokines (including ILs, TNF, and IFNs) function in autocrine, paracrine, and endocrine communication networks to activate other receptors, accessory proteins, kinases, and transcription factors in neighboring or distant cells (C and D). Recommended reviews to seek further details of these pathways have been included in the text. (C) IFN signaling. Binding of IFNs to their cognate receptors, IFNAR-1 and -2 (for type I IFN) and IFNGR1 and 2 (for type II IFN), transduces signals into kinases and transcription factors (yellow boxes) to promote expression of ISGs via ISREs or γ IFN activation sites (GASs) in their promoters. Proteins such as IRFs, PRRs, and viperin are expressed in response to IFNs (Schneider et al., 2014). LD proteins such as Plin2 and Plin5 are also regulated by IFN (Bosch et al., 2020b). (D) Cytokine signaling. Two representative examples of cytokines are included. IL-1 and TNFα bind to their specific receptors on the plasma membrane to activate the signaling machinery and transcription factors, for inflammatory gene expression.
Box 2. Mechanisms of innate immunity
Toll-like receptors (TLRs) are evolutionarily conserved PRRs expressed by a variety of professional immune cells (leukocytes) such as monocytes, macrophages, dendritic cells, neutrophils, and mast cells, as well as a variety of nonimmune cells including hepatocytes, adipocytes, myocytes, and cardiomyocytes. TLR1, 2, 4, 5, and 6 are initially located on the cell membrane, whereas TLR3, 7, 8, and 9 are located on endolysosomal compartments (Iwasaki and Medzhitov, 2015; Fig. 3). Intracellular PRR pathways sensing and responding to viral RNAs and DNAs include (i) the axis formed by the retinoic acid inducible gene-I (RIG-I)/melanoma differentiation associated gene 5 (MDA5)–mitochondrial antiviral-signaling protein (MAVS, located in in the outer mitochondrial membrane, peroxisomes, and the ER), (ii) those formed by cyclic GMP–AMP synthase (cGAS)—stimulator of IFN genes (STING, located in the ER; Ni et al., 2018), and (iii) those absent in melanoma-2 (AIM2)/caspase-1/inflammasome pathway, which is located in the cytosol (Lugrin and Martinon, 2018). When activated by its specific PAMP(s), each PRR recruits a set of accessory proteins and transduces danger signals into signaling molecules that relay responses through a series of intricate post-translational modifications (Fig. 2 B; Iwasaki and Medzhitov, 2015; Paludan et al., 2021). Different classes of PRR generate biological responses through distinct molecular mechanisms. For example, inflammasomes initiate proteolytic cleavage of substrates to enable cytokine release and/or cell death, whereas TLRs primarily function through regulated gene expression. In the case of TLRs, activated signaling molecules in turn activate transcription factors, such as NF-κB, AP-1, IRF3, and IRF7, promoting expression of inflammatory cytokines, type I IFN, and a range of antimicrobial defense genes such as AMPs (Stocks et al., 2018). Cytokines (including ILs, TNF, and IFNs) function in autocrine, paracrine, and endocrine communication networks to activate other receptors, accessory proteins, kinases, and transcription factors in neighboring or distant cells (Fig. 2, C and D). This cytokine network must be tightly regulated to kill pathogens but to avoid host damage. Resolution of this response is a key task of innate immunity, also mediated by cytokines, transcription factors, and specific signaling lipids.
During infection, the intensity of each immune branch relies on multiple factors such as the cell type, the nature of the PAMP, and the affected organ(s). Importantly, these PRRs have the capacity to distinguish not only danger signals of invaders, but also the host's own components that are presented when homeostasis is dysregulated. These are often referred to as damage-associated molecular patterns. For example, circulating fatty acids and oxidized lipids are damage-associated molecular patterns that have been widely implicated in PRR activation, thus connecting metabolic diseases with inflammatory phenotypes (Saltiel and Olefsky, 2017).
Are LDs part of innate immunity?
It is still debatable whether the accumulation of LDs in infected cells reflects the induction of LDs by invaders for nutritional purposes and/or to subvert immune responses, or on the contrary, that LDs actually contribute to effective host defense. With much of the research pursuing the hijacking of LDs (Roingeard and Melo, 2017), it is increasingly evident that innate immunity strongly depends on different aspects of lipid metabolism (Jarc and Petan, 2020).
LD formation follows the fundamental premises of innate immunity being a rapid, generic, and conserved reaction triggered by local and paracrine signals, suggesting that it is an integral part of the defense program. This hypothesis is supported by a number of observations including the following: (i) infected cells accumulate LDs within minutes after infection (Monson et al., 2020; Rabhi et al., 2016); (ii) LDs are formed in response to numerous PAMPs and by activation of many PRRs, including those of insects (Barletta et al., 2016); (iii) the accumulation of LDs also occurs in cells treated with killed pathogens (Chen et al., 2008; Nicolaou et al., 2012); (iv) LDs are formed not only in infected cells but also in uninfected neighboring cells (Chen et al., 2020; Rabhi et al., 2016); and finally, (v) LDs recruit a plethora of innate immune antimicrobial proteins when cells are activated with PAMPs, IFNs, or pathogens (Bosch et al., 2020b; Hinson and Cresswell, 2009a).
The proteomic characterization of LDs formed in response to Gram-negative bacterial lipopolysaccharide (LPS, a TLR4 agonist), demonstrated that at least 30% of the LD proteome is regulated by LPS (Bosch et al., 2020b). Thus, innate immunity has developed a defense program that includes an extensive remodeling of the organelle. The in silico analysis of these LPS-regulated proteins suggested that LDs are innate immune hubs involved in several intra- and extracellular immune responses including (i) acting as signaling platforms, (ii) direct killing of pathogens, (iii) regulating immunometabolism, and (iv) providing fatty acids for pro- and anti-inflammatory molecules.
Innate immunity rewires lipid metabolism to form LDs
As part of their defense mechanisms, infected cells need to rapidly rewire lipid metabolism, accumulate lipids, and form LDs. Obviously, not all cells have the same lipogenic abilities and regulatory circuits. Thus, the capacity for, and nature of, LD formation relies on the cell type. While LDs in macrophages are enriched in cholesterol esters, LDs in hepatocytes mainly contain triacylglycerols (Olzmann and Carvalho, 2019). Furthermore, triacylglycerols in LDs in mastocytes are highly enriched in arachidonic acid (Dichlberger et al., 2011). The described mechanisms by which innate immunity rewires lipid metabolism in response to danger are (i) reduced cholesterol biosynthesis, (ii) reduced cholesterol efflux and increased cholesterol uptake, (iii) increased de novo synthesis and desaturation of fatty acids, (iv) increased fatty acid uptake, and (v) expression of key enzymes and LD proteins to administrate the resources. Below we highlight some key research to support these general statements.
Mouse bone marrow–derived macrophages infected with murine γ herpesvirus-68 decrease expression of genes encoding enzymes of the cholesterol biosynthetic pathway and increase expression of genes encoding lipid importers (York et al., 2015). This transcriptional signature was reproduced when these cells were treated with type I IFN or the synthetic TLR3 agonist Poly:IC. Despite the reduction in cholesterol synthesis, infected macrophages accumulate LDs. This is partially caused because type I IFN both increases cholesterol influx via scavenger receptor-A (SR-A) and decreases ATP binding cassette subfamily A member 1 (ABCA1)–mediated cholesterol efflux (Boshuizen et al., 2016). The transcriptional signature of L. major–infected bone marrow–derived macrophages confirmed that cells infected with parasites also decrease cholesterol efflux while increasing cholesterol uptake and de novo synthesis of fatty acids. Heat-killed promastigotes identically trigger the transcriptional program, indicating that rewiring of lipid metabolism is a host-driven process (Rabhi et al., 2012; Rabhi et al., 2016).
Different studies have showed that LDs are formed in response to type I and II IFNs and by the action of hypoxia-inducible transcription factor-1 α (HIF-1α), which is induced by NF-κB (Knight et al., 2018; Mylonis et al., 2019). Considerable differences in the lipogenic genes involved have been described, with this depending on the system and the nature of the challenge (e.g., different PAMPS and pathogens). Nonetheless, expression of enzymes related to fatty acid synthesis (FASN), desaturation (stearoyl-CoA desaturase-1 [SCD]), uptake and transport (fatty acid transport protein [FATP] or fatty acid translocase CD36), and esterification (ACSL, ACAT, and DGAT), together with the expression of LD-resident proteins (Plins), have been consistently observed (Castoldi et al., 2020; Nicolaou et al., 2012; Nolan et al., 2017; Rabhi et al., 2012). Accumulation of LDs could also be the result of the reduction in fatty acid oxidation described in LPS- and type II IFN–treated macrophages, hepatocytes, and pancreatic β cells (Bosch et al., 2020b; Castoldi et al., 2020; Truong et al., 2020). This metabolic reprogramming could be the consequence of the physical disconnection of LDs and mitochondria observed in proteomic analysis of LDs in LPS-treated hepatocytes and C. trachomatis–infected epithelial cells (Bosch et al., 2020b; Saka et al., 2015).
Rewiring of lipid metabolism is needed for effective innate immunity
The rewiring of lipid metabolism in innate immune cells generates a positive signaling feedback. Type I IFN signaling reduces cholesterol biosynthesis, which in turn enhances the production of type I IFN to potentiate antiviral defense (York et al., 2015). STING is somehow able to detect cholesterol levels in ER membranes, such that the level must be low for STING to signal to TBK1 (O’Neill, 2015). Thus, low cholesterol levels in the ER heighten the activation of the cGAS-STING-TBK1-IRF3 axis to drive type I IFN production (Fig. 2; York et al., 2015). Genetic approaches to inhibit cholesterol biosynthesis in macrophages resulted in enhanced resistance to murine γ herpesvirus-68 in mouse macrophages, as well as influenza A and human immunodeficiency virus 1 (York et al., 2015). In addition, intermediate molecules of the cholesterol biosynthetic pathway, such as lanosterol, cooperate to confer resistance to bacterial infections by regulation of an axis involving the transcription factors STAT1/STAT2, which activate type I IFN–stimulated gene expression (Araldi et al., 2017).
LD formation seems to be needed for some innate immune signaling responses. Hepatic cells with a reduced content of LD had a concomitant reduction in type I and III IFN production in response to double-stranded (ds) DNA, dsRNA, and Sendai virus (Monson et al., 2018). The presence of LDs correlates with IFN production and resistance to Herpes simplex virus 1 and Zika virus (Monson et al., 2020). Inhibitors of de novo lipogenesis markedly attenuated expression of anti-viral gene driven by type II IFN in pancreatic β cells (Truong et al., 2020). In macrophages, SREBP-1a activates not only genes required for lipogenesis but also the gene encoding NLRP1a, which is a core inflammasome component (Im et al., 2011). The possibility that LDs are functioning as signaling platforms that recruit and coordinate adaptors and kinases downstream of different PRRs is increasingly accepted (Monson et al., 2021). Accumulation of many IFN-inducible proteins on LDs supports the notion that LDs are tightly regulated by IFNs (Bosch et al., 2020b). Some examples of IFN-stimulated genes (ISGs) functioning on LDs will be detailed later.
LDs are hubs of innate immunity
The state of the conflict
The studies we have discussed demonstrate that LDs are produced in response to defined pathogenic stimuli to function as a conserved antimicrobial hub that has been heightened during eukaryotic evolution, from the histones on Drosophila LDs protecting embryos and adults to complex clusters of immune proteins described on hepatic LDs (Fig. 3 A; details in Box 3; Anand et al., 2012; Bosch et al., 2020b). In macrophages, LD-mediated defense involves association of LDs with phagolysosomal membranes and, presumably, transfer of antibacterial agents to contact bacterial membranes (Fig. 3 D). Relocation of LDs to interact with intracellular Mycobacterium has been described as early as 10 min after phagocytosis (Barisch and Soldati, 2017). This possibility, observed for bacteria and parasites (Cocchiaro et al., 2008; Nolan et al., 2017; Peyron et al., 2008; Rabhi et al., 2016; Soupene et al., 2012), suggests a specific docking between the two structures, but the mechanisms involved await dissection. In oxidative cells, such as hepatocytes, Plin5 is down-regulated during inflammation to decrease LD–mitochondrial contacts and oxidative metabolism while increasing LD–bacteria interactions (Bosch et al., 2020b). However, not only direct killing but also additional LD-centered defenses are in place.
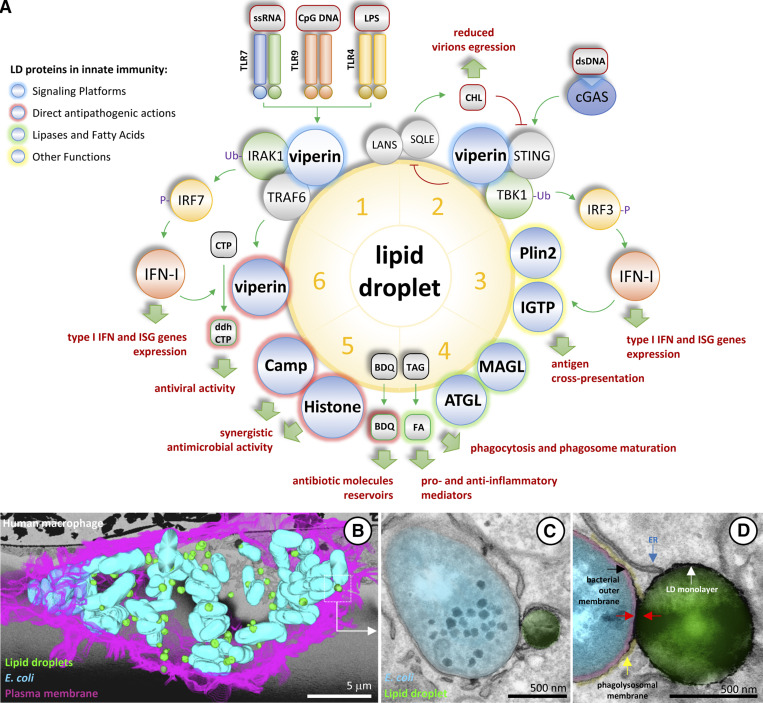
Figure 3.
LDs and innate immunity. (A) Six examples highlighting key roles of LDs in innate immunity. LDs are emerging as key organelles in innate immunity. The figure includes selected examples of the multifaceted roles of LDs during host defense. Details and references are in Box 3. (B–D) LDs interact with bacteria in infected cells. HMD macrophages from healthy donors were infected with E. coli for 4 h and analyzed by transmission electron microscopy. B–D are unpublished images from Robert Parton. (B) Serial blockface scanning electron microscopy data reconstruction showing an infected macrophage. The plasma membrane (pink), bacteria (blue), and LDs (green) in the 3D dataset have been colored and projected onto a single image. (C and D) Representative images of the LD–bacteria interaction are pseudocolored blue (bacteria), green (LD), red (E. coli outer and inner membranes and periplasm), and yellow (phagolysosomal membrane). The analysis reveals numerous contacts between LDs and E. coli. In the LD–E. coli contact sites, the LD monolayer (containing LD proteins) produced an apparent discontinuity in the phagolysosomal membrane (yellow) and probably interacted with the bacterial outer membrane (contacts are indicated with red arrows). FA, fatty acid; ss, single-stranded.
Box 3. Highlighting key roles of LDs in innate immunity
As summarized in Fig. 3, LDs are emerging as key organelles in innate immunity. (1) Although constitutively expressed in many cell types, viperin is strongly expressed in response to different PRRs, including TLR7/9 sensing of nucleic acids in endosomal membranes (Saitoh et al., 2011) and TLR4 recognizing LPS on the plasma membrane (Bosch et al., 2020b). After expression, viperin accumulates on LDs and recruits TRAF6 to facilitate IRAK1 ubiquitination and activation (Saitoh et al., 2011). IRAK1 phosphorylates IRF7, which migrates into the nucleus to induce expression of type I IFN. Type I IFN functions in an autocrine and paracrine fashion to activate the expression of ISGs for host defense. (2) Viperin also recruits signaling molecules in the cGAS sensing pathway, directly binding STING to mediate TBK1 polyubiquitination (Crosse et al., 2020). TBK1 phosphorylates IRF3, which migrates into the nucleus to promote expression of type I IFN. In parallel, the capacity of viperin to inhibit cholesterol biosynthesis by interaction with lanosterol synthase and squalene monooxygenase (Grunkemeyer et al., 2021) could function synergistically to activate STING (York et al., 2015) and to inhibit virus replication and virion release (Wang et al., 2007). (3) In addition to viperin, numerous IFN-inducible proteins accumulate on LDs (Bosch et al., 2020b). Among them, IGTP (IRGM3) on LDs regulates cross-presentation of phagocytosed antigens by MHC class I (Bougnères et al., 2009). (4) The fatty acids produced by lipases on LDs (ATGL, HSL, and MAGL) play crucial roles in phagocytosis, phagosome maturation, and synthesis of key pro- and anti-inflammatory mediators (Jarc and Petan, 2020). (5) LDs have been implicated in direct killing of bacteria by several mechanisms. The highly lipophilic anti-tubercular antibiotic bedaquiline (BDQ) accumulates within LDs (Greenwood et al., 2019). LDs are a transferable reservoir that enhances BDQ antibacterial efficacy. In addition, LDs accumulate antimicrobial molecules such as CAMP and histones, both with direct and synergistic antimicrobial capacities (Bosch et al., 2020b; Doolin et al., 2020). (6) Viperin is an S-adenosyl-L-methionine (SAM) enzyme that produces 3′-deoxy-3′,4'-didehydro-CTP (ddhCTP), a ribonucleotide that functions as a chain terminator of viral RNA synthesis by viral polymerases (Gizzi et al., 2018). Interaction of viperin with IRAK1 and TRAF6 increases viperin activity to produce ddhCTP (Dumbrepatil et al., 2019).
Viperin and the formation of immune signaling platforms on LDs
Viperin is considered to be an ancient core factor of IFN-mediated innate immunity in vertebrates (Rivera-Serrano et al., 2020). Viperin (RSDA2) is a broad-spectrum antiviral protein and a key transducer of the type I IFN–mediated response. Viperin is constitutively expressed in many cell types such as hepatocytes, cardiomyocytes, adipocytes, and macrophages. Its expression is also further induced by viruses, bacteria, IFNs, and different PAMPs such as LPS and nucleotide analogues (Rivera-Serrano et al., 2020). The viperin promoter contains two sequential IFN-stimulated response element (ISRE) sites, thus enabling inducible expression when the IFNAR-JAK-STAT-IRF9 axis is activated (Fig. 2 C; Severa et al., 2006).
After expression, viperin is targeted to the cytoplasmic face of the ER and to LDs via an N-terminal amphipathic α-helix (Hinson and Cresswell, 2009b). The C-terminal domain of viperin contributes to its antiviral activity (Van der Hoek et al., 2017) and facilitates viperin binding to iron (Upadhyay et al., 2014). Viperin on LDs interacts with viral components such as the HCV NS5A protein and the NS3 and DENV-2 capsid, and with viral RNAs (Helbig et al., 2013; Helbig et al., 2011). Viperin inhibits replication of a variety of highly pathogenic viruses including HCV, DENV, Zika virus, West Nile virus, and influenza A virus (Rivera-Serrano et al., 2020). In the case of HCV, the antiviral activity of viperin strongly relies on its location on LDs (Helbig et al., 2011).
The antiviral activity of viperin is multifaceted. Viperin is a radical S-adenosyl-L-methionine (SAM) enzyme that catalyzes conversion of CTP to ddhCTP, a ribonucleotide that functions as a chain terminator of viral RNA synthesis by viral polymerases (Gizzi et al., 2018). In addition, viperin is able to inhibit protein secretion in the ER (Hinson and Cresswell, 2009b) and to reduce the release and replication of the influenza virus by reducing isoprenoid biosynthesis (Wang et al., 2007).
During the last decade, studies centered on viperin functions have led to the hypothesis that LDs are important signaling platforms that regulate and amplify the IFN-mediated immune response. When dendritic cells are challenged with different insults that activate TLR7 and TLR9, cells robustly expressed viperin via IRF3 and IRF7 activation (Fig. 2 B). Loss of viperin reduces TLR7- and TLR9-promoted production of type I IFN, indicating a positive feedback mechanism by which newly synthesized viperin contributes to signal transduction (Saitoh et al., 2011). On LDs, viperin recruits IRAK1 and TRAF6 to facilitate IRAK1 ubiquitination and to induce the nuclear translocation of IRF7 (Fig. 2 B). Further, interaction with IRAK1 and TRAF6 increases the activity of viperin to produce ddhCTP (Dumbrepatil et al., 2019). Thus, LDs coordinate signals arriving from TLR7 and TLR9 to increase the type I IFN–mediated response and locally produce antiviral nucleotides.
In addition, following activation with dsDNA, viperin on LDs binds STING to enhance ubiquitination of TBK1 and the type I IFN response that limits HBV viability (Fig. 2 B; Crosse et al., 2020). The capacity of viperin to inhibit cholesterol biosynthesis by interacting with key enzymes of the biosynthetic pathway that reside on LDs (Grunkemeyer et al., 2021) could function synergistically to activate STING (York et al., 2015) or to reduce the viral replication compartment formation (Ilnytska et al., 2013; Strating et al., 2015). In conclusion, these studies indicate that viperin on LDs nucleates signaling platforms, produces antiviral molecules, and reduces cholesterol biosynthesis to enhance the synthesis of type I IFN and antiviral defenses. Similar regulatory mechanisms for other signaling pathways have been suggested by manipulation of LDs and concomitant reduction in the production of several cytokines (Jaisinghani et al., 2018; Knight et al., 2018; Monson et al., 2018; Truong et al., 2020).
CAMP, histones, and the coordination of direct antibacterial molecules on LDs
In addition to viperin, other defense proteins accumulate on the LDs of challenged cells (Bosch et al., 2020b). AMPs are a large family of evolutionarily conserved cationic molecules generally functioning as antipathogenic compounds by disorganizing pathogens membranes (Lazzaro et al., 2020). AMPs are effective against Gram-negative and -positive bacteria, enveloped viruses, fungi, and even transformed or cancerous cells. AMPs are synthesized as pro-proteins that follow the exocytic pathway to be cleaved and extracellularly released. Cathelicidin (CAMP) is a broad-spectrum AMP with chemotactic and immunomodulatory properties (Fabisiak et al., 2016). Proteolysis of CAMP generates LL-37, the active peptide. Adipocytes secrete CAMP to protect the skin during Staphylococcus aureus infection (Zhang et al., 2015). However, a pool of CAMP is also intracellularly retained for accumulation on LDs, at least in macrophages and hepatocytes (Bosch et al., 2020b). Like viperin, CAMP is bound to LDs by a hydrophobic N-terminal domain with the C-terminal containing LL-37 facing the cytosol. Cells expressing a genetically engineered LD-associated CAMP are more resistant to different bacterial species including Escherichia coli, methicillin-resistant S. aureus, and Listeria monocytogenes. Silencing of CAMP in human monocyte–derived (HMD) macrophages significantly impaired LD-mediated defense against intracellular E. coli (Bosch et al., 2020b). However, Pseudomonas aeruginosa was not affected by LD-CAMP, suggesting evasion mechanisms. In this regard, C. trachomatis, an obligate intracellular bacterium that targets LDs (Kumar et al., 2006), produces PGP3, a virulence factor that neutralizes the anti-chlamydial activity of CAMP (Hou et al., 2015).
Originally identified as antibacterial proteins (Hirsch, 1958; Miller et al., 1942), histones participate in extracellular-mediated antimicrobial and inflammatory responses (Brinkmann et al., 2004). For example, extracellular histones are important components of neutrophil extracellular traps (Tan et al., 2020) and have also been implicated in activation of TLR2 and TLR4 (Xu et al., 2011). However, similarly to Camp, a pool of histones accumulates on cytosolic LDs of Drosophila embryos (Li et al., 2012) and mammalian cells (Anand et al., 2012; Bersuker et al., 2018; Bosch et al., 2020b). In Drosophila, histones are recruited on LDs by interaction with Jabba (Li et al., 2012). Although no Jabba homologue has yet been found in mammals, histones have been described in hepatic LDs when mice are challenged with LPS (Anand et al., 2012; Bosch et al., 2020b). In experiments performed in vitro with LDs purified from Drosophila embryos, histones are released from LDs in the presence of bacterial LPS or lipoteichoic acid to kill both Gram-negative E. coli and Gram-positive S. epidermidis. Furthermore, Jabba mutant embryos and adults are susceptible to a variety of bacteria (Anand et al., 2012).
Histones and CAMP can function synergistically, with CAMP forming pores on bacterial membranes and histones depolarizing the membrane potential and reorganizing bacterial chromosomal DNA to inhibit transcription of E. coli and S. aureus (Doolin et al., 2020; Duong et al., 2021). The physiological relevance of this synergy is perhaps illustrated by the fact that important human pathogens, such as strains of the group A Streptococcus belonging to the hypervirulent M1T1 serogroup, have developed virulence factors to specifically bind and neutralize histones and CAMP (Döhrmann et al., 2017; LaRock et al., 2015). Although crucial during infection, histones and CAMP can be quite harmful for the host and promote cell damage and inflammation (Martin et al., 2015; Yamasaki et al., 2007). Thus, compartmentalization of toxic antibacterial proteins on LDs is likely an important mechanism to limit indiscriminate and dangerous cellular damage.
LDs are reservoirs and suppliers of bioactive lipids
Lipases on LDs have key roles in immunity. Macrophages lacking ATGL show reduced phagocytosis and impaired migration (Aflaki et al., 2011; Chandak et al., 2010), which is only partially restored by glucose supplementation, suggesting a role for ATGL beyond the simple provision of substrates for energy production (Chandak et al., 2010). The ATGL gene promoter has a putative IFN-γ–activated sequence (GAS) that likely interacts with Stat family members (Fig. 2 C; Truong et al., 2020). Further, NF-κB represses ATGL expression in Drosophila fat bodies by inhibition of the transcription factor FOXO-1 (Molaei et al., 2019), indicating that lipolysis is linked to innate immunity.
Triacylglycerol within LDs is a source of arachidonic acid, a precursor for eicosanoid formation (Jarc and Petan, 2020). Arachidonic acid accumulates in LDs during mastocyte maturation (Dichlberger et al., 2011). Key enzymes for eicosanoid biosynthesis are located on LDs, and thus, synthesis of prostaglandins occurs locally on LDs, at least partially (Bozza et al., 2011). Indeed, genetic or pharmacological inhibition of ATGL results in impaired production of eicosanoids in adipocytes, endothelia, and immune cells (Dichlberger et al., 2014; Gartung et al., 2016; Kuo et al., 2018). The fatty acids produced by MAGL (Fig. 1 A) have also been related to eicosanoid formation and neuroinflammation (Nomura et al., 2011). The likely role of LDs in providing polyunsaturated fatty acids, such as eicosapentaenoic acid and docosahexaenoic acid, for the synthesis of pro-resolution lipoxins, resolvins, and protectins (Russell and Schwarze, 2014) or fatty acids to activate key signaling molecules, such as Sirt1, that link metabolism and inflammation (Chen et al., 2015; Najt et al., 2020) deserves further investigation.
Targeting LDs for defense; LDs as antibiotic reservoirs
BDQ is a highly lipophilic antibiotic reducing M. tuberculosis viability by interacting with ATP synthase. In infected HMD macrophages, BDQ accumulates within LDs (Greenwood et al., 2019). LDs do not immobilize the antibiotic but constituted a transferable reservoir to enhance its antibacterial efficacy (Greenwood et al., 2019). Similarly, the broad-spectrum antiviral compound ST-669 accumulates within LDs to restrict chlamydial inclusion development and C. burnetii growth (Sandoz et al., 2014). These studies clearly illustrate the potential of LDs to deliver toxic molecules to invaders.
The capacity for sequestration in LDs of toxic molecules, including lipophilic molecules, lipids, or harmful proteins is conserved through evolution (Geltinger et al., 2020). For example, toxin trapping in LDs confers resistance to fungi, being a self-resistance mechanism for the toxin producer and a resistance mechanism for the toxin recipient (Chang et al., 2015). In an ACSL5- and DGAT2-dependent mechanism, LDs store acyl-ceramides to reduce the pool of de novo ceramides and ceramide-mediated apoptosis (Senkal et al., 2017). This capacity could be beneficial or detrimental when cells are pharmacologically treated with hydrophobic drugs. For example, lasonolide A is a macrolide with antitumoral, antifungal, and antibacterial capacity. It accumulates in LDs from where it is progressively cleaved by a LD-resident serine hydrolase, with this contributing to drug efficacy (Dubey et al., 2020). In contrast, accumulation of anti-tumoral drugs in LDs confers chemoresistance to different cancer cells (Bacci et al., 2021).
Conclusions and future perspectives
The numerous studies discussed add the LD to the repertoire of defense mechanisms of mammalian cells (Fig. 3 A and Box 3). Different tissues vary in their LD content, with the adipose tissue representing an extreme example. Animal adipose tissue has acquired a central role in the regulation of systemic innate immunity, reinforcing the idea that eukaryotes organize a front-line defense around nutrients (Schäffler and Schölmerich, 2010). Lack (lipodystrophy) or malfunctioning (obesity) of adipose tissue are both risk factors for infectious diseases (Falagas and Kompoti, 2006; Lima et al., 2018). The nutritional status of infected animals tightly determines the response to infection by yet to be identified pathways (Wang et al., 2016). Dermal adipocytes, a front line of the body’s defense, protect against invasive S. aureus skin infection by secretion of CAMP, with its expression being integrated within the adipogenesis program (Zhang et al., 2015; additional reading in the Supplemental box).
The evidence discussed highlights that LDs can be antimicrobial vehicles, directly functioning to face and kill invaders. However, it is yet unclear how association of LDs with phagolysosomal membranes is driven and executed (Fig. 3 B) and why LDs are recruited to certain bacteria (such as E. coli) but not others (such as Salmonella; Bosch et al., 2020b). By electron microscopy, a distinct junction between the LD and enclosing membrane is evident (Fig. 3, C and D), but the nature of this junction and the mechanism of any protein, peptide, or lipid transfer to the bacteria is completely unknown. In the case of cytosolic bacteria, additional mechanisms could be operative on LDs. Mysterin (RNF213) is an LD protein (Sugihara et al., 2019) up-regulated in response to LPS (Bosch et al., 2020b) that participates in the cellular defense against cytosolic Salmonella by mediating LPS ubiquitination and triggering antibacterial autophagy (Otten et al., 2021). Thus, the use of LDs as a first defensive line could provide innate immunity with some biological advantages: (i) it is a strategic location to attract a plethora of pathogens, (ii) it allows synergy between different immune systems operating simultaneously or coordinately against invaders, and (iii) it provides safety for the rest of the cellular organelles by sequestering potentially cytotoxic compounds.
Furthermore, the remarkable plasticity of LD formation, distribution, and consumption could support immunity by organizing platforms of functionally connected immune enzymes, adaptors, and kinases to timely boost signaling. Molecular crowding on LDs, known to be important to control lipolysis (Box 1), allows efficient regulation of protein–protein interactions to rapidly switch on and off signaling cascades responding to environmental fluctuations.. In parallel, the key role of LDs in regulating key homeostatic cellular processes such as the ER stress, cholesterol synthesis, autophagy, and oxidative metabolism might contribute to locally generate the environment conducive to host defense. Finally, lipases, Plins, and ACSLs on LDs could not only determine production of pro-inflammatory lipids but also supply fatty acids species needed to generate mitigation mediators or to activate key metabolic cascades to control inflammation. Whether these mechanisms, only tested for some pathogens, could be generic branches of innate immunity deserves further investigation.
Indeed, the diversity of pathogens and experimental systems used to study LDs is vast, complicating the formulation of a general model. The evolutionary arms race between hosts and microbes is likely to lead to distinct scenarios, with LDs defeating the invader in some circumstances, and the pathogen surpassing and/or exploiting this defense system in others. In both cases, elucidating mechanisms evolved by eukaryotes to protect valuable nutrients as well as understanding microbial strategies for exploiting the resources for evasion and survival will undoubtedly reveal strategic choke points to be enhanced or corrected in future therapeutic interventions.
Online supplemental material
Table S1 lists viruses related to LD formation and interaction. Table S2 lists bacteria related to LD formation and interaction. Table S3 lists protozoans related to LD formation and interaction. A supplemental box lists selected publications to illustrate the multifaceted relationship of host lipids with pathogens.
Supplementary Material
lists viruses related to LD formation and interaction.
lists bacteria related to LD formation and interaction.
lists protozoans related to LD formation and interaction.
lists selected publications to illustrate the multifaceted relationship of host lipids with pathogens.
Acknowledgments
M. Bosch acknowledges support from Fundació la Marató de TV3 (31/U/2016). M.J. Sweet is supported by a National Health and Medical Research Council Investigator Grant (APP1194406). A. Pol and R.G. Parton have been supported by the Human Frontier Science Program (RGP0020/2015). A. Pol is supported by the Ministerio de Ciencia e Innovación (RTI2018-098593-B-I00), Fundació la Marató de TV3 (31/U/2016), and the Institució de Centres de Recerca de Catalunya (CERCA) Programme/Generalitat de Catalunya. R.G. Parton is supported by the National Health and Medical Research Council of Australia (program grant APP1037320 and Senior Principal Research Fellowship 569452) and the Australian Research Council Centre of Excellence in Convergent Bio-Nanoscience and Technology (CE140100036).
The authors declare no competing financial interests.
Author contributions: M. Bosch, M.J. Sweet, R.G. Parton, and A. Pol conceived and designed the manuscript. M. Bosch, M.J. Sweet, R.G. Parton, and A. Pol wrote the manuscript. R.G. Parton, and A. Pol secured funding.
References
- Aflaki, E., Balenga N.A., Luschnig-Schratl P., Wolinski H., Povoden S., Chandak P.G., Bogner-Strauss J.G., Eder S., Konya V., Kohlwein S.D., et al. 2011. Impaired Rho GTPase activation abrogates cell polarization and migration in macrophages with defective lipolysis. Cell. Mol. Life Sci. 68:3933–3947. 10.1007/s00018-011-0688-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand, P., Cermelli S., Li Z., Kassan A., Bosch M., Sigua R., Huang L., Ouellette A.J., Pol A., Welte M.A., and Gross S.P.. 2012. A novel role for lipid droplets in the organismal antibacterial response. eLife. 1:e00003. 10.7554/eLife.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araldi, E., Fernández-Fuertes M., Canfrán-Duque A., Tang W., Cline G.W., Madrigal-Matute J., Pober J.S., Lasunción M.A., Wu D., Fernández-Hernando C., and Suárez Y.. 2017. Lanosterol Modulates TLR4-Mediated Innate Immune Responses in Macrophages. Cell Rep. 19:2743–2755. 10.1016/j.celrep.2017.05.093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacci, M., Lorito N., Smiriglia A., and Morandi A.. 2021. Fat and Furious: Lipid Metabolism in Antitumoral Therapy Response and Resistance. Trends Cancer. 7:198–213. 10.1016/j.trecan.2020.10.004 [DOI] [PubMed] [Google Scholar]
- Barisch, C., and Soldati T.. 2017. Mycobacterium marinum Degrades Both Triacylglycerols and Phospholipids from Its Dictyostelium Host to Synthesise Its Own Triacylglycerols and Generate Lipid Inclusions. PLoS Pathog. 13:e1006095. 10.1371/journal.ppat.1006095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barletta, A.B., Alves L.R., Silva M.C., Sim S., Dimopoulos G., Liechocki S., Maya-Monteiro C.M., and Sorgine M.H.. 2016. Emerging role of lipid droplets in Aedes aegypti immune response against bacteria and Dengue virus. Sci. Rep. 6:19928. 10.1038/srep19928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersuker, K., Peterson C.W.H., To M., Sahl S.J., Savikhin V., Grossman E.A., Nomura D.K., and Olzmann J.A.. 2018. A Proximity Labeling Strategy Provides Insights into the Composition and Dynamics of Lipid Droplet Proteomes. Dev. Cell. 44:97–112.e7. 10.1016/j.devcel.2017.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi, K., He Z., Gao Z., Zhao Y., Fu Y., Cheng J., Xie J., Jiang D., and Chen T.. 2016. Integrated omics study of lipid droplets from Plasmodiophora brassicae. Sci. Rep. 6:36965. 10.1038/srep36965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch, M., Parton R.G., and Pol A.. 2020a. Lipid droplets, bioenergetic fluxes, and metabolic flexibility. Semin. Cell Dev. Biol. 108:33–46. 10.1016/j.semcdb.2020.02.010 [DOI] [PubMed] [Google Scholar]
- Bosch, M., Sánchez-Álvarez M., Fajardo A., Kapetanovic R., Steiner B., Dutra F., Moreira L., López J.A., Campo R., Marí M., et al. 2020b. Mammalian lipid droplets are innate immune hubs integrating cell metabolism and host defense. Science. 370:eaay8085. 10.1126/science.aay8085 [DOI] [PubMed] [Google Scholar]
- Boshuizen, M.C., Hoeksema M.A., Neele A.E., van der Velden S., Hamers A.A., Van den Bossche J., Lutgens E., and de Winther M.P.. 2016. Interferon-β promotes macrophage foam cell formation by altering both cholesterol influx and efflux mechanisms. Cytokine. 77:220–226. 10.1016/j.cyto.2015.09.016 [DOI] [PubMed] [Google Scholar]
- Bougnères, L., Helft J., Tiwari S., Vargas P., Chang B.H., Chan L., Campisi L., Lauvau G., Hugues S., Kumar P., et al. 2009. A role for lipid bodies in the cross-presentation of phagocytosed antigens by MHC class I in dendritic cells. Immunity. 31:232–244. 10.1016/j.immuni.2009.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutant, M., Kulkarni S.S., Joffraud M., Ratajczak J., Valera-Alberni M., Combe R., Zorzano A., and Cantó C.. 2017. Mfn2 is critical for brown adipose tissue thermogenic function. EMBO J. 36:1543–1558. 10.15252/embj.201694914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozza, P.T., Bakker-Abreu I., Navarro-Xavier R.A., and Bandeira-Melo C.. 2011. Lipid body function in eicosanoid synthesis: an update. Prostaglandins Leukot. Essent. Fatty Acids. 85:205–213. 10.1016/j.plefa.2011.04.020 [DOI] [PubMed] [Google Scholar]
- Brinkmann, V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., and Zychlinsky A.. 2004. Neutrophil extracellular traps kill bacteria. Science. 303:1532–1535. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Camus, G., Herker E., Modi A.A., Haas J.T., Ramage H.R., Farese R.V. Jr., and Ott M.. 2013. Diacylglycerol acyltransferase-1 localizes hepatitis C virus NS5A protein to lipid droplets and enhances NS5A interaction with the viral capsid core. J. Biol. Chem. 288:9915–9923. 10.1074/jbc.M112.434910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castoldi, A., Monteiro L.B., van Teijlingen Bakker N., Sanin D.E., Rana N., Corrado M., Cameron A.M., Hässler F., Matsushita M., Caputa G., et al. 2020. Triacylglycerol synthesis enhances macrophage inflammatory function. Nat. Commun. 11:4107. 10.1038/s41467-020-17881-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandak, P.G., Radovic B., Aflaki E., Kolb D., Buchebner M., Fröhlich E., Magnes C., Sinner F., Haemmerle G., Zechner R., et al. 2010. Efficient phagocytosis requires triacylglycerol hydrolysis by adipose triglyceride lipase. J. Biol. Chem. 285:20192–20201. 10.1074/jbc.M110.107854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, W., Zhang M., Zheng S., Li Y., Li X., Li W., Li G., Lin Z., Xie Z., Zhao Z., and Lou H.. 2015. Trapping toxins within lipid droplets is a resistance mechanism in fungi. Sci. Rep. 5:15133. 10.1038/srep15133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charron, A.J., and Sibley L.D.. 2002. Host cells: mobilizable lipid resources for the intracellular parasite Toxoplasma gondii. J. Cell Sci. 115:3049–3059. 10.1242/jcs.115.15.3049 [DOI] [PubMed] [Google Scholar]
- Chen, S., Sorrentino R., Shimada K., Bulut Y., Doherty T.M., Crother T.R., and Arditi M.. 2008. Chlamydia pneumoniae-induced foam cell formation requires MyD88-dependent and -independent signaling and is reciprocally modulated by liver X receptor activation. J. Immunol. 181:7186–7193. 10.4049/jimmunol.181.10.7186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, X., Lu Y., Zhang Z., Wang J., Yang H., and Liu G.. 2015. Intercellular interplay between Sirt1 signalling and cell metabolism in immune cell biology. Immunology. 145:455–467. 10.1111/imm.12473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q., Gouilly J., Ferrat Y.J., Espino A., Glaziou Q., Cartron G., El Costa H., Al-Daccak R., and Jabrane-Ferrat N.. 2020. Metabolic reprogramming by Zika virus provokes inflammation in human placenta. Nat. Commun. 11:2967. 10.1038/s41467-020-16754-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocchiaro, J.L., Kumar Y., Fischer E.R., Hackstadt T., and Valdivia R.H.. 2008. Cytoplasmic lipid droplets are translocated into the lumen of the Chlamydia trachomatis parasitophorous vacuole. Proc. Natl. Acad. Sci. USA. 105:9379–9384. 10.1073/pnas.0712241105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppens, I.2006. Contribution of host lipids to Toxoplasma pathogenesis. Cell. Microbiol. 8:1–9. 10.1111/j.1462-5822.2005.00647.x [DOI] [PubMed] [Google Scholar]
- Crosse, K.M., Monson E.A., Dumbrepatil A.B., Smith M., Tseng Y.Y., Van der Hoek K.H., Revill P.A., Saker S., Tscharke D.C., Marsh E.N.G. , et al. 2020. Viperin binds STING and enhances the type-I interferon response following dsDNA detection. Immunol. Cell Biol. [DOI] [PubMed] [Google Scholar]
- Daniel, J., Maamar H., Deb C., Sirakova T.D., and Kolattukudy P.E.. 2011. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 7:e1002093. 10.1371/journal.ppat.1002093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidi, L., Levin Y., Ben-Dor S., and Pick U.. 2015. Proteome analysis of cytoplasmatic and plastidic β-carotene lipid droplets in Dunaliella bardawil. Plant Physiol. 167:60–79. 10.1104/pp.114.248450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Mattos, K.A., Sarno E.N., Pessolani M.C., and Bozza P.T.. 2012. Deciphering the contribution of lipid droplets in leprosy: multifunctional organelles with roles in Mycobacterium leprae pathogenesis. Mem. Inst. Oswaldo Cruz. 107(suppl 1):156–166. 10.1590/S0074-02762012000900023 [DOI] [PubMed] [Google Scholar]
- Dhiman, R., Caesar S., Thiam A.R., and Schrul B.. 2020. Mechanisms of protein targeting to lipid droplets: A unified cell biological and biophysical perspective. Semin. Cell Dev. Biol. 108:4–13. 10.1016/j.semcdb.2020.03.004 [DOI] [PubMed] [Google Scholar]
- Dichlberger, A., Schlager S., Lappalainen J., Käkelä R., Hattula K., Butcher S.J., Schneider W.J., and Kovanen P.T.. 2011. Lipid body formation during maturation of human mast cells. J. Lipid Res. 52:2198–2208. 10.1194/jlr.M019737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichlberger, A., Schlager S., Maaninka K., Schneider W.J., and Kovanen P.T.. 2014. Adipose triglyceride lipase regulates eicosanoid production in activated human mast cells. J. Lipid Res. 55:2471–2478. 10.1194/jlr.M048553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhrmann, S., LaRock C.N., Anderson E.L., Cole J.N., Ryali B., Stewart C., Nonejuie P., Pogliano J., Corriden R., Ghosh P., and Nizet V.. 2017. Group A Streptococcal M1 Protein Provides Resistance against the Antimicrobial Activity of Histones. Sci. Rep. 7:43039. 10.1038/srep43039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolin, T., Amir H.M., Duong L., Rosenzweig R., Urban L.A., Bosch M., Pol A., Gross S.P., and Siryaporn A.. 2020. Mammalian histones facilitate antimicrobial synergy by disrupting the bacterial proton gradient and chromosome organization. Nat. Commun. 11:3888. 10.1038/s41467-020-17699-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey, R., Stivala C.E., Nguyen H.Q., Goo Y.H., Paul A., Carette J.E., Trost B.M., and Rohatgi R.. 2020. Lipid droplets can promote drug accumulation and activation. Nat. Chem. Biol. 16:206–213. 10.1038/s41589-019-0447-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumbrepatil, A.B., Ghosh S., Zegalia K.A., Malec P.A., Hoff J.D., Kennedy R.T., and Marsh E.N.G.. 2019. Viperin interacts with the kinase IRAK1 and the E3 ubiquitin ligase TRAF6, coupling innate immune signaling to antiviral ribonucleotide synthesis. J. Biol. Chem. 294:6888–6898. 10.1074/jbc.RA119.007719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duong, L., Gross S.P., and Siryaporn A.. 2021. Developing Antimicrobial Synergy With AMPs. Frontiers in Medical Technology. 3. [DOI] [PMC free article] [PubMed]
- Dvorak, A.M., Morgan E.S., and Weller P.F.. 2003. RNA is closely associated with human mast cell lipid bodies. Histol. Histopathol. 18:943–968. [DOI] [PubMed] [Google Scholar]
- Egan, J.J., Greenberg A.S., Chang M.K., Wek S.A., Moos M.C. Jr., and Londos C.. 1992. Mechanism of hormone-stimulated lipolysis in adipocytes: translocation of hormone-sensitive lipase to the lipid storage droplet. Proc. Natl. Acad. Sci. USA. 89:8537–8541. 10.1073/pnas.89.18.8537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers, S., and Schaible U.E.. 2013. The granuloma in tuberculosis: dynamics of a host-pathogen collusion. Front. Immunol. 3:411. 10.3389/fimmu.2012.00411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelmann, I., and Pujol N.. 2010. Innate immunity in C. elegans. Adv. Exp. Med. Biol. 708:105–121. 10.1007/978-1-4419-8059-5_6 [DOI] [PubMed] [Google Scholar]
- Fabisiak, A., Murawska N., and Fichna J.. 2016. LL-37: Cathelicidin-related antimicrobial peptide with pleiotropic activity. Pharmacol. Rep. 68:802–808. 10.1016/j.pharep.2016.03.015 [DOI] [PubMed] [Google Scholar]
- Falagas, M.E., and Kompoti M.. 2006. Obesity and infection. Lancet Infect. Dis. 6:438–446. 10.1016/S1473-3099(06)70523-0 [DOI] [PubMed] [Google Scholar]
- Faustino, A.F., Carvalho F.A., Martins I.C., Castanho M.A., Mohana-Borges R., Almeida F.C., Da Poian A.T., and Santos N.C.. 2014. Dengue virus capsid protein interacts specifically with very low-density lipoproteins. Nanomedicine (Lond.). 10:247–255. 10.1016/j.nano.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Fernández, M.A., Albor C., Ingelmo-Torres M., Nixon S.J., Ferguson C., Kurzchalia T., Tebar F., Enrich C., Parton R.G., and Pol A.. 2006. Caveolin-1 is essential for liver regeneration. Science. 313:1628–1632. 10.1126/science.1130773 [DOI] [PubMed] [Google Scholar]
- Filipe, A., and McLauchlan J.. 2015. Hepatitis C virus and lipid droplets: finding a niche. Trends Mol. Med. 21:34–42. 10.1016/j.molmed.2014.11.003 [DOI] [PubMed] [Google Scholar]
- Fukuda, E.Y., Lad S.P., Mikolon D.P., Iacobelli-Martinez M., and Li E.. 2005. Activation of lipid metabolism contributes to interleukin-8 production during Chlamydia trachomatis infection of cervical epithelial cells. Infect. Immun. 73:4017–4024. 10.1128/IAI.73.7.4017-4024.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo-Montejano, V.I., Saxena G., Kusminski C.M., Yang C., McAfee J.L., Hahner L., Hoch K., Dubinsky W., Narkar V.A., and Bickel P.E.. 2016. Nuclear Perilipin 5 integrates lipid droplet lipolysis with PGC-1α/SIRT1-dependent transcriptional regulation of mitochondrial function. Nat. Commun. 7:12723. 10.1038/ncomms12723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartung, A., Zhao J., Chen S., Mottillo E., VanHecke G.C., Ahn Y.H., Maddipati K.R., Sorokin A., Granneman J., and Lee M.J.. 2016. Characterization of Eicosanoids Produced by Adipocyte Lipolysis: IMPLICATION OF CYCLOOXYGENASE-2 IN ADIPOSE INFLAMMATION. J. Biol. Chem. 291:16001–16010. 10.1074/jbc.M116.725937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassen, N.C., Niemeyer D., Muth D., Corman V.M., Martinelli S., Gassen A., Hafner K., Papies J., Mösbauer K., Zellner A., et al. 2019. SKP2 attenuates autophagy through Beclin1-ubiquitination and its inhibition reduces MERS-Coronavirus infection. Nat. Commun. 10:5770. 10.1038/s41467-019-13659-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geltinger, F., Schartel L., Wiederstein M., Tevini J., Aigner E., Felder T.K., and Rinnerthaler M.. 2020. Friend or Foe: Lipid Droplets as Organelles for Protein and Lipid Storage in Cellular Stress Response, Aging and Disease. Molecules. 25:5053. 10.3390/molecules25215053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gizzi, A.S., Grove T.L., Arnold J.J., Jose J., Jangra R.K., Garforth S.J., Du Q., Cahill S.M., Dulyaninova N.G., Love J.D., et al. 2018. A naturally occurring antiviral ribonucleotide encoded by the human genome. Nature. 558:610–614. 10.1038/s41586-018-0238-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, A.S., Egan J.J., Wek S.A., Garty N.B., Blanchette-Mackie E.J., and Londos C.. 1991. Perilipin, a major hormonally regulated adipocyte-specific phosphoprotein associated with the periphery of lipid storage droplets. J. Biol. Chem. 266:11341–11346. 10.1016/S0021-9258(18)99168-4 [DOI] [PubMed] [Google Scholar]
- Greenwood, D.J., Dos Santos M.S., Huang S., Russell M.R.G., Collinson L.M., MacRae J.I., West A., Jiang H., and Gutierrez M.G.. 2019. Subcellular antibiotic visualization reveals a dynamic drug reservoir in infected macrophages. Science. 364:1279–1282. 10.1126/science.aat9689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunkemeyer, T.J., Ghosh S., Patel A.M., Sajja K., Windak J., Basrur V., Kim Y., Nesvizhskii A.I., Kennedy R.T., and Marsh E.N.G.. 2021. Viperin inhibits cholesterol biosynthesis and interacts with enzymes in the cholesterol biosynthetic pathway. bioRxiv. 10.1101/2021.02.19.431989 (Preprint posted February 19, 2021) [DOI]
- Heaton, N.S., and Randall G.. 2010. Dengue virus-induced autophagy regulates lipid metabolism. Cell Host Microbe. 8:422–432. 10.1016/j.chom.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton, N.S., Perera R., Berger K.L., Khadka S., Lacount D.J., Kuhn R.J., and Randall G.. 2010. Dengue virus nonstructural protein 3 redistributes fatty acid synthase to sites of viral replication and increases cellular fatty acid synthesis. Proc. Natl. Acad. Sci. USA. 107:17345–17350. 10.1073/pnas.1010811107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig, K.J., Eyre N.S., Yip E., Narayana S., Li K., Fiches G., McCartney E.M., Jangra R.K., Lemon S.M., and Beard M.R.. 2011. The antiviral protein viperin inhibits hepatitis C virus replication via interaction with nonstructural protein 5A. Hepatology. 54:1506–1517. 10.1002/hep.24542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helbig, K.J., Carr J.M., Calvert J.K., Wati S., Clarke J.N., Eyre N.S., Narayana S.K., Fiches G.N., McCartney E.M., and Beard M.R.. 2013. Viperin is induced following dengue virus type-2 (DENV-2) infection and has anti-viral actions requiring the C-terminal end of viperin. PLoS Negl. Trop. Dis. 7:e2178. 10.1371/journal.pntd.0002178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne, W.M., Reese M.L., and Goodman J.M.. 2018. The assembly of lipid droplets and their roles in challenged cells. EMBO J. 37:e98947. 10.15252/embj.201898947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herker, E., Harris C., Hernandez C., Carpentier A., Kaehlcke K., Rosenberg A.R., Farese R.V. Jr., and Ott M.. 2010. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat. Med. 16:1295–1298. 10.1038/nm.2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson, E.R., and Cresswell P.. 2009a. The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic alpha-helix. Proc. Natl. Acad. Sci. USA. 106:20452–20457. 10.1073/pnas.0911679106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinson, E.R., and Cresswell P.. 2009b. The N-terminal amphipathic alpha-helix of viperin mediates localization to the cytosolic face of the endoplasmic reticulum and inhibits protein secretion. J. Biol. Chem. 284:4705–4712. 10.1074/jbc.M807261200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, J.G.1958. Bactericidal action of histone. J. Exp. Med. 108:925–944. 10.1084/jem.108.6.925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, S., Dong X., Yang Z., Li Z., Liu Q., and Zhong G.. 2015. Chlamydial plasmid-encoded virulence factor Pgp3 neutralizes the antichlamydial activity of human cathelicidin LL-37. Infect. Immun. 83:4701–4709. 10.1128/IAI.00746-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, N.L., Huang M.D., Chen T.L., and Huang A.H.. 2013. Oleosin of subcellular lipid droplets evolved in green algae. Plant Physiol. 161:1862–1874. 10.1104/pp.112.212514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilnytska, O., Santiana M., Hsu N.Y., Du W.L., Chen Y.H., Viktorova E.G., Belov G., Brinker A., Storch J., Moore C., et al. 2013. Enteroviruses harness the cellular endocytic machinery to remodel the host cell cholesterol landscape for effective viral replication. Cell Host Microbe. 14:281–293. 10.1016/j.chom.2013.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im, S.S., Yousef L., Blaschitz C., Liu J.Z., Edwards R.A., Young S.G., Raffatellu M., and Osborne T.F.. 2011. Linking lipid metabolism to the innate immune response in macrophages through sterol regulatory element binding protein-1a. Cell Metab. 13:540–549. 10.1016/j.cmet.2011.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki, A., and Medzhitov R.. 2015. Control of adaptive immunity by the innate immune system. Nat. Immunol. 16:343–353. 10.1038/ni.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaisinghani, N., Dawa S., Singh K., Nandy A., Menon D., Bhandari P.D., Khare G., Tyagi A., and Gandotra S.. 2018. Necrosis Driven Triglyceride Synthesis Primes Macrophages for Inflammation During Mycobacterium tuberculosis Infection. Front. Immunol. 9:1490. 10.3389/fimmu.2018.01490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jallet, D., Caballero M.A., Gallina A.A., Youngblood M., and Peers G.. 2016. Photosynthetic physiology and biomass partitioning in the model diatom Phaeodactylum tricornutum grown in a sinusoidal light regime. Algal Res. 18:51–60. 10.1016/j.algal.2016.05.014 [DOI] [Google Scholar]
- Jarc, E., and Petan T.. 2020. A twist of FATe: Lipid droplets and inflammatory lipid mediators. Biochimie. 169:69–87. 10.1016/j.biochi.2019.11.016 [DOI] [PubMed] [Google Scholar]
- Jaussaud, A., Lupette J., Salvaing J., Jouhet J., Bastien O., Gromova M., and Maréchal E.. 2020. Stepwise Biogenesis of Subpopulations of Lipid Droplets in Nitrogen Starved Phaeodactylum tricornutum Cells. Front Plant Sci. 11:48. 10.3389/fpls.2020.00048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassan, A., Herms A., Fernández-Vidal A., Bosch M., Schieber N.L., Reddy B.J., Fajardo A., Gelabert-Baldrich M., Tebar F., Enrich C., et al. 2013. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J. Cell Biol. 203:985–1001. 10.1083/jcb.201305142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik, S., and Cuervo A.M.. 2015. Degradation of lipid droplet-associated proteins by chaperone-mediated autophagy facilitates lipolysis. Nat. Cell Biol. 17:759–770. 10.1038/ncb3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushik, S., and Cuervo A.M.. 2016. AMPK-dependent phosphorylation of lipid droplet protein PLIN2 triggers its degradation by CMA. Autophagy. 12:432–438. 10.1080/15548627.2015.1124226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketter, E., and Randall G.. 2019. Virus Impact on Lipids and Membranes. Annu. Rev. Virol. 6:319–340. 10.1146/annurev-virology-092818-015748 [DOI] [PubMed] [Google Scholar]
- Kim, M.J., Wainwright H.C., Locketz M., Bekker L.G., Walther G.B., Dittrich C., Visser A., Wang W., Hsu F.F., Wiehart U., et al. 2010. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol. Med. 2:258–274. 10.1002/emmm.201000079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.S., Lee H.M., Kim J.K., Yang C.S., Kim T.S., Jung M., Jin H.S., Kim S., Jang J., Oh G.T., et al. 2017. PPAR-α Activation Mediates Innate Host Defense through Induction of TFEB and Lipid Catabolism. J. Immunol. 198:3283–3295. 10.4049/jimmunol.1601920 [DOI] [PubMed] [Google Scholar]
- Kimmel, A.R., and Sztalryd C.. 2016. The Perilipins: Major Cytosolic Lipid Droplet-Associated Proteins and Their Roles in Cellular Lipid Storage, Mobilization, and Systemic Homeostasis. Annu. Rev. Nutr. 36:471–509. 10.1146/annurev-nutr-071813-105410 [DOI] [PubMed] [Google Scholar]
- Klemm, R.W., and Ikonen E.. 2020. The cell biology of lipid droplets: More than just a phase. Semin. Cell Dev. Biol. 108:1–3. 10.1016/j.semcdb.2020.06.016 [DOI] [PubMed] [Google Scholar]
- Knight, M., Braverman J., Asfaha K., Gronert K., and Stanley S.. 2018. Lipid droplet formation in Mycobacterium tuberculosis infected macrophages requires IFN-γ/HIF-1α signaling and supports host defense. PLoS Pathog. 14:e1006874. 10.1371/journal.ppat.1006874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, Y., Cocchiaro J., and Valdivia R.H.. 2006. The obligate intracellular pathogen Chlamydia trachomatis targets host lipid droplets. Curr. Biol. 16:1646–1651. 10.1016/j.cub.2006.06.060 [DOI] [PubMed] [Google Scholar]
- Kuo, A., Lee M.Y., Yang K., Gross R.W., and Sessa W.C.. 2018. Caveolin-1 regulates lipid droplet metabolism in endothelial cells via autocrine prostacyclin-stimulated, cAMP-mediated lipolysis. J. Biol. Chem. 293:973–983. 10.1074/jbc.RA117.000980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock, C.N., Döhrmann S., Todd J., Corriden R., Olson J., Johannssen T., Lepenies B., Gallo R.L., Ghosh P., and Nizet V.. 2015. Group A Streptococcal M1 Protein Sequesters Cathelicidin to Evade Innate Immune Killing. Cell Host Microbe. 18:471–477. 10.1016/j.chom.2015.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lass, A., Zimmermann R., Haemmerle G., Riederer M., Schoiswohl G., Schweiger M., Kienesberger P., Strauss J.G., Gorkiewicz G., and Zechner R.. 2006. Adipose triglyceride lipase-mediated lipolysis of cellular fat stores is activated by CGI-58 and defective in Chanarin-Dorfman Syndrome. Cell Metab. 3:309–319. 10.1016/j.cmet.2006.03.005 [DOI] [PubMed] [Google Scholar]
- Laufman, O., Perrino J., and Andino R.. 2019. Viral Generated Inter-Organelle Contacts Redirect Lipid Flux for Genome Replication. Cell. 178:275–289.e16. 10.1016/j.cell.2019.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laval, T., Chaumont L., and Demangel C.. 2021. Not too fat to fight: The emerging role of macrophage fatty acid metabolism in immunity to Mycobacterium tuberculosis. Immunol. Rev. 301:84–97. 10.1111/imr.12952 [DOI] [PubMed] [Google Scholar]
- Lazzaro, B.P., Zasloff M., and Rolff J.. 2020. Antimicrobial peptides: Application informed by evolution. Science. 368:eaau5480. 10.1126/science.aau5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, J.Y., Cortese M., Haselmann U., Tabata K., Romero-Brey I., Funaya C., Schieber N.L., Qiang Y., Bartenschlager M., Kallis S., et al. 2019. Spatiotemporal Coupling of the Hepatitis C Virus Replication Cycle by Creating a Lipid Droplet- Proximal Membranous Replication Compartment. Cell Rep. 27:3602–3617.e5. 10.1016/j.celrep.2019.05.063 [DOI] [PubMed] [Google Scholar]
- Leyland, B., Boussiba S., and Khozin-Goldberg I.. 2020. A Review of Diatom Lipid Droplets. Biology (Basel). 9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Thiel K., Thul P.J., Beller M., Kühnlein R.P., and Welte M.A.. 2012. Lipid droplets control the maternal histone supply of Drosophila embryos. Curr. Biol. 22:2104–2113. 10.1016/j.cub.2012.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbing, C.L., McDevitt A.R., Azcueta R.P., Ahila A., and Mulye M.. 2019. Lipid Droplets: A Significant but Understudied Contributor of Host−Bacterial Interactions. Cells. 8:354. 10.3390/cells8040354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lige, B., Sampels V., and Coppens I.. 2013. Characterization of a second sterol-esterifying enzyme in Toxoplasma highlights the importance of cholesterol storage pathways for the parasite. Mol. Microbiol. 87:951–967. 10.1111/mmi.12142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima, J.G., Nobrega L.H.C., Lima N.N., Dos Santos M.C.F., Silva P.H.D., Baracho M.F.P., Lima D.N., de Melo Campos J.T.A., Ferreira L.C., Freire Neto F.P., et al. 2018. Causes of death in patients with Berardinelli-Seip congenital generalized lipodystrophy. PLoS One. 13:e0199052. 10.1371/journal.pone.0199052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen, C.J., Gaspar M.L., Jesch S.A., Delon C., Ktistakis N.T., Henry S.A., and Levine T.P.. 2004. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 304:1644–1647. 10.1126/science.1096083 [DOI] [PubMed] [Google Scholar]
- Lugrin, J., and Martinon F.. 2018. The AIM2 inflammasome: Sensor of pathogens and cellular perturbations. Immunol. Rev. 281:99–114. 10.1111/imr.12618 [DOI] [PubMed] [Google Scholar]
- Lundquist, P.K., Shivaiah K.K., and Espinoza-Corral R.. 2020. Lipid droplets throughout the evolutionary tree. Prog. Lipid Res. 78:101029. 10.1016/j.plipres.2020.101029 [DOI] [PubMed] [Google Scholar]
- Martin, L., van Meegern A., Doemming S., and Schuerholz T.. 2015. Antimicrobial Peptides in Human Sepsis. Front. Immunol. 6:404. 10.3389/fimmu.2015.00404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins, A.S., Carvalho F.A., Faustino A.F., Martins I.C., and Santos N.C.. 2019. West Nile Virus Capsid Protein Interacts With Biologically Relevant Host Lipid Systems. Front. Cell. Infect. Microbiol. 9:8. 10.3389/fcimb.2019.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, B.F., Abrams R., Dorfman A., and Klein M.. 1942. Antibacterial Properties of Protamine and Histone. Science. 96:428–430. 10.1126/science.96.2497.428 [DOI] [PubMed] [Google Scholar]
- Miyanari, Y., Atsuzawa K., Usuda N., Watashi K., Hishiki T., Zayas M., Bartenschlager R., Wakita T., Hijikata M., and Shimotohno K.. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097. 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- Molaei, M., Vandehoef C., and Karpac J.. 2019. NF-κB Shapes Metabolic Adaptation by Attenuating Foxo-Mediated Lipolysis in Drosophila. Dev. Cell. 49:802–810.e6. 10.1016/j.devcel.2019.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson, E.A., Crosse K.M., Das M., and Helbig K.J.. 2018. Lipid droplet density alters the early innate immune response to viral infection. PLoS One. 13:e0190597. 10.1371/journal.pone.0190597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monson, E., Crosse K., Duan M., Chen W., O’Shea R., Wakim L., Whelan D., and Helbig K.. 2020. Intracellular Lipid Droplet Accumulation Occurs Early Following Viral Infection and Is Required for an Efficient Interferon Response. bioRxiv. 2020.02.12.946749. 10.1101/2020.02.12.946749 [DOI] [PMC free article] [PubMed]
- Monson, E.A., Trenerry A.M., Laws J.L., Mackenzie J.M., and Helbig K.J.. 2021. Lipid droplets and lipid mediators in viral infection and immunity. FEMS Microbiol. Rev. 10.1093/femsre/fuaa066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulye, M., Zapata B., and Gilk S.D.. 2018. Altering lipid droplet homeostasis affects Coxiella burnetii intracellular growth. PLoS One. 13:e0192215. 10.1371/journal.pone.0192215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro, E., Atilla-Gokcumen G.E., and Eggert U.S.. 2014. Lipids in cell biology: how can we understand them better? Mol. Biol. Cell. 25:1819–1823. 10.1091/mbc.e13-09-0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mylonis, I., Simos G., and Paraskeva E.. 2019. Hypoxia-Inducible Factors and the Regulation of Lipid Metabolism. Cells. 8:214. 10.3390/cells8030214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najt, C.P., Khan S.A., Heden T.D., Witthuhn B.A., Perez M., Heier J.L., Mead L.E., Franklin M.P., Karanja K.K., Graham M.J., et al. 2020. Lipid Droplet-Derived Monounsaturated Fatty Acids Traffic via PLIN5 to Allosterically Activate SIRT1. Mol. Cell. 77:810–824.e8. 10.1016/j.molcel.2019.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, G., Ma Z., and Damania B.. 2018. cGAS and STING: At the intersection of DNA and RNA virus-sensing networks. PLoS Pathog. 14:e1007148. 10.1371/journal.ppat.1007148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolaou, G., Goodall A.H., and Erridge C.. 2012. Diverse bacteria promote macrophage foam cell formation via Toll-like receptor-dependent lipid body biosynthesis. J. Atheroscler. Thromb. 19:137–148. 10.5551/jat.10249 [DOI] [PubMed] [Google Scholar]
- Nolan, S.J., Romano J.D., and Coppens I.. 2017. Host lipid droplets: An important source of lipids salvaged by the intracellular parasite Toxoplasma gondii. PLoS Pathog. 13:e1006362. 10.1371/journal.ppat.1006362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura, D.K., Morrison B.E., Blankman J.L., Long J.Z., Kinsey S.G., Marcondes M.C., Ward A.M., Hahn Y.K., Lichtman A.H., Conti B., and Cravatt B.F.. 2011. Endocannabinoid hydrolysis generates brain prostaglandins that promote neuroinflammation. Science. 334:809–813. 10.1126/science.1209200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill, L.A.2015. How Low Cholesterol Is Good for Anti-viral Immunity. Cell. 163:1572–1574. 10.1016/j.cell.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Olzmann, J.A., and Carvalho P.. 2019. Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol. 20:137–155. 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten, E.G., Werner E., Crespillo-Casado A., Boyle K.B., Dharamdasani V., Pathe C., Santhanam B., and Randow F.. 2021. Ubiquitylation of lipopolysaccharide by RNF213 during bacterial infection. Nature. 594:111–116. 10.1038/s41586-021-03566-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnon, J., Matzaris M., Stark R., Meex R.C., Macaulay S.L., Brown W., O’Brien P.E., Tiganis T., and Watt M.J.. 2012. Identification and functional characterization of protein kinase A phosphorylation sites in the major lipolytic protein, adipose triglyceride lipase. Endocrinology. 153:4278–4289. 10.1210/en.2012-1127 [DOI] [PubMed] [Google Scholar]
- Paludan, S.R., Pradeu T., Masters S.L., and Mogensen T.H.. 2021. Constitutive immune mechanisms: mediators of host defence and immune regulation. Nat. Rev. Immunol. 21:137–150. 10.1038/s41577-020-0391-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, A.K., and Sassetti C.M.. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc. Natl. Acad. Sci. USA. 105:4376–4380. 10.1073/pnas.0711159105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, J., and Byrne G.I.. 2015. Chlamydia trachomatis growth depends on eukaryotic cholesterol esterification and is affected by Acyl-CoA:cholesterol acyltransferase inhibition. Pathog. Dis. 73:ftv028. 10.1093/femspd/ftv028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyron, P., Vaubourgeix J., Poquet Y., Levillain F., Botanch C., Bardou F., Daffé M., Emile J.F., Marchou B., Cardona P.J., et al. 2008. Foamy macrophages from tuberculous patients’ granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 4:e1000204. 10.1371/journal.ppat.1000204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol, A., Gross S.P., and Parton R.G.. 2014. Review: biogenesis of the multifunctional lipid droplet: lipids, proteins, and sites. J. Cell Biol. 204:635–646. 10.1083/jcb.201311051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak, N.M., Schweiger M., Jaeger D., Kolb D., Kumari M., Schreiber R., Kolleritsch S., Markolin P., Grabner G.F., Heier C., et al. 2013. Cardiac-specific overexpression of perilipin 5 provokes severe cardiac steatosis via the formation of a lipolytic barrier. J. Lipid Res. 54:1092–1102. 10.1194/jlr.M034710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabhi, I., Rabhi S., Ben-Othman R., Rasche A., Daskalaki A., Trentin B., Piquemal D., Regnault B., Descoteaux A., Guizani-Tabbane L., and Sysco C.. Sysco Consortium . 2012. Transcriptomic signature of Leishmania infected mice macrophages: a metabolic point of view. PLoS Negl. Trop. Dis. 6:e1763. 10.1371/journal.pntd.0001763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabhi, S., Rabhi I., Trentin B., Piquemal D., Regnault B., Goyard S., Lang T., Descoteaux A., Enninga J., and Guizani-Tabbane L.. 2016. Lipid Droplet Formation, Their Localization and Dynamics during Leishmania major Macrophage Infection. PLoS One. 11:e0148640. 10.1371/journal.pone.0148640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rank, R.G., Whittimore J., Bowlin A.K., and Wyrick P.B.. 2011. In vivo ultrastructural analysis of the intimate relationship between polymorphonuclear leukocytes and the chlamydial developmental cycle. Infect. Immun. 79:3291–3301. 10.1128/IAI.00200-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recuero-Checa, M.A., Sharma M., Lau C., Watkins P.A., Gaydos C.A., and Dean D.. 2016. Chlamydia trachomatis growth and development requires the activity of host Long-chain Acyl-CoA Synthetases (ACSLs). Sci. Rep. 6:23148. 10.1038/srep23148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renne, M.F., Klug Y.A., and Carvalho P.. 2020. Lipid droplet biogenesis: A mystery “unmixing”? Semin. Cell Dev. Biol. 108:14–23. 10.1016/j.semcdb.2020.03.001 [DOI] [PubMed] [Google Scholar]
- Rivera-Serrano, E.E., Gizzi A.S., Arnold J.J., Grove T.L., Almo S.C., and Cameron C.E.. 2020. Viperin Reveals Its True Function. Annu. Rev. Virol. 7:421–446. 10.1146/annurev-virology-011720-095930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogne, M., Chu D.T., Küntziger T.M., Mylonakou M.N., Collas P., and Tasken K.. 2018. OPA1-anchored PKA phosphorylates perilipin 1 on S522 and S497 in adipocytes differentiated from human adipose stem cells. Mol. Biol. Cell. 29:1487–1501. 10.1091/mbc.E17-09-0538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roingeard, P., and Melo R.C.. 2017. Lipid droplet hijacking by intracellular pathogens. Cell. Microbiol. 19:e12688. 10.1111/cmi.12688 [DOI] [PubMed] [Google Scholar]
- Russell, C.D., and Schwarze J.. 2014. The role of pro-resolution lipid mediators in infectious disease. Immunology. 141:166–173. 10.1111/imm.12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu-Osen, A., Montero-Moran G., Schittmayer M., Fritz K., Dinh A., Chang Y.F., McMahon D., Boeszoermenyi A., Cornaciu I., Russell D., et al. 2015. CGI-58/ABHD5 is phosphorylated on Ser239 by protein kinase A: control of subcellular localization. J. Lipid Res. 56:109–121. 10.1194/jlr.M055004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh, T., Satoh T., Yamamoto N., Uematsu S., Takeuchi O., Kawai T., and Akira S.. 2011. Antiviral protein Viperin promotes Toll-like receptor 7- and Toll-like receptor 9-mediated type I interferon production in plasmacytoid dendritic cells. Immunity. 34:352–363. 10.1016/j.immuni.2011.03.010 [DOI] [PubMed] [Google Scholar]
- Saka, H.A., Thompson J.W., Chen Y.S., Dubois L.G., Haas J.T., Moseley A., and Valdivia R.H.. 2015. Chlamydia trachomatis Infection Leads to Defined Alterations to the Lipid Droplet Proteome in Epithelial Cells. PLoS One. 10:e0124630. 10.1371/journal.pone.0124630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salloum, S., Wang H., Ferguson C., Parton R.G., and Tai A.W.. 2013. Rab18 binds to hepatitis C virus NS5A and promotes interaction between sites of viral replication and lipid droplets. PLoS Pathog. 9:e1003513. 10.1371/journal.ppat.1003513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen, T.S., and Vale P.F.. 2020. Drosophila as a Model System to Investigate the Effects of Mitochondrial Variation on Innate Immunity. Front. Immunol. 11:521. 10.3389/fimmu.2020.00521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo, V.T., Li S., Vihinen H., Hölttä-Vuori M., Szkalisity A., Horvath P., Belevich I., Peränen J., Thiele C., Somerharju P., et al. 2019. Seipin Facilitates Triglyceride Flow to Lipid Droplet and Counteracts Droplet Ripening via Endoplasmic Reticulum Contact. Dev. Cell. 50:478–493.e9. 10.1016/j.devcel.2019.05.016 [DOI] [PubMed] [Google Scholar]
- Saltiel, A.R., and Olefsky J.M.. 2017. Inflammatory mechanisms linking obesity and metabolic disease. J. Clin. Invest. 127:1–4. 10.1172/JCI92035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoz, K.M., Valiant W.G., Eriksen S.G., Hruby D.E., Allen R.D. III, and Rockey D.D.. 2014. The broad-spectrum antiviral compound ST-669 restricts chlamydial inclusion development and bacterial growth and localizes to host cell lipid droplets within treated cells. Antimicrob. Agents Chemother. 58:3860–3866. 10.1128/AAC.02064-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sant’Anna, C., Nakayasu E.S., Pereira M.G., Lourenço D., de Souza W., Almeida I.C., and Cunha-E-Silva N.L.. 2009. Subcellular proteomics of Trypanosoma cruzi reservosomes. Proteomics. 9:1782–1794. 10.1002/pmic.200800730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffler, A., and Schölmerich J.. 2010. Innate immunity and adipose tissue biology. Trends Immunol. 31:228–235. 10.1016/j.it.2010.03.001 [DOI] [PubMed] [Google Scholar]
- Schneider, W.M., Chevillotte M.D., and Rice C.M.. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32:513–545. 10.1146/annurev-immunol-032713-120231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze, R.J., Sathyanarayan A., and Mashek D.G.. 2017. Breaking fat: The regulation and mechanisms of lipophagy. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862(10 pt B):1178–1187. 10.1016/j.bbalip.2017.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkal, C.E., Salama M.F., Snider A.J., Allopenna J.J., Rana N.A., Koller A., Hannun Y.A., and Obeid L.M.. 2017. Ceramide Is Metabolized to Acylceramide and Stored in Lipid Droplets. Cell Metab. 25:686–697. 10.1016/j.cmet.2017.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severa, M., Coccia E.M., and Fitzgerald K.A.. 2006. Toll-like receptor-dependent and -independent viperin gene expression and counter-regulation by PRDI-binding factor-1/BLIMP1. J. Biol. Chem. 281:26188–26195. 10.1074/jbc.M604516200 [DOI] [PubMed] [Google Scholar]
- Sharma, M., Recuero-Checa M.A., Fan F.Y., and Dean D.. 2018. Chlamydia trachomatis regulates growth and development in response to host cell fatty acid availability in the absence of lipid droplets. Cell. Microbiol. 20:e12801. 10.1111/cmi.12801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn, J.H., Lee Y.K., Han J.S., Jeon Y.G., Kim J.I., Choe S.S., Kim S.J., Yoo H.J., and Kim J.B.. 2018. Perilipin 1 (Plin1) deficiency promotes inflammatory responses in lean adipose tissue through lipid dysregulation. J. Biol. Chem. 293:13974–13988. 10.1074/jbc.RA118.003541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene, E., and Kuypers F.A.. 2017. Phosphatidylserine decarboxylase CT699, lysophospholipid acyltransferase CT775, and acyl-ACP synthase CT776 provide membrane lipid diversity to Chlamydia trachomatis. Sci. Rep. 7:15767. 10.1038/s41598-017-16116-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene, E., Rothschild J., Kuypers F.A., and Dean D.. 2012. Eukaryotic protein recruitment into the Chlamydia inclusion: implications for survival and growth. PLoS One. 7:e36843. 10.1371/journal.pone.0036843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soupene, E., Wang D., and Kuypers F.A.. 2015. Remodeling of host phosphatidylcholine by Chlamydia acyltransferase is regulated by acyl-CoA binding protein ACBD6 associated with lipid droplets. MicrobiologyOpen. 4:235–251. 10.1002/mbo3.234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks, C.J., Schembri M.A., Sweet M.J., and Kapetanovic R.. 2018. For when bacterial infections persist: Toll-like receptor-inducible direct antimicrobial pathways in macrophages. J. Leukoc. Biol. 103:35–51. 10.1002/JLB.4RI0917-358R [DOI] [PubMed] [Google Scholar]
- Strating, J.R., and van Kuppeveld F.J.. 2017. Viral rewiring of cellular lipid metabolism to create membranous replication compartments. Curr. Opin. Cell Biol. 47:24–33. 10.1016/j.ceb.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strating, J.R., van der Linden L., Albulescu L., Bigay J., Arita M., Delang L., Leyssen P., van der Schaar H.M., Lanke K.H., Thibaut H.J., et al. 2015. Itraconazole inhibits enterovirus replication by targeting the oxysterol-binding protein. Cell Rep. 10:600–615. 10.1016/j.celrep.2014.12.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian, V., Rothenberg A., Gomez C., Cohen A.W., Garcia A., Bhattacharyya S., Shapiro L., Dolios G., Wang R., Lisanti M.P., and Brasaemle D.L.. 2004. Perilipin A mediates the reversible binding of CGI-58 to lipid droplets in 3T3-L1 adipocytes. J. Biol. Chem. 279:42062–42071. 10.1074/jbc.M407462200 [DOI] [PubMed] [Google Scholar]
- Sugihara, M., Morito D., Ainuki S., Hirano Y., Ogino K., Kitamura A., Hirata H., and Nagata K.. 2019. The AAA+ ATPase/ubiquitin ligase mysterin stabilizes cytoplasmic lipid droplets. J. Cell Biol. 218:949–960. 10.1083/jcb.201712120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztalryd, C., and Brasaemle D.L.. 2017. The perilipin family of lipid droplet proteins: Gatekeepers of intracellular lipolysis. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862(10 pt B):1221–1232. 10.1016/j.bbalip.2017.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztalryd, C., Xu G., Dorward H., Tansey J.T., Contreras J.A., Kimmel A.R., and Londos C.. 2003. Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J. Cell Biol. 161:1093–1103. 10.1083/jcb.200210169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, C., Aziz M., and Wang P.. 2020. The vitals of NETs. J. Leukoc. Biol. 10.1002/JLB.3RU0620-375R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, W.C., Lin R.J., Liao C.L., and Lin Y.L.. 2014. Rab18 facilitates dengue virus infection by targeting fatty acid synthase to sites of viral replication. J. Virol. 88:6793–6804. 10.1128/JVI.00045-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tansey, J.T., Huml A.M., Vogt R., Davis K.E., Jones J.M., Fraser K.A., Brasaemle D.L., Kimmel A.R., and Londos C.. 2003. Functional studies on native and mutated forms of perilipins. A role in protein kinase A-mediated lipolysis of triacylglycerols. J. Biol. Chem. 278:8401–8406. 10.1074/jbc.M211005200 [DOI] [PubMed] [Google Scholar]
- Taurino, M., Costantini S., De Domenico S., Stefanelli F., Ruano G., Delgadillo M.O., Sánchez-Serrano J.J., Sanmartín M., Santino A., and Rojo E.. 2018. SEIPIN Proteins Mediate Lipid Droplet Biogenesis to Promote Pollen Transmission and Reduce Seed Dormancy. Plant Physiol. 176:1531–1546. 10.1104/pp.17.01430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazi, A., Araujo J.R., Mulet C., Arena E.T., Nigro G., Pédron T., and Sansonetti P.J.. 2018. Disentangling Host-Microbiota Regulation of Lipid Secretion by Enterocytes: Insights from Commensals Lactobacillus paracasei and Escherichia coli. MBio. 9:e01493-18. 10.1128/mBio.01493-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong, N.T.T., Lydic T.A., Bazil J.N., Suryadevara A., and Olson L.K.. 2020. Regulation of lipid metabolism in pancreatic beta cells by interferon gamma: A link to anti-viral function. Cytokine. 133:155147. 10.1016/j.cyto.2020.155147 [DOI] [PubMed] [Google Scholar]
- Upadhyay, A.S., Vonderstein K., Pichlmair A., Stehling O., Bennett K.L., Dobler G., Guo J.T., Superti-Furga G., Lill R., Överby A.K., and Weber F.. 2014. Viperin is an iron-sulfur protein that inhibits genome synthesis of tick-borne encephalitis virus via radical SAM domain activity. Cell. Microbiol. 16:834–848. 10.1111/cmi.12241 [DOI] [PubMed] [Google Scholar]
- Vallochi, A.L., Teixeira L., Oliveira K.D.S., Maya-Monteiro C.M., and Bozza P.T.. 2018. Lipid Droplet, a Key Player in Host-Parasite Interactions. Front. Immunol. 9:1022. 10.3389/fimmu.2018.01022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoek, K.H., Eyre N.S., Shue B., Khantisitthiporn O., Glab-Ampi K., Carr J.M., Gartner M.J., Jolly L.A., Thomas P.Q., Adikusuma F., et al. 2017. Viperin is an important host restriction factor in control of Zika virus infection. Sci. Rep. 7:4475. 10.1038/s41598-017-04138-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese, M., Kimler V.A., Ghazi F.R., Rathore G.K., Perkins G.A., Ellisman M.H., and Granneman J.G.. 2019. Adipocyte lipolysis affects Perilipin 5 and cristae organization at the cardiac lipid droplet-mitochondrial interface. Sci. Rep. 9:4734. 10.1038/s41598-019-41329-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielemeyer, O., McIntosh M.T., Joiner K.A., and Coppens I.. 2004. Neutral lipid synthesis and storage in the intraerythrocytic stages of Plasmodium falciparum. Mol. Biochem. Parasitol. 135:197–209. 10.1016/j.molbiopara.2003.08.017 [DOI] [PubMed] [Google Scholar]
- Vieyres, G., and Pietschmann T.. 2019. HCV Pit Stop at the Lipid Droplet: Refuel Lipids and Put on a Lipoprotein Coat before Exit. Cells. 8:233. 10.3390/cells8030233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virchow, R.1867. Die krankhaften Geschwülste: dreissig Vorlesungen, gehalten während des Wintersemesters 1862-1863 an der Universität zu Berlin. Verlag von August Hirschwald, Berlin. [Google Scholar]
- Vogt, D.A., Camus G., Herker E., Webster B.R., Tsou C.L., Greene W.C., Yen T.S., and Ott M.. 2013. Lipid droplet-binding protein TIP47 regulates hepatitis C Virus RNA replication through interaction with the viral NS5A protein. PLoS Pathog. 9:e1003302. 10.1371/journal.ppat.1003302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wake, K.1974. Development of vitamin A-rich lipid droplets in multivesicular bodies of rat liver stellate cells. J. Cell Biol. 63:683–691. 10.1083/jcb.63.2.683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Hinson E.R., and Cresswell P.. 2007. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2:96–105. 10.1016/j.chom.2007.06.009 [DOI] [PubMed] [Google Scholar]
- Wang, H., Sreenivasan U., Hu H., Saladino A., Polster B.M., Lund L.M., Gong D.W., Stanley W.C., and Sztalryd C.. 2011. Perilipin 5, a lipid droplet-associated protein, provides physical and metabolic linkage to mitochondria. J. Lipid Res. 52:2159–2168. 10.1194/jlr.M017939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C., Zhao Y., Gao X., Li L., Yuan Y., Liu F., Zhang L., Wu J., Hu P., Zhang X., et al. 2015. Perilipin 5 improves hepatic lipotoxicity by inhibiting lipolysis. Hepatology. 61:870–882. 10.1002/hep.27409 [DOI] [PubMed] [Google Scholar]
- Wang, A., Huen S.C., Luan H.H., Yu S., Zhang C., Gallezot J.D., Booth C.J., and Medzhitov R.. 2016. Opposing Effects of Fasting Metabolism on Tissue Tolerance in Bacterial and Viral Inflammation. Cell. 166:1512–1525.e12. 10.1016/j.cell.2016.07.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte, M.A., and Gould A.P.. 2017. Lipid droplet functions beyond energy storage. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 1862(10 pt B):1260–1272. 10.1016/j.bbalip.2017.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whytock, K.L., Shepherd S.O., Wagenmakers A.J.M., and Strauss J.A.. 2018. Hormone-sensitive lipase preferentially redistributes to lipid droplets associated with perilipin-5 in human skeletal muscle during moderate-intensity exercise. J. Physiol. 596:2077–2090. 10.1113/JP275502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J., Zhang X., Monestier M., Esmon N.L., and Esmon C.T.. 2011. Extracellular histones are mediators of death through TLR2 and TLR4 in mouse fatal liver injury. J. Immunol. 187:2626–2631. 10.4049/jimmunol.1003930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, T., Omatsu N., Matsushita S., and Osumi T.. 2004. CGI-58 interacts with perilipin and is localized to lipid droplets. Possible involvement of CGI-58 mislocalization in Chanarin-Dorfman syndrome. J. Biol. Chem. 279:30490–30497. 10.1074/jbc.M403920200 [DOI] [PubMed] [Google Scholar]
- Yamasaki, K., Di Nardo A., Bardan A., Murakami M., Ohtake T., Coda A., Dorschner R.A., Bonnart C., Descargues P., Hovnanian A., et al. 2007. Increased serine protease activity and cathelicidin promotes skin inflammation in rosacea. Nat. Med. 13:975–980. 10.1038/nm1616 [DOI] [PubMed] [Google Scholar]
- Yao, J., Dodson V.J., Frank M.W., and Rock C.O.. 2015. Chlamydia trachomatis Scavenges Host Fatty Acids for Phospholipid Synthesis via an Acyl-Acyl Carrier Protein Synthetase. J. Biol. Chem. 290:22163–22173. 10.1074/jbc.M115.671008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- York, A.G., Williams K.J., Argus J.P., Zhou Q.D., Brar G., Vergnes L., Gray E.E., Zhen A., Wu N.C., Yamada D.H., et al. 2015. Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling. Cell. 163:1716–1729. 10.1016/j.cell.2015.11.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, C., and Liu P.. 2017. The lipid droplet: A conserved cellular organelle. Protein Cell. 8:796–800. 10.1007/s13238-017-0467-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L.J., Guerrero-Juarez C.F., Hata T., Bapat S.P., Ramos R., Plikus M.V., and Gallo R.L.. 2015. Innate immunity. Dermal adipocytes protect against invasive Staphylococcus aureus skin infection. Science. 347:67–71. 10.1126/science.1260972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, J., Lan Y., Li M.Y., Lamers M.M., Fusade-Boyer M., Klemm E., Thiele C., Ashour J., and Sanyal S.. 2018. Flaviviruses Exploit the Lipid Droplet Protein AUP1 to Trigger Lipophagy and Drive Virus Production. Cell Host Microbe. 23:819–831.e5. 10.1016/j.chom.2018.05.005 [DOI] [PubMed] [Google Scholar]
- Zhang, E., Najt C.P., Hu H., and Mashek D.G.. 2019. Hepatic perilipin 5 promotes lipophagy and alters lipid droplet and mitochondrial dynamics. FASEB J. 33:490.19-490.19.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists viruses related to LD formation and interaction.
lists bacteria related to LD formation and interaction.
lists protozoans related to LD formation and interaction.
lists selected publications to illustrate the multifaceted relationship of host lipids with pathogens.