Abstract
Bioengineers have designed numerous instructive brain extracellular matrix (ECM) environments with tailored and tunable protein compositions and biomechanical properties in vitro to study astrocyte reactivity during trauma and inflammation. However, a major limitation of both protein-based and synthetic model microenvironments is that astrocytes within fail to retain their characteristic stellate morphology and quiescent state without becoming activated under “normal” culture conditions. Here, a synthetic hydrogel is introduced, which for the first time demonstrates maintenance of astrocyte quiescence and activation on demand. With this synthetic brain hydrogel, the brain-specific integrin-binding and matrix metalloprotease-degradable domains of proteins are shown to control astrocyte star-shaped morphologies, and an ECM condition that maintains astrocyte quiescence with minimal activation can be achieved. In addition, activation can be induced in a dose-dependent manner via both defined cytokine cocktails and low molecular weight hyaluronic acid. This synthetic brain hydrogel is envisioned as a new tool to study the physiological role of astrocytes in health and disease.
Keywords: biomaterials, hydrogels, mass spectrometry, peptides, poly(ethylene glycol), tissue engineering
1. Introduction
Astrocytes constitute ≈30%[1] of the cells within the mammalian brain and function as key producers and maintainers of the brain extracellular matrix (ECM) during brain tissue homeostasis.[2] During brain trauma[3] and inflammation,[1,4] changes in the ECM composition,[5] ECM stiffness,[6] and the introduction of cytokine molecules[7] transform astrocytes from a quiescent to a reactive state. This reactive state is typically characterized by the upregulation of the intermediate filament proteins glial fibrillary acidic protein (GFAP)[1,2] and vimentin.[8] Recent studies[1] have sought to understand the profile and origination of reactive and quiescent astrocytes[7,9] toward developing therapeutics that inhibit astrocyte activation.[1] Yet, these studies are hindered due to the large complexity and lack of control over the in vivo astrocyte microenvironment. Thus, researchers have increasingly sought in vitro models in which to study astrocyte activation. However, a major limitation to the study of normal and healthy astrocytes in vitro is that there is no reproducible system that can maintain astrocytes in a quiescent state to study activation.
For this reason, bioengineers have developed defined and controllable cell culture environments in which to study cells in more native-like conditions. Relevant to the brain, astrocytes grown as 3D organoids[10] provide aspects of their native phenotype, such as a stellate morphology and astrocyte cell-to-cell heterogeneity. However, the close cell–cell contact in this organoid culture is an important drawback, as astrocyte processes only overlap in vivo during their reactive state.[2a,11] Additionally, culturing cells as organoids is time consuming[10a,12] and does not allow for the customization of ECM cues like stiffness[5a,13] and ligand density[5b,13b] present in real tissue. Protein and glycosaminoglycan-based 3D hydrogels, e.g., Type I collagen,[5b,d] hyaluronic acid (HA),[14] or defined mixtures of the two,[5b,d] have been popularized as in vitro models of brain ECM as they are naturally occurring in the brain, are biocompatible, and have shown lower upregulation of GFAP[5d] compared to astrocytes grown as a 2D monolayer. These environments still sacrifice a stellate astrocyte morphology even in cases where GFAP expression is low, cause significant astrocyte activation compared to in vivo, and a major drawback of protein and sugar-based ECM hydrogels is that their constituent proteins,[5a,c–e] and stiffnesses[5a] (which likely influence astrocyte activation) cannot be independently controlled without major chemical modifications.
Synthetic hydrogels, in contrast, provide a tremendous opportunity to design tissue-specific scaffolds[13b] with tight control over environmental parameters, and there is an engineering opportunity to incorporate bioactive molecules to represent any microenvironment of interest.[15] However, astrocytes either become reactive when cultured in synthetic hydrogels,[15b] or in cases where activation was reduced, their characteristic stellate morphology was sacrificed.[14b,c] Currently there is no in vitro model that can maintain and control astrocyte quiescence, and therefore further the study of how astrocytes activate during central nervous system (CNS) diseases or other injuries/trauma. This highlights the imminent need for ECM environments that can retain astrocyte physiological quiescence in vitro.
We sought to develop such an in vitro model that would enable us and others to study the specific extracellular factors that control astrocyte quiescence and activation. To do this, we first characterized the human brain ECM via mechanical indentation, mass spectrometry, and Protein Atlas histology[16] in order to incorporate the appropriate stiffness and most prevalent ECM proteins responsible for integrin-mediated adhesion and matrix metalloprotease (MMP)-mediated degradation in the brain. We synthesized peptides to represent these proteins, and combined them with a modulus-tunable poly(ethylene glycol) (PEG) network to create a carefully designed, yet very simple synthetic hydrogel brain-like ECM. With this hydrogel we overcome the current challenge of retaining primary human astrocyte quiescence in vitro and, therefore, controlling activation. We demonstrate modulation of astrocyte activation via tuning of the integrin-binding and MMP-degradable profile of the hydrogel, in combination with dosing with cytokine molecules and low molecular weight HA, currently not possible in other in vitro systems where astrocytes remain in a permanently activated state.
2. Results and Discussion
2.1. Characterization of the Human Brain ECM
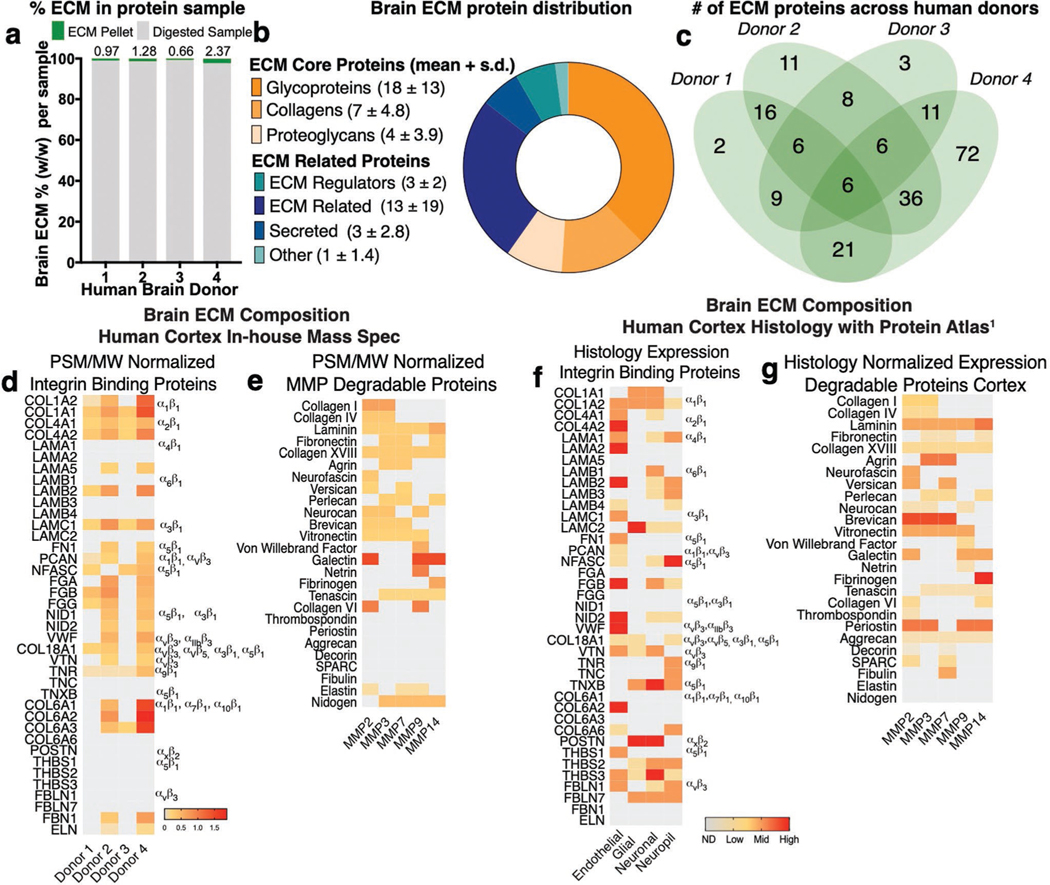
Astrocytes are responsive to ECM proteins in vitro. For example, ECM composition[5a] determines astrocyte responses to substrate stiffness and other inflammatory stimuli. These authors observed arrested astrocyte migration on fibronectin, and rapid migration on tenascin or laminin. Others have shown astrocyte attachment to fibronectin, laminin, and fibrillin-1 regulates IL-1-induced activation in an integrin-dependent manner.[17] Integrin heterodimers are the transmembrane receptors that mediate tissue-specific cell binding to the ECM. We thus hypothesized that a brain-specific ECM with defined integrin-binding interactions would allow guide astrocyte quiescence and activation in vitro. To define the ECM components of real brain tissue, we acquired four healthy human frontal cortex samples (Figure 1a). These samples were decellularized and enriched for ECM proteins following an ECM enrichment protocol recently introduced by Naba et al.,[18] which resulted in an insoluble pellet of 1–2 wt% ECM, which we then analyzed via liquid chromatography-mass spectrometry (LC-MS/MS) (Figure 1a). Protein hits were compared to the Human Matrisome Database[19] and classified into ECM-core or ECM -related proteins (Figure 1b). Glycoproteins, collagens, and proteoglycans, which are known to interact with integrin heterodimers, compose the core ECM proteins (Figure 1b). From this list, and limiting to hits found in at least two donors (Figure 1c), proteins were further stratified by affinity to integrin binding and enzymatic degradation via MMPs (Figure 1d,e; Table S2, Supporting Information). Peptide spectrum match (PSM) normalized by the protein molecular weight was used to quantify a relative abundance of selected proteins (Figure 1d,e). At the culmination of this process, we found 116 ECM proteins, 18 of which with known integrin interactions, and 21 known to be degraded by MMPs (Figure 1d,e; Tables S1 and S2, Supporting Information).
Figure 1.
Characterization of the human brain cortex extracellular matrix. a) Right frontal cortex samples from four healthy human donors were decellularized and enriched for ECM proteins—resulting in an insoluble pellet that was solubilized, reduced, and digested into peptides identified via liquid chromatography-mass spectrometry (LC-MS). b) Protein hits were compared to the Human Matrisome Database and classified into ECM-core or ECM-related proteins. Data shown are average number of proteins found across the four donors + standard deviation (s.d.). c) Distribution of brain ECM signature proteins among donors. d) Proteins identified in at least two donors were screened for affinity to integrin binding and e) enzymatic degradation via matrix metalloproteinases (MMPs). Heat map depicts protein abundance based on peptide spectrum match (PSM) normalized to protein molecular weight (MW) for integrin-binding and MMP-degradable proteins. f) The Human Protein Atlas was screened for ECM proteins in the human brain cortex as identified via the provided histology. Heat map depicts protein abundance via histology expression of integrin binding and g) MMP-degradable proteins in the human brain cortex. Degree of expression denoted as ND (not detected), low, medium, or high.
To ensure that the proteins we identified were in areas of the brain both near astrocytes and in the extracellular space (not intracellular), we complemented our LC-MS/MS findings with a published histological screen of brain proteins by the Human Protein Atlas,[16] where we found 147 brain ECM proteins in the cortex (Table S3, Supporting Information). We similarly reduced this list to core proteins known from the Matrisome Database, resulting in 17 proteins with known integrin binding sites and 24 that are degraded by MMPs (Figure 1f,g; Table S4, Supporting Information). Spatial location of proteins within the brain cortex was assessed via histology images from the Protein Atlas to differentiate proteins located at the basement membrane (endothelial cells) or the interstitium (astrocytes, neurons, neurophils; Figure 1f,g).[20] We used the grade of histology reported by the Protein Atlas (ND, low, medium, or high) to assess protein abundance (Tables S3 and S4, Supporting Information). Both approaches, in-house mass spectrometry and Protein Atlas histology, were combined to determine the relative amounts of proteins present in the brain cortex (Figure 2a; Table S5, Supporting Information).
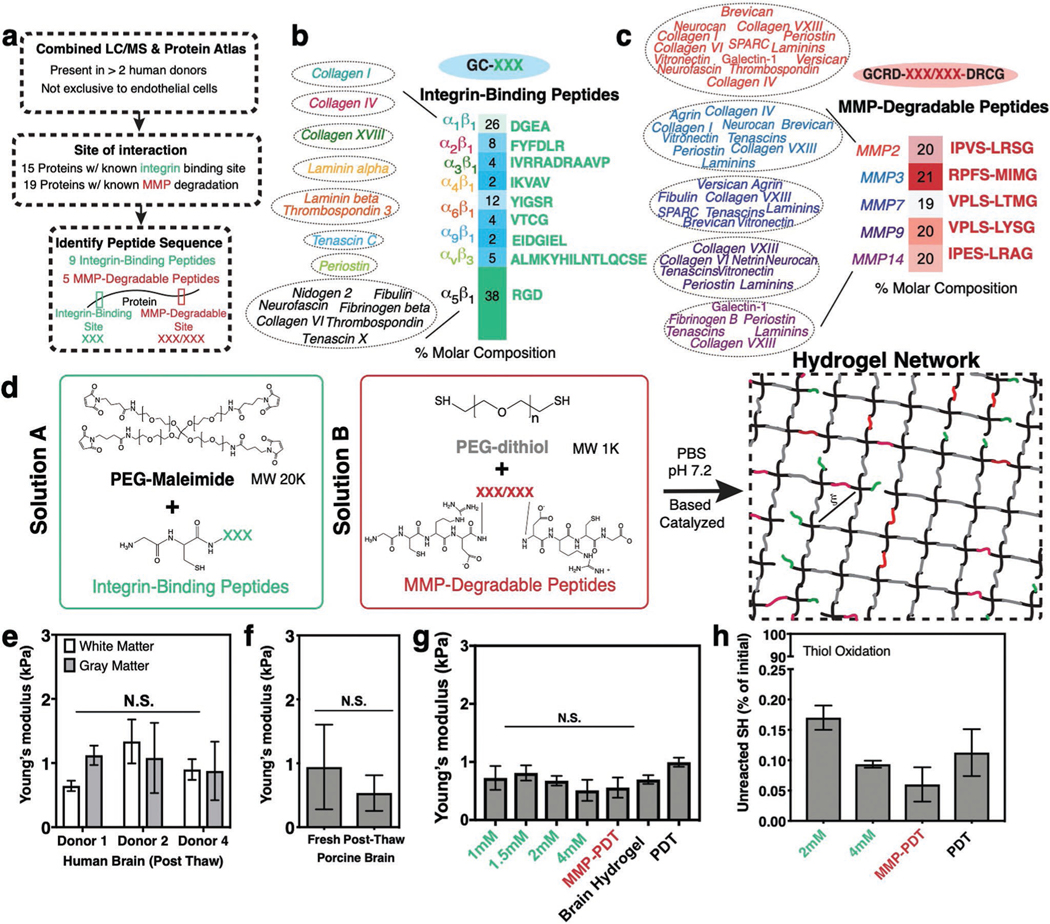
Figure 2.
Design of a synthetic brain ECM hydrogel. a) Proteins identified via in-house proteomics (LC/MS, present in >2 donors) and histology from the Protein Atlas (to exclude proteins specific to endothelial cells) were screened by integrin-binding and matrix metalloproteinase (MMP)-degradable peptide sequences. b) Integrin-binding peptides include 15 proteins, incorporated into the hydrogel via single cysteine. Heat map depicts molar concentration of each peptide in the final design. c) MMP-degradable peptides include 19 proteins, and were incorporated into the hydrogel by a cysteine at each end. Heat map depicts molar concentration of each peptide in the final design. d) A Michael-addition reaction is used to combine these peptides with PEG-maleimide to form a hydrogel network via two solutions: solution A containing integrin-binding peptides and the PEG polymer, and solution B with the MMP-degradable peptides and a nondegradable crosslinker. ξ is the mesh size of the hydrogel (≈20 nm). e) Young’s modulus of previously frozen human brain (3 donors), f) fresh and previously frozen porcine brain n = 6, and g) hydrogel tuned to the same Young’s modulus as brain tissue as measured via indentation. Modulus is maintained after incorporation of integrin-binding (green) or MMP-degradable (red) peptides. n = 8. h) Percent of unreacted thiols after hydrogel is formed, and then reduced with NaBH4, indicating high network formation efficiency. n = 8. All data are mean ± s.d. Data in (e)–(g) were analyzed using an ANOVA followed by a Tukey’s multiple comparison test with 95% confidence interval. N.S. is not significant.
2.2. Design of a Synthetic Brain ECM Hydrogel
To create a brain ECM-mimicking hydrogel, we narrowed the ECM Matrisome quantification results to proteins with known interactions with integrin heterodimers.[21] From the pool of proteins present in Figure 1d,f, integrin-binding sequences were matched via literature mining to their integrin heterodimer pairs (Tables S2, S4, and S5, Supporting Information). Proteins identified were screened to those present in at least two donors, not present only near endothelial cells, and with known integrin-binding amino acid sequences (Figure 2a,b). Seven of these 15 proteins interact with integrins via the RGD site while eight proteins had unique amino acid sequences— resulting in a total of 9 integrin-binding peptides that represent the integrin-binding ECM protein landscape of the synthetic brain ECM (Figure 2a,b). These integrin-binding peptides are represented in the hydrogel as shown in Figure 2b, with mole percents derived from the LC-MS/MS PSM performed on human brain tissue.
MMPs are a family of enzymes that cleave ECM proteins.[22] Since the PEG-maleimide synthetic hydrogel system has mesh sizes on the order of tens of nanometers,[23] cells must degrade the biomaterial in order to extend processes, proliferate, and migrate.[24] We screened brain ECM proteins with the same criteria established for integrins as described above (Figure 2a) and identified those susceptible to MMP degradation. These were grouped based on the MMP via literature mining (19 proteins; Table S2 and S4, Supporting Information; Figure 2a,c). Within this group, we identified the specific peptide sequences known to be degraded by MMPs (Figure 2a,c).[25] This process resulted in 5-degradable peptides that comprise the degradable landscape of the synthetic brain ECM (Figure 2c). The MMP-degradable peptides are represented in the hydrogel with the molar concentrations depicted in Figure 2c based on the same strategy described for the integrin-binding peptides (LC-MS/MS).
Both integrin-binding and MMP-degradable peptides can be easily incorporated in the hydrogel during crosslinking via the Michael addition reaction.[26] Integrin-binding peptides contain a single cysteine functionalization, and MMP-degradable peptides contain cysteines near each end to serve as degradable crosslinks (Figure 2d). We form the hydrogel by creating two solutions that contain a 1) maleimide functionalized PEG with the integrin-binding peptides, and 2) a PEG-dithiol with the MMP-degradable peptides (Figure 2d). When the two solutions are combined, the hydrogel network forms in under a minute. We previously showed that this reaction speed can be tuned by manipulating the ionic strength of the buffer, the pH, and the electronegativity of the amino acids flanking the MMP- degradable peptides.[27]
Since brain cell phenotype is sensitive to the modulus of the ECM,[28,39] we needed to account for this in our design of an in vitro brain ECM. Unfortunately, we quickly discovered that reported values for Young’s moduli of the brain ranged from 100s of Pa to 10s of kPa, and vary considerably among experimental techniques.[29] We therefore independently characterized the modulus of human brain, focusing on the cortex, via an indentation technique previously published by our group,[30] and we found the Young’s modulus to be ≈1 kPa (Figure 2e). Due to our inability to acquire fresh human brain tissue, and to confirm there was no difference in modulus of fresh tissue versus frozen, we also characterized the modulus of fresh and frozen-then-thawed porcine brain cortex.[31] We were surprised to find a similar Young’s modulus when comparing porcine and human brain, and no statistically significant difference between fresh and previously frozen porcine tissue (Figure 2e,f). To incorporate this modulus into our brain hydrogel, and taking a similar approach to work by others[32] that have modulated a biomaterial’s Young’s modulus, we adjusted the macromer concentration until reaching a similar Young’s modulus to that of brain (Figure S1, Supporting Information). We also confirmed that the Young’s modulus was not affected by the incorporation of a range of concentrations of integrin-binding peptides, or by crosslinking the gel with either PEG-dithiol (PDT) versus the desired combination of PDT and the MMP-degradable peptides (Figure 2g). Finally, these peptides can be incorporated efficiently into the PEG-maleimide system as confirmed by an Ellman’s assay and LC/MS (Figure 2h; Figure S1, Supporting Information).
2.3. Human Brain-Specific Integrin-Binding Peptides Validation via Cell Adhesion
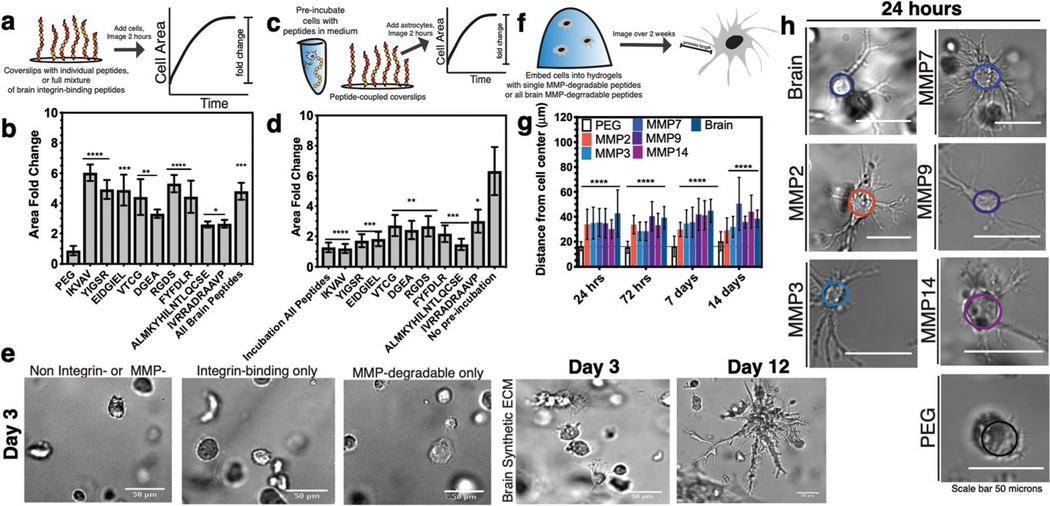
To validate the function of the integrin-binding peptides in the brain-specific hydrogel, we performed a 2D adhesion assay with a human neuroblastoma cell line SK-N-AS. Neuroblastoma cell area increased significantly when cells were seeded on surfaces functionalized with integrin-binding peptides compared to negative control surfaces (PEG; Figure 3a,b; Figure S2, Supporting Information). Experiments with the breast cancer cell line MDA-MB-231 showed similar results (Figure S2 and Tables S1–S4, Supporting Information).[33] We conversely performed a competitive binding assay where cells were preincubated with soluble, individual integrin-binding peptides before seeding onto a surface with the full cocktail of brain-specific integrin-binding peptides (Figure 3c). Compared to those cells preincubated with peptide, control cells fully spread and adhered to the brain peptide sur- faces (Figure 3d; Figure S2, Supporting Information). We used Pubmed to compare the sequence homology of these peptides found in human ECM proteins to murine (Figure S3a–i, Supporting Information). Murine cell lines Neuro2A and mHippoE18 did not adhere as strongly to the human peptide sequences (Figure S3c,d, Supporting Information), and the competitive binding assay did not inhibit mouse cell adhesion to coverslips, particularly in comparison to the human SK-N-AS cells (Figure S3g,h, Supporting Information).
Figure 3.
Biological validation of brain integrin-binding and MMP-degradable peptides. a) Schematic depicting coverslip functionalization with integrin-binding peptides and cell seeding. b) Cell area fold change after seeding onto integrin-binding functionalized coverslips in comparison to a negative control (PEG). N = 2, n = 3. c) Schematic of competitive binding assay of integrin-binding peptides. d) Change in cell area after preincubation with peptides in solution, before seeding onto peptide-functionalized coverslips. Cell area is depicted as a fold change compared to a positive control (no preincubation). N = 2, n = 3. e) Representative human primary astrocyte morphology in the different hydrogel conditions. f) Schematic depicting encapsulation of cells in 3D hydrogels functionalized with MMP-degradable peptides and measurement of process length originating from the cell center. g) Astrocyte process length for individual and all brain MMP-degradable peptides in comparison to a PEG gel with no degradable crosslinks after 24 h, 72 h, 7 days, and 14 days of encapsulation. N = 2, n = 4. h) Representative images of astrocytes after 24 h of encapsulation. All data are mean + s.d. Statistical analyses were performed using Prism (GraphPad). Data in (b), (d), and (f) were analyzed using a one-way analysis of variance followed by a Dunnett’s multiple comparison test with 95% confidence interval. *, **, ***, and **** indicate P < 0.05, P < 0.01, P < 0.001, and P < 0.0001.
We then encapsulated human primary astrocytes in hydrogels containing a single integrin-binding peptide for 5 min prior to fixing and staining for the integrin heterodimers receptors expected to bind the peptide ligand, based on an assay developed by Li et al. (Table S5, Supporting Information).[34] This assay only stains integrins at the cell membrane, and positive staining correlates with ECM binding. We positively identified integrin heterodimers via immunofluorescence for the predicted integrin ligand-receptor pairs in contrast to culture in PEG alone (Figure S4, Supporting Information). Together, this data illustrates that the integrin-binding peptides are human brain specific, and appropriate for incorporation into the hydrogel.
2.4. Human Astrocytes Must Cleave MMP-Degradable Peptides to Achieve Star-Like Morphologies
In the brain, astrocytes are visually phenotypically characterized by extensive ramification and star-like morphologies.[2a,b] Without MMP-degradable crosslinks, hydrogels of a 20–25 nm mesh size would yield rounded astrocytes with little or no ramification. We encapsulated human primary astrocytes in 3D hydrogels that were either blank (PEG-only, negative control), hydrogels with integrin-binding peptides only, or containing both brain integrin-binding and MMP-degradable peptides (Figure 3e). We optimized astrocyte cell density in the brain hydrogel to prevent cell clustering over long culture periods (Figure S5, Supporting Information). We found that astrocytes extended processes in 3D as quantified via Sholl Analysis (distance of cell protrusions from the cell center), in conditions that incorporated both integrin-binding and MMP peptides, and astrocytes remained viable in the brain hydrogel for extended culture times (12 days) (Figure 3e). A timepoint of 3 days was chosen to illustrate astrocytes remained round unless both integrin-binding and MMP peptide cocktails were included. We verified that each MMP-degradable peptide was susceptible to degradation by the astrocytes by encapsulating cells and measuring process length compared to a PEG hydrogel crosslinked with PEG-dithiol (Figure 3h; Figure S6, Supporting Information). 24 h of culture was sufficient to allow for cells to extend processes in each MMP condition, and agrees with work by others for other cell types.[25]
2.5. Astrocyte Activation Can Be Controlled with Hydrogel Composition
Recent work by Placone et al.[5d] illustrated that modifying the concentration of components in a protein-based hydrogel composed of Type I collagen, HA, and Matrigel reduced the expression of GFAP, an astrocyte-specific marker indicative of activation, in primary fetal human astrocytes as compared to culture in 3D collagen alone. Similarly, Hara et al.[5c] demonstrated that astrocytes become reactive when cultured in a collagen-coated substrate mediated by the integrin-N-cadherin pathway. We thus hypothesized that tuning the composition of the brain hydrogel would have an impact on astrocyte activation as measured by GFAP expression.
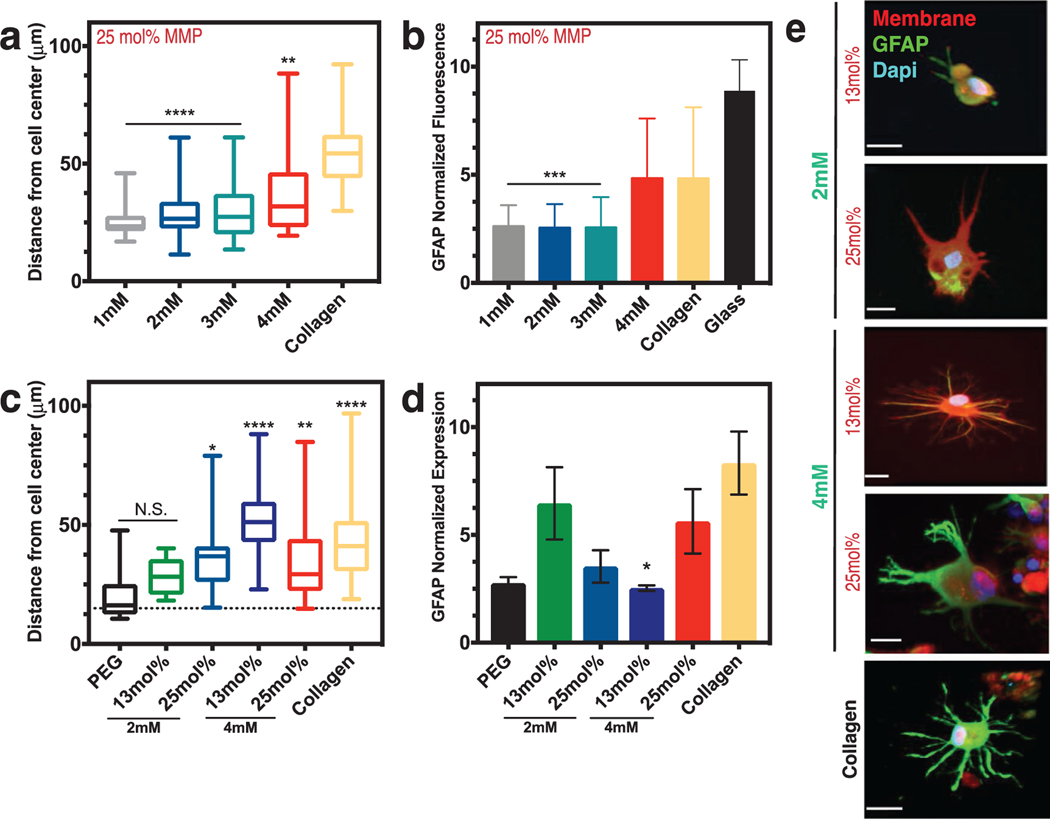
While keeping the MMP peptide concentration constant, we varied the integrin-binding peptide concentrations in the brain hydrogel and quantified cell spreading (Figure S7, Supporting Information). We found that increasing the concentration of adhesive peptides beyond a concentration of 2 × 10−3 m resulted in astrocytes with long processes similar to those in a collagen-based gel, but above 3 × 10−3 m yielded astrocytes with high levels of GFAP fluorescence similar to that in collagen (Figure 4a,b).[5d] We thus decided to test if modifying the concentration of MMP-degradable peptides would also have an effect in activation. We found that combining a 4 × 10−3 m concentration of integrin-binding peptides with a 13 mol% concentration of MMP-degradable peptides yielded significantly low activation of the astrocytes compared to collagen (Figure 4c,d; Figures S7 and S8, Supporting Information). This result agrees with that of others upon ECM deposition in vivo, where, for example, increase in protein secretion is correlated with astrocyte activation.[5c,35] Figure 4e shows representative morphologies and GFAP fluorescence in astrocytes encapsulated across these different conditions. We found a particular brain hydrogel formulation that resulted in both low activation (GFAP expression) while maintaining long, stellate morphologies: a 1 kPa stiffness with 13 mol% MMP-degradable peptides and 2 × 10−3 m integrin-binding peptides.
Figure 4.
Astrocyte activation can be controlled via integrin-binding and MMP-degradable peptides in the brain hydrogel. a) Box and whisker plots show distance of process length from the cell center as a function of the integrin-binding peptide concentration (in mm) in the hydrogel, with collagen gels as a comparison. N = 2, n = 3. b) Normalized fluorescence intensity of glial fibrillary acidic protein (GFAP) as a function of integrin-specific peptide concentration, with both collagen gels and a glass coverslip for comparison (N = 3). All hydrogels in (a) and (b) had 25 mol% MMP-degradable peptides and the time point is 48 h after encapsulation. c) Box and whisker plots showing distance of process length from the cell center as a function of both integrin-binding peptide concentration and concentration of the MMP-degradable peptides. Statistics are in comparison to a PEG hydrogel with no peptides included (negative control). N = 2, n = 4. d) Normalized GFAP fluorescent intensity as a function of hydrogel integrin-binding and MMP-degradable peptide concentrations. N = 2, n = 4. Data in (c) and (d) are after 72 h of encapsulation. e) Representative images of astrocytes encapsulated in different hydrogel conditions after 72 h. Scale bar is 20 μm. Data in (a)–(c) are mean + s.d. Data in (d) are mean + SEM. Data in (a)–(d) were analyzed using a one-way analysis of variance (ANOVA) followed by a Dunnett’s multiple comparison test with 95% confidence interval. *, **, ***, and **** indicate P < 0.05, P < 0.01, P < 0.001, and P < 0.0001. N.S. is not significant.
2.6. Astrocyte Quiescence In Vitro Is Maintained in Synthetic Brain ECM Hydrogels
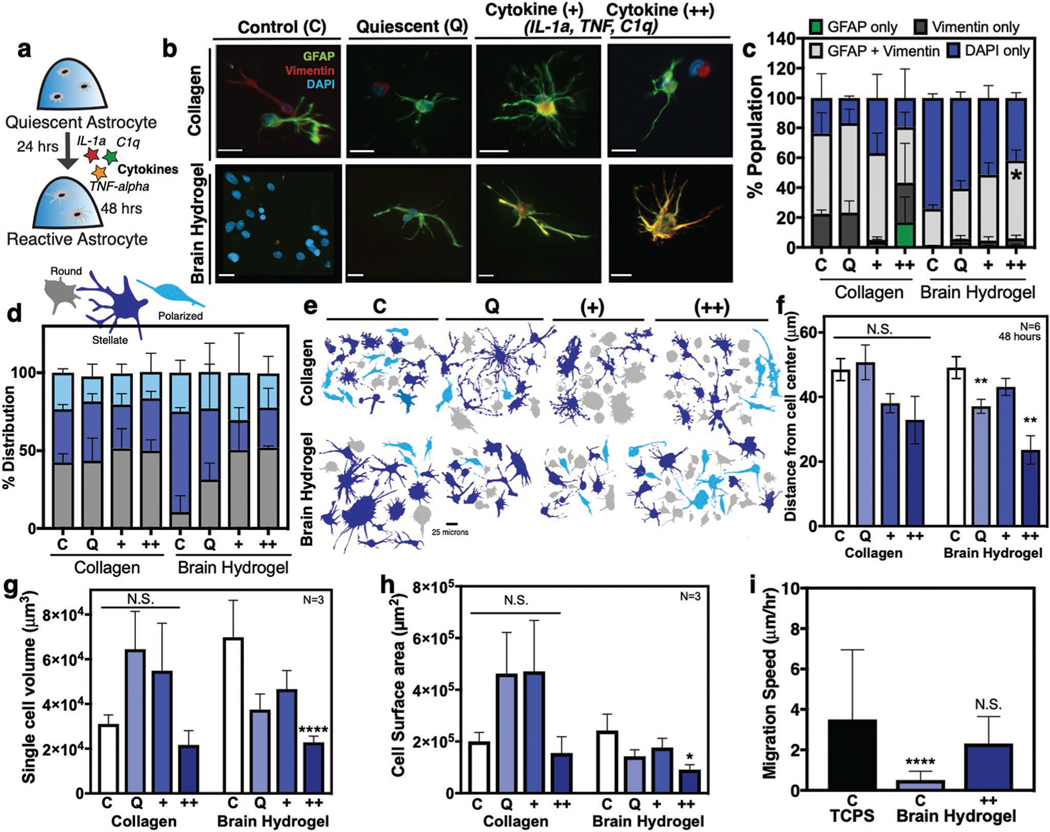
Recent work by Liddelow et al.[7] described astrocyte activation via three cytokines produced by microglia: interleukin 1 alpha (IL-1alpha), tumor necrosis factor alpha (TNF-alpha), and complement component 1, subcomponent q (C1q), resulting in high expression of GFAP.[7] Similarly, work by Hara et al.,[5c] identified gfap and vimentin as genes highly expressed by spinal cord injury-activated astrocytes. To confirm that the quiescent astrocytes can undergo activation in the brain hydrogel, we dosed them with these three cytokines after 24 h of culture, following the established protocols by Liddelow et al. (Figure 5a). Under the standard culture medium conditions, astrocytes expressed low levels of activation markers GFAP and Vimentin in the brain hydrogel, in contrast to collagen (Figure 5b,c).[5c] Similarly, Liddelow et al.[7] defined a quiescent serum-free medium that is supplemented with heparin-binding EGF-like growth factor (HBEGF). We used this serum-free medium composed of 50% neurobasal, 50% DMEM, 1× SATO, supplemented with HBEGF, penicillin, streptomycin, sodium pyruvate, l-glutamine, and N-acetyl cysteine (marked Q for “quiescent”). In this condition, brain hydrogel-cultured astrocytes were largely quiescent with a small percentage of cells positive for GFAP, while collagen still showed signs of promoting astrocyte activation (Figure 5b,c; Figure S9, Supporting Information). In the brain hydrogel, stimulation with cytokines in increasing doses resulted in increased GFAP expression in the astrocyte population (Figure 5b,c). In contrast, astrocytes cultured in collagen retained a high GFAP expression across all cytokine concentrations.
Figure 5.
Astrocyte activation can be controlled in the brain hydrogel. a) Human primary astrocytes were encapsulated in hydrogels for 24 h and subsequently dosed with cytokines IL-1α, TNF-α, and C1q for an additional 24 h. b) Representative images of astrocytes after 48 h in brain hydrogels and collagen and incubated with either standard culture medium (control), defined quiescent medium, dosed with cytokines (+) IL-1α (3 ng mL−1), TNF-α (30 ng mL−1), and C1q (400 ng mL−1), or dosed with a 2× dose of cytokines (++) IL-1α (6 ng mL−1), TNF-α (60 ng mL−1), and C1q (800 ng mL−1). Scale bar is 20 μm. c) Quantification of cells from (b) stained for GFAP, vimentin, and DAPI. N = 2, n = 250 cells per condition. d) Distribution of cell morphologies and e) representative cell morphologies in the different populations identified as round (gray), stellate (blue), or polarized (light blue). N = 3, n = 100 cells per condition. f) Quantification of astrocyte process length N = 6, n = 3. g) Single cell volume and h) single cell surface area for astrocytes encapsulated for 48 h. N = 3, n = 3. i) Comparison of migration speed astrocytes seeded on plastic (TCPS), or in the brain hydrogels with and without cytokines. N = 2. All plots show mean + SEM. Data in (c) were analyzed using a one-way analysis of variance (ANOVA). Data in (f)–(h) were analyzed using a two-way ANOVA followed by a Tukey’s multiple comparison test with 95% confidence interval. Data in (i) were analyzed using a one-way ANOVA followed by a Dunnett’s multiple comparison test with 95% confidence interval. *, **, and **** indicate P < 0.05, P < 0.01, and P < 0.0001, respectively; N.S., not significant.
Work by others has demonstrated that activated astrocytes become significantly smaller compared to quiescent astrocytes in vivo.[7] The aberrant morphologies coincide with high expression of gfap during activation.[36] In our brain hydrogel, astrocytes retain a predominantly stellate morphology in both standard and quiescent medium, and shift to a mix of rounded, polarized, and stellate cells in cytokine medium (Figure 5d,e). In collagen, astrocytes exhibited a heterogeneous range of morphologies as expected in activated populations (Figure 5d,e).[36] Similar to A1 activated astrocytes described by Liddelow et al.,[7] we found astrocyte process length, volume, and surface area decreased significantly in the highest cytokine concentration in the brain hydrogel, while there was no significant difference across collagen cultured conditions (Figure 5f–h). Astrocytes have also been shown to become highly migratory upon activation in vivo.[4,37] We expectedly found highly migratory astrocytes on 2D TCPS,[38] and that cells migrated significantly more in brain hydrogels with cytokine medium, compared to the standard culture in the control brain hydrogel (Figure 5i).
HA is a polysaccharide in brain tissue that has been commonly incorporated into in vitro ECM models of the brain.[14b,28]c,[39] Others have shown correlation between concentration of low-[43] and high-[40] MW HA in a biomaterial with disease progression (e.g., glioblastoma) and the activation of other cells (macrophages).[40] We found that low MW HA induced astrocyte activation in our brain hydrogel. We incorporated thiolated low MW HA (10 kDa) in the brain hydrogel and stained for GFAP after 48 h (Figure S10, Supporting Information). We observed a trend where GFAP expression increased in a dose-dependent manner, with 0.1 × 10−3 m HA being the lowest dose at which astrocytes remained quiescent (Figure S10, Supporting Information). This in part agrees with observed increases in glioma malignancy in an HA concentration-dependent manner.[28c,39] Together these data demonstrate that astrocytes cultured in the brain hydrogel remain quiescent in vitro and can undergo cytokine- and HA-mediated activation as defined by morphology, expression of GFAP and vimentin, and a migratory phenotype. Further, quiescent astrocyte populations were observed in the synthetic brain-mimicking hydrogel, and not in collagen.
3. Conclusion
Bioengineers have designed in vitro models that can recapitulate the brain ECM.[5,15a] However, there is no tunable in vitro model that can maintain astrocyte quiescence. The instructive design of this brain ECM specific hydrogel allows for tunable control of astrocyte activation, in contrast to protein- and sugarbased models like collagen and hyaluronic acid.
An exciting recent finding illustrated how proteins secreted by cells within synthetic hydrogels regulate cell phenotype,[41] potentially putting into question the need for inclusion of ECM binding and degradation sites into gels. However, we found here that these initial signals are very important in driving astrocyte quiescence and activation. For example, astrocytes encapsulated in hydrogels with either integrin-binding or MMP-degradable peptides did not show a stellate morphology (Figure 3). The need for these tissue-specific hydrogels is of particular importance to instruct cells that do not produce their own matrix, for example oligodendrocytes and Schwann cells,[42] and it has been shown many times that these cues can impact cell differentiation[42,43] and viability.[27] Similarly, specific ECM cues provided by integrin-binding sites,[5c,17,44] inflammatory molecules,[7,45] and mechanical properties,[28a] have been shown to influence the deposition of ECM proteins by astrocytes.
What we still do not know is whether all these peptides are necessary to drive the observed astrocyte quiescence. A future useful study would be to perform an N-1 factorial design across all 14 peptides, and across multiple concentrations for each peptide to determine the simplest possible hydrogel design needed to instruct the desired astrocyte phenotype. Regardless, we foresee the use of this synthetic brain hydrogel to understand extracellular mechanisms of astrocyte activation, as well as individual contributions of quiescent and reactive astrocytes with other CNS cells in neurodegenerative diseases. In fact, recent studies demonstrate the need to control for and study the dynamic cell-to-cell interactions, especially in the context of neurons, astrocytes, and microglia[46] to better study Alzheimer’s disease in vitro. Rational biomaterial design is also critical for to accelerate and/or support proper brain tissue repair after stroke.[47]
Future studies investigating the functional interactions among these various neural cell types in our tunable system will allow for precise and controlled manipulation, experimental schemes to tease apart the complex signaling mechanisms underlying cell-to-cell communication in the context of neurodevelopmental and neurodegenerative diseases, and potentially offer synergistic therapeutic value.
4. Experimental Section
Cell Culture:
Normal human astrocytes of cortical origin were purchased from ScienCell Research Laboratories (San Diego, CA, USA) and maintained in Astrocyte medium (ScienCell, San Diego, CA, USA). Primary cells were used between passages 1 to 3 for all experiments. MDA-MB-231-BrM2a cells were a generous gift from Joan Massague, and MDA-MB-231 cells were a generous gift from Dr. Sallie Schnedier lines. Both 231 cell lines were cultured in DMEM with 10% fetal bovine serum (FBS, Thermo). The Neuro2A, mHippoE18, and SK-N-AS were cultured in DMEM supplemented with 10% FBS, 0.1 × 10−3 m NEAA, 1% l-glutamine, and 1 × 10−3 m sodium pyruvate.
Acquisition of Human Brain Tissue:
Deidentified human brain tissues were obtained from the NIH NeuroBioBank at Harvard Medical School. All tissues had been flash-frozen from four postmortem neurologically normal individuals: three females, and one male from the ages of 30–60.
Mass Spectrometry:
The CNMCS Compartment Protein Extraction Kit (MilliporeSigma) was used to decellularize all human brain tissue samples as described in ref. [18]. Briefly, intracellular soluble proteins were extracted with sequential incubation in the appropriate buffers according to manufacturer’s instructions. This resulted in an insoluble ECM-rich pellet. The ECM-rich pellet of four brain tissue samples was solubilized and reduced in 8 m urea, 100 × 10−3 m ammonium bicarbonate, and 10 × 10−3 m dithiothreitol (DTT, Thermo Fisher Scientific, Waltham, MA) for 30 min at pH 8 and 37 °C. Samples were then alkylated with 25 × 10−3 m iodoacetamide (Sigma-Aldrich) in the dark at RT for 30 min. Samples were subsequently quenched with 5 × 10−3 m DTT and the solution was diluted to 2 m urea with 100 × 10−3 m ammonium bicarbonate (pH 8). Proteins were digested via trypsin (Thermo) and Lys-C endoproteinase (Promega, Madison, WI) at a weight ratio of 1:50 enzyme to protein overnight (12–16 h) at 37 °C. Finally, samples were cleaned and concentrated using a C18 column (Thermo).
Digested peptides were separated by reverse phase LC gradient prior to mass spectrometry analysis with an Orbitrap Fusion Tribid (Thermo). Identified peptides were aligned against the Matrisome using the Thermo Proteome Discoverer 1.41.x. Parameters for analysis used trypsin as a protease, with four missed cleavages per peptide, a precursor mass tolerance of 10 ppm, and fragment tolerance of 0.6 Da.
Determination of Young’s Modulus for Brain Tissue and Hydrogels:
The Young’s modulus was measured using an indentation custom built instrument used as previously described.[30] Briefly, a flat cylindrical punch of 1.5 mm diameter was brought into contact with the hydrogel at a fixed displacement rate of 15 μm s−1, for a maximum load of 1.5 mN. The linear response of the low strain regime was analyzed using a Hertzian model accounting for dimensional confinement between the contact radius (a) and the sample height (h) (0.5 < a/h < 2) as described previously.[48] To tune the hydrogel Young’s modulus to match the brain, PEG-maleimide hydrogels were formed at different macromer concentrations containing the integrin-binding and MMP-degradable peptides and swollen in PBS buffer overnight. Finally, the Young’s modulus was similarly obtained for six individual porcine brains (n ≥ 5) and three human brain donors (n = 2). All samples had a thickness of 1 mm to ensure the dimensional confinement remained within 0.5 < a/h < 2.
Solid-Phase Peptide Synthesis:
Peptides (GRGDSPCG, GCALMKYH ILNTLQCSE, GCDPGIVRRADRAAVP, GCDPGIKVAV, GCDPGYISGR, GCGDGEA, GCGFYFDLR, CSVTCG, CGGAEIDGIEL, GCRDIPVSLRSGDRCG, GCRDRPFSMIMGDRCG, GCRDVPLSLTMGDRCG, GCRDVPLSLYSGDRCG, and GCRDIPESLRAGDRCG) were synthesized on a CEM Liberty Blue automated solid phase peptide synthesizer (CEM, Mathews, NC) using FMOC protected amino acids (Iris Biotech GMBH, Germany). The peptide was cleaved from the resin by sparging-nitrogen gas through a solution of trifluoroacetic acid (TFA), triisopropylsilane (TIPS), 2,2′(ethylenedioxy)diethanethiol (DODT), and water at a ratio of 92.5:2.5:2.5:2.5 vol%, respectively (Sigma-Aldrich) for 2–3 h at RT in a reactor vessel (ChemGlass, Vineland, NJ). After reaction, the solution was filtered, and the peptide was precipitated using ethyl ether at −80 °C (Thermo). The molecular weight of the peptide was validated using a MicroFlex MALDI-TOF (Bruker, Billerica, MA) using alpha-cyano-4-hydroxycinnamic acid as the matrix (Sigma-Aldrich). Peptides were analyzed and purified to >95% on a VYDAC reversed-phase C18 column attached to a Waters 2487 dual (lambda) adsorbable detector and 1525 binary HPLC pump (Waters, Milford, MA).
Cell Adhesion Assay to Brain ECM Peptide Surfaces:
Integrin-binding peptides were attached to 15 mm glass coverslips by adapting a previous published approach.[49] Coverslips were treated with UV/ozone at 1 atm (UV/Ozone ProCleaner, 180 μg m−3 ozone level in chamber, 4.6 mW cm−2 peak UV intensity, 10.15 W UV lamp power requirement, BioForce Nanosciences, Salt Lake City, UT) for 10 min to expose −OH groups. (3-aminopropyl)triethoxysilane (APTES, Sigma-Aldrich, St. Louis, MO) was reacted with the glass surfaces in a 90 °C oven via vapor deposition overnight, wrapped in foil. The glass coverslips were sequentially rinsed three times in toluene (Thermo), 95% ethanol (Thermo), and distilled water. The glass was allowed to dry in the oven at 90 °C for 1 h, and then functionalized with 10 g L−1 N,N-disuccinimidyl carbonate (DSC, Sigma-Aldrich) and 5 vol% N,N-diisopropylethylamine (DIEA, Sigma-Aldrich) in acetone (Thermo), sequentially, for 2 h each. Coverslips were then rinsed with acetone, air-dried for 10 min, and either used immediately or stored in a desiccator overnight.
Each glass coverslips surface was coated with 70 μL of integrinbinding peptide solution, allowed to react for 2 h, rinsed three times with pH 7.4 PBS, and then blocked with 10 μg cm−2 MA(PEG)24 (Thermo) for 2 h to prevent nonspecific protein adsorption. The coverslips were glued to the surfaces of each well in a 24-well plate with epoxy (Thermo), rinsed three times with PBS, and UV sterilized for 1 h before cell seeding. Cells were seeded at 10 000 cells cm−2 to functionalized glass coverslip surfaces in serum free DMEM and imaged by a controlled Zeiss Axio Observer Z1 microscope (Carl Zeiss, Oberkochen, Germany) using an AxioCam MRm camera and an EC Plan-Neofluar 20 × 04 NA air objective. Images were taken every 5 min for an incubation period of 1, 2, and 12 h. Cell area was quantified by manual tracing in ImageJ (NIH, Bethesda, MD).
Competitive Binding Assay:
Cells were seeded at 10 000 cells cm−2 onto coverslips functionalized with integrin-binding peptides after 30 min of pretreatment with individual peptides or the complete brain-ECM peptide mixture. Cells were seeded at 10 000 cells cm−2 onto glass coverslips (15 mm, Thermo) functionalized with the individual peptides or a 500 μg mL−1 (35 μg cm−2) complete brain-ECM peptide mixture: 35 wt% RGD, 8 wt% LRE, 5 wt% IKVAV, 2 wt% PHSRN-RGDS, 13 wt% DGEA, 10 wt% FYFDLR, 4 wt% VTCG, 1 wt% AEIDGIEL, 3 wt% ALMKYHILNTLQCSE, 3 wt% IVRRADRAAVP, 5 wt% TWSKVGGHLRPGIVQSG, 2 wt% GRKRK, 7 wt% YIGSR, and 1 wt% GWTVFQKRLDGS. Conditions with individual peptides were at the same concentration as the peptide is represented in the full mixture. Cells were imaged beginning at 5 min after seeding in an environment-controlled Zeiss Axio Observer Z1 microscope. Images were taken at 5 min intervals for 2 h, or every 15 min for 12 h, and cell areas were manually traced in ImageJ.
Synthesis of 3D Brain Hydrogels:
A 10 wt% solution of a 20K 4-arm PEG-maleimide (Jenkem Technology, Plano, TX) was reacted with 4 × 10−3 m integrin-binding peptides for 5 min in serum-free medium at pH 7.4 (solution A). The crosslinker solution (solution B) was composed of 77 molar% of 1 K linear PEG-dithiol (Jenkem) and 13 molar% of the MMP-degradable peptide cocktail. These two solutions were combined at a 1:1 molar ratio of thiol to maleimide in TEA at pH 7.4 for a total final gel volume of 10 μL. For experiments with cells, cells were added to “solution A” by using solution A as the fluid to resuspend a cell pellet prior to gelation with “solution B.” Gels were allowed to polymerize for 10 min prior to addition of cell culture medium.
To ensure that the brain hydrogels stayed in a fixed location during imaging, coverslips that would covalently crosslink to the gels during network polymerization were prepared, as previously described.[50] Glass coverslips were UV/ozone treated for 10 min and functionalized with 2 vol% solution of 3-mercaptopropyl-trimethoxysilane (MPT, Thermo) in 95% ethanol (adjusted to pH 5.0 with glacial acetic acid) for 1 h. The wells were rinsed three times with 100% ethanol and allowed to air dry for 10 min before addition of the brain hydrogel solutions.
For experiments where integrin-binding peptides were varied, the composition of the cocktail was maintained at a constant mole percent, as shown in Figure 2b. The overall number of moles of peptide were stoichiometrically tuned to the number of moles of available maleimide groups from the star-PEG polymer at a 1:4 ratio (i.e., the star PEG has 4 moles of maleimide for each mole of PEG, and the integrin-binding peptides have 1 mole of cysteines for every 1 mole of peptide). The same approach was taken when varying the MMP-peptide concentration, where each MMP peptide had 2 moles of cysteines per mole of polymer. When describing the % MMP-degradable peptides included in the gels, we reported how much of the total crosslinker is MMP-degradable, relative to the nondegradable PEG-dithiol crosslinker. All hydrogels were synthesized such that all hydrogels were 100% crosslinked (with some reported ratio of those crosslinks being MMP-degradable or not).
Validation of Peptide Incorporation:
The Measure-iT thiol kit was used to quantify unreacted thiols (Thermo) as previously described.[27] Integrin-binding peptides were incorporated into gels at 2 × 10−3 and 4 × 10−3 m concentrations in a 100 μL volume of PEG-maleimide for 10 min before reacting hydrogels with 100 μL of the Measure-iT kit working solution. Separately, dithiol terminated crosslinkers (PDT at an equimolar ratio of thiol to maleimide, or MMP-degradable peptide (13 mol%) with 87 mol% PDT) were reacted with the PEG-maleimide in 10 μL aliquots for 10 min before reacting with 100 μL of the kit working solution. These hydrogels were incubated in sodium borohydride (NaBH4, Sigma) in water at a molar ratio of 4:1 NaBH4 to thiol for 4 h before adding the thiol kit working solution. Hydrogel supernatant and solutions were acquired and read at an excitation of 494 nm and emission of 517 nm, as was previously reported.[51] To detect any peptides that did not incorporate into the hydrogel network, the supernatant was removed and it was analyzed using a MicroFlex MALDI-TOF (Bruker) with alphacyano-4-hydroxy cinnamic acid (Sigma) as the matrix.
Human Astrocytes Spreading and Variation of Integrin-Binding Peptide Concentration:
Human astrocytes were encapsulated into the brain customized hydrogel with integrin-binding peptide concentrations varying from 0 × 10−3 to 4 × 10−3 m at a density of 5000 cells μL−1. After 24 h, hydrogels were fixed with 4% formaldehyde (Acros) for 10 min and stained with GFAP (Abcam ab7260), CellMask Membrane stain (Thermo C10046), and DAPI (Sigma). Cells were imaged on a Zeiss Spinning Disc microscope (Zeiss) using an HRm AxioCam. Images were processed with Zen software (Zeiss) and cell areas were manually traced with ImageJ (NIH).
Collagen Hydrogel Synthesis:
Type I collagen (rat tail, Corning, NY, lot 7079004) were formed by combining with NaOH, 10× DMEM (Sigma), and cells in cold, serum-free medium. Gels were allowed to polymerize at 37 °C for 30 min before adding cell culture medium.
Control, Quiescent, and Cytokine-Containing Astrocyte Media:
Serumfree base medium containing 50% neurobasal (Thermo), 50% DMEM, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin, 1 × 10−3 m sodium pyruvate (Thermo), 292 μg mL−1 l-glutamine (Thermo), and 5 μg mL−1 of N-acetyl cysteine (Sigma) were supplemented with HBEGF (Peprotech, Rocky Hill, NJ Cat #100–47) at 5 ng mL−1 as previously described.[7] 1× SATO was incorporated from 100× SATO aliquots (10 mg mL−1 BSA (Thermo), 10 mg mL−1 transferrin (Sigma), 1.6 mg mL−1 putrescine (Sigma), 6 μg mL−1 progesterone (Sigma), and 4 μg mL−1 sodium selenite (Sigma)) at the moment of the experiment.
Astrocytes were cultured in either this quiescent medium or control human astrocyte medium from ScienCell (ScienCell, San Diego, CA, USA). For dosing with cytokines, quiescent medium was treated with IL-1α (3 ng mL−1, Sigma, #I3901), TNF-α (30 ng mL−1, Cell Signaling Technology, Danvers, MA #8902SF), and C1q (400 ng mL−1, MyBioSource, San Diego, CA #MBS143105) for the single dose cytokine conditions (+). Double cytokine conditions (++) were dosed with double the cytokine concentrations listed above.
Passage 1–3 primary human astrocytes were cultured in the brain hydrogel for 24 h in either control or quiescent astrocyte medium. After 24 h, the medium was changed to that of control, quiescent, single dose-, or double dose-cytokine medium. Following 24 h after cytokine dosing, the medium was removed, and the hydrogels were fixed in 4% PFA for 10 min at 37 °C before immunocytochemistry.
Astrocyte Culture in Synthetic Brain Hydrogels Containing Ultralow MW HA:
To functionalize the brain hydrogel with low MW hyaluronate thiol (MW 10 kDa, Creative PEG Works, HA-371), HA containing thiol groups was reconstituted in PBS and incorporated with the crosslinker mixture composed of 77 molar% of 1 K linear PEG-dithiol (Jenkem) and 13 molar% of the MMP-degradable peptide cocktail where HA was added to a final concentration of 0.1 × 10−3, 0.5 × 10−3, and 1 × 10−3 m HA in a 10 μL hydrogel. Astrocytes were encapsulated in these HA-containing brain hydrogel as described earlier. Hydrogel contained 4 × 10−3 m integrin-binding peptides and were crosslinked at a 1:1 molar ratio of thiol to maleimide in TEA at pH 7.4. Gels were allowed to polymerize in 10 μL volumes with 5000 cells μL−1, and cell culture media was added after 10 min. Astrocytes were cultured for 24, 48, and 72 h in the HA containing hydrogels where medium was removed and replaced with 4% PFA to prepare the hydrogels for immunocytochemistry.
Immunocytochemistry:
Hydrogels were fixed in 4% paraformaldehyde solution for 10 min at room temperature, and then washed 3× with ice-cold PBS. Gel-embedded cells were blocked with 5% bovine serum albumin (BSA, Thermo) for 30 min, followed by incubation with the CellMask membrane stain (Thermo C10046) for 1 h in the dark at RT, and then washed 3× with PBS. To quantify activation, gel-embedded cells were permeabilized with 0.25% Triton X-100 (Sigma) for 10 min, rinsed with 0.1% Triton X-100 for 3 min, washed with 3× PBS over 5 min, blocked with 5% BSA at room temperature for 30 min, and incubated with primary antibodies to GFAP (1:1000 ab7260, Abcam), vimentin (1:200 ab24525, Abcam), and Cell Mask membrane stain (1:1000), overnight at 4 °C. Finally, gels were washed and incubated in secondary antibody solutions (1:500 ab96883, Abcam, Goat antirabbit, green and 1:500 ab96950, Abcam, Goat antichicken, red, respectively) for 1 h at RT. Cells were imaged on a Zeiss Spinning Disc confocol (Zeiss) using an HRm AxioCam. Images were taken using Zen software (Zeiss) and cell morphology was analyzed in ImageJ (NIH) and Imaris (BitPlane, Belfast, UK). Normalized GFAP expression was determined quantitatively from the ratio of the average fluorescence per pixel in the cell body and processes to the average intensity of the background as quantified with ImageJ (NIH). The number of GFAP-positive cells was counted using the 3D cell counter plug-in in ImageJ.
3D reconstructions and filament traces of the astrocytes were generated from confocal Z-stacks of the CellMask membrane stain using Imaris (Bitplane) and ImageJ (NIH) to obtain quantitative morphological data. Sholl[52] analysis was performed to quantify astrocyte process number and length. Total additive process length, degree of branching (# process ends/# primary processes), and cell diameter were calculated for each cell.
Statistical Analysis:
Statistical analysis was performed with GraphPad Prism (7.0d) (GraphPad Software, Inc., La Jolla, CA). Statistical significance was evaluated using a one-way analysis of variance (ANOVA) followed by a Tukey’s post-test for pairwise comparisons. P-values <0.05 were considered significant, where P < 0.05 is denoted with *, P < 0.01 with **, P < 0.001 with ***, and P < 0.0001 with ****.
Supplementary Material
Acknowledgements
The authors would like to thank Sarah Perry for providing peptide synthesizer equipment, the NIH NeuroBioBank for providing human brain tissue, and support from Lauren Jansen on hydrogel design workflow. The authors thank Dr. Stephen Eyles and the Mass Spectrometry Core Facility at the Institute of Applied Life Sciences for support with LC-MS/MS. This work was supported by NIH Supplement Award 3DP2CA186573-01S1 to S.G., NIH New Innovator Award to S.R.P. (1DP2CA186573-01), both from the National Cancer Institute, an National Science Foundation CAREER to S.R.P. (DMR1454806), a grant from the Office of Naval Research N00014-17-1-2056 to A.J.C. and S.R.P., a Northeast Alliance for the Graduate Education and Professoriate (NEAGEP) fellowship to S.G., and a Spaulding-Smith Fellowship to S.G. from the UMass Amherst Graduate School. This work was also funded by the National Institute of General Medical Sciences (R25GM099649). S.R.P. is a Biomedical Scholar funded by the Pew Foundation.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Sualyneth Galarza, Department of Chemical Engineering, University of Massachusetts, Amherst, MA 01003, USA.
Alfred J. Crosby, Department of Polymer Science and Engineering, University of Massachusetts Amherst, MA 01003, USA
ChangHui Pak, Department of Biochemistry and Molecular Biology, University of Massachusetts, Amherst, MA 01003, USA
Shelly R. Peyton, Department of Chemical Engineering, University of Massachusetts, Amherst, MA 01003, USA
References
- [1].Liddelow SA, Barres BA, Immunity 2017, 46, 957. [DOI] [PubMed] [Google Scholar]
- [2]a).Oberheim NA, Goldman SA, Nedergaard M, in Heterogeneity of astrocytic form and function, Astrocytes, Humana Press; 2012, pp. 23–45; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Oberheim NA, Takano T, Han X, He W, Lin JH, Wang F, Xu Q, Wyatt JD, Pilcher W, Ojemann JG, J. Neurosci 2009, 29, 3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Okada S, Hara M, Kobayakawa K, Matsumoto Y, Nakashima Y, Neurosci. Res 2018, 126, 39. [DOI] [PubMed] [Google Scholar]
- [4].Farina C, Aloisi F, Meinl E, Trends Immunol. 2007, 28, 138. [DOI] [PubMed] [Google Scholar]
- [5]a).Johnson KM, Milner R, Crocker SJ, Neurosci. Lett 2015, 600, 104; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hsiao TW, Tresco PA, Hlady V, Biomaterials 2015, 39, 124; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Hara M, Kobayakawa K, Ohkawa Y, Kumamaru H, Yokota K, Saito T, Kijima K, Yoshizaki S, Harimaya K, Nakashima Y, Nat. Med 2017, 23, 818; [DOI] [PubMed] [Google Scholar]; d) Placone AL, McGuiggan PM, Bergles DE, Guerrero-Cazares H, Quiñones-Hinojosa A, Searson PC, Biomaterials 2015, 42, 134; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Struve J, Maher PC, Li Y.-q, Kinney S, Fehlings MG, Kuntz Iv C, Sherman LS, Glia 2005, 52, 16. [DOI] [PubMed] [Google Scholar]
- [6].Pogoda K, Chin L, Georges PC, Byfield FJ, Bucki R, Kim R, Weaver M, Wells RG, Marcinkiewicz C, Janmey PA, New J Phys. 2014, 16, 075002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Münch AE, Chung W-S, Peterson TC, Nature 2017, 541, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Calvo J, Carbonell A, Boya J, Brain Res. 1991, 566, 333. [DOI] [PubMed] [Google Scholar]
- [9].Zamanian JL, Xu L, Foo LC, Nouri N, Zhou L, Giffard RG, Barres BA, J. Neurosci 2012, 32, 6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]a).Sloan SA, Darmanis S, Huber N, Khan TA, Birey F, Caneda C, Reimer R, Quake SR, Barres BA, Pasça SP, Neuron 2017, 95, 779; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Dezonne RS, Sartore RC, Nascimento JM, Saia-Cereda VM, Romão LF, Alves-Leon SV, De Souza JM, Martins-de-Souza D, Rehen SK, Gomes FCA, Sci. Rep 2017, 7, 45091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sofroniew MV, Trends Neurosci. 2009, 32, 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pasça SP, Nat. Neurosci 2018, https://www.stemcell.com/media/files/wallchart/WA27077-Building_Three-Dimensional_Human_Brain_Organoids.pdf. [DOI] [PubMed] [Google Scholar]
- [13]a).DeForest CA, Anseth KS, Annu. Rev. Chem. Biomol. Eng 2012, 3, 421; [DOI] [PubMed] [Google Scholar]; b) Tibbitt MW, Anseth KS, Biotechnol. Bioeng 2009, 103, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]a).Seidlits SK, Khaing ZZ, Petersen RR, Nickels JD, Vanscoy JE, Shear JB, Schmidt CE, Biomaterials 2010, 31, 3930; [DOI] [PubMed] [Google Scholar]; b) Jeffery AF, Churchward MA, Mushahwar VK, Todd KG, Elias AL, Biomacromolecules 2014, 15, 2157; [DOI] [PubMed] [Google Scholar]; c) Seidlits SK, Liang J, Bierman RD, Sohrabi A, Karam J, Holley SM, Cepeda C, Walthers CM, J. Biomed. Mater. Res., Part A 2019, 107, 704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15]a).Puschmann TB, Zandén C, De Pablo Y, Kirchhoff F, Pekna M, Liu J, Pekny M, Glia 2013, 61, 432; [DOI] [PubMed] [Google Scholar]; b) Papadimitriou C, Celikkaya H, Cosacak MI, Mashkaryan V, Bray, Bhattarai P, Brandt K, Hollak H, Chen X, He S, Dev. Cell 2018, 46, 85; [DOI] [PubMed] [Google Scholar]; c) Celikkaya H, Cosacak MI, Papadimitriou, Popova S, Bhattarai, Biswas SN, Siddiqui T, Wistorf, Nevado-Alcalde I, Naumann, Front. Cell. Neurosci 2019, 13, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C, Sjöstedt E, Asplund A, Science 2015, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- [17].Summers L, Kangwantas K, Nguyen L, Kielty C, Pinteaux E, Mol. Cell. Neurosci 2010, 44, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Naba A, Clauser KR, Hynes RO, Visualized Exp J. 2015, 101, e53057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Naba A, Clauser KR, Ding H, Whittaker CA, Carr SA, Hynes RO, Matrix Biol. 2016, 49, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dityatev A, Seidenbecher CI, Schachner M, Trends Neurosci. 2010, 33, 503. [DOI] [PubMed] [Google Scholar]
- [21]a).Peng H, Ong YM, Shah WA, Holland PC, Carbonetto S, Dev. Neurobiol 2013, 73, 333; [DOI] [PubMed] [Google Scholar]; b) Zaidel-Bar R, Geiger B, J. Cell Sci 2010, 123, 1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sternlicht MD, Werb Z, Annu. Rev. Cell Dev. Biol 2001, 17, 463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Barney LE, Jansen LE, Polio SR, Galarza S, Lynch ME, Peyton SR, Curr. Opin. Chem. Eng 2016, 11, 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Raeber GP, Lutolf MP, Hubbell JA, Biophys. J 2005, 89, 1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Patterson J, Hubbell JA, Biomaterials 2010, 31, 7836. [DOI] [PubMed] [Google Scholar]
- [26].Phelps EA, Enemchukwu NO, Fiore VF, Sy JC, Murthy N, Sulchek TA, Barker TH, García AJ, Adv. Mater 2012, 24, 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Jansen LE, Negrón-Piñeiro LJ, Galarza S, Peyton SR, Acta Biomater. 2018, 70, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]a).Tyler WJ, Nat. Rev. Neurosci 2012, 13, 867; [DOI] [PubMed] [Google Scholar]; b) Ulrich TA, de Juan Pardo EM, Kumar S, Cancer Res. 2009, 69, 4167; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Ananthanarayanan B, Kim Y, Kumar S, Biomaterials 2011, 32, 7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chatelin S, Constantinesco A, Willinger R, Biorheology 2010, 47, 255. [DOI] [PubMed] [Google Scholar]
- [30].Jansen LE, Birch NP, Schiffman JD, Crosby AJ, Peyton SR, J. Mech. Behav. Biomed. Mater 2015, 50, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Budday S, Nay R, de Rooij R, Steinmann P, Wyrobek T, Ovaert TC, Kuhl E, J. Mech. Behav. Biomed. Mater 2015, 46, 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Cruz-Acuña R, Quirós M, Farkas AE, Dedhia PH, Huang S, Siuda D, García-Hernández V, Miller AJ, Spence JR, Nusrat A, Nat. Cell Biol 2017, 19, 1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]a).Chandrasekaran S, Guo N.-h., Rodrigues RG, Kaiser J, Roberts DD, J. Biol. Chem 1999, 274, 11408; [DOI] [PubMed] [Google Scholar]; b) Lichtner RB, Howlett AR, Lerch M, Xuan J-A, Brink J, Langton-Webster B, Schneider MR, Exp. Cell Res 1998, 240, 368. [DOI] [PubMed] [Google Scholar]
- [34].Li S, Nih LR, Bachman H, Fei P, Li Y, Nam E, Dimatteo R, Carmichael ST, Barker TH, Segura T, Nat. Mater 2017, 16, 953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Valiente M, Obenauf AC, Jin X, Chen Q, Zhang XH-F, Lee DJ, Chaft JE, Kris MG, Huse JT, Brogi E, Cell 2014, 156, 1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Sosunov AA, Guilfoyle E, Wu X, McKhann GM, Goldman JE, J. Neurosci 2013, 33, 7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Renault-Mihara F, Okada S, Shibata S, Nakamura M, Toyama Y, Okano H, Int. J. Biochem. Cell Biol 2008, 40, 1649. [DOI] [PubMed] [Google Scholar]
- [38].Etienne-Manneville S, Hall A, Cell 2001, 106, 489. [DOI] [PubMed] [Google Scholar]
- [39].Pedron S, Becka E, Harley BAC, Biomaterials 2013, 34, 7408. [DOI] [PubMed] [Google Scholar]
- [40].Rayahin JE, Buhrman JS, Zhang Y, Koh TJ, Gemeinhart RA, ACS Biomater. Sci. Eng 2015, 1, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Loebel C, Mauck RL, Burdick JA, Nat. Mater 2019, 18, 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Colognato H, Tzvetanova ID, Dev. Neurobiol 2011, 71, 924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pang Y, Fan LW, Tien LT, Dai X, Zheng B, Cai Z, Lin RCS, Bhatt A, Brain Behav. 2013, 3, 503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Bonneh-Barkay D, Wiley CA, Brain Pathol. 2009, 19, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Nishio T, Kawaguchi S, Iseda T, Kawasaki T, Hase T, Brain Res. 2003, 990, 129. [DOI] [PubMed] [Google Scholar]
- [46]a).Park J, Wetzel I, Marriott I, Dréau D, D’Avanzo C, Kim DY, Tanzi RE, Cho H, Nat. Neurosci 2018, 21, 941; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lin Y-T, Seo J, Gao F, Feldman HM, Wen H-L, Penney J, Cam HP, Gjoneska E, Raja WK, Cheng J, Neuron 2018, 98, 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Nih LR, Gojgini S, Carmichael ST, Segura T, Nat. Mater 2018, 17, 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Shull KR, Ahn D, Chen WL, Flanigan CM, Crosby AJ, Macromol. Chem. Phys 1998, 199, 489. [Google Scholar]
- [49].Barney LE, Jansen LE, Peyton SR, Dandley EC, Reich NG, Mercurio AM, Integr. Biol 2014, 7, 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Brooks EA, Jansen LE, Gencoglu MF, Yurkevicz AM, Peyton SR, ACS Biomater. Sci. Eng 2018, 4, 707. [DOI] [PubMed] [Google Scholar]
- [51].Jansen LE, Negron-Pineiro LJ, Galarza S, Peyton SR, Acta Biomater. 2018, 70, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Ferreira TA, Blackman AV, Oyrer J, Jayabal S, Chung AJ, Watt AJ, Sjöström PJ, Van Meyel DJ, Nat. Methods 2014, 11, 982. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.