Abstract
Liver malignancies are among the tumor types that are resistant to immune checkpoint inhibition therapy. Tumor-associated macrophages (TAMs) are highly enriched and play a major role in inducing immunosuppression in liver malignancies. Herein, CCL2 and CCL5 are screened as two major chemokines responsible for attracting TAM infiltration and inducing their polarization towards cancer-promoting M2-phenotype. To reverse this immunosuppressive process, we directly evolve an innovative single-domain antibody that bispecifically binds and neutralizes CCL2 and CCL5 (BisCCL2/5i) with high potency and specificity. mRNA encoding BisCCL2/5i was encapsulated in a clinically approved lipid nanoparticle platform, resulting in a liver-homing biomaterial that allows transient yet efficient expression of BisCCL2/5i in the diseased organ in a multiple dosage manner. This BisCCL2/5i mRNA nanoplatform significantly induces the polarization of TAMs toward the antitumoral M1 phenotype and reduces immunosuppression in the tumor microenvironment. The combination of BisCCL2/5i with PD-1 ligand inhibitor (PD-Li) achieves long-term survival in mouse models of primary liver cancer and liver metastasis of colorectal and pancreatic cancers. Our work provides an effective bispecific targeting strategy that could broaden the PD-Li therapy to multiple types of malignancies in the human liver.
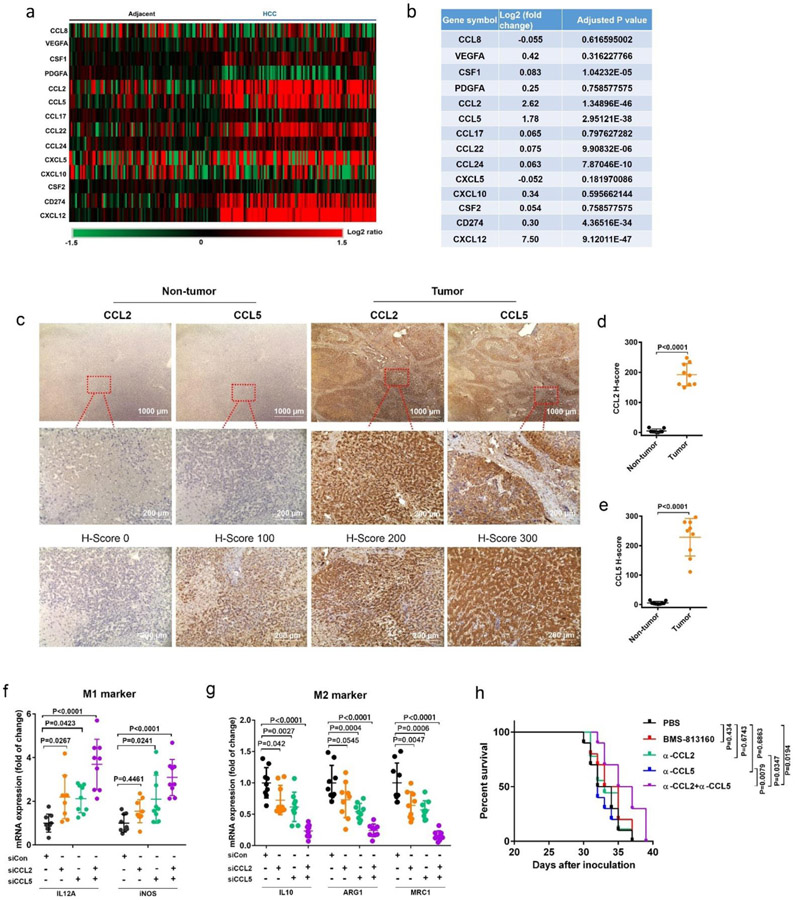
Tumors non-responsive to immune checkpoint inhibition are primed by a high density of immunosuppressive cells, including tumor-associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs) and regulatory T cells (Tregs), with little T cell infiltration in the tumor microenvironment (TME), a characteristic that emerged as a major barrier to the effectiveness of immune checkpoint inhibition therapy[1]. As the first extraintestinal organ, the liver is constantly at risk of attack from various harmful factors including bacterial endotoxins and virus infection, resulting in an immunosuppressive microenvironment that is particularly attractive for malignancy development and metastasis[2]. A large number of macrophages were reported to be presented in the peritumor and intratumor tissues (38.6%) of hepatocellular carcinoma (HCC) patients, and the level of FOXP3+ Treg cells in tumor tissues was much higher than that in normal liver tissues (3.9% vs. 0.3%; P<0.0001)[3]. Furthermore, compared to that in normal liver tissues, the frequency of MDSCs in HCC tumors was significantly increased and correlated with tumor size, burden and stage[4]. In an attempt to assess the top candidates of monocyte-associated genes that prime immunosuppression in liver malignancy, we analyzed the gene expression profiles (data extracted from the Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) database under the accession number GSE25097) of HCC liver tumor lesions and their matched adjacent normal liver samples from 197 patients. The distribution of differentially expressed probes and expression levels of monocyte-associated genes were displayed in Figure 1a and b. Comparison of the gene expression profiles of monocyte attractants showed profound changes in the TME. Both CCL2 and CCL5 were significantly upregulated (Log2 fold change >1.5, P<0.0001, FDR<0.05) in the HCC tumor sites. In contrast, no obvious differences in CXCL5, CXCL10, and CSF2, which are known to promote M1-phenotype polarization of macrophages[5], were found between malignant HCC and normal liver tissues (Figure 1a, b). Another monocytes attractant, CXCL12, was also upregulated at HCC tumor sites compared to HCC-free adjacent sites (Log2 fold change >5.0, P<0.0001, FDR<0.05), supporting the strategy of targeting CXCL12 as we demonstrated previously in the liver metastasis tumor models[6]. Consistent with the gene signature analysis, pronounced upregulation of CCL2 and CCL5 was observed via immunohistochemistry (IHC) in human HCC tumor tissues relative to non-tumor liver tissues (Figure 1c-e). Therefore, CCL2 and CCL5 appear to be the two top-ranked genes that trigger the tumor-infiltrating monocytes in liver cancer patients. Previous clinical trials used small molecular antagonists or monoclonal antibodies to block either CCL2/CCR2 or CCL5/CCR5 signaling pathways for the treatment of solid tumors such as colorectal cancer/liver metastasis, advanced prostate cancer, and pancreatic cancer/liver metastasis[7]. However, the therapeutic effects by blocking these signaling pathways have not been fully realized in clinical studies. More evidence suggests that TAMs differentiated from monocytes are a major component of the immune cells recruited to the TME during cancer progression[8]. To assess the impact of overexpression of CCL2 and CCL5 in TAMs, gene expression correlation analysis in the human HCC tumor sites was performed. As shown in Figure S1, gene expression of CCL2 and CCL5 was positively correlated with the expression of M2-phenotype macrophage markers (MRC1 and IL10), suggesting the indispensable role of CCL2 and CCL5 in the M2 polarization of TAMs during HCC progression. To evaluate the crosstalk between TAMs and tumor cells mediated by CCL2 and CCL5, murine bone-marrow-derived macrophages (BMDMs) were exposed to the culture medium of tumor cells to mimic the interactions between TAMs and cancer cells in the TME. We first knocked down CCL2 and CCL5 in Hepa1-6 cells, a mouse HCC cell line with pathological features similar to those of human HCC[9], and then cocultured BMDMs with the conditioned medium from Hepa1-6 tumor cells to track macrophage polarization in the presence or absence of a microenvironment with secreted CCL2 and CCL5. qRT-PCR analyses confirmed that silencing either CCL2 or CCL5 suppressed the gene expression of M2 markers and increased the expression of M1 markers to some extent. However, compared to mono-silencing, the combination of CCL2 and CCL5 silencing was much more effective in priming macrophages toward the M1 phenotype (Figure 1f, g). Consistent with the findings from BMDMs, the coculture of RAW 264.7 cells, a murine macrophage cell line, with the conditioned medium from Hepa1-6 cells also showed significant inhibition of M2 polarization (Figure S2c, d). To explore possible therapeutic strategy, we first investigated antitumor efficacy when CCL2 or/and CCL5 signaling was blocked by using a neutralizing antibody or a CCR2/CCR5 dual antagonist (BMS-813160) in HCC tumor-bearing mice (Figure 1h). Compared to the PBS treatment, mono-blockade of CCL2 or CCL5 did not result in significantly prolonged survival. Moreover, dual blockade of CCR2 and CCR5 via BMS-813160 showed negligible survival benefit, which might be caused by the poor pharmacokinetic profile of small molecule through systemic administration. The existence of other cognate receptors for CCL2 and CCL5 (e.g., CCR1 and CCR4) may also account for CCL2- and CCL5-driven chemotaxis[10], leading to the unsatisfactory antitumor effect of BMS-813160 in our mouse models. In contrast, a significant survival benefit was observed when CCL2 and CCL5 were simultaneously blocked using a combination of two neutralizing antibodies. These results support our hypothesis that both CCL2 and CCL5 are indispensable in driving macrophage polarization towards tumor-promoting M2 phenotype and mono-blockade of CCL2 or CCL5 only confers limited survival benefit in treating primary HCC.
Figure 1. Identification of the top-ranked monocytes-related genes that are associated with HCC cancer progression.
(a-b) Heat map (a) and comparison of relative gene expression levels (log2 fold change and adjusted p value) (b) between HCC-free sites (Adjacent) and tumor sites (HCC) in the diseased samples from liver cancer patients. In heat map, columns represent 197 patient samples from gene expression omnibus (GEO) database; rows represent monocytes-related genes. Values represent the log2 ratio over control (gene expression in adjacent samples). Data processing were performed using the R software package (version 4.0.2; https://cran.r-project.org/). (c) Upper panel: Representative IHC staining images of CCL2 and CCL5 (10×) and its regional magnification (40×) in the human liver cancer tissues and paired adjacent non-tumor liver tissues. Lower panel: Representative scores of IHC staining. Positive staining is indicated by brown color. (d-e) Staining score analysis of CCL2 (d) and CCL5 (e) expression in tumor samples from 9 HCC patients; data were analyzed by unpaired two-tailed Student’s t-test. (f-g) mRNA expression of classic M1 (f) and M2 (g) markers in BMDMs 24 hr post the addition of the conditioned medium from Hepa1-6 tumor cells. n = 9 biologically independent samples; data were analyzed by one-way ANOVA and Tukey’s multiple comparisons test. (h) Kaplan-Meier survival curves of orthotopic HCC tumor-bearing mice treated with PBS, BMS-813160 (25 mg/kg/day, i.p., 5 doses, 1 day apart), CCL2 neutralizing antibody alone (α-CCL2: 10 mg/kg, i.p., 3 doses, 3 days apart), CCL5 neutralizing antibody alone (α-CCL5: 5 mg/kg, i.p., 3 doses, 3 days apart), and α-CCL2 plus α-CCL5 antibodies, using 30% weight loss as the endpoint criteria. Each line represents one survival curve for each group of ten mice; Log-rank (Mantel-Cox) test. Data are represented as the mean ± s.d.
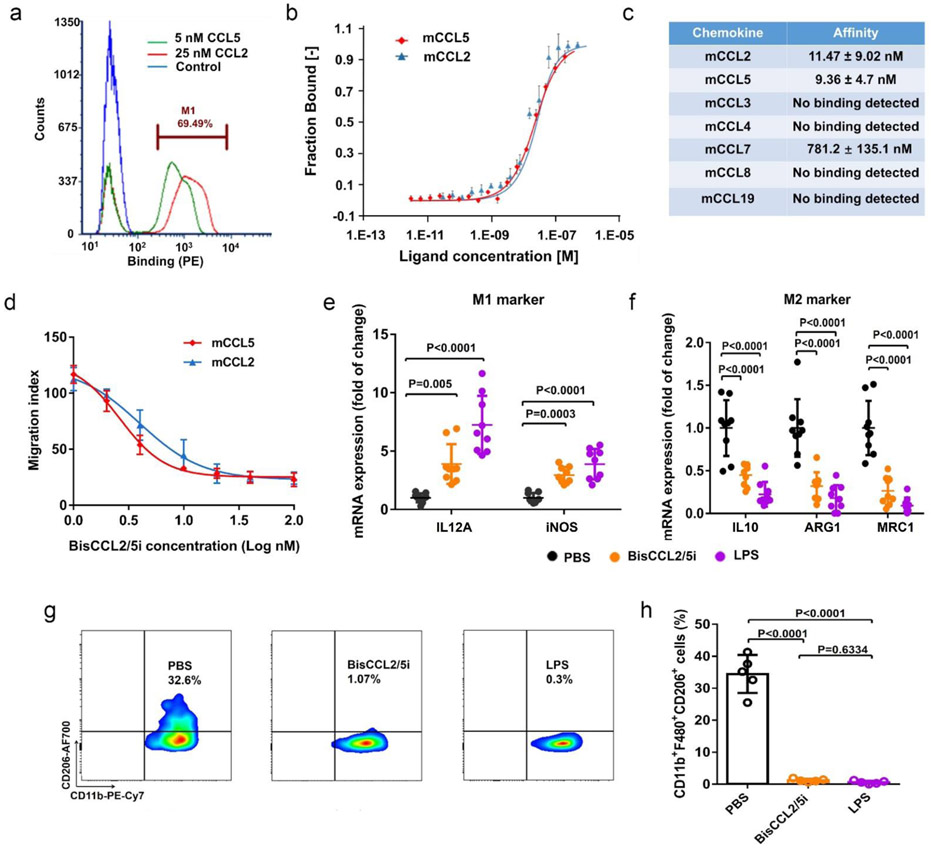
To effectively reverse immunosuppression without disrupting the signaling pathways mediated by other cognate chemokines that share the same receptors as CCL2 or CCL5, we directly evolved a highly specific CCL2/CCL5 dual inhibitor (BisCCL2/5i) from a single-domain antibody (VH) library displayed on the yeast cell surface (~108 variants) (Figure 2a). The evolved BisCCL2/5i was found to have a binding affinity of ~11.5 nM and ~9.4 nM for murine CCL2 and CCL5, respectively (Figure 2b). Moreover, BisCCL2/5i does not bind to other chemokines we tested except weakly to CCL7 (with an affinity around 780 nM, which is 70 times less compared to CCL2 and CCL5) (Figure 2c). The migration inhibition assay showed that BisCCL2/5i potently inhibited CCL2- or CCL5-mediated migration of macrophages, with IC50 values of approximately 4.0 nM and 2.6 nM, respectively (Figure 2d), similar to those of anti-CCL2 and anti-CCL5 neutralizing antibodies (α-CCL2: IC50 1.2 nM; α-CCL5: IC50 2.2 nM, Figure S3). Treatment of BMDMs with BisCCL2/5i significantly increased the expression of M1 markers while suppressed the expression of M2 markers (Figure 2e, f). Flow cytometric analysis confirmed a significant decline in the percentage of M2-phenotype macrophages after BisCCL2/5i or LPS (a classic M1 inducer) treatment, revealing a pronounced shift of macrophage polarization toward the M1 phenotype (Figure 2g, h). These results demonstrated that BisCCL2/5i can simultaneously block CCL2 and CCL5 signaling and effectively promote macrophage polarization toward the cancer-inhibitory M1 phenotype.
Figure 2. A unique single domain antibody that binds both CCL2 and CCL5 and blocks their biological activities.
(a) Demonstration of dual specificity of BisCCL2/5i by flow cytometry. (b-c) Binding affinity of BisCCL2/5i to mCCL2, mCCL5 or other related chemokine family members, measured by MST. (d) In Vitro inhibition of chemotaxis of monocytes in the Transwell assays. (n = 3 biologically independent samples). (e-f) mRNA expression of M1 and M2 markers in BMDMs 24 hr after the treatment with PBS, BisCCL2/5i protein, and LPS, respectively. n = 9 biologically independent samples; data were analyzed by one-way ANOVA and Tukey’s multiple comparisons test. (g-h) M2 macrophages sorted from IL4 stimulated BMDMs were stained with F4/80, CD11b, and CD206 16 hr post the incubation with PBS, BisCCL2/5i protein, and LPS. The percentage of M2 macrophage (CD11b+F4/80+CD206+) in the total sorted cells was shown in Figure 2h. The representative flow dots and FACS quantification showed BisCCL2/5i and LPS promoted M1 polarization of macrophages (n = 5 biologically independent samples; data were analyzed by one-way ANOVA and Tukey’s multiple comparisons test). Data are represented as the mean ± s.d.
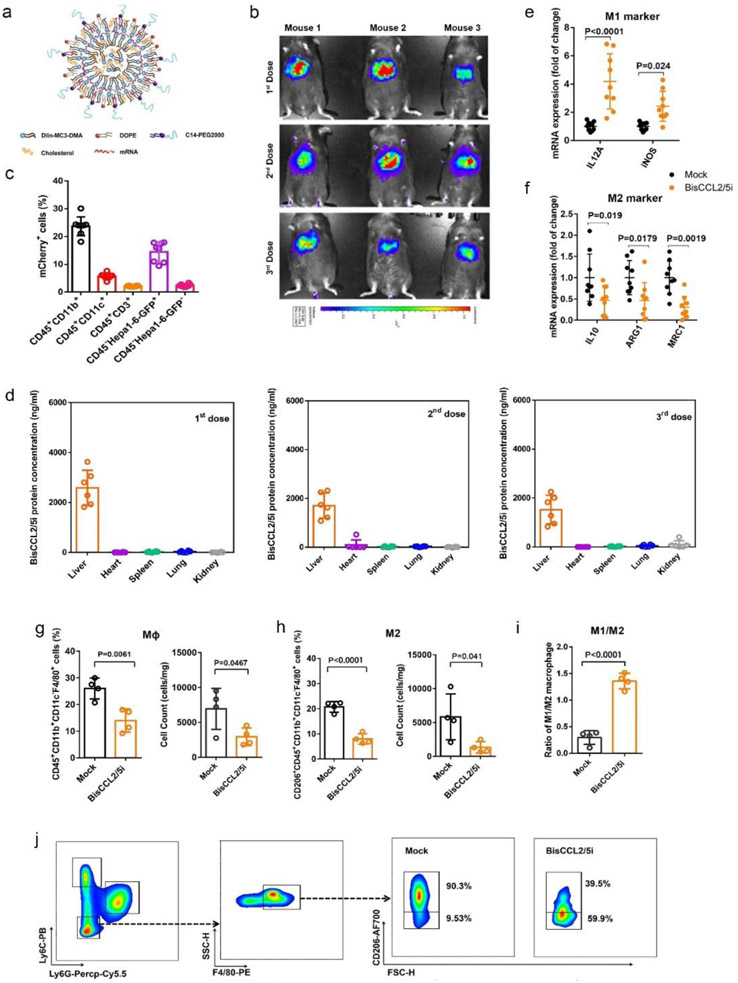
However, use of full-length antibodies during early stage of translational studies remains a challenge due to their complexity of posttranslational modification[11] and long circulatory half-life that could increase toxicities especially used at a high doses or in a combination[12]. During the past decade, an alternative approach emerged in cancer immunotherapy by biomaterial-delivery of the mRNA encoding a therapeutic protein such as single-domain antibody[13]. When appropriately delivered in vivo, an mRNA drug induces rapid and efficient production of the corresponding therapeutic protein by taking advantage of the translational machinery of the host cells, making the strategy safe and controllable[11a]. The first FDA-approved siRNA drug (ONPATTRO®) relies on Dlin-MC3-DMA-based lipid nanoparticles (MC3 LNPs) for siRNA delivery to hepatocytes[14]. In addition to their use in the delivery of siRNAs, MC3 LNPs have been explored for the delivery of therapeutic mRNA[15], in particular those that encode vaccines including recent ones against SARS-CoV-2[16]. Mechanistically, ionizable lipids such as MC3 have a pKa around 6.4 and are able to encapsulate mRNA during LNP production at low pH, ensuring the LNPs’ neutral surface charge in the circulation at physiological pH, and promote the quick release of the mRNA cargo from maturing endosomes into cytosol for protein synthesis following cell internalization[17]. Therefore, in this study, the mRNA encoding BisCCL2/5i was delivered to the liver malignancy based on the liver-homing MC3 LNP (Figure 3a). It should be noted that a highly potent signaling peptide (from albumin) was engineered at the very N-terminus of BisCCL2/5i, resulting in efficient secretion from cytoplasm into the local TME.
Figure 3. Dual blockade of CCL2 and CCL5 via LNP-mediated mRNA delivery of BisCCL2/5i polarizes macrophage M1 phenotype and reduces the immunosuppression in the TME.
(a) Schematic of the mRNA-loaded LNPs. (b) In vivo transfection of Luc mRNA-LNPs after repeated administration (i.v., every 4 days, in total 3 doses). The luciferase was injected intraperitoneally into the mice 6 hr post the administration of Luc mRNA-LNPs, followed by measuring the luc bioluminescence signal using IVIS imaging. n = 3 biologically independent samples. (c) The quantification of mCherry-positive cells expressed in murine orthotopic HCC tumor tissue 6 hr after injection of mCherry mRNA-LNPs (mCherry mRNA: 0.5 mg/kg). mRNA is mainly expressed in monocytes (CD45+CD11b+) and tumor cells (Hepa1-6-GFP+) (n = 8 biologically independent mice per group). (d) BisCCL2/5i expression in different organs 6 hr after each administration of BisCCL2/5i mRNA-LNPs (mRNA: 1 mg/kg, i.v., 3 days apart). n = 6 biologically independent samples. The BisCCL2/5i mRNA was mainly expressed in the liver tissue and the repeated administration resulted in comparable protein level. (e-f) mRNA expression of classic M1 (d) and M2 (e) markers in the HCC tumor tissues 48 hr after systemic administration of formulated LNPs as a dose corresponding to 1 mg/kg mRNA (Mock, HcRed mRNA). Each data point is an individual sample (n = 9); one-way ANOVA and Tukey’s multiple comparisons test. (g-j) Change of the immunocellular composition in the HCC TME 48 hr following Mock mRNA-LNPs and BisCCL2/5i mRNA-LNPs treatments (mRNA: 1 mg/kg), measured by flow cytometry (n = 4 biologically independent samples; unpaired two-tailed Student’s t-test; the experiment was conducted three times independently with similar results). (g-h) The percentage and cell counts of macrophages (g) and their M2 subtype (h) in the total immune cells. (i-j) Representative flow dots of M1- and M2-phenotype macrophages (j) and the ratio of M1/M2 (i). MΦ, macrophages (CD45+CD11b+CD11c−Ly6C−Ly6G−F4/80+); M2, M2-phenotype macrophages (CD206+). Data are represented as the mean ± s.d.
The mRNA-loaded LNPs with a diameter of approximately 100-120 nm showed a high transfection efficacy in the liver tissue but drastically less in other organs, as demonstrated by using luciferase (Luc) mRNA as a reporter gene (Figure S4 and Figure 3b). To determine which cell types within the liver tumor were transfected, we delivered mCherry mRNA-LNPs to an orthotopic HCC tumor model in which Hepa1-6 tumor cells were stably transfected with a vector carrying GFP. Confocal images and flow cytometric analysis demonstrated that the mRNA-LNPs were preferentially internalized by Hepa1-6 tumor cells (GFP+) and myeloid cells (CD11b+), resulting in efficient expression of mCherry protein in these cells (Figure 3c, Figure S5 and Figure S6). When BisCCL2/5i mRNA-LNP was used, the corresponding BisCCL2/5i protein showed highest expression in the liver compared with other major organs (Figure 3d), consistent with the results from use of Luc mRNA. BisCCL2/5i protein levels in the plasma were evaluated after a single injection into mice. The maximal protein concentration in the plasma (2,086 ± 927 ng/mL) was observed 6 hr after injection, followed by a decrease yet detectable measurement until day 3. The total protein level over time (area under the curve (AUC)) was approximately 34,403 ng·hr/mL. The decay phase (λz) in the serum suggests a distribution process between central blood and peripheral tissues. The BisCCL2/5i protein generated by the expression of delivered mRNA has a MW around 13 kDa, which presumably will be quickly cleared by kidney during circulation. Nevertheless, the half-life of the protein expression after each administration of BisCCL2/5i mRNA-LNPs is around 87 hr (Figure S8a, b), indicating the continuous translation of the mRNA inside the transfected cells prior to its degradation, consistent with the results from other LNP-based mRNA delivery in the literature[12b, 18]. Unlike viral gene delivery that is often limited to single dosage, repeated dosage is possible for non-viral approach. It has been reported that repeated dosage of mRNA therapy substantially improved survival relative to a single dose in the MC38-R tumor model[12b]. We confirmed that repeated administration of mRNA-LNPs induced comparable protein expression by quantifying the luciferase activity and BisCCL2/5i levels in different organs (Figure S4c and Figure 3d), supporting the feasibility of long-term treatment with our mRNA therapeutics. We further evaluated antitumor efficacy in the orthotopic HCC tumor model after treatment with either Mock (HcRed) mRNA-LNPs as control, BisCCL2/5i mRNA-LNPs, or combined CCL2- and CCL5-neutralizing antibodies, respectively. As shown in Figure S9a and b, both BisCCL2/5i and neutralizing antibodies resulted in prolonged survival relative to the Mock group. Notably, the BisCCL2/5i mRNA delivered by LNPs showed a greater survival benefit than a combination of neutralizing antibodies, indicating a clear advantage of using the BisCCL2/5i mRNA-LNPs over the sophisticated administration of anti-CCL2 and anti-CCL5 antibodies simultaneously. qRT-PCR results confirmed the BisCCL2/5i mRNA-LNPs were able to suppress the M2-TAM polarization (Figure 3e, f). We further evaluated the effect of BisCCL2/5i mRNA-LNPs on the immunocellular composition of the HCC TME. Treatment with BisCCL2/5i mRNA-LNPs decreased the percentage and cell number of tumor-infiltrating macrophages relative to Mock mRNA-LNPs (Figure 3g), indicating the blockage of CCL2 and CCL5 reduced intratumoral trafficking of macrophages. It is noteworthy that BisCCL2/5i treatment led to more than 50% reduction of M2 fraction in total macrophage population (20.8% vs. 8.0% of M2 fraction in total macrophages in Mock vs. BisCCL2/5i groups) (Figure 3h) and 4.4-fold increase of M1/M2 ratio (0.3 vs. 1.31 in Mock vs. BisCCL2/5i groups) (Figure 3i, j), suggesting that BisCCl2/5i not only inhibited the further infiltration of macrophages, but also drove the polarization of existing M2 macrophage towards M1 subtype. No significant change in monocytic MDSCs was observed between the Mock group and BisCCL2/5i group (Figure S10a, b). However, the proportion of granulocytic MDSCs, a predominate composition of MDSCs in most cancer types[19], was decreased after BisCCL2/5i treatment, suggesting the existence of chemotaxis-mediated reduction of g-MDSCs by blocking CCL2 and CCL5. It should be noted that there are many different TAM/MDSC subtypes. The investigation of detailed effect of BisCCL2/5i on other TAM/MDSC subtypes is necessary in the future studies. The reduced proportion of Tregs was also observed in the BisCCL2/5i treated group (Figure S10c). The suppression of M2-phenotype macrophage polarization, g-MDSCs, and Tregs after BisCCL2/5i treatment indicated reduced immunosuppression in the TME, which was confirmed by increased levels of CD8+ T cells and NK cells (Figure S10d, e). These results demonstrated that the liver-homing delivery of BisCCL2/5i mRNA via LNP efficiently promoted the polarization of TAMs from the cancer-promoting M2 phenotype to the cancer-inhibitory M1 phenotype and shifted the immunocellular composition of TME into antitumor immunity.
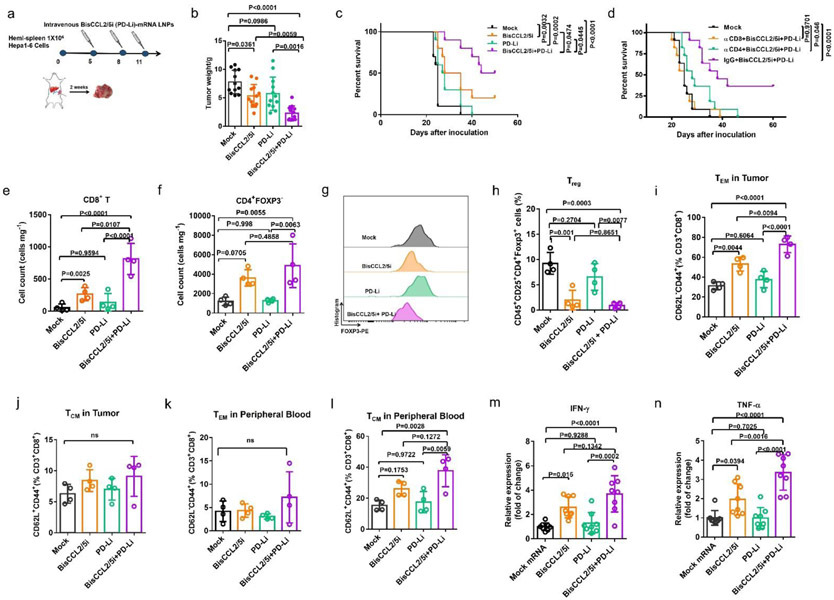
To evaluate whether the reduced immunosuppression in the TME by BisCCL2/5i treatment could synergistically improve the immunotherapy, we used a trimeric PD-1 ligand inhibitor (PD-Li) that was developed in our group[20] (Figure S11), by delivering its mRNA encapsulated in LNPs in the identical manner as that for BisCCL2/5i, allowing the same approach of drug administration and almost exclusive uptake by the liver where the malignancies are located. We therefore evaluate the therapeutic efficacy of BisCCL2/5i in sensitizing tumors to the blockade of PD-1/PDL-1 in the large orthotopic HCC tumor model (Figure S12a). Tumor weight measurements and survival rates showed a modest therapeutic benefit with the monotherapies (Figure S12b). Systemic administration of BisCCL2/5i mRNA-LNPs significantly improved the response to PD-Li inhibition therapy, with Kaplan-Meier survival estimates at 32 days (Mock), 37 days (BisCCL2/5i therapy), 35 days (PD-Li therapy), and 49 days (BisCCL2/5i plus PD-Li combination therapy), respectively, demonstrating that BisCCL2/5i therapy sensitized the orthotopic HCC tumors to the blockade of PD-1/PD-L1 therapy (Figure S12c). We further adopted a hemi-spleen approach that allows for efficient establishment of uniform and diffuse HCC in the liver, which better mirrors HCC in humans. Hepa1-6 tumor cells were inoculated specifically to the liver via a hemi-spleen injection, and the treatment was initiated in mice bearing diffuse tumors (~5 days) (Figure 4a). Notably, approximately 58% of mice (7 out of 12) administered BisCCL2/5i in combination with PD-Li exhibited complete antitumor responses, without evidence of residual tumor burden at least 50 days after tumor cell inoculation (Figure 4b, c). These results clearly demonstrated that the combination therapy can confer a significant survival benefit and promote tumor eradication in diffuse liver cancers. The important role of CD4+ and CD8+ T cells in the treatments was further verified by a depletion study in which anti-CD4 or anti-CD8 mAbs significantly compromised therapeutic efficacy compared to that of the IgG isotype control, while CD8+ T cells appeared to play a more important role in antitumor immunity than CD4+ T cells (Figure 4d). As expected, significant upregulation of intratumoral CD8+ T cells were observed 4 days after the last treatment of BisCCL2/5i and PD-Li (Figure 4e). The counts of CD4+FOXP3− T cells were also elevated while the proportion of Tregs was decreased in the combination therapy (Figure 4f-h), suggesting the reduced immunosuppression in the TME. Due to the long-term survival in this hemi-spleen tumor model, memory CD8+ T cells (effector memory CD8+ T cell (TEM) and central memory CD8+ T cell (TCM)) within tumor site and in peripheral blood were also characterized after different treatments (Figure 4i-l). The frequency of both CD8+ TEM and CD8+ TCM cells was measured 4 days after the final treatment. Negligible change of TEM cells (CD44+CD62L− gated from CD3+CD8+ T cells) was observed after PD-Li treatment compared to the Mock group. However, BisCCL2/5i mRNA-LNPs treatment showed increased fraction of CD8+ TEM cells. Notably, the combination treatment considerably increased the level of CD8+ TEM cells relative to Mock and monotherapies. Although the fraction of CD8+ TCM (CD44+CD62L+ gated from CD3+CD8+ T cells) was not elevated in the tumor site, this fraction remarkably increased in the systemic circulation after combined treatment (Figure 4l). These results indicated that memory CD8+ T cells are mainly responsible for the long-term survival after BisCCL2/5i and PD-Li combination therapy. Additionally, the combination treatment caused a remarkable increase in mRNA expression level of TNF-α and IFN-γ relative to Mock or monotherapies in the TME (Figure 4m, n), confirming that the combination therapies resulted in the activation of T cells and revoked the immunosuppression.
Figure 4. Dual blockade of CCL2 and CCL5 sensitizes HCC tumors to the PD-1/PD-L1 inhibition therapy.
(a) Treatment scheme for HCC tumors established by hemi-spleen approach. (b) Average tumor weight of different treated groups. n = 12 biologically independent samples; one-way ANOVA and Tukey’s multiple comparisons test. (c) Kaplan-Meier survival curve in different treated groups. n = 10 biologically independent samples; Log-rank (Mantel-Cox) test. (d) Kaplan-Meier survival analysis of HCC tumor bearing-mice (for CD4+ or CD8+ T-cell depletion and BisCCL2/5i plus PD-Li treatments). n = 11 biologically independent samples; Log-rank (Mantel-Cox) test. (e-l) Change of the immunocellular composition in the HCC TME 4 days following Mock mRNA-LNPs and BisCCL2/5i mRNA-LNPs treatments (mRNA: 1 mg/kg), measured by flow cytometry (n = 4 biologically independent samples; one-way ANOVA and Tukey’s multiple comparisons test). (e-f) The counts of total CD8+ T (e) cells and CD4+FOXP3− T cells (f) in the HCC tumor site. (g-h) Representative flow cytometry histograms of FOXP3 expression (g) and the percentage of Treg cells (h) in the CD4+ T cells. (i-j) The percentages of effector memory CD8+ T cells (TEM) (i) and central memory CD8+ T cells (TCM) (j) in the CD8+ T population in HCC TME. (k-l) The percentages of CD8+ TEM cells (i) and CD8+ TCM cells (j) in the CD8+ T population in peripheral blood. (m-n) Relative expressions of IFN-γ and TNF-α cytokines in HCC TME 4 days following indicated treatments, detected by quantitative RT-PCR (n = 8 biologically independent samples; one-way ANOVA and Tukey’s multiple comparisons test). CD8+ T (CD45+CD3+CD8+); CD4+ T (CD45+CD3+CD4+); Treg (CD45+CD3+CD4+CD25+FOXP3+), CD8+ TEM (CD44+CD62L− gated from CD3+CD8+ T cells), CD8+ TCM (CD44+CD62L+gated from CD3+CD8+ T cells). Data are represented as the mean ± s.d.
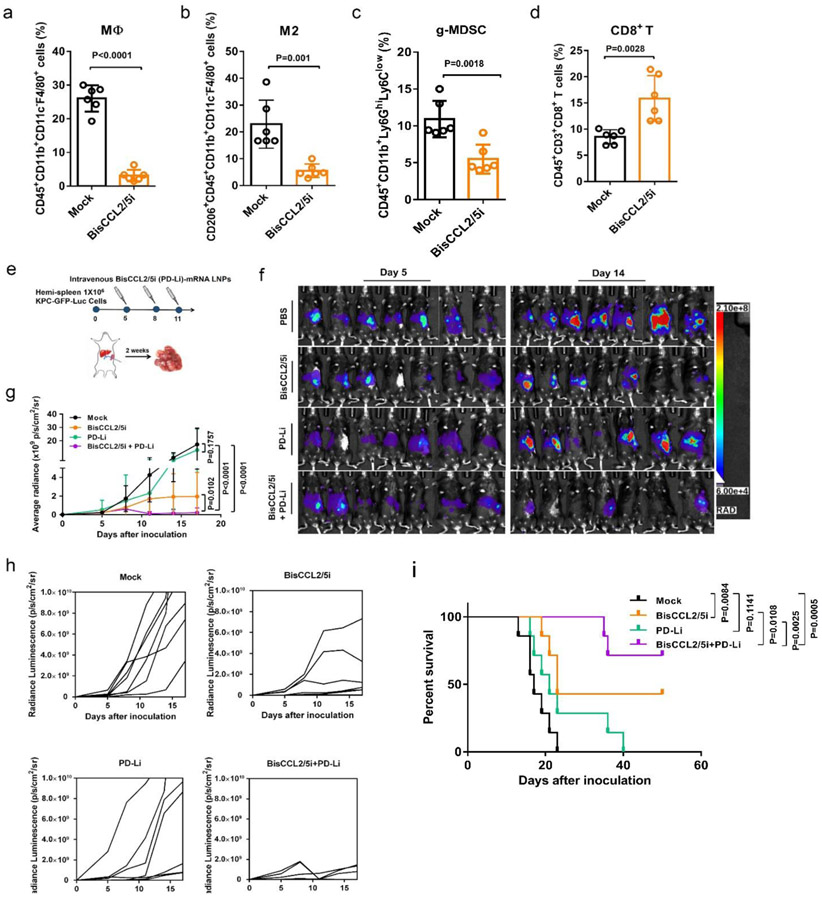
Clinically, the high recurrence rate in cancer patients is typically caused by residual metastases after surgery, and liver metastasis is the major cause of death in patients with digestive tract malignancies, in particular colorectal and pancreatic cancers[21]. To evaluate whether BisCCL2/5i therapy can sensitize liver metastatic tumors to the inhibition of PD-1/PD-L1 therapy, pancreatic cancer liver metastasis mouse model was established using a KPC-GFP-Luc cell line. Enriched macrophage infiltration (~25%) (Figure 5a) and upregulation of CCL2 and CCL5 as shown by IHC staining (Figure S7) in KPC liver metastatic tumors implied the suitability of BisCCL2/5i treatment. Similar to the HCC tumor, treatment with BiCCL2/5i mRNA-LNP decreased M2-polarized macrophage and MDSC populations inside KPC liver metastases (Figure 5b, c), presumably leading to increased intratumoral infiltration of CD3+ T cells (Figure S13f) and in particular CD8+ T cells (Figure 5d) compared to the Mock group. BisCCL2/5i monotherapy significantly mitigated the progression of the liver metastasis of KPC tumor, whereas PD-Li monotherapy showed negligible tumor inhibition compared to the Mock treatment (Figure 5f-i). Relative to the monotherapies, the combination of BisCCL2/5i with PD-Li most effectively controlled the liver metastatic KPC tumor growth and prolonged survival benefit, inducing a complete response in approximately 57% of KPC-bearing mice. The therapeutic efficacy of BisCCL2/5i was further tested in a CT26 colorectal cancer liver metastasis model, which also shows signatures including high expression levels of CCL2 and CCL5 and enriched macrophages (~30%) within the metastatic lesions (Figure S7 and Figure S14b). BisCCL2/5i-mediated modulation of immune microenvironment in this CRC liver metastatic model was confirmed by upregulated CD3+ T cell infiltration, downregulated macrophage accumulation and decreased M2-phenotype inside CT26 liver metastases (Figure S14a and Figure S14c). Similar to that observed in the KPC liver metastasis model, BisCCL2/5i plus PD-Li exhibited significant tumor inhibition and conferred a prolonged survival benefit relative to Mock and monotherapies (Figure S14e-g). These results clearly demonstrated that dual-blockade of CCL2/CCL5 via BisCCL2/5i mRNA-LNPs and its synergistic potential combined with PD-Li therapy can be expanded to a variety of secondary liver malignancies.
Figure 5. Dual blockade of CCL2 and CCL5 sensitized KPC liver metastasis tumor to PD-1/PD-L1 inhibition therapy.
(a-d) Change of the immunocellular composition in the KPC liver metastatic TME 48 hr following Mock mRNA and BisCCL2/5i mRNA-LNPs treatment (mRNA: 1 mg/kg, i.v.), measured by flow cytometry (n = 6 biologically independent samples; unpaired two-tailed Student’s t-test). The percentage of macrophages (a), M2-phenotype macrophages (b), and g-MDSC (c) in the total immune cells. The percentage of CD8+ T cells in the CD3+ cells (d). (e) Treatment scheme for KPC liver metastasis tumors administered intravenously with various formulations (mRNA: 1 mg/kg). (f-g) In vivo bioluminescence imaging (f) and tumor growth burden (g) of mice bearing KPC liver metastasis receiving various treatments (n = 7 biologically independent samples; two-way ANOVA with multiple comparisons). The experiments were conducted twice independently with similar results. (h) Spider plots of individual tumor growth curves (n = 7 biologically independent samples in each group). (i) Kaplan-Meier survival analysis of KPC liver metastasis tumor bearing-mice after indicated treatments. n = 8 biologically independent samples; Log-rank (Mantel-Cox) test. Data are represented as the mean ± s.d.
Systemic side effects are major concerns for anti-cancer therapies. We tested the safety of the BisCCL2/5i mRNA-LNP and PD-Li mRNA-LNP delivery strategy in orthotopic HCC tumor models. Blood was collected and subjected to blood panel analysis. Although the level of red blood cell (RBC) tends to decrease after the combination treatment compared with the Mock control, the cell counts for RBC are not out of normal range (Figure S15a), suggesting it is not biologically relevant. Additionally, no abnormal changes in white blood cell (WBC) counts were observed in each group, indicating the low immunogenicity of different indicated treatments (Figure S15b). The lack of detectable systemic toxicity was further confirmed by normal liver function (ALT and AST levels) and kidney function (blood urea nitrogen (BUN) and creatinine (CREAT)) across all the treatment groups (Figure S15c-f). Moreover, body weight was consistent across all groups tested (Figure S15g). It should be noted that immunotherapy-related adverse events (irAEs) are the common complications of systemic administration of immunotherapeutics. Th17 cells have been reported to be highly upregulated in inflammatory tissues of autoimmune diseases[22]. Our previous study confirmed the elevation of Th17 cell numbers in the spleen after systemic treatment using anti-PD-L1 monoclonal antibody [23]. Therefore, we measured the proportion of Th17 cells as a parameter to monitor irAEs during mRNA-LNP immunotherapy. No obvious upregulation of Th17 cells was observed in the spleen in the treated groups, indicating that this liver-homing biomaterial approach to deliver the mRNAs encoding BisCCL2/5i and PD-Li protein may mitigate irAEs (Figure S15h). Our work supports the notion that directed molecular evolution allows to generate innovative multi-functional biological molecules that are otherwise difficult to achieve by rational design. It is challenging to develop a small protein domain that tightly and specifically binds to both CCL2 and CCL5, two chemokines that share low homology. We suspect that BisCCL2/5i recognizes these two chemokines at a motif or region that is structurally conserved at a three-dimensional level. It will be of great interest to investigate the elimination and the interaction of BisCCL2/5i protein with their target proteins in future studies.
In summary, we have demonstrated the indispensable role of both CCL2 and CCL5 in the development of liver malignancies and bioengineered a single domain antibody by directed molecular evolution to bispecifically block both CCL2 and CCL5 simultaneously. By delivering the mRNA encoding BisCCL2/5i, we confirmed the clear advantage of using liver-homing therapeutic mRNA-LNPs over the administration of a combination of anti-CCL2 and anti-CCL5 antibodies or of small molecule antagonists against CCR2/CCR5 that have numerous endogenous ligands other than CCL2/CCL5. This advanced biomaterial based on mRNA/LNP system could be an ideal approach to fill the gap between small molecules and full-length mAbs, and facilitate the deeper exploration of combination immunotherapy with great clinical potential. The combination of BisCCL2/5i with PD-Li therapy produced a robust anti-cancer response and long-term survival in the syngeneic mouse models of three major liver malignancies, including primary HCC, liver metastasis of colorectal cancer and liver metastasis of pancreatic cancer. Since the same nanoplatform was used to deliver different therapeutic mRNAs, it allows for the same approach of drug administration, almost exclusive uptake by the liver as the diseased organ, and the potential to study other mRNA combinations in the future if modulators other than immune checkpoint blockade are of interest. The expression level achieved via LNP-mediated mRNA delivery allows the immunotherapeutic proteins to take effect at very low doses of mRNA administration, making it possible to further reduce the systemic toxicity. We measured the proportion of Th17 cells as a parameter to monitor the irAEs of immunotherapy. No obvious changes in the proportion of Th17 cells were observed in the spleen across different treatment groups, indicating that the mRNA-LNP delivery and transient expression system could be a promising approach to mitigate irAEs. It should be noted that the murine tumor models used in this study do not allow us to recapitulate the irAEs occurred in human patients. Hopefully, future tumor models will better simulate the development of tumors and irAE phenotypes in human patients, allowing more accurate evaluation of potential immunogenicity of therapeutics based on mRNA-LNP nanoplatforms. High expression of CCL2 and CCL5 have been observed in human HCC, suggesting clinical feasibility of the CCL2/CCL5-dual blockade strategy. The combination strategy based on mRNA-LNPs delivery system reported here has the potential to be applied to other cancer types, especially those that contain enriched TAMs. It will be of great interest to investigate the effect of BisCCL2/5i in other TAM enriched preclinical murine tumor models. Our study provides a strong rationale for combining PD-Li therapy with co-blockade of CCL2/CCL5 in treating primary and metastatic liver malignancies aiming to achieve a meaningful impact on patient survival and broaden PD-Li inhibition therapy to other cancer types.
Statistical analysis
Data were expressed as the mean ± standard deviation (s.d.). Statistical analysis was performed by unpaired two-tailed Student’s t-test when only two value sets were compared or ANOVA comparison between multiple groups. Log-rank Mantel-Cox test was used for survival curves. All statistical analyses were done using GraphPad Prism 7.0. No exclusion criteria were incorporated in the design of the experiments for this study.
Supplementary Material
Acknowledgement
Funding: The work was supported by an R01 grant CA157738 (to R.L.) from the NCI and the innovation grants (RX03812125, RX03612118, and RX03202109) from the Eshelman Institute for Innovation (to R.L.).
Contributor Information
Ying Wang, Division of Chemical Biology and Medicinal Chemistry, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Karthik Tiruthani, Division of Chemical Biology and Medicinal Chemistry, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Sirui Li, Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, NC 27599, USA.
Mengying Hu, Division of Pharmacoengineering and Molecular Pharmaceutics, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Guojie Zhong, Department of Systems Biology, Columbia University, New York, NY, 10032, USA.
Yu Tang, Division of Pharmacotherapy and Experimental Therapeutics, Eshelman School of Pharmacy, University of North Carolina, Chapel Hill, NC, 27599, USA.
Sourav Roy, Division of Chemical Biology and Medicinal Chemistry, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Lillian Zhang, Division of Chemical Biology and Medicinal Chemistry, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Jun Tan, Department of Hepatology, HwaMei Hospital, University of Chinese Academy of Sciences, Ningbo, Zhejiang 315010, P.R. China..
Chengheng Liao, Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, NC 27599, USA.
Rihe Liu, Division of Chemical Biology and Medicinal Chemistry, Eshelman School of Pharmacy, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; Lineberger Comprehensive Cancer Center, University of North Carolina School of Medicine, Chapel Hill, NC 27599, USA; Carolina Center for Genome Sciences, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Reference
- [1].a) Zhou J, Liu M, Sun H, Feng Y, Xu L, Chan AW, Tong JH, Wong J, Chong CCN, Lai PB, Gut 2018, 67, 931; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Zamarin D, Holmgaard RB, Subudhi SK, Park JS, Mansour M, Palese P, Merghoub T, Wolchok JD, Allison JP, Science translational medicine 2014, 6, 226ra32; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kong L-Q, Zhu X-D, Xu H-X, Zhang J-B, Lu L, Wang W-Q, Zhang Q-B, Wu W-Z, Wang L, Fan J, PloS one 2013, 8, e59771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Yan J, Li S, Li S, International reviews of immunology 2014, 33, 498; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Abu-Shanab A, Quigley EM, Nature reviews Gastroenterology & hepatology 2010, 7, 691; [DOI] [PubMed] [Google Scholar]; c) Schnabl B, Brenner DA, Gastroenterology 2014, 146, 1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Tu J-F, Ding Y-H, Ying X-H, Wu F-Z, Zhou X-M, Zhang D-K, Zou H, Ji J-S, Scientific reports 2016, 6, 35056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Wang D, An G, Xie S, Yao Y, Feng G, Tumor Biology 2016, 37, 10427. [DOI] [PubMed] [Google Scholar]
- [5].Jaguin M, Houlbert N, Fardel O, Lecureur V, Cellular immunology 2013, 281, 51. [DOI] [PubMed] [Google Scholar]
- [6].Goodwin TJ, Zhou Y, Musetti SN, Liu R, Huang L, Science translational medicine 2016, 8, 364ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].a) Marques RE, Guabiraba R, Russo RC, Teixeira MM, Expert opinion on therapeutic targets 2013, 17, 1439; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hardaway AL, Herroon MK, Rajagurubandara E, Podgorski I, Cancer and Metastasis Reviews 2014, 33, 527; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Klarenbeek A, Maussang D, Blanchetot C, Saunders M, van der Woning S, Smit M, de Haard H, Hofman E, Drug Discovery Today: Technologies 2012, 9, e237. [Google Scholar]
- [8].a) Devalaraja S, To TKJ, Folkert IW, Natesan R, Alam MZ, Li M, Tada Y, Budagyan K, Dang MT, Zhai L, Cell 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Cho H, Seo Y, Loke KM, Kim S-W, Oh S-M, Kim J-H, Soh J, Kim HS, Lee H, Kim J, Clinical Cancer Research 2018, 24, 5407; [DOI] [PubMed] [Google Scholar]; c) Movahedi K, Laoui D, Gysemans C, Baeten M, Stangé G, Van den Bossche J, Mack M, Pipeleers D, In't Veld P, De Baetselier P, Cancer research 2010, 70, 5728. [DOI] [PubMed] [Google Scholar]
- [9].a) Bernhard H, Darlington G, Ruddle F, Developmental biology 1973, 35, 83; [DOI] [PubMed] [Google Scholar]; b) Li X, Yao W, Yuan Y, Chen P, Li B, Li J, Chu R, Song H, Xie D, Jiang X, Gut 2017, 66, 157. [DOI] [PubMed] [Google Scholar]
- [10].a) Mantovani A, Sozzani S, Locati M, Allavena P, Sica A, Trends in immunology 2002, 23, 549; [DOI] [PubMed] [Google Scholar]; b) Ruytinx P, Proost P, Van Damme J, Struyf S, Frontiers in immunology 2018, 9, 1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Pardi N, Secreto AJ, Shan X, Debonera F, Glover J, Yi Y, Muramatsu H, Ni H, Mui BL, Tam YK, Nature communications 2017, 8, 1; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sifniotis V, Cruz E, Eroglu B, Kayser V, Antibodies 2019, 8, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].a) Hassel JC, Heinzerling L, Aberle J, Bähr O, Eigentler TK, Grimm M-O, Grünwald V, Leipe J, Reinmuth N, Tietze JK, Cancer treatment reviews 2017, 57, 36; [DOI] [PubMed] [Google Scholar]; b) Hewitt SL, Bai A, Bailey D, Ichikawa K, Zielinski J, Karp R, Apte A, Arnold K, Zacharek SJ, Iliou MS, Science Translational Medicine 2019, 11. [DOI] [PubMed] [Google Scholar]
- [13].a) Miao L, Li L, Huang Y, Delcassian D, Chahal J, Han J, Shi Y, Sadtler K, Gao W, Lin J, Nature biotechnology 2019, 37, 1174; [DOI] [PubMed] [Google Scholar]; b) Billingsley MM, Singh N, Ravikumar P, Zhang R, June CH, Mitchell MJ, Nano Letters 2020, 20, 1578; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Rybakova Y, Kowalski PS, Huang Y, Gonzalez JT, Heartlein MW, DeRosa F, Delcassian D, Anderson DG, Molecular Therapy 2019, 27, 1415; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Krasniqi A, Bialkowska M, Xavier C, Van der Jeught K, Muyldermans S, Devoogdt N, D’Huyvetter M, New biotechnology 2018, 45, 69; [DOI] [PubMed] [Google Scholar]; e) Peterson EC, Laurenzana EM, Atchley WT, Hendrickson HP, Owens SM, Journal of Pharmacology and Experimental Therapeutics 2008, 325, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Cheng Q, Wei T, Farbiak L, Johnson LT, Dilliard SA, Siegwart DJ, Nature Nanotechnology 2020, 15, 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].a) Sayers EJ, Peel SE, Schantz A, England RM, Beano M, Bates SM, Desai AS, Puri S, Ashford MB, Jones AT, Molecular Therapy 2019, 27, 1950; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hajj KA, Melamed JR, Chaudhary N, Lamson NG, Ball RL, Yerneni SS, Whitehead KA, Nano Letters 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Kong N, Tao W, Ling X, Wang J, Xiao Y, Shi S, Ji X, Shajii A, Gan ST, Kim NY, Science Translational Medicine 2019, 11; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Islam MA, Xu Y, Tao W, Ubellacker JM, Lim M, Aum D, Lee GY, Zhou K, Zope H, Yu M, Nature biomedical engineering 2018, 2, 850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].a) Zhang N-N, Li X-F, Deng Y-Q, Zhao H, Huang Y-J, Yang G, Huang W-J, Gao P, Zhou C, Zhang R-R, Cell 2020, 182, 1271; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tai W, Zhang X, Drelich A, Shi J, Hsu JC, Luchsinger L, Hillyer CD, Tseng C-TK, Jiang S, Du L, Cell research 2020, 30, 932. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Moderna. https://www.modernatx.com/.; d) Pfizer. https://www.pfizer.com/health/coronavirus
- [17].Maugeri M, Nawaz M, Papadimitriou A, Angerfors A, Camponeschi A, Na M, Hölttä M, Skantze P, Johansson S, Sundqvist M, Nature communications 2019, 10, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kose N, Fox JM, Sapparapu G, Bombardi R, Tennekoon RN, De Silva AD, Elbashir SM, Theisen MA, Humphris-Narayanan E, Ciaramella G, Science immunology 2019, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].a) Youn JI, Collazo M, Shalova IN, Biswas SK, Gabrilovich DI, Journal of leukocyte biology 2012, 91, 167; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Lu L-C, Chang C-J, Hsu C-H, Journal of hepatocellular carcinoma 2019, 6, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Song W, Shen L, Wang Y, Liu Q, Goodwin TJ, Li J, Dorosheva O, Liu T, Liu R, Huang L, Nature communications 2018, 9, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].a) Malik I, Hussein F, Bush D, Alqaisi M, Bernal P, Byrd J, Garberoglio C, American journal of clinical oncology 2010, 33, 242; [DOI] [PubMed] [Google Scholar]; b) Zheng D-X, Meng S-C, Liu Q-J, Li C-T, Shang X-D, Zhu Y-S, Bai T-J, Xu S-M, World Journal of Gastroenterology 2016, 22, 3031; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Brodt P, Clinical Cancer Research 2016, 22, 5971. [DOI] [PubMed] [Google Scholar]
- [22].a) Yamada H, Journal of inflammation research 2010, 3, 33; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Hirota K, Hashimoto M, Ito Y, Matsuura M, Ito H, Tanaka M, Watanabe H, Kondoh G, Tanaka A, Yasuda K, Immunity 2018, 48, 1220; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Zhu S, Qian Y, Clinical science 2012, 122, 487. [DOI] [PubMed] [Google Scholar]
- [23].Song W, Shen L, Wang Y, Liu Q, Goodwin TJ, Li J, Dorosheva O, Liu T, Liu R, Huang L, Nature communications 2018, 9, 2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.