Abstract
Space radiation is characterized by high-linear energy transfer (LET) ionizing radiation. The relationships between the early biological effects of space radiation and the probability of cancer in humans are poorly understood. Bcl2 not only functions as a potent antiapoptotic molecule but also as an oncogenic protein that induces DNA replication stress. To test the role and mechanism of Bcl2 in high-LET space radiation-induced lung carcinogenesis, we created lung-targeting Bcl2 transgenic C57BL/6 mice using the CC10 promoter to drive Bcl2 expression selectively in lung tissues. Intriguingly, lung-targeting transgenic Bcl2 inhibits ribonucleotide reductase activity, reduces dNTP pool size and retards DNA replication fork progression in mouse bronchial epithelial cells. After exposure of mice to space radiation derived from 56iron, 28silicon or protons, the incidence of lung cancer was significantly higher in lung-targeting Bcl2 transgenic mice than in wild-type mice, indicating that Bcl2-induced DNA replication stress promotes lung carcinogenesis in response to space radiation. The findings provide some evidence for the relative effectiveness of space radiation and Bcl-2 at inducing lung cancer in mice.
Here we have discovered that Bcl2 potently promotes lung cancer development in response to space radiation. Our findings contribute to the development of new strategies for the prevention and/or treatment of space radiation-induced lung cancer by disrupting the oncogenic activity of Bcl2.
Introduction
Space radiation is comprised of high energy protons and high charge (Z) and energy (E) nuclei, whose ionization patterns in molecules, cells and tissues, and the resulting initial biological insults, are distinct from those of typical low-linear energy transfer (low-LET) radiation. Space radiation is characterized by high-LET ionizing radiation (high-LET IR) (1). High-LET IR is induced by high charge (Z) and energy (E) particles or high energy ions. Protons are characterized by low-LET although protons have slightly higher-LET than X-rays (2). Major radiation-induced solid cancer sites include lung, breast, thyroid and colon (3). Lung cancer makes up about one-third of the cancers attributable to radiation observed in atomic-bomb survivors (3) and nuclear-reactor workers (4). The relationships between the early biological effects of high-LET IR and the probability of cancer in human males and females are poorly understood. It is this missing knowledge that leads to large uncertainties in projecting cancer risk during space exploration.
We have previously demonstrated that high-LET IR derived from 56Fe or 28Si particles induced DNA double-strand break foci along ion tracks (i.e. clustered DSBs) in normal human bronchial epithelial cells (5). In contrast, low-LET IR derived from X-rays induced DSB foci with sparse ionization, and DNA breaks were scattered more evenly throughout the nucleus (5). The high-LET IR-induced clustered DSBs could be repaired by homologous recombination and non-homologous end-joining pathways, probably mainly via homologous recombination because high-LET IR has been reported to induce more small fragments of DNA DSBs (<40 base pairs) that prevent Ku binding to two ends of one DSB fragment at the same time, thus delaying Ku-dependent repair (6). Failure to repair, or inaccurate repair, could lead to either cell death (7), or gene mutations and the accumulation of genomic rearrangements that potentially promote tumorigenesis (8,9). It has recently been reported that exposure to 1 Gy (single or fractionated dose as 0.2 Gy × 5) of high-LET IR derived from 56Fe, 28Si and 16O induced a higher incidence of lung tumorigenesis than X-ray at the same dose in wild-type C57BL/6 mice (8).
DNA replication is a fundamental process of the cell that ensures accurate duplication of the genetic information and subsequent transfer to daughter cells. Various perturbations, originating from endogenous or exogenous sources, can interfere with proper progression and completion of the replication process, thus threatening genome integrity (10). DNA replication stress is an inefficient DNA synthesis process that leads replication forks to progress slowly or stall (11). DNA replication stress-induced fork collapse can also generate DSBs (12). Importantly, DNA replication stress has emerged as a major source of the deletions, translocations, amplifications and aneuploidies that are abundant in cancer (13). Oncogene activation alters DNA replication leading to increased replication stress, DNA breaks and activation of the DNA damage response (14). Bcl2 not only functions as a potent antiapoptotic molecule but also as an oncogenic protein that can enhance susceptibility to carcinogenesis (15). We have recently discovered that Bcl2 negatively regulates the homologous recombination DNA repair pathway in response to space radiation (5). Bcl2 also retards DNA replication fork progression via suppression of ribonucleotide reductase (RNR) activity (11). These properties of Bcl2 may synergize with space radiation to facilitate carcinogenesis. Here we report that selective expression of Bcl2 in lung tissues via the CC10 promoter causes DNA replication stress in mouse bronchial epithelial (MBE) cells leading to a significant increase in the incidence of lung cancer in response to high-LET space radiation or protons.
Materials and methods
Materials
Bcl2 and CC10 antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Mouse anti-BrdU clone B44 was purchased from Becton Dickinson (San Jose, CA). Rat anti-BrdU BU1/75 (ICR1) was purchased from Abcam (Cambridge, MA). Alexa Fluor 488 (green) goat anti-mouse, Alexa Fluor (red) 555 goat anti-rabbit and Alexa Fluor (red) 555 goat anti-rat were purchased from Invitrogen Life Technologies (Carlsbad, CA). The R.T.U. Vectastain kit, 3,3′-diaminobenzidine and hematoxylin were purchased from Vector Laboratories (Burlingame, CA). EpCAM (CD326) magnetic microbeads were obtained from Miltenyi Biotec (Auburn, CA). Keratinocyte serum-free medium was purchased from Invitrogen Life Technologies (Carlsbad, CA). C14-CDP and dC14-CDP were obtained from Moravek Biochemicals (Brea, CA). Adenylyl-imidodiphosphate, cytidine diphosphate (CDP), cytidine monophosphate, deoxycytidine monophosphate, deoxyuridine monophosphate, collagenase A and DNase I were purchased from Sigma Aldrich (St. Louis, MO). TLC Silica Plate was purchased from EMD Chemicals (Gibbstown, NJ). Bcl2T1F and Bcl2T1R primers were synthesized by Integrated DNA Technologies (Coralville, IA). The plasmids mcc10-BG1-rSPD and h3.7SPC/SV40 were kindly provided by Dr Jeffrey A. Whitsett (Cincinnati Children’s Hospital Medical Center, OH). The mBcl2/pCIneo construct was constructed by our laboratory (16). All of the reagents used were obtained from commercial sources unless otherwise stated.
Lung-targeting Bcl2 construct, transgenic mice and genotyping
Bcl2 transgenic construct was derived from mcc10-BG1-rSPD by removal of the Rat Pulmonary Surfactant Associated Protein D (rSPD) gene and ligation of a 0.8-kb EcoRI fragment containing the coding sequence for the mouse Bcl2 gene from h3.7SPC-mBcl2. Briefly, pCI-neo-Bcl2 consisting of the XhoI–XbaI fragment of the mouse Bcl2 gene was cloned into the SalI to XbaI site of h3.7SPC/SV40 by using the isoschizmers SalI and XboI. Then, the EcoRI fragment of the Bcl2 gene from h3.7SPC-mBcl2 was cloned into the EcoRI site in a mCC10 expression vector consisting of the HindIII–Hph1 fragment of the mCC10 promoter cloned into the BamHI site of pBSKS (17). Cloned into this plasmid were rabbit globin gene intron sequences (into a BamHI to EcoRI site) and the bovine growth hormone polyadenylation signal (XbaI to XhoI fragment from pGKneo cloned into the EcoRV site). The NotI–KpnI fragment containing the mcc10-Bcl2 transgene was excised, purified and introduced into the inbred C57BL/6 mouse genome by pronuclear microinjection as described previously (18–20). C57BL/6 mouse is a resistant strain with very clean background, which is not common to spontaneously develop cancers and has been widely used for studying exogenous carcinogen(s) or radiation-induced tumorigenesis (8,21–23). Transgenic founders positive for the transgene and transgenic pups were identified by PCR of tail DNA with a forward mcc10-Bcl2 vector primer (Bcl2T1F: 5′-CGT GCT GGT TAT TGT GCT GTC TC-3′) and a reverse Bcl2 complementary DNA (cDNA) primer (Bcl2T1R: 5′-CAT CCC ACT CGT AGC CCC TCT G-3′) as described previously (24). PCR was performed as follows: denaturation at 95°C for 5 min, 30 cycles of denaturation at 95°C for 30 s, annealing at 54°C for 30 s and extension at 72°C for 30 s, followed by a 5 min extension at 72°C. The primers were used at 0.4 µM and produced an 860-bp PCR product after 30 cycles. Bcl2 transgenic positive mice were bred and the colony was expanded.
High-LET radiation treatment
All animal studies were performed according to protocols approved by the Institutional Animal Care Use Committees (IACUC) of both Emory University and Brookhaven National Laboratory (BNL). Mice were housed in a temperature- and humidity-controlled environment in filter top cages with ad libitum access to food and water in an animal facility at Emory University. High-LET irradiation was performed in the Alternating Gradient Synchrotron Booster in the NASA Space Radiation Laboratory (NSRL) at the BNL. Mice were shipped from Emory directly to BNL. At the NSRL, each mouse was placed in a small transparent rectangular Lucite box (7.6 cm × 3.8 cm × 3.8 cm) with multiple holes, and 10 of these boxes were slid into a sample holder made of low-density foam. The sample holder was placed in the path of the beam (20 × 20 cm). The average age of mice at time of exposure was approximately 6 weeks. Mice were exposed to 0.5 Gy of X-ray, 56Fe, 28Si or proton once a day for 5 days continuously. After irradiation at the NSRL, all mice were shipped back to Emory by courier for same day delivery in a temperature-controlled truck. Control untreated mice were kept in the same conditions. X-ray radiation was performed in the Department of Radiation Oncology at Emory. All mice were housed in the Emory animal care facility for observation of tumor development for up to 18 months.
Cell lines and transfection
H1299 and MCF7 cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) in 2012. Two months after receipt, these cell lines were employed for the described experiments without further authentication by authors. H1299 cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). MCF7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) medium supplemented with 10% FBS. Various Bcl2 constructs or empty vectors were transfected into cells using NanoJuice (EMD Millipore, Billerica, MA) according to the manufacturer’s instruction.
MBE cells isolation and culture
MBE cells were isolated from the bronchi of wild-type C57BL/6 mice and CC10-Bcl2 transgenic mice using a magnetic cell separation method and cultured as described previously (25–27). Briefly, after euthanasia, three mouse lungs with the same genotype were subjected to thoracotomy and the lungs were rinsed with 1× phosphate-buffered saline (PBS) via the right cardiac ventricle and excised. After removal of the larger blood vessels, the lungs were minced with scissors under sterile conditions and placed in a 60-mm dish containing DMEM. Tissues were digested in Hanks’ balanced salt solution supplemented with 1 mg/ml collagenase A and 20 µg/ml DNase I at 37°C for 45 min with occasional agitation. The cellular digest was filtered through a 40 µm cell strainer (BD Bioscience, San Jose, CA), centrifuged at 400g for 10 min, and washed twice in DMEM with 10% FBS. The cell pellet was resuspended in 4 ml of DMEM with 10% FBS. Cells were incubated with EpCAM (CD326) magnetic microbeads (Miltenyi Biotec Auburn, CA) and separated in a magnetic field. After washing away the unbound cells, the beads were released by trypsin/ethylenediaminetetraacetic acid (EDTA) treatment. Cells were seeded with the microbeads. The cells were immortalized after continuous culture in keratinocyte serum-free medium (Invitrogen Life Technologies, Carlsbad, CA) with 20% FBS. Confluent cells were passed routinely at a split ratio of 1–3 after trypsin/EDTA digestion and cultured under the same conditions.
Preparation of cell lysate and western blot
Cells were washed with cold 1× PBS and resuspended in ice-cold EBC buffer (0.5% Nonidet P-40, 50 mM Tris, pH 7.6, 120 mM NaCl, 1 mM EDTA and 1 mM β-mercaptoethanol) containing protease inhibitor mixture set I. Following cell lysis by sonication and centrifugation at 14 000g for 15 min at 4°C, the resulting supernatant was collected as the total cell lysate. Protein expression was analyzed by western blot as described previously (28).
Immunofluorescence staining
Staining of tissue sections was performed using the R.T.U. Vectastain kit following the manufacturer’s standard protocol (Vector Laboratories, Burlingame, CA). Tissue sections were incubated with Bcl2 primary antibody from mouse and CC10 primary antibody from rabbit (dilution 1:50) overnight at 4°C. After washing with 1× PBS, tissue sections were incubated with Alexa 488 (green)-conjugated anti-mouse or Alexa 555 (red)-conjugated anti-rabbit secondary antibodies or 4′,6-diamidino-2-phenylindole for 60 min at room temperature. Tissue sections were observed under a fluorescence confocal microscope (Zeiss, Sweden).
DNA molecular combing
DNA fiber spreads were performed as described (11,29,30). Briefly, MBE cells were first labeled with 5′ chlorodeoxyuridine (CIdu, 100 µM) 20 min. After washing, cells were then labeled with iododeoxyuridine (IdU, 100 µM) for another 20 min. 2 µl of cell suspension (1 × 106 cells/ml) in cold PBS were spotted onto a microscope slide and mixed with 12 µl lysis buffer (0.5% sodium dodecyl sulfate, 50 mM EDTA and 200 mM Tris–Cl) for 10 min at room temperature. Samples were carefully tilted at a 15° angle to allow spreading of the genomic DNA into single-molecule DNA fibers by gravity. Fibers were then fixed with methanol and acetic acid (3:1) buffer. Fibers were subsequently acid treated with HCl (2.5 M) to denature the DNA fibers. Slides were neutralized and washed four times with 1× PBS (pH 8.0 PBS one time, pH 7.4 PBS three times). Slides were blocked with 10% goat serum and 0.1% Triton-X in PBS for at least 1 h, and then incubated with primary antibodies against IdU (mouse anti-BrdU clone B44) and 5′ chlorodeoxyuridine [rat anti-BrdU BU1/75(ICR1)] and secondary antibodies [Alexa Fluor 488 (green) goat anti-mouse and Alexa Fluor (red) 555 goat anti-rat] for 1 h. Slides were mounted in Prolong with 4′,6-diamidino-2-phenylindole and scanned with a Zeiss Axioplan2 upright microscope (Axioplan, Zeiss). Images were analyzed by Zeiss AxioVision software. The lengths of CIdU (red) and IdU (green) labeled patches were analyzed using the ImageJ software (31). The fork rate (kb/min) was calculated from the length of fluorescent signal (kb) divided by the time of the pulse. The median replication rates, interorigin distances and P values derived from Mann–Whitney U-test were computed with Prism v5.0. Representative images of DNA fibers were cropped from different fields of view and assembled with Adobe Photoshop, as described previously (32).
Measurement of intracellular dNTPs
Cellular deoxynucleoside triphosphate (dNTP) levels were analyzed as we described previously (11,33). Briefly, cells were harvested, and cellular nucleotides were extracted with 6% trichloroacetic acid followed by neutralization with the addition of 5 M K2CO3 just prior to high-performance liquid chromatography analysis. The dNTPs were separated from nucleoside triphosphates using a boronic acid resin column (Thermo Fisher Scientific, Waltham, MA). Then, chromatographic separations of dNTPs were performed using a Symmetry C (18) 3.5 µM (150 × 4.6 mm) column equipped with fluorescence detector (Waters Corporation, Milford, MA). All data were plotted using the GraphPad Prism v 5.0 program (GraphPad Prism software).
RNR activity assay
RNR activity was analyzed as described previously (11,34). Briefly, MBE cells were harvested and washed with 1× PBS. Low salt homogenization buffer (10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, 2 mM dithiothreitol, pH 7.2) was added to the cell pellet. After homogenization with a 27G1/2 syringe needle, cell debris were removed by centrifugation at 16 000g at 4°C for 20 min. The supernatant was passed through a sephadex G25 spin column. 600 µg of protein was added to a 40 µl reaction mixture (50 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid buffer, pH 7.2, 10 mM dithiothreitol, 4 mM adenylyl-imidodiphosphate, 20 µl FeCl3, 2 mM magnesium acetate, 50 µl CDP and 100 µl C14-CDP) and incubated at 37°C for 1 h. Then, 4 µl of 10 M perchloric acid was added for 15 min on ice. After centrifugation, the supernatant was transferred to a new tube and boiled for 20 min. 4 µl of a marker solution (60 mM cytidine monophosphate, 60 mM deoxycytidine monophosphate and 60 mM deoxyuridine monophosphate plus 12 µl 5 M KOH) was added and then incubated on ice for 15 min. Samples were centrifuged at 14 000 rpm for 5 min. The resulting supernatant containing nucleotides was spotted on a thin-layer chromatography (TLC) plate and separated by TLC. TLC plates were analyzed with quantification using the variable scanner Typhoon 9210 (GE Health). All data were plotted using the GraphPad Prism v 5.0 program. RNR activity was calculated by C14-dCDP/(C14-CDP + C14-dCDP).
Immunohistochemistry analysis
Mouse lung tissues were fixed in 10% formalin, embedded in paraffin and cut into sections. After deparaffinization, rehydration, inactivation of endogenous peroxidase and antigen retrieval, immunohistochemistry staining was performed using R.T.U. Vectastain Kit (Vector Laboratories) according to the manufacturer’s instructions. Tissue section slides were incubated with Bcl2 antibody (dilution 1:50) overnight at 4°C, followed by staining with 3,3′-diaminobenzidine and counterstained with hematoxylin, dehydrated, treated with xylene and mounted. All slides were examined, and representative pictures were taken using an Olympus BX41 microscope (Olympus America, Melville, NY).
Analysis of mouse survival and lung tumor incidence
Mouse survival and lung tumor incidence were observed up to 18 months after irradiation exposure. At the end of experiments, the entire lung tissue of each mouse was removed, fixed in 10% formalin and embedded in paraffin. Three parts of lung tissues representing different regions of the lung were put vertically into paraffin blocks for hematoxylin and eosin (H&E) staining. Lung tissue samples were sectioned at 3 mm three times for placement of slides and stained with H&E, followed by scanning analysis using an Aperio Whole Slide Scanner (ScanScope XT; Aperio Technologies), which digitizes whole microscope slides at ×20 or ×40 magnification with high-resolution images (0.5 µm/pixel for ×20 and 0.25 µm/pixel for ×40 scans). Images were analyzed using ImageScope viewing software (Leica Biosystems). Lung cancer classification was determined by a certified pathologist in the Department of Pathology and Laboratory Medicine, Emory University. Tumor numbers were counted under a microscope, and tumor area was quantified using Openlab modular imaging software (PerkinElmer). Tumor incidence of each experimental group was counted accurately.
Statistical analysis
The sample size of mice (n = 30–54) was chosen to detect a minimum effect size of 1.5 with at least 80% power and a type I error of 0.05 for each comparison. Significant differences between two groups were analyzed using 2-tailed Mann–Whitney test or 2-tailed t-test. A P value <0.05 was considered statistically significant. Mouse survival data were analyzed using Kaplan–Meier survival curve.
Results
Generation of the lung-targeting Bcl2 transgenic mice under CC10 promoter
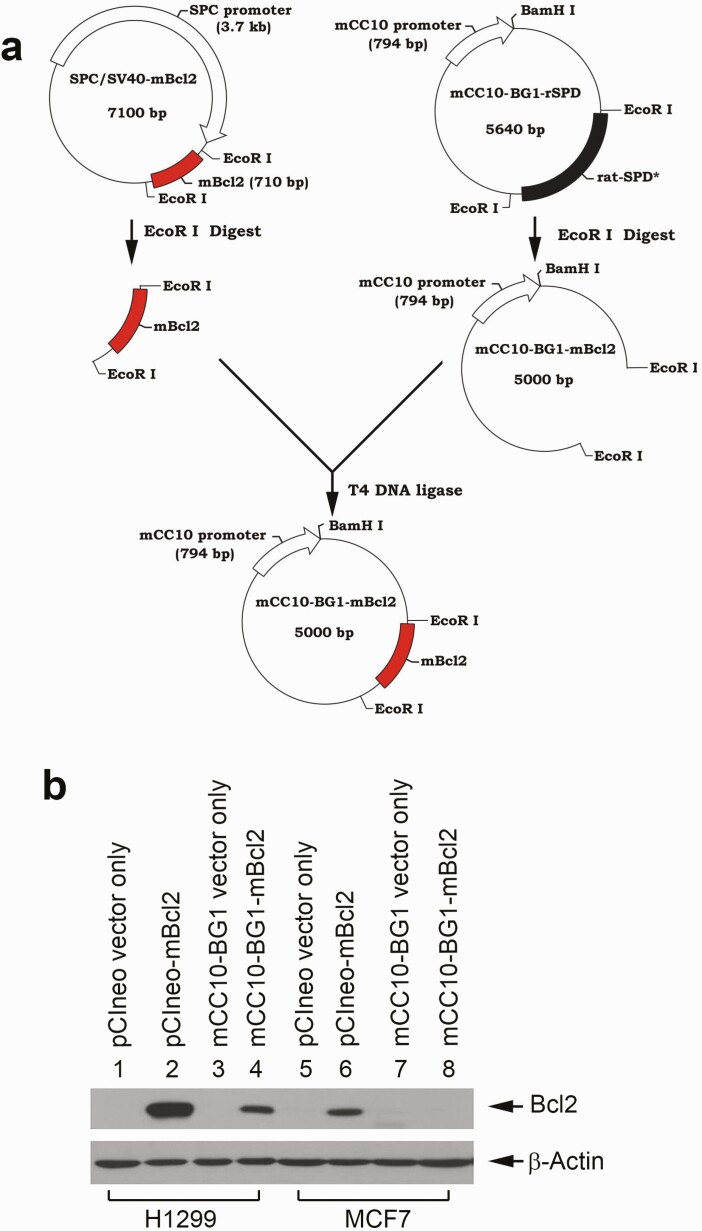
The CC10 promoter has been well characterized to drive expression of a selected gene specifically in airway epithelial cells in the bronchioles of the lung (35–39), which are common origins of tobacco smoke-related lung cancer. To create CC10-promoter-driven Bcl2 transgenic mice, we obtained the mCC10-BG1-rSPD construct from Dr Jeffrey A. Whitsett (Cincinnati Children’s Hospital Medical Center, Cincinnati, OH). WT mouse Bcl2 cDNA (mBcl2) was excised from SPC/SV40-mBcl2 using EcoRI and subsequently cloned into the pCC10-BG1-rSPD construct after removal of the rSPD insert using EcoRI, leading to generation of the mCC10-BG1-mBcl2 construct as shown in Figure 1a. To confirm whether the CC10 promoter can drive the selective expression of Bcl2 in lung cancer cells, we compared Bcl2 expression using the pCIneo/CMV promoter mammalian expression vector and mCC10-BG1 vector with or without Bcl2 cDNA in non-small cell lung cancer H1299 and breast cancer MCF7 cells. Results indicated that both H1299 and MCF7 cells expressed exogenous Bcl2 when transfected with pCIneo-mBcl2 constructs with the CMV promoter (Figure 1b, lane 2 versus lane 6). In contrast, Bcl2 expression was observed only in H1299 but not in MCF7 cells after transfection of mCC10-BG1-mBcl2 (Figure 1b, lanes 4 versus lane 8). Bcl2 expression was not observed in any of these two cell lines after transfection with empty vectors (Figure 1b, lanes 1, 3, 5 and 7). These results indicate that the CC10 promoter drives the selective expression of Bcl2 in non-small cell lung cancer H1299 cells, but not in breast cancer cells.
Figure 1.
CC10 promoter drives selective expression of Bcl-2 in a lung cancer cell line but not a breast cancer cell line in vitro. (a) Schematic diagram of cloning of mCC10-BG1-mBcl2 construct. (b) Various constructs were transfected into non-small cell lung cancer H1299 cells and breast cancer MCF7 cells, followed by western blot analysis of Bcl2 expression.
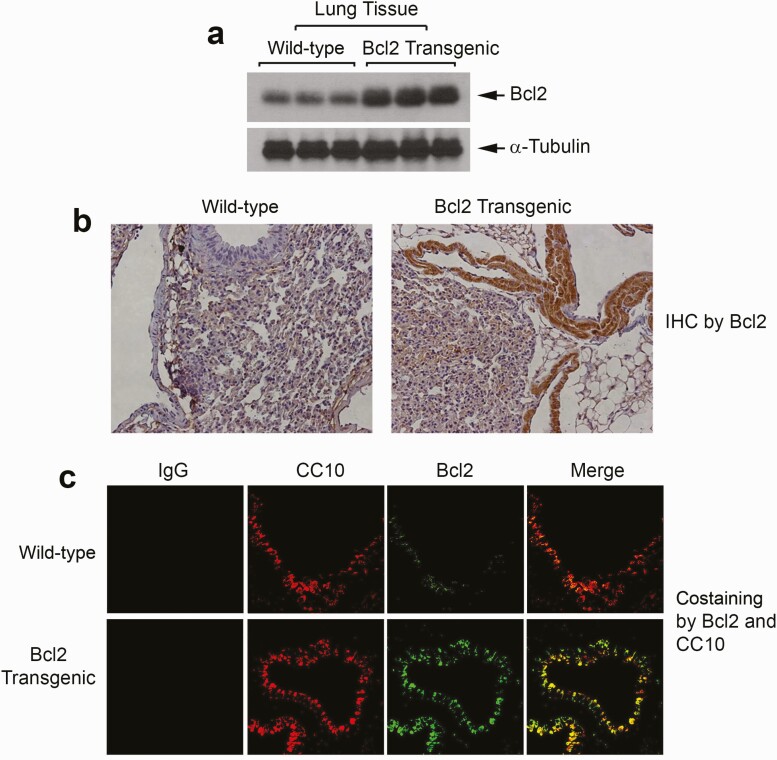
After confirming the expected expression of Bcl2 under the CC10 promoter, the mCC10-BG1-mBcl2 construct was used to create lung-targeting Bcl2 transgenic mice. Generation of lung-targeting Bcl2 transgenic mice was performed by the Transgenic Mouse/Gene Targeting Core Facility at Emory University. The purified linear DNA was microinjected into the pronuclei of fertilized eggs isolated from C57BL/6 mice by standard methods (40). Viable oocytes were implanted into the oviducts of pseudopregnant C57BL/6 recipient mice. Four positive founder (F0) transgenic mice (M1, M3, M5 and M8) were identified by PCR using tail DNA (Supplementary Figure 1, available at Carcinogenesis Online). Founder F0 mice bearing the transgene were then bred to wild-type C57BL/6 mice to generate hemizygous F1 offspring. F1 mice from each F0 line were genotyped for germline transmission of the transgenes. The established transgenic F1 mice were bred as necessary to maintain and expand the colony. The lung-targeting Bcl2 transgenic mice were maintained in compliance with the Emory University Institutional Review Board procedures. Bcl2 transgene-positive mice from subsequent generations were identified by PCR-based genotyping using tail DNA, a forward mcc10-Bcl2 vector primer and a reverse Bcl2 cDNA primer as described in ‘Methods’. Representative genotyping data are shown in Supplementary Figure 2, available at Carcinogenesis Online. Bcl2 expression in lung tissues was confirmed by western blot or immunohistochemistry staining using Bcl2 antibody (Figure 2a and b). As expected, Bcl2 is extensively colocalized with CC10 protein in the lung tissue from lung-targeting Bcl2 transgenic mice (Figure 2c). Intriguingly, colocalization of Bcl2 and CC10 protein mainly occurs in airway epithelial cells of the bronchioles (Figure 2c, lower panel), which are common origins of tobacco smoke-related lung cancer.
Figure 2.
Selective expression of Bcl-2 in lung tissues in Bcl2 transgenic mice. (a and b) Bcl2 expression was compared in lung tissues from wild-type C57BL/6 and CC10-promoter-WT Bcl2 transgenic mice by western blot or immunohistochemistry using Bcl2 antibody. (c) Colocalization of Bcl2 and CC10 was analyzed by immunofluorescence using CC10 and Bcl2 antibodies in lung tissues from wild-type control and CC10-promoter-Bcl2 transgenic mice.
Lung-targeting transgenic Bcl2 reduces RNR activity and intracellular dNTP pool size leading to retardation of DNA replication fork progression in bronchial epithelial cells
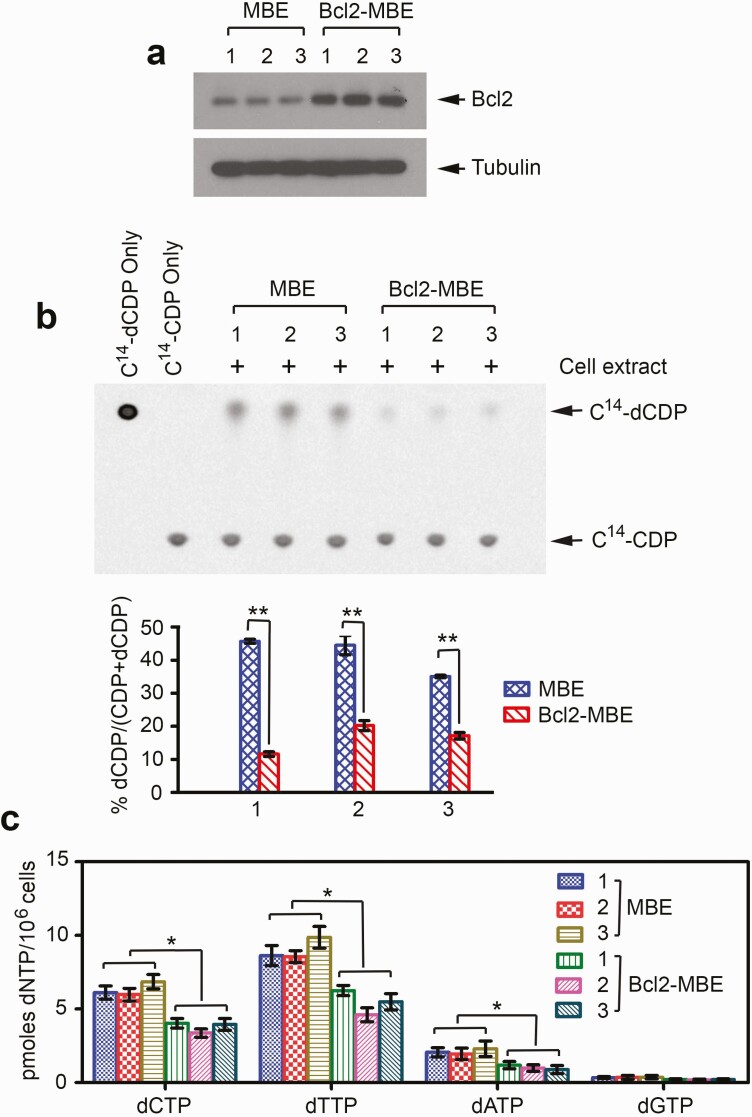
We have recently discovered that Bcl2 interacts with RRM2 leading to suppression of RNR activity, decreased intracellular dNTP levels and retardation of DNA replication fork progression at the cellular level in vitro (11). To test whether Bcl2 plays such a role in vivo, MBE cells were isolated from lung tissues of the lung-targeting Bcl2 transgenic C57BL/6 or wild-type control mice as described (41), followed by measurement of RNR activity and intracellular dNTPs, and analysis of DNA fibers as we described previously (11). Decreased RNR activity and reduced dNTP pool size were observed in MBE cells isolated from three representative lung-targeting Bcl2 transgenic mice (Bcl2-MBE) compared with MBE cells from three representative wild-type mice (Figure 3).
Figure 3.
Expression of Bcl2 results in decreased RNR activity and reduced dNTP pool size in MBE cells isolated from Bcl2 lung-targeting transgenic mice. (a) MBE cells or Bcl2-MBE cells were isolated from lung tissues of three representative wild-type mice or three representative Bcl2 transgenic mice, respectively. Bcl2 expression was analyzed by western blot. (b) Extracts from MBE cells isolated from three representative wild-type mice or Bcl2-MBE cells isolated from three representative Bcl2 transgenic mice were incubated with 14C-CDP. The conversion from 14C-CDP to 14C-dCDP was analyzed by TLC. RNR activity was calculated as 14C-dCDP/(14C-CDP+ 14C-dCDP). The error bars indicate ± SD of three separate experiments. **P < 0.01, by 2-tailed t-test. (c) Intracellular dNTP levels were measured by high-performance liquid chromatography analysis in MBE and Bcl2 MBE cells. The error bars indicate ± SD of three separate experiments. *P < 0.05, by 2-tailed t-test.
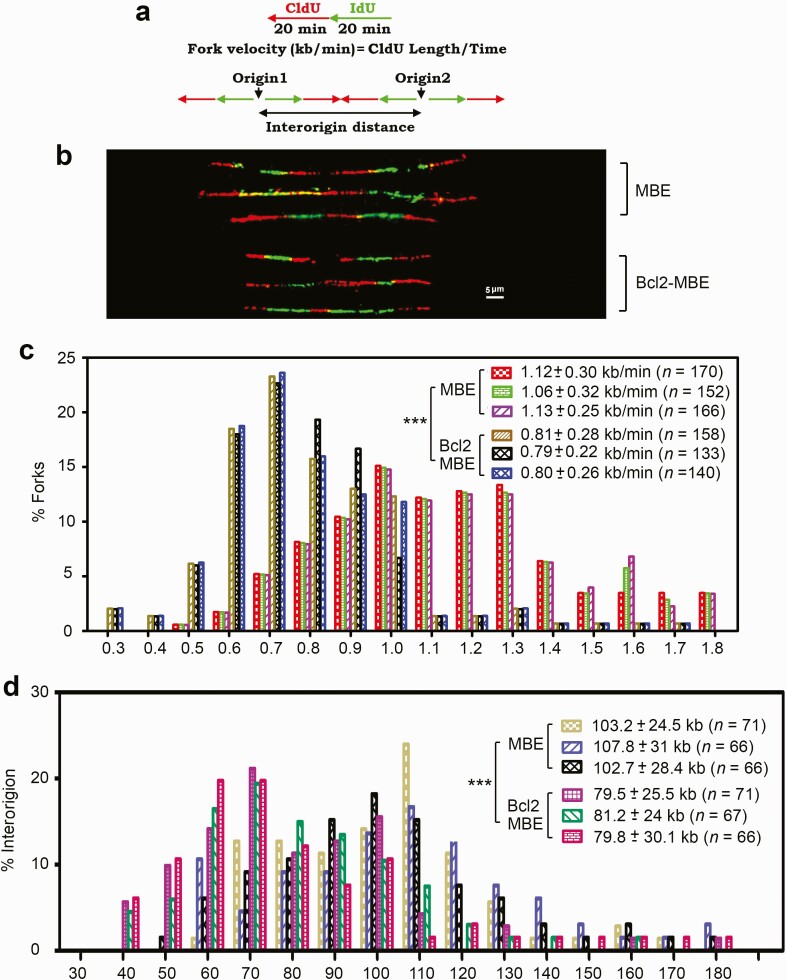
Because the dNTP pool size determines fork progression and origin usage (42,43), Bcl2-reduced RNR activity and dNTP pool size in MBE cells possibly affects DNA replication fork progression. Replication dynamics was evaluated using single-molecule DNA fiber analysis. This technique can measure the speed of individual replication fork progression as well as the frequency of replication initiation events. The rate of individual replication fork progression was calculated by dividing the length of each fluorescent segment (1 µm = 2.59 kb) by the time of pulse (20 min), as described previously (44) (Figure 4a). Newly synthesized DNA labeled with IdU (green) and 5′ chlorodeoxyuridine (CIdU, red) was detected with fluorescent antibodies (Figure 4b). A pronounced and reproducible decrease in the mean replication fork progression rate was observed in Bcl-2-MBE cells isolated from three representative lung-targeting Bcl2 transgenic mice (0.79 ± 0.22 to 0.81 ± 0.28 kb/min) compared with MBE cells isolated from three representative wild-type mice (1.06 ± 0.32 to 1.13 ± 0.25 kb/min) (P < 0.001; Figure 4b and c). A dramatic increase in the percentage of slowly progressing forks was observed in the Bcl2-expressing MBE cells (Figure 4c). These results reveal that expression of Bcl2 in MBE cells significantly reduces the rate of DNA replication fork progression in lung-targeting Bcl2 transgenic mice.
Figure 4.
Bcl2 retards DNA replication fork progression in MBE cells isolated from Bcl2 lung-targeting transgenic mice. (a) Schematic diagram for calculation of the fork rate (top) and inter-origin distance (bottom) of replication fork progression. (b) MBE cells isolated from three representative wild-type mice or Bcl2-MBE cells isolated from three representative Bcl2 transgenic mice were pulse-labeled with 100 µM CldU for 20 min and 100 µM IdU for another 20 min. The labeled cells were processed for DNA combing. Representative pairs of sister replication forks are shown. Red, CldU; green, IdU, scale bar, 5 µm (1 µm = 2.59 kb). (c) Distribution of fork rate (kb/min) in MBE and MBE-Bcl2 cells. The mean ± SD for fork rate and number of scores are summarized. The error bars indicate ± SD, n = 150 fibers. ***P < 0.001, by a 2-tailed Mann–Whitney test. (d) Distribution of inter-origin distances (kb) was compared in MBE and MBE-Bcl2 cells. The error bars indicate ± SD, n = 150 fibers. ***P < 0.001, by a 2-tailed Mann–Whitney test. CldU, 5′ chlorodeoxyuridine.
To test the effect of Bcl2 on the origin density, we measured the distance between two adjacent origins as described (43). The mean origin distance in MBE cells isolated from three representative wild-type mice was 102.7 ± 28.4 to 107.8 ± 31 kb, whereas in Bcl2-expressing MBE cells isolated from three representative lung-targeting Bcl2 transgenic mice, it was significantly shorter, only 79.5 ± 25.5 to 81.2 ± 24 kb (P < 0.001; Figure 4d). This decrease is expected from the observed decreased fork rate, as fork rate and interorigin distance show a linear correlation (44,45). A dramatically increased population with short origin distance was observed in the Bcl2-expressing MBE cells (Figure 4d). These results indicate that Bcl2 expression leads to activation of an increased number of origins in MBE cells in vivo.
Lung-targeting transgenic Bcl2 promotes lung tumorigenesis in response to high-LET radiation or protons
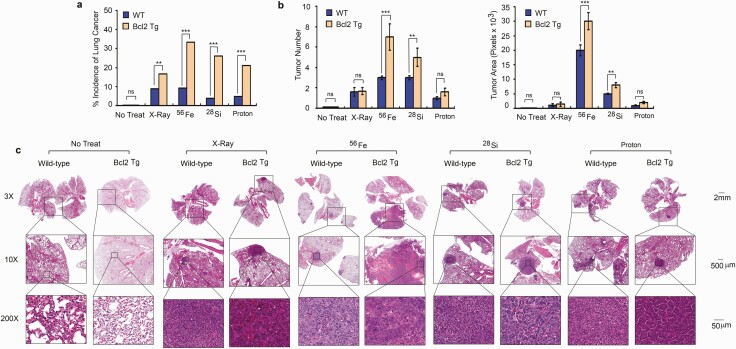
To test the effect of high-LET radiation, protons versus X-ray radiation on lung carcinogenesis, 56Fe, 28Si, X-ray and protons were chosen for radiation of mice as shown in Supplementary Table 1, available at Carcinogenesis Online. Wild-type C57BL/6 and lung-targeting Bcl2 transgenic mice were randomized into five treatment groups as indicated in Supplementary Table 2, available at Carcinogenesis Online. Group 1 → no treatment Ctrl; Group 2 → X-ray; Group 3 → 56Fe; Group 4 → 28Si; Group 5 → protons. Mice were irradiated with 0.5 Gy of X-ray, 56Fe, 28Si or protons once a day for continuous 5 days. After various radiation treatments, lung tumor formation and survival of mice in different groups were observed for up to18 months. At the end of experiments, mice were euthanized by CO2 asphyxiation. Lung tissues were collected for pathological analysis. No tumors were observed in the lungs of wild-type or lung-targeting Bcl2 transgenic mice receiving no treatment. Treatment with X-ray, 56Fe, 28Si and protons induced a very low incidence of lung cancer in wild-type mice (Supplementary Table 2, Figure 5a and Supplementary Figure 3, available at Carcinogenesis Online). However, high-LET radiation derived from 56Fe and 28Si or radiation derived from protons induced a significantly higher incidence of lung cancer than low-LET X-ray in lung-targeting Bcl2 transgenic mice (Supplementary Table 2, Figure 5a and Supplementary Figure 3, available at Carcinogenesis Online), indicating that the incidence of lung cancer in response to high-LET radiation or protons was greater in lung-targeting Bcl2 transgenic mice than in wild-type mice. In addition to tumor incidence, tumor burden including tumor number and tumor area in each lung was quantified as described in ‘Methods’. Exposure of wild-type or lung-targeting Bcl2 transgenic mice to 56Fe or 28Si resulted in greater tumor burden than X-ray or proton exposure among lung cancer positive mice (Figure 5b and c). Intriguingly, lung-targeting Bcl2 transgenic mice had greater tumor burden than wild-type mice in response to 56Fe or 28Si (Figure 5b and c). Although the major tumor type induced by high-LET and low-LET radiation was lung adenocarcinoma (Supplementary Table 2, available at Carcinogenesis Online and Figure 5c), lung squamous cell carcinomas were observed in five mice (two wild-type and three lung-targeting Bcl2 transgenic mice) and occurred only among mice exposed to 56Fe as indicated in Supplementary Table 2, available at Carcinogenesis Online. Our findings suggest that Bcl2 significantly promotes high-LET radiation- or protons-induced lung tumorigenesis, but has less effect on X-ray-induced lung carcinogenesis.
Figure 5.
Bcl2 enhances high-LET irradiation-induced lung tumorigenesis. (a) Wild-type C57BL/6 mice and lung-targeting Bcl2 transgenic mice were exposed to 0.5 Gy of X-ray, 56Fe, 28Si or proton once a day for 5 days continuously, followed by monitoring lung tumorigenesis for 18 months. Percentage of lung cancer incidence was calculated for all groups. Number of mice (n) for each group is indicated in Supplementary Table 2, **P < 0.01, ***P < 0.001, ns, no significance, by 2-tailed t-test. (b) Tumor numbers were counted under the microscope and tumor area was quantified using Openlab modular imaging software. The error bars indicate ± SD, number of mice (n) for each group is indicated in Supplementary Table 2. **P < 0.01, ***P < 0.001, ns, no significance, by 2-tailed t-test. (c) Representative H&E images from no treatment or treatment groups for wild-type C57BL/6 and lung-targeting Bcl2 transgenic mice.
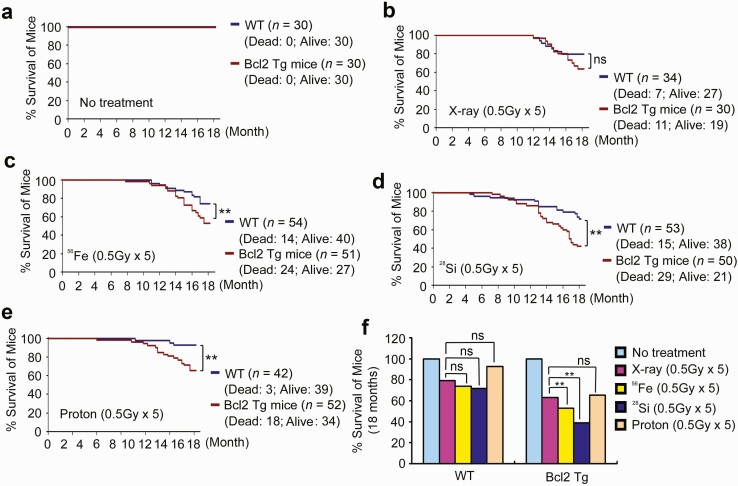
Lung-targeting transgenic Bcl2 reduces survival of mice in response to high-LET radiation
Survival of mice was observed for 18 months following their exposure to fractionated X-ray, 56Fe, 28Si or protons (0.5 Gy/day ×5). No deaths occurred in the no treatment Ctrl group among both wild-type and lung-targeting Bcl2 transgenic mice (Figure 6a). Low-LET X-ray radiation induced more deaths in lung-targeting Bcl2 transgenic mice than in wild-type mice but the difference did not reach statistical significance (P = 0.19, Figure 6b). A significantly higher death rate was observed in lung-targeting Bcl2 transgenic mice compared with wild-type mice following exposure of mice to 56Fe, 28Si or protons (P < 0.01, Figure 6c–e). Intriguingly, high-LET radiation derived from 56Fe or 28Si induced a significantly higher death rate compared with protons or X-ray among lung-targeting Bcl2 mice, but there was no significant difference in wild-type mice (Figure 6f).
Figure 6.
Bcl2 reduces survival of mice following high-LET irradiation exposure. (a–f) Survival of mice was analyzed up to 18 months after exposure of wild-type C57BL/6 and lung-targeting Bcl2 transgenic mice to X-ray, 56Fe, 28Si, proton or no treatment Ctrl. **P < 0.01, ns, no significance, by 2-tailed t-test.
Discussion
Space radiation consists primarily of IR, which exists in the form of energetic protons and charged heavy nuclei particles, and has been shown to produce distinct biological damage and cancer risk compared with radiation on Earth. Deep-space radiation may have health risks and be dangerous for astronauts. The study of heavy ion radiation-induced effects on mice could provide insight into the human health risks of space radiation exposure (46). In the present report, animal models were employed to acquire critical knowledge regarding space radiation-induced lung cancer initiation, promotion and progression. It is not common for C57BL/6 mice to spontaneously develop lung cancer without any treatment (8). Therefore, the C57BL/6 mouse provides a very clean background and an ideal mouse model to study space radiation carcinogenesis.
The Bcl2 oncoprotein not only functions as a potent antiapoptotic molecule but also causes DNA replication stress via induction of inefficient DNA synthesis by inhibiting RNR activity (11,47). However, the role and molecular mechanism(s) of Bcl2-mediated DNA replication stress in space radiation-induced lung carcinogenesis remain currently unknown. To test this, we utilized C57BL/6 mice to generate lung-targeting Bcl2 transgenic mouse models to evaluate lung tumorigenesis in response to space radiation. Intriguingly, fractionated exposure to high-LET radiation derived from 56Fe and 28Si or protons at 0.5 Gy ×5, but not low-LET radiation derived from X-ray at the same dose, induced a significantly higher incidence of lung cancer and rate of death in lung-targeting Bcl2 transgenic mice than in wild-type mice (Figures 5 and 6, and Supplementary Table 2, available at Carcinogenesis Online). Mechanistically, decreased RNR activity, reduced dNTP pool size and stalled DNA replication forks were observed in MBE cells isolated from lung-targeting Bcl2 transgenic mice compared with wild-type mice (Figures 3 and 4). This suggests that Bcl2-mediated DNA replication stress promotes lung carcinogenesis in response to high-LET radiation or protons in vivo.
Intriguingly, 56Fe is the most potent in the induction of lung cancer in lung-targeting Bcl2 transgenic mice, followed by 28Si and then protons (Figure 5, Supplementary Table 2 and Supplementary Figure 3, available at Carcinogenesis Online). This may be due to 56Fe having the highest energy (i.e. 56Fe → 600 MeV/u, 28Si → 300 MeV/u and proton → 150 MeV/u). Additionally, X-ray, 28Si and protons only induced adenocarcinoma while 56Fe can induce both squamous cell carcinoma and adenocarcinoma (Supplementary Table 2, available at Carcinogenesis Online). The exact mechanism(s) underlying this difference is not clear. Protons induced the lowest death rate compared with 56Fe and 28Si, similar to that observed with low-LET X-ray (Figure 6). This indicates that protons induce less in vivo toxicity than 56Fe and 28Si.
In summary, we have demonstrated that selective expression of Bcl2 in lung tissues via the CC10 promoter suppresses RNR activity, reduces dNTP pool size and retards DNA replication fork progression in MBE cells, which enhances susceptibility to space radiation-induced lung carcinogenesis. The findings provide some evidence for the relative effectiveness of space radiation and Bcl-2 at inducing lung cancer in mice.
Funding
This work was supported by National Aeronautics and Space Administration (NASA) grant NNX12AC30G (to X. Deng), by The National Institutes of Health (NIH)/The National Cancer Institute (NCI) grants R01CA193828 and R01CA200905 (to X. Deng), by the Winship Research Pathology and Integrated Cellular Imaging shared resource supported by the Winship Cancer Institute of Emory University (P30CAJ 38292), by the Winship Fashion a Cure Research Scholar Award (to X. Deng), a philanthropic award provided by the Winship Cancer Institute of Emory University, and by Winship Endowment Fund (to X. Deng).
Conflict of Interest Statement: The authors disclose no potential conflicts of interest.
Supplementary Material
Acknowledgements
We are grateful to Dr Jeffrey A. Whitsett (Cincinnati Children’s Hospital Medical Center, Cincinnati, OH) for kindly providing the mCC10-BG1-rSPD construct. We thank Anthea Hammond for editing of the manuscript.
Glossary
Abbreviations
- DMEM
Dulbecco’s modified Eagle’s medium
- DSB
double-strand break
- EDTA
ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- IR
ionizing radiation
- LET
linear energy transfer
- MBE
mouse bronchial epithelial
- PBS
phosphate-buffered saline
- RNR
ribonucleotide reductase
References
- 1. Durante, M. et al. (2008) Heavy ion carcinogenesis and human space exploration. Nat. Rev. Cancer, 8, 465–472. [DOI] [PubMed] [Google Scholar]
- 2. Beheshti, A. et al. (2019) GeneLab database analyses suggest long-term impact of space radiation on the cardiovascular system by the activation of FYN through reactive oxygen species. Int. J. Mol. Sci., 20, E661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Preston, D.L. et al. (2007) Solid cancer incidence in atomic bomb survivors: 1958–1998. Radiat. Res., 168, 1–64. [DOI] [PubMed] [Google Scholar]
- 4. Cardis, E. et al. (2007) The 15-country collaborative study of cancer risk among radiation workers in the nuclear industry: estimates of radiation-related cancer risks. Radiat. Res., 167, 396–416. [DOI] [PubMed] [Google Scholar]
- 5. Xie, M. et al. (2015) Bcl2 inhibits recruitment of Mre11 complex to DNA double-strand breaks in response to high-linear energy transfer radiation. Nucleic Acids Res., 43, 960–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang, H. et al. (2008) The Ku-dependent non-homologous end-joining but not other repair pathway is inhibited by high linear energy transfer ionizing radiation. DNA Repair (Amst)., 7, 725–733. [DOI] [PubMed] [Google Scholar]
- 7. Wang, H. et al. (2010) Characteristics of DNA-binding proteins determine the biological sensitivity to high-linear energy transfer radiation. Nucleic Acids Res., 38, 3245–3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang, X. et al. (2015) Relative effectiveness at 1 Gy after acute and fractionated exposures of heavy ions with different linear energy transfer for lung tumorigenesis. Radiat. Res., 183, 233–239. [DOI] [PubMed] [Google Scholar]
- 9. Khanna, K.K. et al. (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat. Genet., 27, 247–254. [DOI] [PubMed] [Google Scholar]
- 10. Mazouzi, A. et al. (2014) DNA replication stress: causes, resolution and disease. Exp. Cell Res., 329, 85–93. [DOI] [PubMed] [Google Scholar]
- 11. Xie, M. et al. (2014) Bcl2 induces DNA replication stress by inhibiting ribonucleotide reductase. Cancer Res., 74, 212–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Toledo, L.I. et al. (2013) ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell, 155, 1088–1103. [DOI] [PubMed] [Google Scholar]
- 13. Lecona, E. et al. (2014) Replication stress and cancer: it takes two to tango. Exp. Cell Res., 329, 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Halazonetis, T.D. et al. (2008) An oncogene-induced DNA damage model for cancer development. Science, 319, 1352–1355. [DOI] [PubMed] [Google Scholar]
- 15. Linette, G.P. et al. (1995) Peripheral T-cell lymphoma in lckpr-bcl-2 transgenic mice. Blood, 86, 1255–1260. [PubMed] [Google Scholar]
- 16. Deng, X. et al. (2000) Survival function of ERK1/2 as IL-3-activated, staurosporine-resistant Bcl2 kinases. Proc. Natl. Acad. Sci. USA, 97, 1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Linnoila, R.I. et al. (2000) Constitutive achaete-scute homologue-1 promotes airway dysplasia and lung neuroendocrine tumors in transgenic mice. Cancer Res., 60, 4005–4009. [PubMed] [Google Scholar]
- 18. Plück, A. et al. (2009) Generation of chimeras by microinjection. Methods Mol. Biol., 561, 199–217. [DOI] [PubMed] [Google Scholar]
- 19. Tchou-Wong, K.M. et al. (2002) Lung-specific expression of dominant-negative mutant p53 in transgenic mice increases spontaneous and benzo(a)pyrene-induced lung cancer. Am. J. Respir. Cell Mol. Biol., 27, 186–193. [DOI] [PubMed] [Google Scholar]
- 20. DiCosmo, B.F. et al. (1994) Airway epithelial cell expression of interleukin-6 in transgenic mice. Uncoupling of airway inflammation and bronchial hyperreactivity. J. Clin. Invest., 94, 2028–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miller, Y.E. et al. (2003) Induction of a high incidence of lung tumors in C57BL/6 mice with multiple ethyl carbamate injections. Cancer Lett., 198, 139–144. [DOI] [PubMed] [Google Scholar]
- 22. Rivera, J. et al. (2008) Genetic background and the dilemma of translating mouse studies to humans. Immunity, 28, 1–4. [DOI] [PubMed] [Google Scholar]
- 23. Klopstock, N. et al. (2009) HCV tumor promoting effect is dependent on host genetic background. PLoS One, 4, e5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perl, A.K. et al. (2002) Conditional gene expression in the respiratory epithelium of the mouse. Transgenic Res., 11, 21–29. [DOI] [PubMed] [Google Scholar]
- 25. Dong, Q.G. et al. (1997) A general strategy for isolation of endothelial cells from murine tissues. Characterization of two endothelial cell lines from the murine lung and subcutaneous sponge implants. Arterioscler. Thromb. Vasc. Biol., 17, 1599–1604. [DOI] [PubMed] [Google Scholar]
- 26. Licht, A.H. et al. (2006) JunB is required for endothelial cell morphogenesis by regulating core-binding factor beta. J. Cell Biol., 175, 981–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang, X.Y. et al. (2012) Novel method for isolation of murine Clara cell secretory protein-expressing cells with traces of stemness. PLoS One, 7, e43008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Han, B. et al. (2015) Small-molecule Bcl2 BH4 antagonist for lung cancer therapy. Cancer Cell, 27, 852–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schlacher, K. et al. (2011) Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell, 145, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jackson, D.A. et al. (1998) Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J. Cell Biol., 140, 1285–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Henry-Mowatt, J. et al. (2003) XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol. Cell, 11, 1109–1117. [DOI] [PubMed] [Google Scholar]
- 32. Pasero, P. et al. (2002) Single-molecule analysis reveals clustering and epigenetic regulation of replication origins at the yeast rDNA locus. Genes Dev., 16, 2479–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Huang, D. et al. (2003) Analysis of intracellular nucleoside triphosphate levels in normal and tumor cell lines by high-performance liquid chromatography. J. Chromatogr. B: Analyt. Technol. Biomed. Life Sci., 784, 101–109. [DOI] [PubMed] [Google Scholar]
- 34. Chimploy, K. et al. (2009) E2F4 and ribonucleotide reductase mediate S-phase arrest in colon cancer cells treated with chlorophyllin. Int. J. Cancer, 125, 2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Temann, U.A. et al. (1998) Expression of interleukin 9 in the lungs of transgenic mice causes airway inflammation, mast cell hyperplasia, and bronchial hyperresponsiveness. J. Exp. Med., 188, 1307–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhu, Z. et al. (1999) Pulmonary expression of interleukin-13 causes inflammation, mucus hypersecretion, subepithelial fibrosis, physiologic abnormalities, and eotaxin production. J. Clin. Invest., 103, 779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stripp, B.R. et al. (1992) cis-acting elements that confer lung epithelial cell expression of the CC10 gene. J. Biol. Chem., 267, 14703–14712. [PubMed] [Google Scholar]
- 38. Ray, M.K. et al. (1996) Immunohistochemical localization of mouse Clara cell 10-KD protein using antibodies raised against the recombinant protein. J. Histochem. Cytochem., 44, 919–927. [DOI] [PubMed] [Google Scholar]
- 39. Castro, C.M. et al. (2000) Attenuation of pulmonary neuroendocrine differentiation in mice lacking Clara cell secretory protein. Lab. Invest., 80, 1533–1540. [DOI] [PubMed] [Google Scholar]
- 40. Brown, G.A. et al. (2002) Oocyte injection in the mouse. Methods Mol. Biol., 180, 39–70. [DOI] [PubMed] [Google Scholar]
- 41. Ginzkey, C. et al. (2012) Analysis of nicotine-induced DNA damage in cells of the human respiratory tract. Toxicol. Lett., 208, 23–29. [DOI] [PubMed] [Google Scholar]
- 42. Poli, J. et al. (2012) dNTP pools determine fork progression and origin usage under replication stress. EMBO J., 31, 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bester, A.C. et al. (2011) Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell, 145, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Conti, C. et al. (2007) Replication fork velocities at adjacent replication origins are coordinately modified during DNA replication in human cells. Mol. Biol. Cell, 18, 3059–3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Courbet, S. et al. (2008) Replication fork movement sets chromatin loop size and origin choice in mammalian cells. Nature, 455, 557–560. [DOI] [PubMed] [Google Scholar]
- 46. Suman, S. et al. (2012) Relative biological effectiveness of 12C and 28Si radiation in C57BL/6J mice. Radiat. Environ. Biophys., 51, 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deng, X. et al. (2004) Mono- and multisite phosphorylation enhances Bcl2’s antiapoptotic function and inhibition of cell cycle entry functions. Proc. Natl. Acad. Sci. USA, 101, 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.