Abstract
Summary
The web platform 3DBionotes-WS integrates multiple web services and an interactive web viewer to provide a unified environment in which biological annotations can be analyzed in their structural context. Since the COVID-19 outbreak, new structural data from many viral proteins have been provided at a very fast pace. This effort includes many cryogenic electron microscopy (cryo-EM) studies, together with more traditional ones (X-rays, NMR), using several modeling approaches and complemented with structural predictions. At the same time, a plethora of new genomics and interactomics information (including fragment screening and structure-based virtual screening efforts) have been made available from different servers. In this context, we have developed 3DBionotes-COVID-19 as an answer to: (i) the need to explore multiomics data in a unified context with a special focus on structural information and (ii) the drive to incorporate quality measurements, especially in the form of advanced validation metrics for cryo-EM.
Availability and implementation
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The 3DBionotes-WS web platform has been operational for several years as part of the online offer of the Spanish Institute of Bioinformatics, the Spanish Node of the Research Infrastructure (RI) ELIXIR and Instruct-ERIC RI (Segura et al., 2019). It is, in fact, one of ELIXIR Recommended Interoperability Resources (https://elixir-europe.org/platforms/interoperability/rirs). A major goal is to provide an interactive graphical environment over the web where structural and multiomics data can be intuitively explored, complemented by a powerful API.
The COVID-19 outbreak has changed science: worldwide, scientists came together across disciplines and national borders in order to fight the pandemic. Structural biologists are no exception and the role of cryogenic electron microscopy (cryo-EM) in elucidating key viral structures is paramount [for a brief outline, see (Kearns, 2020)], complementing more traditional approaches, such as X-ray crystallography, NMR and fold predictions. However, SARS-CoV-2 maps did not achieve very high resolution (in most cases close to 3 Å, particularly for cryo-EM), which made it difficult to build atomic models suitable for drug development. Additionally, the pressure to publish these structures as fast as possible has never been so high. Early in the pandemic, specific resources have been created to address structural needs (https://github.com/thorn-lab/coronavirus_structural_task_force), acknowledging the requirement to pay special attention not only to data quantity, but also to data quality. In this way, validation information on cryo-EM maps provided by the Coronavirus Structural Task Force has been integrated into 3DBionotes, which have evolved to supply quality measurements, keeping its orientation toward the web (and its API) and focusing on integrative analysis. Indeed, its COVID-19 edition described here, combines in the same analysis framework key viral genomics, interactomics and structural information, including drug screening approaches [both experimental fragment-based screening (Douangamath et al., 2020) and virtual screening]. In the following, we describe 3DBionotes-COVID-19 edition, illustrating its use and value for users with some case studies in Supplementary Material.
2 Results
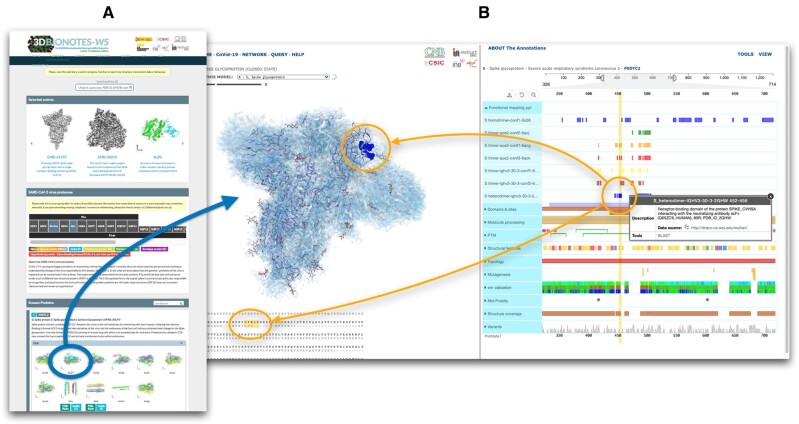
The design of 3DBionotes COVID-19 adds an additional layer of interactive information over the classical design of 3DBionotes. The new edition has a specific landing page to manage multiple sources of structural information that, once selected, launches a new version of 3DBionotes accessing COVID-19 specific information (Fig. 1). We describe this design in the following:
Fig. 1.
3DBionotes COVID-19 application screenshots. (A) Landing page, showing some of the main sections: representative examples, a simplified schema of the virus proteome that serves as index with links to the corresponding subsection for every protein, followed by various panels with the structures. (B) 3D viewer and annotations, showing the example of EMD-21452, corresponding to SARS-CoV-2 spike glycoprotein (closed state). By clicking in any of the symbols representing an annotation, all the residues associated with it will be highlighted in the protein sequence alignment as well as in the atomic structure. At the same time, those residues will also be highlighted with vertical yellow bars so it is easier to locate in relation with other annotations types. Additionally, a panel will pop-up with more detailed information about the annotation, including links to the origin of the data source
2.1 Landing page
This page acts as a structural information organizer, collecting data from SARS-CoV-2 and related coronavirus, as well as their interactions with host proteins. We automatically harvest structures deposited in PDB and EMDB together with predicted models from SWISSModel (Bienert et al., 2017), AlphaFold (Jumper et al., 2020) and BSM-Arc (Hijikata et al., 2020). When validation and quality information are available from PDB-REDO (Joosten et al., 2014) and the Coronavirus Structural Task Force (Croll et al., 2021) special tags are incorporated for every entry, pointing to the rerefined models. Entries are organized into five categories: PDB, EMDB, Interactions with other proteins (PPI) and Ligands, Related (to SARS-CoV or others) and Computational Models. In ‘Ligands’, we initially incorporate experimental information from fragment-based screening (Douangamath et al., 2020) as well as our own structure-based repurposing virtual screening (https://covid19drugrepurposing.cnb.csic.es).
Every entry is displayed with its reference and a static view of the model, when available, that serves the user as a preliminary visual hint of the structure (Fig. 1A). A pop-up panel is displayed with a brief description of the entry and a set of external links when the pointer is placed on the image for a few moments. Upon clicking on the entry, data are transferred to a new instance of 3DBionotes that pays special attention to multiomics and cryo-EM quality information, as detailed in the next section, opening the 3D viewer (Fig. 1B) and starting the annotation collection process. Users can go back to the landing page at any time in their analysis.
2.2 New annotations
3DBionotes-WS, in general, collects and organizes a wide range of annotations of the selected macromolecule. The COVID-19 release includes access to a collection of specifically developed servers adding new functionality, as it can be appreciated in the case studies detailed in Supplemental Material. Among them, we highlight:
Cryo-EM quality information at the amino acid level coloring the map being displayed in the 3D viewer, including:
Local resolution information on cryo-EM maps, calculated using a deep learning approach which does not require half maps (Ramírez-Aportela et al., 2019).
Quantitative validation metrics, such as Q-scores (Pintilie et al., 2020) and FSC-Q-scores (Ramírez-Aportela et al., 2021).
SARS-CoV-2 main protease fragment screening by PanDDA analysis (Pearce et al., 2017).
Genomics variants, with source data from CNCB (https://bigd.big.ac.cn/ncov/variation).
Functional mapping of Protein–Protein Interactions (PPI), with source data from Korbin’s lab (http://draco.cs.wpi.edu/wuhan) (Srinivasan et al., 2020).
2.3 Selected case studies
In Supplementary Material, we present how 3DBionotes can be used in four different use cases, namely:
SARS-CoV-2 spike protein cryo-EM map validation analysis based on local resolution metrics.
Use of improved structural models using new refinement methods collected from the Coronavirus Structural Task Force.
Analysis of SARS-CoV-2 spike variant D614G.
Study of drug screening on SARS-CoV-2 main protease (NSP5).
3 Conclusions
3DBionotes-COVID-19 is fully accessible at https://3dbionotes.cnb.csic.es/ws/covid19, providing a unique analysis environment tailored to COVID-19 information. It has all the advantages of 3DBionotes in terms of complex interactive analysis over the web and API access, offering both the possibility to work with structural data already deposited in public databases and with new user data, plus a series of new services geared toward quality (cryo-EM validation and curated structural models) and information integration. We demonstrate the usefulness of this interactive resource on four selected cases.
Funding
We acknowledge financial support from CSIC [PIE/COVID-19 number 202020E079], the Comunidad de Madrid through grant CAM [S2017/BMD-3817], the Spanish Ministry of Science and Innovation through projects [SEV 2017-0712, FPU-2015/264, PID2019-104757RB-I00/AEI/10.13039/501100011033], the Instituto de Salud Carlos III: PT17/0009/0010 [ISCIII-SGEFI/ERDF] and the European Union and Horizon 2020 through grant: CORBEL [INFRADEV-01-2014-1, Proposal 654248] and EOSC Life [INFRAEOSC-04-2018, Proposal: 824087]. This work was supported by Instruct-ULTRA [Grant 731005], an EU H2020 project to further develop the services of Instruct-ERIC. Contributions from the Coronavirus Structural Task Force were supported by the German Federal Ministry of Education and Research [Grant 05K19WWA] and Deutsche Forschungsgemeinschaft [project TH2135/2-1]. The authors acknowledge the support and the use of resources of Instruct, a Landmark ESFRI project.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Jose Ramon Macias, Spanish National Bioinformatics Institute (INB ELIXIR-ES), Biocomputing Unit, National Centre of Biotechnology (CNB-CSIC), Instruct Image Processing Centre, Campus de Cantoblanco, 28049 Madrid, Spain.
Ruben Sanchez-Garcia, Spanish National Bioinformatics Institute (INB ELIXIR-ES), Biocomputing Unit, National Centre of Biotechnology (CNB-CSIC), Instruct Image Processing Centre, Campus de Cantoblanco, 28049 Madrid, Spain.
Pablo Conesa, Spanish National Bioinformatics Institute (INB ELIXIR-ES), Biocomputing Unit, National Centre of Biotechnology (CNB-CSIC), Instruct Image Processing Centre, Campus de Cantoblanco, 28049 Madrid, Spain.
Erney Ramirez-Aportela, Spanish National Bioinformatics Institute (INB ELIXIR-ES), Biocomputing Unit, National Centre of Biotechnology (CNB-CSIC), Instruct Image Processing Centre, Campus de Cantoblanco, 28049 Madrid, Spain.
Marta Martinez Gonzalez, Spanish National Bioinformatics Institute (INB ELIXIR-ES), Biocomputing Unit, National Centre of Biotechnology (CNB-CSIC), Instruct Image Processing Centre, Campus de Cantoblanco, 28049 Madrid, Spain.
Carlos Wert-Carvajal, Spanish National Bioinformatics Institute (INB ELIXIR-ES), Biocomputing Unit, National Centre of Biotechnology (CNB-CSIC), Instruct Image Processing Centre, Campus de Cantoblanco, 28049 Madrid, Spain.
Alberto M Parra-Perez, Spanish National Bioinformatics Institute (INB ELIXIR-ES), Biocomputing Unit, National Centre of Biotechnology (CNB-CSIC), Instruct Image Processing Centre, Campus de Cantoblanco, 28049 Madrid, Spain.
Joan Segura Mora, Research Collaboratory for Structural Bioinformatics Protein Data Bank, San Diego Supercomputer Center, University of California, San Diego, La Jolla, CA, USA.
Sam Horrell, Diamond Light Source Ltd. (DLS), Oxfordshire, UK.
Andrea Thorn, Institute for Nanostructure and Solid State Physics, HARBOR, Universität Hamburg, Hamburg, Germany.
Carlos O S Sorzano, Spanish National Bioinformatics Institute (INB ELIXIR-ES), Biocomputing Unit, National Centre of Biotechnology (CNB-CSIC), Instruct Image Processing Centre, Campus de Cantoblanco, 28049 Madrid, Spain.
Jose Maria Carazo, Spanish National Bioinformatics Institute (INB ELIXIR-ES), Biocomputing Unit, National Centre of Biotechnology (CNB-CSIC), Instruct Image Processing Centre, Campus de Cantoblanco, 28049 Madrid, Spain.
References
- Bienert S. et al. (2017) The SWISS-MODEL repository-new features and functionality. Nucleic Acids Res., 45, D313–D319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croll,T.I. et al. (2021) Making the invisible enemy visible. Nature Structural & Molecular Biology, 28, 404–408. doi:10.1038/s41594-021-00593-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douangamath A. et al. (2020) Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease. Nat. Commun., 11, 5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hijikata A. et al. (2020) Knowledge‐based structural models of SARS‐CoV‐2 proteins and their complexes with potential drugs. FEBS Lett., 594, 1960–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten R.P. et al. (2014) The PDB_REDO server for macromolecular structure model optimization. IUCrJ, 1, 213–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J. et al. (2020) Computational predictions of protein structures associated with COVID-19. Version 3, DeepMind website. https://deepmind.com/research/open-source/computational-predictions-of-protein-structures-associated-with-COVID-19.
- Kearns S. (2020) Structure of the pandemic. Structure, 28, 874–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce N.M. et al. (2017) A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density. Nat. Commun., 8, 15123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintilie G. et al. (2020) Measurement of atom resolvability in cryo-EM maps with Q-scores. Nat. Methods, 17, 328–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Aportela E. et al. (2019) DeepRes: a new deep-learning- and aspect-based local resolution method for electron-microscopy maps. IUCrJ, 6, 1054–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez-Aportela E. et al. (2021) FSC-Q: A CryoEM Map-to-Atomic Model Quality Validation Based on the Local Fourier Shell Correlation. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura J. et al. (2019) 3DBIONOTES v3.0: crossing molecular and structural biology data with genomic variations. Bioinformatics, 35, 3512–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S. et al. (2020) Structural genomics of SARS-CoV-2 indicates evolutionary conserved functional regions of viral proteins. Viruses, 12, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.