Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), started in 2019 in China and quickly spread into a global pandemic. Nucleocapsid protein (N protein) is highly conserved and is the most abundant protein in coronaviruses and is thus a potential target for both vaccine and point-of-care diagnostics. N Protein has been suggested in the literature as having posttranslational modifications (PTMs), and accurately defining these PTMs is critical for its potential use in medicine. Reports of phosphorylation of N protein have failed to provide detailed site-specific information. We have performed comprehensive glycomics, glycoproteomics and proteomics experiments on two different N protein preparations. Both were expressed in HEK293 cells; one was in-house expressed and purified without a signal peptide (SP) sequence, and the other was commercially produced with a SP channeling it through the secretory pathway. Our results show completely different PTMs on the two N protein preparations. The commercial product contained extensive N- and O-linked glycosylation as well as O-phosphorylation on site Thr393. Conversely, the native N Protein model had O-phosphorylation at Ser176 and no glycosylation, highlighting the importance of knowing the provenance of any commercial protein to be used for scientific or clinical studies. Recent studies have indicated that N protein can serve as an important diagnostic marker for COVID-19 and as a major immunogen by priming protective immune responses. Thus, detailed structural characterization of N protein may provide useful insights for understanding the roles of PTMs on viral pathogenesis, vaccine design and development of point-of-care diagnostics.
Keywords: glycosylation of SARS-CoV-2 N protein, N protein phosphorylation, N protein site-mapping, SARS-CoV-2 nucleocapsid posttranslational modifications, SARS-CoV-2 phosphoproteomics
Introduction
In early December 2019 in Wuhan, China, an outbreak of disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), commonly known as coronavirus disease 2019 (COVID-19), spread rapidly and became a worldwide pandemic (Zhu et al. 2020). According to the World Health Organization (WHO) report, as of mid-April 2021, there have been 137.9 million confirmed cases of COVID-19 globally, including 2 million deaths. In particular, to date, the United States alone has over 31.2 million confirmed cases and over 561,356 deaths due to Centers for Disease Control and Prevention (2021). There are currently more than 80 vaccine candidates in clinical trials, and 2 have been approved for full use in the United States (Zimmer et al. 2021). However, the search for alternative vaccines to protect against SARS-CoV-2 still continues due to recent reports of mutations and new strains of SARS-CoV-2 (Wang et al. 2021). Most of the current vaccine candidates targeting a specific protein are aimed at the virus’s spike protein (Dong et al. 2020). However, the nucleocapsid or nucleocapsid protein (N protein) has also been suggested as a vaccine or therapeutic target as well as a biomarker for the disease (Kumar et al. 2020; Kwarteng et al. 2020; Nikolaev et al. 2020). The N protein is an attractive target because it is highly conserved, strongly antigenic and is the most abundant protein in SARS-CoV-2 (Li et al. 2020; Mu et al. 2020; Yoshimoto 2020). Since no detailed analysis of potential posttranslational modifications (PTMs) of the SARS-CoV-2 N protein has been put forward to date, we decided to undertake this challenging analysis in our laboratory.
SARS-CoV-2 is a virus from the coronavirus family. The coronavirus virion consists of a nucleocapsid surrounded by a lipid envelope in which the membrane glycoprotein (M) and envelop small membrane protein (E) are embedded. Protrusions composed of trimeric glycoproteins (spike protein, S) are anchored in the lipid envelope and extend radially (Figure 1A). The S protein is the most studied protein in SARS-CoV-2 and due to its exposed position on the virus surface and its role in attaching to host cells, it has been chosen as a target for a number of vaccine candidates (Dong et al. 2020). The S protein is highly glycosylated and binds to the human angiotensin-converting enzyme 2 (hACE2) for entry into the host cell (Chen et al. 2020; Shajahan, Archer-Hartmann, et al. 2020; Wang et al. 2020; Zhao et al. 2020). Through a combination of glycoproteomics and glycomics, we and others have recently deduced a detailed glycosylation profile of both S protein and its receptor hACE2 (Shajahan, Supekar, et al. 2020; Watanabe et al. 2020; Zhao et al. 2020). Moreover, a number of studies have highlighted the importance of spike protein glycosylation, which plays a critical role in the mechanism of viral attachment to the hACE2 receptor (Chen et al. 2020; Shajahan, Archer-Hartmann, et al. 2020; Zhao et al. 2020). However, there are reports suggesting that the S protein alone may be an insufficient target for vaccine and therapeutic development (Ferretti et al. 2020;Gouveia et al. 2020; Ihling et al. 2020). A recent clinical study involving SARS-CoV-2 patients identified the majority of immunodominant CD8+ T cell epitopes (that activate CD8+ cells to kill virally infected cells) from virus proteins other than the S protein, leading to the conclusion that more protein targets will need to be included for new and less vulnerable vaccine designs (Ferretti et al. 2020).
Fig. 1 .
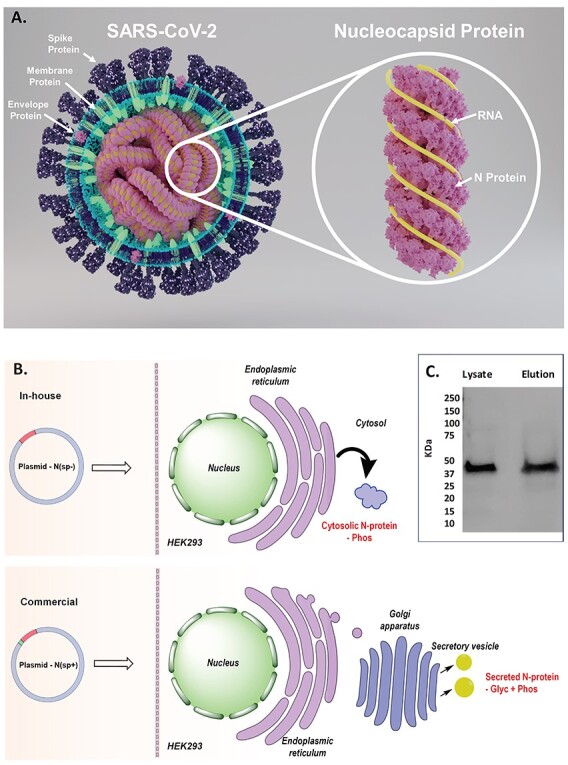
Structural proteins of SARS-CoV-2. (A) Structure of SARS-CoV-2 showing key proteins and structure of N protein. (B) Representation of N protein expression without SP, N(SP-) and with SP, N(SP+). (C) Western blot evaluation of MagneHis™ Protein Purified N(SP-).
In addition to N protein’s role as a potential therapeutic or vaccine target, the N protein can also serve as an important diagnostic marker for COVID-19. Recent mass spectrometry (MS)-based studies of SARS-CoV-2 proteins from gargle-solution and nasal swab samples from COVID-19 patients detected the presence of N protein peptides, making them potential diagnostic biomarkers for COVID-19 (Gouveia et al. 2020; Ihling et al. 2020). The presence of N protein peptides in the gargle and nasopharyngeal swabs could be used to develop a point-of-care high-throughput test for fast detection of SARS-CoV-2. The authors demonstrated in these studies that SARS-CoV-2 peptidome detection through tandem MS can be used as alternative methodologies to polymerase chain reaction (PCR) and immunodiagnostics. The clinical study on gargle solution showed that peptide 41RPQGLPNNTASWFTALTQHGK61 from the N protein could be detected in the saliva of COVID-19-positive patients (Ihling et al. 2020). Another study demonstrated that N protein peptides, 375ADETQALPQR385 and 170GFYAQGSR177, can be detected with intense signal within short retention time in the nasopharyngeal samples from COVID-19 patients (Gouveia et al. 2020). These recent findings on the COVID-19 clinical samples show the presence of the viral N protein in the host body fluids and its potential of the utilization as a diagnostic biomarker at the point-of-care.
The N protein makes up the most abundant source of proteins in the coronavirus (Surjit and Lal 2008). The N protein interacts with viral genome ribonucleicacid (RNA) to form long, flexible, helical ribonucleoprotein (RNP) complexes (Figure 1A) and contributes toward viral genome condensation and packaging (Narayanan et al. 2003; Chen et al. 2007). The C-terminal interactions between the N and M proteins result in specific genome encapsidation during the budding process of the viral particle (He et al. 2004).
Mapping studies of SARS-CoV-2 and other coronaviruses, including the closely related SARS-CoV-1, have revealed the RNA-binding function to a fragment of 55 amino acids located at the N-terminal half that resides in Domain 1, and the RNA-binding, as well as dimerization functions, to the C-terminal half from Domain 3 (Chen et al. 2007). The linker region (LKR) connects the N- and C-terminal domains of the protein and includes a Ser-/Arg-rich (S-/R-rich) region (Chang et al. 2014). Studies have shown that the S-/R-rich LKR contains multiple putative sites of phosphorylation that may play a role in regulating the N protein function and the N–M protein interactions (He et al. 2004; Surjit et al. 2005; Peng et al. 2008; Wu et al. 2009).
Early studies on the N protein from coronaviruses suggested that the N protein is a phosphoprotein and does not bear glycans on its backbone (Parker and Masters 1990; Laude and Masters 1995; Fung and Liu 2018). To discover the PTMs of the SARS-CoV-2 N protein, we explored several high-resolution MS-based approaches, including glycomics, glycoproteomics and phosphoproteomics analyses, using N protein from two different recombinant sources, a commercial source and a recombinant protein produced in our lab.
Results
PTM analysis of commercially produced recombinant N protein N(SP+)
The N protein obtained from the commercial source was expressed in HEK293 with a signal peptide (SP), designed to cause the protein to enter the secretory pathway (Figure 1B). For convenience, we will term this commercial N protein “N(SP+).” Evaluated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS PAGE), N(SP+) had a molecular mass of 60 kDa, which differs from its theoretical protein mass of 47 kDa (Figure 2). The added mass is not due to the SP, which is removed during protein expression and is no longer present in the final commercial product. Rather, the added mass is likely due to PTMs, including glycosylation and phosphorylation (Figure 3) (Masters 2006). In the following, we describe our characterization of the PTMs of N(SP+), focusing primarily on the glycoproteomic analysis. However, a detailed glycomic analysis of the detected glycans can be found in the supplementary material (Supplementary Figures S1–S28 and Supplementary Tables SI and SII).
Fig. 2 .
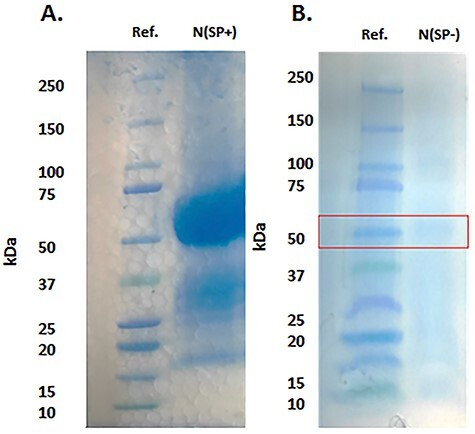
SDS PAGE evaluation of N protein. (A) N protein expressed with SP, N(SP+) (60 kDa). (B) N protein expressed without SP, N(SP-) (47 kDa).
Fig. 3 .
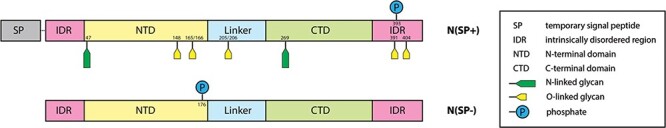
Domain structure and PTMs of the commercial N protein, produced with SP N(SP+) (top) and the N protein, expressed in-house, without the SP N(SP-) (bottom).
N-linked glycosylation on N(SP+)
We used trypsin and elastase proteases, both separately and sequentially, to produce three peptide samples. The Liquid Chromatography (LC) tandem Mass Spectrometry (MS/MS) data of these protease digests were analyzed by using search algorithms in the Byonic software, employing all possible mammalian N-glycans, O-glycans and phosphorylation as possible PTMs.
There are five potential N-glycosylation sites (-NXS/T- with X ≠ P) on the N protein (N47, N77, N192, N196 and N269) (Figure 4). The Byonic N-glycopeptide search suggested that only N47 and N269 were glycosylated in N(SP+). We manually validated this conclusion for accurate precursor mass (5 ppm) and precise glycan neutral loss in the MS/MS experiments (<20 ppm). The data confirmed that the two N-glycosylation sites vary distinctly in their N-glycosylation profiles. The site N47 was part of glycopeptide 41RPQGLPNNTASWF53 and had a glycan occupancy of about 53% (Figure 5 and Supplementary Table SIII) (Supplementary Figures S29–S64). We observed only complex-type glycan structures at this site. Based on the common biosynthetic pathway (Varki et al. 2015-2017) and glycan neutral loss pattern in MS/MS experiments from glycopeptide, the most abundant glycoform with an m/z 3590.51 was identified as glycan NeuAc1GalNAc1Gal1GlcNAc2Man3GlcNAc2Fuc1, which showed 6.56% glycan occupancy relative to other glycoforms at site N47. Moreover, all the glycans detected at this site featured core fucosylation. We also observed multiple doubly fucosylated glycans (~16%) and a minor amount of triply fucosylated structure (0.45%) (Supplementary Table SIII).
Fig. 6 .
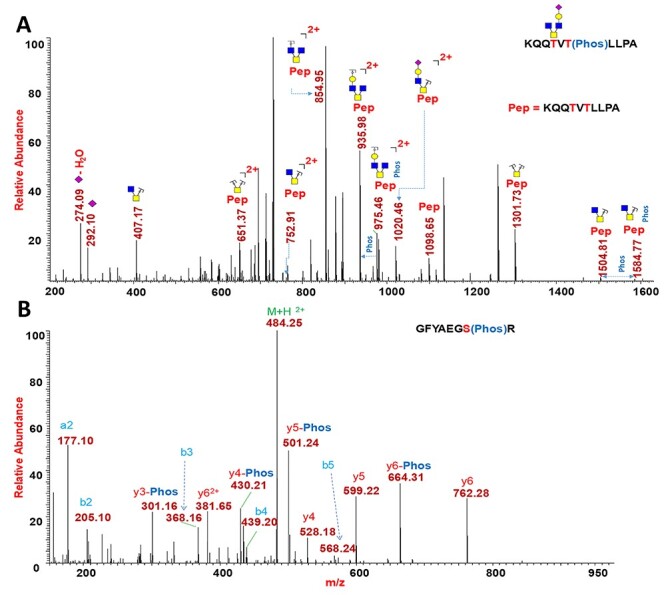
PTM analysis of N(SP+) and N(SP-). (A) CID MS2 of N(SP+) phosphorylated O-glycopeptide 388KQQTVTLLPA397 indicating phosphate at Thr393. (B) HCD MS2 of N(SP-) phosphorylated peptide 170GFYAEGSR177 showing phosphate at Ser176.
Fig. 5 .
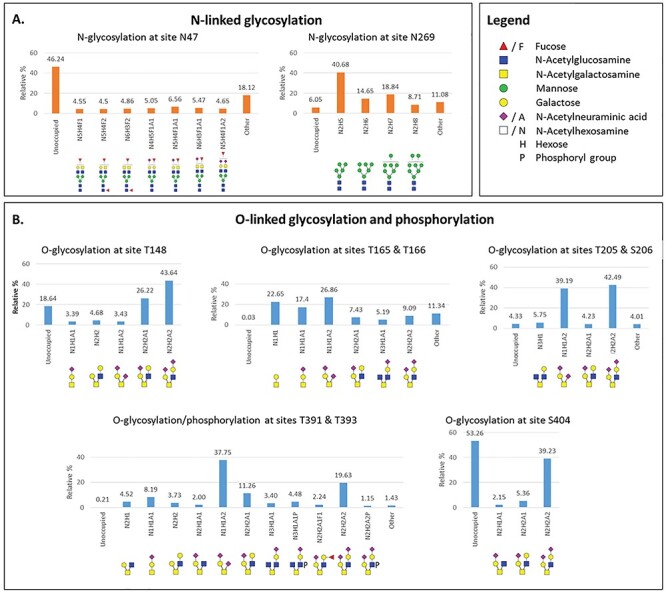
Site-specific relative quantification of N- and O-linked glycosylation of N(SP+). (A) Relative percentage of N-glycans at sites N47 (complex-type glycans only) and N269 (mainly high-mannose type glycans, Man5 structure as the most abundant). (B) Relative percentage of O-glycans at sites T148, T165, T166, T205, S206, T391 and S404. Phosphorylation is detected on T393. These sites showed major glycan occupancy ranging from 47–99%. Minor O-glycosylated site distribution is shown in the SI. Minor glycoforms with less than 2% abundance are referred to as “other.”
Fig. 4 .

SARS-CoV-2 N protein sequence coverage and detected PTM sites for N(SP+) and N(SP-).
The Byonic N-glycopeptide and manual search also resulted in the detection of N-glycan on peptide 267AYNVTQAFGR276 at site N269. The glycan occupancy at site N269 was found to be higher (94%) than that of site N47 (53%). At site N269, we detected mostly high-mannose type glycosylation making up ~85% of total glycan (Figure 5A). The relative percentage of the complex- (4.5%) and hybrid-type N-glycans was lower (~3.6%). Fucosylation and sialylated glycans were identified at trace levels (0.63% and 0.76%, respectively) (Supplementary Table SIII). The tryptic peptide 69GQGVPINTNSSPDDQIGYYR88 was detected only in nonglycosylated form (Supplementary Figure S48), indicating that N77 is not occupied.
The N-glycosylation sites N192 and N196 occur in the LKR, which is rich in serine and arginine (SR-rich). Investigation of the N-glycosylation at these sites proved difficult because the two N-glycan sequons are located next to each other and are separated by only one arginine residue (Figure 4). For site N192, the trypsin digest possibly generated the tripeptide 192NSS195 (Figure 4), which is too small to efficiently fragment and detect in the mass spectra. For site N196, in silico trypsin digestion generated the peptide 196NSTPGSSR203, which also contains four potential O-glycan sites (Figure 4). The software searches for the possible N-glycan combined with four O-glycans were not successful due to software limitations and were too complicated to hand-calculate. Similarly, in silico analysis of the elastase digest of N protein produced the longer peptide 183SSRSSSRSRNSSRNSTPGSSRGTSPA208, comprising two potential N-glycosylation sites and 14 potential O-glycosylation or phosphorylation sites, presenting a formidable challenge to software-based detection (Figure 4). All these factors made the PTM characterization at this SR-rich LKR extremely difficult. To overcome this obstacle, we set out to locate the O-glycans on this peptide first, using the de-N-glycosylated N protein. We reasoned that searching for O-glycosylation in the SR-rich region would later be helpful in finding N-glycans since confirmed O-glycan information from this de-N-glycosylation experiment would enable manual-/software-assisted search for N-glycosylation in non-de-N-glycosylation experiments. To investigate O-glycosylation in the SR-rich region, we first removed the N-glycans from the N protein by the PNGase F treatment and subsequently digested the protein by trypsin/elastase. The resulting O-glycopeptides were subjected to LC–MS/MS analysis, followed by manual- and software-assisted searches for O-glycans. This eliminated interference of N-glycans while searching for O-glycans and reduced the number of possible combinations for manual-/software-based glycan search. We detected the O-glycosylated 204GTSPAR209 glycopeptide from the LKR and observed several O-glycans on the peptide (see detection of O-linked glycosylation of N(SP+)). However, even with this stepwise approach, we could not identify any N-glycans at sites N192 and N196.
To further investigate the possibility of glycosylation at sites N192 and N196, we performed an 18O labeling experiment where N-glycans are removed from glycoproteins with PNGase F in the presence of H218O, followed by enzymatic digestion with trypsin/elastase (Supplementary Figure S1). In the process of de-glycosylation, the N-glycan-bearing Asn is converted to 18O-Asp (N → D*), which can be detected by MS. In this experiment, we observed conversion of N47 and N269 to 18O-labeled Asp, confirming the presence of N-glycans in these positions. LC–MS/MS analysis of the trypsin and elastase digests detected the peptides, 41RPQGLPD*NTA50 and 268YD*VTQAFGR276 with 18O-Asp (D*) at positions 47 and at 269, respectively (Supplementary Figures S30 and S50). However, in either digest, we did not observe any 18O-labels at site N196 but detected the unlabeled peptide 196NSTPGSSRGTSPA208, confirming absence of an N-glycan at N196 (Supplementary Figure S49). For site N192, we observed neither 18O-labeled nor unlabeled peptide in any of the enzymatic digests.
O-linked glycosylation of N(SP+)
For the detection of O-glycosylation, we performed LC–MS/MS experiments on tryptic and elastase digests. The samples were prepared by separate and sequential enzymatic digestion. We also confirmed O-glycosylation on peptides by removing N-glycans by treatment of PNGase F, followed by enzymatic digestion with trypsin and elastase. We identified the O-glycosylation on the N protein by performing higher-energy collisional dissociation (HCD) and collision-induced dissociation (CID) fragmentations and identified the O-glycosylation site occupancy by a targeted electron-transfer dissociation (ETD) MS2 fragmentation on O-glycopeptides. ETD is a chemical dissociation technique that induces fragmentation along the glycopeptide backbone generating c and z ions with the intact glycan, which helps to pinpoint the position of the otherwise labile glycan in the peptide (Shajahan et al. 2017). We were able to confirm O-glycosylation on 11 O-glycan sites within eight distinct peptides and determined the sites of O-glycan attachment (Supplementary Figures S65–S135). The glycan occupancy on four O-glycosylation sites (S23, T245, T247 and T379; found on three glycopeptides) in total was very low (<10% for each site) (Supplementary Figures S136–S138).
We observed ions with the mass corresponding to the peptide 15ITFGGPSDSTGSNQNGER32 with multiple glycans, such as m/z 2771.1381 for peptide+HexNAc1Hex1NeuAc2 and m/z 3137.2547 for peptide+HexNAc2Hex2NeuAc2. In HCD and CID MS experiments, oxonium ions for HexNAc (m/z 204.0865) and NeuAc (m/z 292.1026) and respective b and y peptide fragment ions confirmed the identity of the glycopeptide (Supplementary Figures S66–S69). The ETD experiments suggested that glycosylation is present at site S23 (fragments c8, c9, z9 and c10) (Supplementary Figure S65). We observed that the glycan occupancy at S23 was only about 2% as shown in Supplementary Figure S136. HCD and CID MS2 analysis of glycopeptide 144DHIGTR149 confirmed the O-glycosylation at T148 by the presence of glycan oxonium ions, peptide fragments (b and y ions) and glycan neutral losses (Supplementary Figures S70–S74). The overall glycan occupancy at site T148 was determined at ~81%, out of which mono- and di-sialyl core 2-type glycans (~70%) were predominant (Figure 5B and Supplementary Table SIV).
The Byonic glycopeptide search showed a strong indication of glycosylation on the peptide 159LQLPQGTTLPK169, which contains two potential O-glycosylation sites (Supplementary Figures S75–S92). To further evaluate the glycosylation in this peptide, we manually validated full mass, HCD and CID MS2 spectra that showed oxonium ions, b and y peptide fragments, along with peaks corresponding to the masses of the peptide and peptide+HexNAcHex (Supplementary Figure S76). The ETD experiment for site-mapping of T165 or T166 in the peptide 159LQLPQGTTLPK169 detected two spectra that showed c and z ions, indicating that some of these peptides are O-glycosylated on T165 and others on T166 (Supplementary Figures S91 and S92). The ETD MS2 spectra showed the fragment peaks z5 at m/z 908.57 and c7 at m/z 1120.57, diagnostic for the presence of glycans at site T165 (Supplementary Figure S91), as well as the fragment peaks z4 at m/z 807.36 and c8 at m/z 1221.65, diagnostic for the presence of glycans at site T166 (Supplementary Figure S92). The data demonstrated that the peptide 159LQLPQGTTLPK169 is almost fully occupied with glycans. Moreover, core 1-type structures were predominant, accounting for 77% of total glycans at these sites (Figure 5B). However, we also detected core 2- to core 4-type glycans, including extended core 2 glycans with fucose and sialic acid on the peptide 159LQLPQGTTLPK169 (Supplementary Figures S75–S92).
The peptide 204GTSPAR209, located in the LKR region, showed O-glycans at both sites, T205 and S206. We were not able to deduce site-specific information at these sites. However, based on the glycosylation pattern of the peptide, it was confirmed that both sites contained O-glycans. This peptide was about 85% occupied with sialylated core 1- and core 2-type glycans in total (Figure 5B and Supplementary Figures S93–S99).
The Byonic and manual glycoproteomics search also found the peptide 238GQQQQGQTVTK248, which contains sites T245 and T247 at the C-terminal dimerization domain (CTD) region. A corresponding, minor glycopeptide with the glycan HexNAcHexNeuAc was identified at m/z 1858.8428; however, due to the low abundance of this glycopeptide, we were unable to obtain conclusive MSn data and so the site of O-glycosylation on this glycopeptide remains unknown (Supplementary Figures S100–S103). Based on full mass and fragment ions, another peptide 376ADETQALPQR385 that contains O-glycosylation site T379 indicated glycans (Supplementary Figures S104–S113). Nonetheless, the peptides 238GQQQQGQTVTK248 and 376ADETQALPQR385 were mostly unoccupied (>90%, Supplementary Figures S137 and S138).
Toward the C-terminus end of N protein, the peptide 388KQQTVTLLPA397, which includes O-glycosylation sites T391 and T393, was found with attached glycans. The ETD experiments detected corresponding c and z peptide fragments with intact glycan, suggesting that the site T391 was glycosylated (Supplementary Figure S132). At site T391, di-sialylated core 1- to core 2-type glycans were predominant over other glycans (Figure 5B and Supplementary Figure S114–S128 and Supplementary Table SIV). Interestingly, site S404 was found to contain only mono- and di-sialylated core 2-type structures as shown in Supplementary Figures S129–S131. We confirmed the O-glycan structures and glycosylation sites through both Byonic software search and manual analysis.
In summary, as shown in Figure 5B, the sites T148, T165, T166, T205, S206, T391 and S404 comprised significant levels of O-glycosylation (47–90%), and the sites S23, T245, T247 and T379 indicated a lower level (1–9%) of O-glycosylation. We identified core 1-, core 2-, core 3- and core 4-type O-glycans at these sites, confirming the glycomics analysis findings (see supplementary material and Supplementary Figures S2 and S5 and Supplementary Tables SII and SIV).
O-phosphorylation
We digested the N protein with elastase/trypsin and conducted a Byonic-based phosphate search in the elastase digest. In this experiment, we identified phosphorylation on glycopeptide 388KQQTVTLLPA397, which contains two potential sites—T391 and T393—for glycosylation or phosphorylation on N(SP+). As shown in the HCD MS2 spectrum (Supplementary Figure S133), the glycopeptide identity was confirmed by validating the b and y ions and glycan neutral losses. The presence of a phosphate in the peptide was confirmed by CID MS2 experiments using the high-resolution precursor mass and neutral losses of phosphate (loss of m/z 79.9663) from the glycopeptide (Supplementary Figure S134). As shown in the deconvoluted CID MS2 spectrum in Figure 6A, the peak at m/z 1584.77 represents the glycopeptide fragment 388KQQTVTLLPA397 with a phosphate group and a glycan, and the peak at m/z 1504.81 represents the same peptide with glycosylation but without phosphorylation (neutral loss of one phosphate group of 79.9663 Da). Similarly, peaks at m/z 975.45 and at 935.98 show loss of a phosphate group from the peptide fragment bearing a HexNAc3Hex glycan. In the site-mapping ETD experiments of this peptide, we observed characteristic c4, c6 and z5, z7 ions at m/z 1565.76, 1846.15, 578.26 and 1840.85, respectively, to unambiguously assign the site of phosphorylation at site T393 and glycan at site T391 (Supplementary Figure S132).
PTM analysis of recombinant N protein without SP N(SP-)
Since the extensive glycosylation of the commercial N protein N(SP+) was likely an artifact resulting from the presence of the SP during expression, we wanted to also determine the PTMs of properly expressed (i.e. without the SP) N protein N(SP-) for comparison. We expressed N(SP-) in our laboratory in HEK293 cells and confirmed it by western blot using HRP anti-His Tag Antibody from Biolegend (Figure 1C). N(SP-) was purified from cell lysate using the MagneHis™ Protein Purification kit from Promega (V8500). The SDS PAGE showed that, in contrast to N(SP+), whose mass was higher due to glycosylation, N(SP-) had a molecular mass of 47 kDa, as expected from the known protein sequence. N(SP-) was subjected to proteolysis and the enzymatic digests analyzed by LC–MS/MS for the identification of PTM and peptide mass fingerprinting.
Protein databases suggest that SARS-CoV-2 N protein is a phosphoprotein. We analyzed protease-digested N(SP-) via LC–MS/MS analysis, followed by manual- and software-assisted searches for proteomic analysis. For software-aided searches, we used Byonic software, employing all possible mammalian phosphorylation, N-glycans and O-glycans as possible PTMs. We performed a Byonic search for the detection of phosphorylation, followed by manual validation for accurate precursor mass (5 ppm) and precise b and y ions in the MS/MS experiments (<20 ppm) (Supplementary Figures S139– S165). In the tryptic digest of N(SP-), the peptide 170GFYAEGSR177 was detected with phosphate modification at Ser176. As shown in Figure 6B, the precise b and y ions in the MS2 spectrum uncovered the site of phosphorylation. For this site, the peptide without phosphate modification was also found, suggesting that the site Ser176 is not fully phosphorylated. Our analysis found that the N(SP-) at site S176 is 44.4% occupied with the phosphate modification. There are five potential N-glycosylation sites (-NXS/T- with X ≠ P) on the N protein. Our Byonic N-glycopeptide search, followed by manual validation for accurate precursor mass (5 ppm) and precise glycan neutral loss in MS/MS experiments (<20 ppm), did not detect N-glycosylation on the sites N47, N77 and N269 in the tryptic digest of N(SP-). For these three sites, we were able to detect only peptides, 41RPQGLPNNTASWFTALTQHGK61, 69GQGVPINTNSSPDDQIGYYR88 and 267AYNVTQAF274, confirming that this region is covered by the tryptic digest. We did not obtain sequence coverage for the N-glycosylation sites N192 and N196 that occur in the LKR. Surprisingly, we did not detect the peptides of the SR-rich LKR between amino acids G178–R209, possibly due to the presence of multiple PTMs, although no PTMs were detected in this region other than phosphorylation on S176. Further, we conducted Byonic-assisted and manual O-glycan search on the tryptic peptides of N(SP-). However, we did not identify peptides with O-glycans.
We observed cleavage of methionine at the N-terminal end and acetylation of the subsequent serine residue (Supplementary Figures S139 and S140). The cleavage of N-terminal methionine and acetylation of the N-terminus are the most common protein PTM (Bonissone et al. 2013). Two-thirds of the proteins in any proteome are suggested to be potential substrates for N-terminal methionine excision (NME), and methionine aminopeptidases (MetAPs) are expressed in all organisms from bacteria to eukaryotes (Giglione et al. 2004). For the methionine excision, the second or P2, amino acid in protein substrates is important as MetAP preferentially excises the N-terminal Met when the second residue is Gly, Ala, Ser, Thr, Cys, Pro or Val. The substrates for N-acetyltransferase (NAT), which acetylates the N-terminus, are usually the four smallest residues (Gly, Ala, Ser and Thr). In the case of the N protein, the P2 amino acid is Ser, and thus our observation of PTM at the N-terminus of the N protein is following the conventional N-terminal processing of proteins (Bonissone et al. 2013). Since the NME and NTA activity is necessary for protein function and stability, such processing of the N protein could be critical for its activity in viral replication and infection.
Discussion
The coronavirus N protein consists of three distinct but highly conserved domains, including an RNA-binding N-terminal domain (NTD), a CTD and a central S-/R-rich linker domain, which display an intrinsically disordered structure and facilitate molecular movements to aid interactions (Figure 2). The NTD is reportedly responsible for RNA-binding, CTD for oligomerization, and the SR-rich linker is generally known to be primarily involved in phosphorylation events (Wootton et al. 2002; Chang et al. 2006; Kang et al. 2020). However, no site-specific PTMs have been reported in any of these domains in the current literature on SARS-CoV-2. Our LC–MS/MS studies have shown that the commercial preparation of the recombinant SARS-CoV-2 N protein expressed with an N-terminal SP N(SP+) targeted it to the host cell secretory pathway and resulted in both N- and O-glycosylations, in addition to phosphorylation. By contrast, recombinant N protein expressed without the leader sequence N(SP-) has no detectable N- or O-glycosylation, and we identified the phosphorylation at a different site (S176) than that found in N(SP+) (T393). The addition of nonnatural PTMs carries the risk of generating artifacts when using such protein in diagnostic or research settings, potentially complicating the interpretation of results. Our data indicate that N protein expressed without the SP does not contain glycan structures but is phosphorylated. This form is much more likely to represent characteristics of the in vivo N protein expressed during the course of a SARS-CoV-2 viral infection.
A recent report used the same commercial glycosylated version of N protein and studied the binding antibodies isolated from the plasma of human Covid-19 patients (Rudberg et al. 2020). The authors compared the serological titer of antibodies with three antigens of SARS-CoV-2, including spike (S) protein and nucleocapsid (N) protein, and found that 235 out of 243 positive cases had antibodies that bound to this glycosylated version of the N protein. That study shows that the glycosylated N protein can bind antibodies generated by Covid-19 patients, possibly by binding only to the nonglycosylated regions of this N protein (Rudberg et al. 2020). However, additional epitopes might be revealed using an N protein without such PTMs.
Recently, several enzyme-linked immunosorbent assay (ELISA)-based serological tests have been developed to detect serum immunoglobulins (Igs) against SARS-CoV-2. Serological assays are shown to be critical in assessing the population spectrum that has been exposed to the virus as well as the heterogeneity of antibody response. A dual ELISA test against SARS-CoV-2 N protein showed the critical importance of epitope unmasking by de-glycosylation of the protein produced in a mammalian system (Rump et al. 2020).
The phosphorylation sites in the SARS-CoV-2 N protein were investigated in a recent study and were reported as occurring within the N-terminal portion of the protein, at or near the RNA-binding region, but not at the CTD (Bouhaddou et al. 2020). There are clusters of phosphorylation sites within the arginine/serine (RS)- dipeptide-rich region, which is C-terminal to the RNA-binding region, and such region is a conserved sequence across N proteins within the coronavirus family. In SARS-CoV-1, this region is phosphorylated by serine–arginine (SR) protein kinases and modulates the role of SARS-CoV-1 N protein in host translation inhibition (Peng et al. 2008). Phosphorylation of this same region can possibly play a similar role in SARS-CoV-2.
To our knowledge, the coronavirus N protein is the only phosphorylated structural protein on the virus, and this phosphorylation has been proposed to play a role in regulating its functions (Calvo et al. 2005; Chen et al. 2005; Lin et al. 2007). Evidence of significant conformational changes in the N protein structure due to phosphorylation has been reported (Stohlman et al. 1983). N protein from SARS-CoV-1 has been shown to elicit a well-defined immunological response, which underscores the importance of the N protein as a potent target for a vaccine against the COVID-19 infection (Blicher et al. 2005). A recent study, aimed to investigate the effect of early SARS-CoV-2-specific humoral immune responses on the disease outcome, found that deceased patients had stronger antibody responses toward N protein, while survivors had a much stronger antibody response to the S protein, highlighting the importance of the N protein in disease outcome (Atyeo et al. 2020). Although several vaccine candidates have exhibited high efficacy, the majority are based on the recognition of the S protein (Zimmer et al. 2021, Dong et al. 2020, Mohan et al. 2020, Polack et al. 2020). There is a concern that the vaccine will no longer be effective if the new SARS-CoV-2 strains with altered S protein sequence emerge (Koyama et al. 2020). The N protein has been reported as less susceptible to mutations and thus merits further consideration as a target for future vaccine development, although another study has claimed that N protein is subject to increased mutations (Dutta et al. 2020; Grifoni et al. 2020; Rahman et al. 2021). A recent study reported 684 amino acid substitutions across 317 (75.66% of total amino acids) unique positions of the SARS-CoV-2 N protein, such as in positions 82, 21 and 83 in the RNA-binding N-terminal domain, SR-rich region and CTD, respectively. In addition, they reported 11 in-frame deletions, mostly (N protein SARS-Cov-2 sequences, n = 10) within the highly flexible LKR and the NTD region (Rahman et al. 2020). Nevertheless, understanding the patterns of the PTMs of the N protein described in this study may ultimately help in new strategies to detect, prevent or treat the SARS-CoV-2 infection.
In summary, we noted that the commercial N protein preparation contained numerous PTMs, including N- and O-linked glycosylation and phosphorylation at an unexpected location (Thr393). By contrast, our recombinant product was not glycosylated and was phosphorylated at a site distinct from the commercial preparation (Ser176). Further inspection revealed that the commercial recombinant N protein preparation was designed as a fusion protein with a proprietary N-terminal SP sequence that caused it to enter the secretory pathway of the HEK293 host cells. On the other hand, our recombinant expression product did not contain such an N-terminal signal sequence causing the N protein to remain in the cytosol (Figure 1B). Our results indicated that targeting to the mammalian host cell secretory pathway led to glycosylation and phosphorylation distinctive from physiologically cytosolic viral N protein. It is expected that these additional PTMs may impact the validity of the experimental conclusions derived from the commercial preparation when studied in the context of virus biology.
Materials and methods
Sequencing-grade modified trypsin and elastase were purchased from Promega (Madison, WI). Peptide-N-Glycosidase F (PNGase F) was purchased from New England Biolabs (Ipswich, MA). All other reagents were purchased from Sigma-Aldrich (St. Louis, MO) unless indicated otherwise. Data analysis was performed using Byonic 2.3 software and was performed manually using Xcalibur 4.2 and GlycoWorkbench 1.1. The purified N protein expressed in HEK293 cells (Cat. No. NUN-C5227) was purchased from AcroBiosystems (Newark, DE).
Expression of N protein in HEK293 cells
The coding region for the N protein gene of SARS-CoV-2 (2019-nCoV) in the expression vector pCMV3ORF-his, was obtained from Sino Biological (Beijing, China) (Cat: VG40588-CH) and transformed into E. coli DH5α (NEB, Cat. No. C2987H). A DNA Maxi prep was generated using the Purelink Hipure Plasmid Maxiprep kit from Invitrogen (Carlsbad, CA) (Cat: K210006) and was resuspended in sterile extra pure water. The recombinant N Protein was expressed by transient transfection of suspension culture in HEK293 cells (FreeStyle™ 293-F cells, ThermoFisher Scientific, Waltham, MA). Cells were maintained at 0.5–3.0 × 106 cells/mL in a humidified CO2 platform shaker incubator at 37°C with 50% humidity and 125 rpm. Transient transfection was performed at a cell density of 2.5–3.0 × 106 cells/mL in an expression medium comprised of a 9:1 ratio of Freestyle™293 expression medium (ThermoFisher Scientific) and EX-Cell expression medium, including Glutamax (Sigma-Aldrich). Transfection was initiated by the addition of plasmid DNA and polyethyleneimine as transfection reagent (linear 25-kDa polyethyleneimine, Polysciences, Inc., Warrington, PA). Twenty-four hours posttransfection, the cell cultures were diluted with an equal volume of fresh media supplemented with valproic acid (2.2 mM of final concentration), and protein production was continued for an additional 5 d at 37°C with 125 rpm shaking. The cell cultures were harvested, clarified by sequential centrifugation at 1200 rpm for 10 min and 3500 rpm for 15 min at 4°C. The cell pellets were further used for the purification of N Protein.
Purification of N protein in HEK293F cells without SP N(SP-)
N protein expressed in HEK293F was extracted and purified using MagneHis™ Protein Purification kit purchased from Promega (Madison, WI) (Catalog number: V8500). Approximately, 4 × 106 cells (1 mL) were lysed using the buffer provided in the kit with adding 1 μL of DNase by passing cells through 1-mL syringe. The cell lysate was centrifuged, and supernatant was used for further purification. Approximately, 200 μL of lysate was diluted to 1 mL using PBS buffer with 20 mM imidazole. To this solution was added 50 μL of magnetic his-Ni-particles and they were incubated at room temp for 90 min. Magnetic particles were settled using magnetic stand. Using a pipette, we carefully removed the supernatant, washed the particles using 500 μL of 0.1% Tween-20, 10 min each time, for a total of 10 times. We washed the particles using 500 μL of 100 mM HEPES, 10 mM of imidazole and 500 mM of NaCl buffer. Total of 10 times, 10 min for each time. The protein was eluted with elution Buffer (pH 7.5), 100 mM HEPES, 500 mM imidazole, by incubating for 5 min for two times. The protein solution was desalted by passing through 10 kDa MWCO filters and was lyophilized. The protein was then suspended in 50-mM ammonium bicarbonate and was digested with sequence grade trypsin and elastase enzymes separately. The enzymatic digests of N protein expressed without the SP was then analyzed by LC–MS/MS for the identification of PTM detection and peptide mass fingerprinting.
N- and O-linked glycan release, purification and permethylation of N(SP+)
The N protein, N(SP+) was purified using SDS PAGE prior to glycomic analysis (Figure 3A). The 45–65 kDa gel band was cut into smaller pieces (1 mm squares approx.) and was transferred to clean micro-centrifuge tubes. The gel pieces were de-stained by adding 500 μL of acetonitrile (ACN):50 mM NH4HCO3 (1:1) and were incubated at room temperature (RT) for about 30 min. Tubes were centrifuged, and the supernatant was discarded. Then 250 μL of ACN was added, and the gel pieces were incubated for 20–30 min. Samples were centrifuged, and the supernatant was discarded. The gel pieces were then suspended in 500 μL of 50 mM NH4HCO3 and 3 μL PNGase F was added. The sample was incubated for 18 h at 37°C. The released N-glycans were extracted out by adding 1:2 H2O:ACN (500 μL) at RT for 15 min, and the supernatant was collected. The released N-glycans were speed-dried, reconstituted in 0.1% formic acid (FA) solution, purified by passing through a C18 SPE cartridge and eluted with 0.1% FA solution (3 mL) before being dried by lyophilization. The gel pieces containing O-glycoprotein were digested by adding sequence grade trypsin in digestion buffer (50 mM NH4HCO3) for 18 h at 37°C. The O-glycopeptides were extracted out from gel by the addition of 1:2, H2O:ACN containing 5% FA (500 μL) and were kept at RT for 15 min and the supernatant was collected. The released O-glycopeptides were speed-dried and used for the release of O-glycans by β-elimination.
The O-glycans were released from the glycopeptide peptide by reductive β-elimination reported elsewhere (Shajahan et al. 2017). Briefly, eluted N protein glycopeptides were treated with a solution of 19 mg/500 μL of sodium borohydride in 50 mM of sodium hydroxide. The reaction mixture was heated to 45°C for 16 h, then neutralized with a solution of 10% acetic acid. The sample was desalted on a hand-packed ion exchange resin (DOWEX H+) by eluting with 5% acetic acid and dried by lyophilization. The borates were removed by the addition of a solution of methanol:acetic acid (9:1) and evaporation under a steam of nitrogen. The released N- and O-linked glycans were then permethylated using NaOH/DMSO–methyl iodide method published previously (Shajahan et al. 2017).
N- and O-linked glycomic profiling by MALDI-MS and ESI-MSn of N(SP+)
The permethylated N- and O-glycans were dissolved in 2 μL of methanol. About 0.5 μL of sample was mixed with equal volume of DHB matrix solution (10 mg/mL in 1:1 methanol–water) and spotted on to a MALDI plate. MALDI–MS spectra were acquired in positive ion and reflector mode using an AB Sciex 5800 MALDI-TOF-TOF mass spectrometer.
Two microliters of permethylated N- and O- glycans dissolved in methanol were mixed with 98 μL of ESI–MS infusion buffer (1:1:1–0.1% FA in water [with 1 mM NaOH]:methanol:ACN) and were infused directly into an Orbitrap Fusion Tribrid mass spectrometer through a nanospray ion source. ESI-MSn spectra of glycans were acquired by both total ion monitoring and manual MSn fragmentation. Original glycoform assignments were made based on full-mass molecular weight. Additional structural details were determined by ESI-MSn and analysis with GlycoWorkbench 1.1 software.
Protease digestion for glycoproteomics of N protein, N(SP+)
The purified N protein, N(SP+) (20 μg), expressed on HEK293 cells was dissolved in 50 mM of ammonium bicarbonate solution and digested with trypsin and elastase separately as well as sequentially by incubating for 18 h at 37°C. The digest was filtered through 0.2-μm filter and was directly analyzed by LC–MS/MS.
18O labeling via de-glycosylation and subsequent protease digestion of N(SP+)
One micrograms of N protein was dissolved in 36 μL of H218O and 2 μL of 10× glycobuffer (NEB). To this solution, 2 μL PNGase F was added, and the reaction mixture was incubated at 37°C for 16 h. Enzymatic activity of PNGase F was deactivated by heating the mixture to 95°C for 5 min followed by lyophilizing the sample. The de-N-glycosylated protein was then suspended in 48 μL 50 mM NH4HCO3 and we added 2 μL elastase/trypsin (0.5 μg/μL). The protein was then digested at 37°C for 16 h. The enzyme was deactivated by heating to 95°C for 5 min and the solvents were removed by speed vacuum. The peptides were reconstituted in 0.1% FA, filtered through a 0.2 μm filter and analyzed via LC–MS/MS.
Data acquisition of protein digests using nano-LC–MS/MS
The glycoprotein digests were analyzed on an Orbitrap Fusion Tribrid mass spectrometer equipped with a nanospray ion source and were connected to a Dionex Ultimate 3000 RSLC nano system (ThermoFisher). A prepacked nano-LC column (Cat. No. 164568, ThermoFisher) of 15 cm length with 75 μm internal diameter (id), filled with 3 μm C18 material (reverse phase), were used for chromatographic separation of samples. The precursor ion scan was acquired at 120,000 resolutions in the Orbitrap analyzer, and precursors at a time frame of 3 s were selected for subsequent MS/MS fragmentation in the Orbitrap analyzer at 15,000 resolution. The LC–MS/MS runs of each digest were conducted for 180 min. About 0.1% FA and 80% ACN–0.1% FA was used as mobile phases A and B, respectively, in order to separate the glycopeptides. The threshold for triggering an MS/MS event was set to 1000 counts, and monoisotopic precursor selection was enabled. MS/MS fragmentation was conducted with stepped HCD product-triggered (pd) CID (collision-induced dissociation) (HCDpdCID) program. The O-linked glycopeptides were also analyzed for site-mapping by targeted ETD and were acquired on both Orbitrap and ion trap (IT) analyzers. Charge state screening was enabled, and precursors with unknown charge state or a charge state of +1 were excluded (positive ion mode). Dynamic exclusion was also enabled (exclusion duration of 30 s).
Data analysis of glycoproteins
The LC–MS/MS spectra of combined tryptic/elastase digest of N protein were searched against the FASTA sequence of N protein using the Byonic software 2.3 by choosing appropriate peptide cleavage sites (semi-specific cleavage option enabled). Oxidation of methionine, deamidation of asparagine and glutamine, protein N-acetylation and possible common mammalian N-glycans, O-glycan and phosphorylation masses were used as variable modifications. The LC–MS/MS spectra were also analyzed manually for the glycopeptides with the support of the ThermoFisher Xcalibur 4.2 software, GlycoMod tool and ProteinProspector v6.2.1. The HCDpdCID and ETD MS2 spectra of glycopeptides were evaluated for the glycan neutral loss pattern, oxonium ions and glycopeptide fragmentations to assign the sequence and the presence of glycans in the glycopeptides. Supplemental glycomics and glycoproteomics workflow figure (Supplementary Figure S1) was created with the help of BioRender.com.
Supplementary Material
Funding
Financial support from the US National Institutes of Health (S10OD018530, P41GM103390, R24GM137782 and R01GM130915) is gratefully acknowledged.
Contributor Information
Nitin T Supekar, Complex Carbohydrate Research Center, The University of Georgia, 315 Riverbend Road, Athens, GA 30602, USA.
Asif Shajahan, Complex Carbohydrate Research Center, The University of Georgia, 315 Riverbend Road, Athens, GA 30602, USA.
Anne S Gleinich, Complex Carbohydrate Research Center, The University of Georgia, 315 Riverbend Road, Athens, GA 30602, USA.
Daniel S Rouhani, Complex Carbohydrate Research Center, The University of Georgia, 315 Riverbend Road, Athens, GA 30602, USA.
Christian Heiss, Complex Carbohydrate Research Center, The University of Georgia, 315 Riverbend Road, Athens, GA 30602, USA.
Digantkumar Gopaldas Chapla, Complex Carbohydrate Research Center, The University of Georgia, 315 Riverbend Road, Athens, GA 30602, USA.
Kelley W Moremen, Complex Carbohydrate Research Center, The University of Georgia, 315 Riverbend Road, Athens, GA 30602, USA.
Parastoo Azadi, Complex Carbohydrate Research Center, The University of Georgia, 315 Riverbend Road, Athens, GA 30602, USA.
Authors’ contributions
P.A., N.T.S. and A.S. conceived the idea of the paper; N.T.S. performed glycoproteomics and glycomics sample processing, A.S. conducted glycomics data acquisition, N.T.S, A.S., A.S.G. and D.R. performed data analysis; everyone contributed toward writing the manuscript; D.G.C. and K.W.M performed the production of N protein at CCRC. P.A. and C.H. monitored the project.
Conflict of interest statement
None declared.
Data availability
The raw data files and search results can be accessed from glycopost repository: https://glycopost.glycosmos.org/preview/519892202600f0148a95a0 PIN CODE 7735.
Abbreviations
- ACN
acetonitrile
- CID
collision-induced dissociation
- COVID-19
coronavirus disease 2019
- CTD
C-terminal dimerization domain
- DTT
dithiothreitol
- E
envelope
- ELISA
enzyme-linked immunosorbent assay
- ETD
electron-transfer dissociation
- FA
formic acid
- hACE2
human angiotensin-converting enzyme 2
- HCD
higher-energy collisional dissociation
- HEK
human embryonic kidney
- id
internal diameter
- Ig
immunoglobulins
- IT-MS
ion trap-mass spectrometry
- M
membrane
- MetAPs
methionine aminopeptidases
- MS
mass spectrometry
- N protein
nucleocapsid protein
- NAT
N-acetyltransferase
- NME
N-terminal methionine excision
- NTD
N-terminal RNA-binding N-terminal domain
- pd
product-triggered
- PNGase F
peptide-N-glycosidase F
- S
spike
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
References
- Centers for Disease Control and Prevention . 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/cases-in-us.html.
- Atyeo C, Fischinger S, Zohar T, Slein MD, Burke J, Loos C, McCulloch DJ, Newman KL, Wolf C, Yu J, et al. 2020. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 53:524–532.e524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blicher T, Kastrup JS, Buus S, Gajhede M. 2005. High-resolution structure of HLA-A*1101 in complex with SARS nucleocapsid peptide. Acta Crystallogr D Biol Crystallogr. 61:1031–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonissone S, Gupta N, Romine M, Bradshaw RA, Pevzner PA. 2013. N-terminal protein processing: a comparative proteogenomic analysis. Mol Cell Proteomics. 12:14–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouhaddou M, Memon D, Meyer B, White KM, Rezelj VV, Correa Marrero M, Polacco BJ, Melnyk JE, Ulferts S, Kaake RM, et al. 2020. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 182:685–712.e619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo E, Escors D, Lopez JA, Gonzalez JM, Alvarez A, Arza E, Enjuanes L. 2005. Phosphorylation and subcellular localization of transmissible gastroenteritis virus nucleocapsid protein in infected cells. J Gen Virol. 86:2255–2267. [DOI] [PubMed] [Google Scholar]
- Chang CK, Hou M-H, Chang C-F, Hsiao C-D, Huang T-h. 2014. The SARS coronavirus nucleocapsid protein—forms and functions. Antiviral Res. 103:39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CK, Sue SC, Yu TH, Hsieh CM, Tsai CK, Chiang YC, Lee SJ, Hsiao HH, Wu WJ, Chang WL, et al. 2006. Modular organization of SARS coronavirus nucleocapsid protein. J Biomed Sci. 13:59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C-Y, Chang C-K, Chang Y-W, Sue S-C, Bai H-I, Riang L, Hsiao C-D, Huang T-H. 2007. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J Mol Biol. 368:1075–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Gill A, Dove BK, Emmett SR, Kemp CF, Ritchie MA, Dee M, Hiscox JA. 2005. Mass spectroscopic characterization of the coronavirus infectious bronchitis virus nucleoprotein and elucidation of the role of phosphorylation in RNA binding by using surface plasmon resonance. J Virol. 79:1164–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Guo Y, Pan Y, Zhao ZJ. 2020. Structure analysis of the receptor binding of 2019-nCoV. Biochem Biophys Res Commun. 525:135–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Dai T, Wei Y, Zhang L, Zheng M, Zhou F. 2020. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct Target Ther. 5:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta NK, Mazumdar K, Gordy JT. 2020. The nucleocapsid protein of SARS-CoV-2: A target for vaccine development. J Virol. 94:e00647–e00620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti AP, Kula T, Wang Y, Nguyen DMV, Weinheimer A, Dunlap GS, Xu Q, Nabilsi N, Perullo CR, Cristofaro AW, et al. 2020. Unbiased screens show CD8 (+) T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 53:1095–1107.e1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung TS, Liu DX. 2018. Post-translational modifications of coronavirus proteins: Roles and function. Future Virol. 13:405–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C, Boularot A, Meinnel T. 2004. Protein N-terminal methionine excision. Cell Mol Life Sci. 61:1455–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouveia D, Miotello G, Gallais F, Gaillard J-C, Debroas S, Bellanger L, Lavigne J-P, Sotto A, Grenga L, Pible O, et al. 2020. Proteotyping SARS-CoV-2 virus from nasopharyngeal swabs: A proof-of-concept focused on a 3 min mass spectrometry window. J Proteome Res. 19:4407–4416. [DOI] [PubMed] [Google Scholar]
- Grifoni A, Sidney J, Zhang Y, Scheuermann RH, Peters B, Sette A. 2020. A sequence homology and bioinformatic approach can predict candidate targets for immune responses to SARS-CoV-2. Cell Host Microbe. 27:671–680.e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He R, Leeson A, Ballantine M, Andonov A, Baker L, Dobie F, Li Y, Bastien N, Feldmann H, Strocher U, et al. 2004. Characterization of protein-protein interactions between the nucleocapsid protein and membrane protein of the SARS coronavirus. Virus Res. 105:121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihling C, Tänzler D, Hagemann S, Kehlen A, Hüttelmaier S, Arlt C, Sinz A. 2020. Mass spectrometric identification of SARS-CoV-2 proteins from gargle solution samples of COVID-19 patients. J Proteome Res. 19:4389–4392. [DOI] [PubMed] [Google Scholar]
- Kang S, Yang M, Hong Z, Zhang L, Huang Z, Chen X, He S, Zhou Z, Zhou Z, Chen Q, et al. 2020. Crystal structure of SARS-CoV-2 nucleocapsid protein RNA binding domain reveals potential unique drug targeting sites. Acta Pharm Sin B. 10:1228–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T, Weeraratne D, Snowdon JL, Parida L. 2020. Emergence of drift variants that may affect COVID-19 vaccine development and antibody treatment. Pathogens. 9:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Qureshi R, Sagurthi SR, Qureshi IA. 2020. Designing of nucleocapsid protein based novel multi-epitope vaccine against SARS-COV-2 using immunoinformatics approach. Int J Pept Res Ther. 27:941–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwarteng A, Asiedu E, Sakyi SA, Asiedu SO. 2020. Targeting the SARS-CoV2 nucleocapsid protein for potential therapeutics using immuno-informatics and structure-based drug discovery techniques. Biomed Pharmacother. 132:110914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laude H, Masters PS. 1995. The coronavirus nucleocapsid protein. In: Siddell SG, editor. The Coronaviridae. Springer US: Boston (MA): Springer US. p. 141–163. [Google Scholar]
- Li JY, Liao CH, Wang Q, Tan YJ, Luo R, Qiu Y, Ge XY. 2020. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 286:198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Shao J, Sun M, Liu J, Xu G, Zhang X, Xu N, Wang R, Liu S. 2007. Identification of phosphorylation sites in the nucleocapsid protein (N protein) of SARS-coronavirus. Int J Mass Spectrom. 268:296–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters PS. 2006. The Molecular Biology of Coronaviruses. Advances in Virus Research. 66:193–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan P, Singhal A, Mangal V. 2020. Novel coronavirus vaccine: An international holy grail. J Mar Med Soc. 22:20–26. [Google Scholar]
- Mu J, Xu J, Zhang L, Shu T, Wu D, Huang M, Ren Y, Li X, Geng Q, Xu Y, et al. 2020. SARS-CoV-2-encoded nucleocapsid protein acts as a viral suppressor of RNA interference in cells. Sci China Life Sci. 63:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan K, Kim KH, Makino S. 2003. Characterization of N protein self-association in coronavirus ribonucleoprotein complexes. Virus Res. 98:131–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaev EN, Indeykina MI, Brzhozovskiy AG, Bugrova AE, Kononikhin AS, Starodubtseva NL, Petrotchenko EV, Kovalev GI, Borchers CH, Sukhikh GT. 2020. Mass-spectrometric detection of SARS-CoV-2 virus in scrapings of the epithelium of the nasopharynx of infected patients via nucleocapsid N protein. J Proteome Res. 19:4393–4397. [DOI] [PubMed] [Google Scholar]
- Parker MM, Masters PS. 1990. Sequence comparison of the N genes of five strains of the coronavirus mouse hepatitis virus suggests a three domain structure for the nucleocapsid protein. Virology. 179:463–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T-Y, Lee K-R, Tarn W-Y. 2008. Phosphorylation of the arginine/serine dipeptide-rich motif of the severe acute respiratory syndrome coronavirus nucleocapsid protein modulates its multimerization, translation inhibitory activity and cellular localization. FEBS J. 275:4152–4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Pérez Marc G, Moreira ED, Zerbini C, et al. 2020. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med. 383:2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MS, Hoque MN, Islam MR, Islam I, Mishu ID, Rahaman MM, Sultana M, Hossain MA. 2021. Mutational insights into the envelope protein of SARS-CoV-2. Gene Rep. 22:100997–100997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MS, Islam MR, Alam ASMRU, Islam I, Hoque MN, Akter S, Rahaman MM, Sultana M, Hossain MA. 2020. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J Med Virol. 93:2177–2195. [DOI] [PubMed] [Google Scholar]
- Rudberg A-S, Havervall S, Månberg A, Jernbom Falk A, Aguilera K, Ng H, Gabrielsson L, Salomonsson A-C, Hanke L, Murrell B, et al. 2020. SARS-CoV-2 exposure, symptoms and seroprevalence in healthcare workers in Sweden. Nat Commun. 11:5064–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rump A, Risti R, Kristal M-L, Reut J, Syritski V, Lookene A, Ruutel Boudinot S. 2020. Dual ELISA using SARS-CoV-2 nucleocapsid protein produced in E. coli and CHO cells reveals epitope masking by N-glycosylation. Biochem Biophys Res Commun. 534:457–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan A, Archer-Hartmann S, Supekar NT, Gleinich AS, Heiss C, Azadi P. 2020. Comprehensive characterization of N- and O- glycosylation of SARS-CoV-2 human receptor angiotensin converting enzyme 2. Glycobiology. 31:410–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan A, Heiss C, Ishihara M, Azadi P. 2017. Glycomic and glycoproteomic analysis of glycoproteins-a tutorial. Anal Bioanal Chem. 409:4483–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shajahan A, Supekar NT, Gleinich AS, Azadi P. 2020. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 30:981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stohlman SA, Fleming JO, Patton CD, Lai MM. 1983. Synthesis and subcellular localization of the murine coronavirus nucleocapsid protein. Virology. 130:527–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M, Kumar R, Mishra RN, Reddy MK, Chow VT, Lal SK. 2005. The severe acute respiratory syndrome coronavirus nucleocapsid protein is phosphorylated and localizes in the cytoplasm by 14-3-3-mediated translocation. J Virol. 79:11476–11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M, Lal SK. 2008. The SARS-CoV nucleocapsid protein: A protein with multifarious activities. Infect Genet Evol. 8:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A, Cummings RD, Esko JD, Stanley P, Hart G W, Aebi M, Darvill AG, Kinoshita T, Packer NH, Prestegard JH, et al. 2015-2017. Essentials of Glycobiology [Internet]. 3rd ed. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press. [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen K-Y, et al. 2020. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 181:894–904.e899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Schmidt F, Weisblum Y, Muecksch F, Barnes CO, Finkin S, Schaefer-Babajew D, Cipolla M, Gaebler C, Lieberman JA, et al. 2021. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 592:616–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. 2020. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 369:330–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wootton SK, Rowland RR, Yoo D. 2002. Phosphorylation of the porcine reproductive and respiratory syndrome virus nucleocapsid protein. J Virol. 76:10569–10576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CH, Yeh SH, Tsay YG, Shieh YH, Kao CL, Chen YS, Wang SH, Kuo TJ, Chen DS, Chen PJ. 2009. Glycogen synthase kinase-3 regulates the phosphorylation of severe acute respiratory syndrome coronavirus nucleocapsid protein and viral replication. J Biol Chem. 284:5229–5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto FK. 2020. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 39:198–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Praissman JL, Grant OC, Cai Y, Xiao T, Rosenbalm KE, Aoki K, Kellman BP, Bridger R, Barouch DH, et al. 2020. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 28:586–601.e586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 382:727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmer C, Corum J, Wee S. 2021. https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data files and search results can be accessed from glycopost repository: https://glycopost.glycosmos.org/preview/519892202600f0148a95a0 PIN CODE 7735.


