Abstract
The ongoing coronavirus disease 19 (COVID-19) pandemic has infected millions of people, claimed hundreds of thousands of lives, and made a worldwide health emergency. Understanding the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) mechanism of infection is crucial in the development of potential therapeutics and vaccines. The infection process is triggered by direct binding of the SARS-CoV-2 receptor-binding domain (RBD) to the host-cell receptor angiotensin-converting enzyme 2 (ACE2). Many efforts have been made to design or repurpose therapeutics to deactivate the RBD or ACE2 and prevent the initial binding. In addition to direct inhibition strategies, small chemical compounds might be able to interfere and destabilize the metastable, prefusion complex of ACE2-RBD. This approach can be employed to prevent the further progress of virus infection at its early stages. In this study, molecular docking was employed to analyze the binding of two chemical compounds, SSAA09E2 and Nilotinib, with the druggable pocket of the ACE2-RBD complex. The structural changes as a result of the interference with the ACE2-RBD complex were analyzed by molecular dynamics simulations. Results show that both Nilotinib and SSAA09E2 can induce significant conformational changes in the ACE2-RBD complex, intervene with the hydrogen bonds, and influence the flexibility of proteins. Moreover, essential dynamics analysis suggests that the presence of small molecules can trigger large-scale conformational changes that may destabilize the ACE2-RBD complex.
Significance
This study aims to shed light on the interference mechanism of small molecules and their destabilization effects on the ACE2-RBD complex. Instead of direct inhibition mechanisms, we focused on the structural analysis of the metastable ACE2-RBD complex targeted by small molecules such as SSAA09E2 and Nilotinib. We showed that in addition to direct inhibition, small molecules can potentially destabilize the early-stage, prefusion, ACE2-RBD complex by inducing conformational changes, changing structural flexibility, and triggering different large-scale motions.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the newest member of coronaviruses, a family of single-stranded, RNA-based viruses able to infect mammals such as bats and humans. Although the SARS-CoV-2 preserves almost 70% of the sequence of the previous SARS-CoV virus, the differences are enough to affect the pathogenicity. Several studies show that the SARS-CoV-2 has a higher binding affinity with its host-cell receptor, angiotensin-converting enzyme 2 (ACE2) (1, 2, 3, 4). The higher risk of virus initial recognition can justify the higher infectivity of the novel coronavirus. However, the overall path of infection is similar to the SARS-CoV virus. The infection process is initialized by virus recognition through binding of the receptor-binding domain (RBD) of spike glycoproteins with ACE2 receptors. The recognition step is followed by the cleavage and activation processes induced by protease enzymes such as TMPRSS2, which paves the way for virus fusion and entry into the cell membrane. Since the initial days of the emergence of the novel coronavirus, huge scientific efforts have been made to target various stages of the infection process from inhibiting the initial host-cell binding to intervene in the virus reproduction pathway. Here, we focus on the early stages of infection and virus recognition process by host-cell receptors.
The crystal structure of the ACE2-RBD complex with the resolution of 2.45 Å (Protein Data Bank (PDB): 6M0J) was determined by Lan et al. (5). Moreover, Yan et al. (6) presented the cryoelectron microscopy structures of full-length ACE2 receptors with and without the RBD (PDB: 6M17). Various strategies have been evaluated to block ACE2-RBD interactions. Monolocal antibodies show promising results in deactivating SARS-CoV-2 spike proteins (7, 8, 9). Moreover, ACE2-based peptides can be designed to compete with real cellular receptors and reduce the risk of infection (10, 11, 12). In the absence of approved drugs and vaccines, screening techniques have been widely employed to repurpose approved drugs and find chemical compounds able to inhibit various proteins that are essential for the infection process. Adedeji et al. (13) carried out the screening of a large library of pharmacologically active small molecules and suggested three compounds with different mechanisms of action to inhibit the infection process of SARS-CoV. Interestingly, they reported that N-[[4-(4-methylpiperazin-1-yl)phenyl]methyl]-1,2-oxazole-5-carboxamide or SSAA09E2 has a novel inhibition mechanism that allows it to interfere with ACE2-RBD interactions and block the early stages of infections. However, details of this novel mechanism and its effectiveness in the case of the SARS-CoV-2 virus are yet to be understood completely. Wei et al. (14) employed virtual screening to evaluate the binding energy of ~15,000 drugs with the RBD. Based on their analysis, among all the drugs listed in the DrugBank database (15), Digitoxin and Nilotinib have the best docking scores with the RBD. In addition to virtual screening techniques, all-atomistic (16,17) and coarse-grained (18, 19, 20) molecular dynamics (MD) methods are effective computational tools for multiscale modeling of virus infection process and computer-aided drug design (21). Deganutti et al. (22) employed structure-based screening and supervised MD to find potential drugs that can bind to the druggable pockets of the RBD and inhibit the binding process. They reported Cefsulodin and Nilotinib as the potential disruptors of the ACE2-RBD interface. Nilotinib is a member of Abelson (Abl) tyrosine-protein kinase (Abl-TK) inhibitor drugs that can potentially affect the infection process through inhibiting the virus fusion before hemifusion (23). In addition, these drugs can reduce the risk of infection through Abl-mediated cytoskeletal rearrangement and interfering with actin dynamics (24). The in vitro observations of positive impacts of Abl-TK inhibitors have recently been strengthened by in vivo studies and medical surveys. For instance, Foà et al. (25) and Breccia et al. (26) reported a lower percentage of COVID-19 infection among the Philadelphia-positive acute lymphoblastic leukemia and chronic myeloid leukemia patients under treatment by Abl-TK inhibitors.
In this study, we focused on the interference mechanism of small molecules that can potentially destabilize the ACE2-RBD complex. The disruption of protein-protein interactions can be a strategy for potential drugs to interfere with the early stages of infection. Based on in vitro and virtual screening studies, we chose SSAA09E2 and Nilotinib to study their potential intervention effects. Instead of direct inhibition of ACE2 or RBD, this study aims to understand the structural changes induced by binding of SSAA09E2 and Nilotinib into the druggable pocket at the central part of the ACE2-RBD interface that may shed light on the mechanisms of action of these drugs. Molecular docking was employed to find the most favorable binding poses. MD was then utilized to analyze the structural changes of the ACE2-RBD complex. We showed that both of these chemical compounds are able to make stable bonds with the ACE2-RBD complex that may potentially reduce the infection risk by interfering with the ACE2-RBD interactions and triggering large-scale conformational changes.
Materials and methods
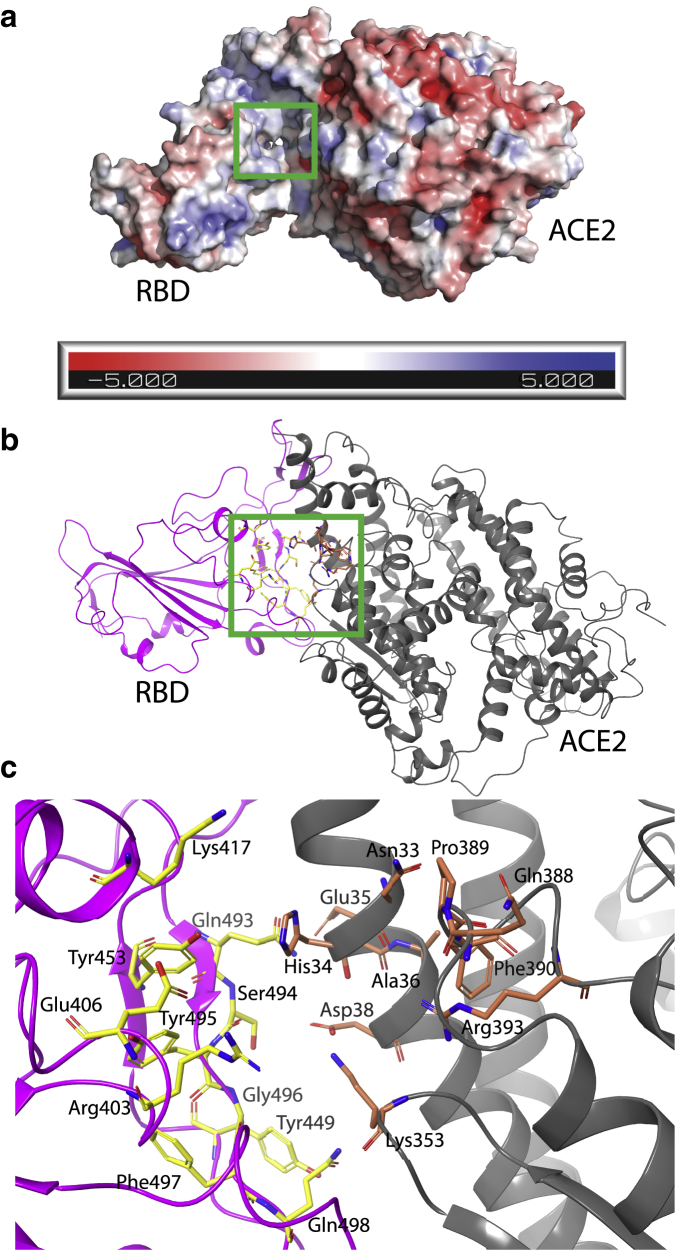
The crystal structure of the SARS-CoV-2 RBD complexed with the ACE2 enzyme was obtained from the protein data bank, PDB: 6M0J (5). The protein structure includes chains A and E, which represent the ACE2 and RBD structures, respectively. The protonation state of proteins was investigated by the H++ server (27). H++ server automates the process of pKa prediction based on the standard continuum solvent methodologies and the environment pH provided by the user. In this work, the protonation state is assigned at the neutral pH of 7.0. Moreover, the salinity, internal dielectric, and external dielectric were set to be 0.15, 10, and 80 M, respectively (28). The chemical structures of Nilotinib and SSAA09E2 were retrieved from the PubChem database (29). Avogadro software was used to prepare the ligand structures and adding hydrogen atoms (30). DoGSiteScorer (31) was used to analyze the binding pockets at the interface of the ACE2-RBD complex. DoGSiteScorer is an online tool that analyzes and ranks potential binding pockets based on their volume, surface area and assigns a drug score to them. Drug score is a number between 0 and 1, the higher the score, the better estimated druggability. The most druggable binding pocket at the ACE2-RBD interface with the volume of 532.4 , surface area of 572.2 , and drug score of 0.69 was chosen for further analysis. This binding pocket is made by residues Asn33, His34, Glu35, Ala36, Asp38, Lys353, Gln388, Pro389, Phe390, Arg393, of the ACE2 protein and residues Arg403, Glu406, Lys417, Tyr449, Tyr453, Gln493, Ser494, Tyr495, Gly496, Phe497, Gln498, Asn501, and Tyr505 of the RBD. Fig. 1 a shows the electrostatic surface (with the unit of ) of the ACE2-RBD complex. The residues that form a druggable binding pocket at the interface of ACE2 enzyme and SARS-CoV-2 RBD are displayed in Fig. 1, b and c.
Figure 1.
(a) The electrostatic surface (with the unit of ) of the ACE2-RBD complex, the binding site is indicated by a green square. (b) Cartoon representation of the ACE2-RBD complex and binding pocket location. (c) Key residues that form the binding pocket. The SARS-CoV-2 RBD and ACE2 enzyme are shown by magenta and gray cartoons, respectively. Key residues are shown by licorice-stick representations. The carbon atoms of the RBD and ACE2 residues are colored yellow and coral, respectively. Red and dark blue sticks show oxygen and nitrogen atoms, respectively. This color scheme is used in figures throughout this article. To see this figure in color, go online.
Ligplot+ package (32) was used to prepare the two-dimensional diagrams of ligand-protein interactions. Protein-Ligand Interaction Profiler (PLIP) web-tool (33) was employed for further analysis of ligand-protein interactions. Molecular docking simulations were performed by Autodocktools 4 (34). The ligand preparation for docking simulations was performed by Autodocktools (34). This process includes adding Gasteiger-Marsili charges to the ligand atoms (35), define torsional degrees of freedom, merge nonpolar hydrogen atoms, and prepare PDBQT files (PDB format with partial charges (Q), and atom types (T)). More details on this standard process can be found in Forli et al. (36) and Morris et al. (37) protocols for molecular docking. A grid with the size of 60 Å 60 Å 60 Å centered on the druggable pocket at the interface of the ACE2-RBD complex was used for docking simulations. The docking process was carried out with the Lamarckian Genetic algorithm (38,39). The best docking pose was used for further MD simulations. The system topology was made by CHARMM-GUI solution builder module by using the CHARMM36(m) all-atom force field (40,41) and TIP3P water model (42). The ligand reader and modeler module of CHARMM-GUI (43) was employed for ligand parameterization and topology preparation. The parameterization was performed based on the CHARMM General Force field (44) approach in which the chemical space that is not covered by the CHARMM force field is parameterized with a rule-based interpretation of existing force field parameters. More details on the automation process of ligand parameterization for the CHARMM force field can be found in the works of Vanommeslaeghe et al. (44,45) and Kim et al. (43). An octahedral computational box with 12 Å distance between the edges was employed for the protein solvation. The system is neutralized by adding potassium (K) and chloride (Cl) ions with the final concentration of 0.15 M. Systems were initially minimized by the steepest descent algorithm with 5000 steps. The minimization was followed by 125 ps of equilibration simulation in the canonical ensemble with the constant number of atoms (N), constant volume (V), and constant temperature (T) (NVT ensemble). Linear Constraint Solver algorithm was employed to constrain the hydrogen bonds (46). The Verlet cutoff scheme with the cutoff radius of 1.2 nm, and the Particle Mesh Ewald method was used to compute long-range electrostatics. All the systems were simulated at the temperature of 310 K that was maintained by the Nosé–Hoover temperature coupling method with the time constant for coupling (τ-t) of 1 ps (47). The 200 ns of production simulation were performed with the constant number of atoms (N), pressure (P), and temperature (T) (NPT ensemble) at the pressure of 1 atm, which was controlled by the Parinello-Rahman method with the time constant for pressure coupling (τ-p) of 5 ps. The production simulations were performed in triplicates with the total simulation time of ns or 1.8 and average results were calculated. GROMACS 5.1 (48) was employed for all the MD simulations and trajectories were visualized by PyMol (49). g_RMS, g_RMSF, g_gyrate, g_hbond, and g_distance packages of GROMACS software were employed to analyze the root mean-square deviations (RMSD), root mean-square fluctuations (RMSF), radius of gyration , number of hydrogen bonds, and distances between residues, respectively. To understand the effects of drug molecules on the essential dynamics of the ACE2-RBD complex, Principal Component Analysis (PCA) was performed on the combined trajectory made by concatenating three trajectory replicates for each case (3 × 200 ns or 600 ns trajectory for each case). The covariance matrix of backbone atoms was diagonalized by the g_covar package of the GROMACS software. The g_anaeig package was employed to project the principal components into essential phase space. Porcupine plots were drawn based on the extreme conformations sampled during the simulation time along each of the first three eigenvectors by using the principal components calculated for the concatenated trajectory of each case. PyMol (49) package was used to prepare the porcupine plots and show direction and strength of movements.
Results and discussion
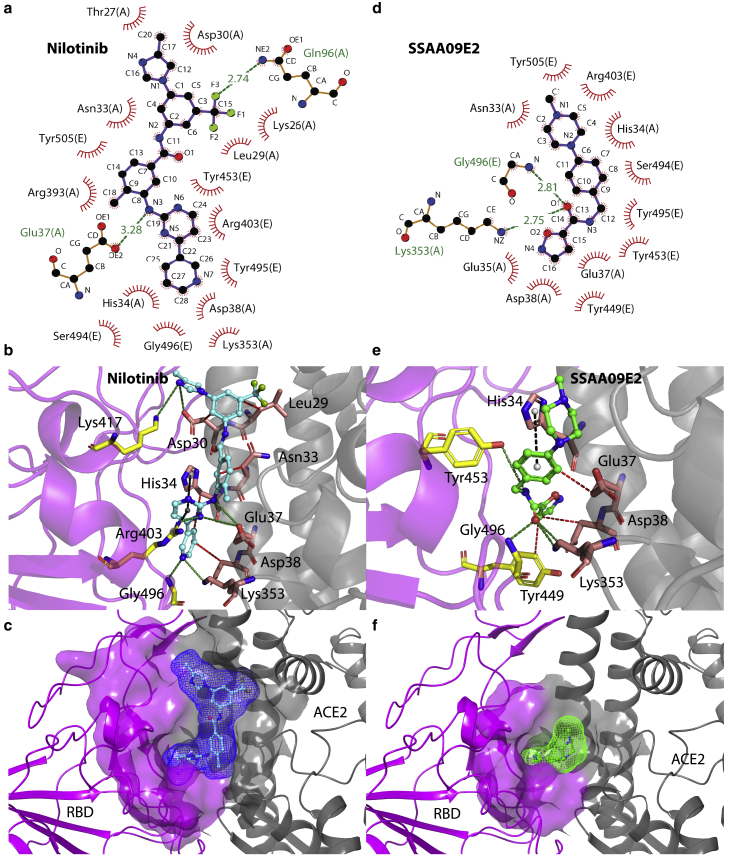
Our docking simulations show that both Nilotinib and SSAA09E2 have high binding affinities with the druggable pocket of the ACE2-RBD complex. Fig. 2 a shows the two-dimensional diagram of potential interactions between the Nilotinib and ACE2-RBD complex. It can be seen that at this binding pocket, Nilotinib has nonbonded interactions with residues Lys26, Thr27, Leu29, Asn33, His34, Glu37, Asp38, Lys353, and Arg393 of the ACE2 protein, which is shown by chain A of the PDB: 6M0J in the diagram. Two residues of the ACE2 enzyme, Glu37 and Gln96, can potentially form hydrogen bonds with Nilotinib. The RBD, chain E of PDB: 6M0J structure, has nonbonded interactions with the Nilotinib through residues Arg 403, Ser494, Tyr495, Gly496, and Tyr505. Moreover, as shown in Fig. 2 b, the PLIP web-tool (33) characterizes two potential cation-π interactions for Nilotinib with His34 and Arg403 residues and more potential hydrogen bonds with Asp30 and Lys 353 of the ACE2 and Lys417 and Gly496 of the RBD. The best docking pose for the Nilotinib is shown in Fig. 2 c with the score of −8.34 kcal/mol. Fig. 2 d shows the interaction diagram of SSAA09E2. Residues Asn33, His34, Glu35, Glu37, Asp38 of the ACE2 enzyme have nonbonded interactions with SSAA09E2. Moreover, Lys353 is able to make a hydrogen bond with the ligand. SSAA09E2 can also form a hydrogen bond with the RBD by interacting with Gly496. Arg403, Tyr449, Tyr453, Ser494, Tyr495, Gly496, and Tyr505 from the RBD have nonbonded interactions with SSAA09E2. As illustrated by Fig. 2 e, the PLIP web-tool predicts one more hydrogen bond between the SSAA09E2 and Tyr453. Moreover, a potential π-stacking interaction is predicted between His34 and SSAA09E2. The best docking pose of SSAA09E2 is displayed in Fig. 2 f with the score of −8.04 kcal/mol, which is close to the case of Nilotinib with the score of −8.34 kcal/mol. Our docking results show that both Nilotinib and SSAA09E2 can potentially interfere with some of the important ACE2-RBD interactions such as His34-Tyr453, Glu35-Gln493, and Asp38-Tyr449 (5,6,50,51).
Figure 2.
(a) Two-dimensional interactions diagram of Nilotinib and ACE2-RBD complex, chains A and E of PDB: 6M0J represent the ACE2 and RBD, respectively. (b) Interactions between the Nilotinib, in the best docking pose, and ACE2-RBD complex characterized by the PLIP web-tool, the dashed black lines show the potential cation-π interactions. (c) The best docking pose of Nilotinib inside the binding pocket. (d) Two-dimensional interactions diagram of SSAA09E2 and the ACE2-RBD complex. (e) Interactions between the SSAA09E2, in the best docking pose, and ACE2-RBD complex characterized by the PLIP web-tool, the dashed black line shows the potential π-stacking interaction, (f) The best docking pose of SSAA09E2 inside the binding pocket. Nonbonded interactions are shown by red-spoked arcs (parts (a) and (d)) and red-dashed lines (parts (b) and (e)). Hydrogen bonds are represented by the green-dashed lines. Nilotinib and SSAA09E2 are shown by the ball-stick representation with light blue and green carbon atoms, respectively. This color scheme for drug molecules is used in figures throughout this article. The mesh surfaces of ligand molecules in parts (c) and (f) show the molecular surfaces of ligands made by rolling a probe ball with the radius of 1.4 Å on the Van der Waals surface of atoms. To see this figure in color, go online.
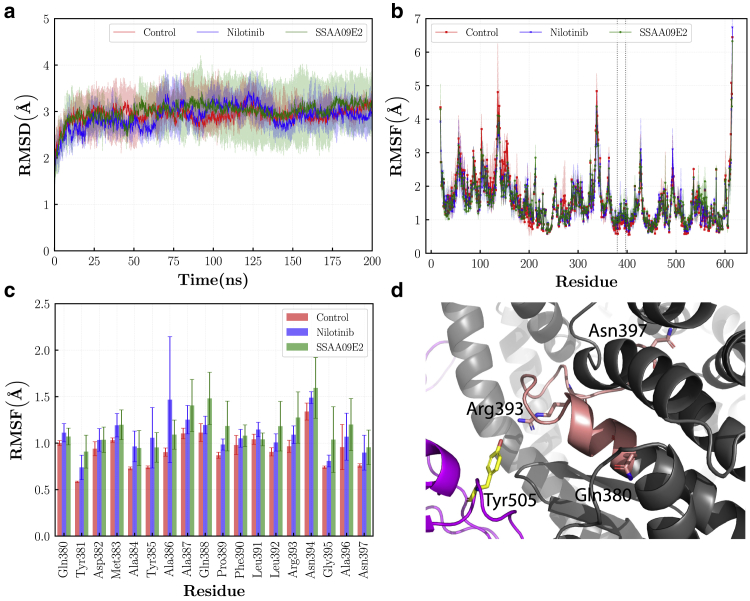
To analyze the stability and conformational changes of protein structures, 200 ns of MD simulation were performed in triplicates for the control group without drug molecules and the ACE2-RBD complex bound with Nilotinib and SSAA09E2. The RMSD is a measure of protein drift from a reference structure. Fig. 3 a shows the RMSD of the ACE2 enzyme in various cases. In all simulations, ACE2 reaches an equilibrium after ~50 ns of simulation. The average RMSD is 3.020.37, 2.890.49, and 2.990.57 Å for the control, Nilotinib, and SSAA09E2 cases, respectively. Therefore, adding drug molecules does not change the ACE2 protein drift from the reference structure significantly. The RMSF shows the time-averaged fluctuations of individual residues. As shown in Fig. 3 b, both drugs can induce changes in the RMSF of some ACE2 regions. For instance, the RMSF of residues Gln380 to Asn397 is increased for both Nilotinib and SSAA09E2 cases (Fig. 3 c). Arg393 is among the key residues that can form a hydrogen bond with the residue Tyr505 of the RBD (52). The higher estimated flexibility of those interface residues can lead to higher entropy penalty that negatively influences the binding energy, especially at higher temperatures (53). Moreover, drug molecules can increase or decrease the average fluctuations in some loop regions that are far from the ACE2-RBD interface such as residues Leu142-Gly147 and Asn194-Tyr202, suggesting that both drugs may have secondary effects on the enzyme activity of ACE2 proteins, which needs to be further studied in experimental works.
Figure 3.
(a) RMSD of the ACE2 protein. (b) RMSF of the ACE2 protein, the region between residues Gln380 and Asn397 is separated by two vertical dashed lines. (c) Means and standard deviations of the RMSF for residues Gln380-Asn397. (d) Snapshot of the Gln380-Asn397 region of the ACE2 protein shown by the coral cartoon. In parts (a) and (b), means and standard deviations are shown by solid lines and filled regions, respectively. To see this figure in color, go online.
The average RMSF over all residues is 1.570.79, 1.520.71, and 1.540.69 Å for the control, Nilotinib, and SSAA09E2 cases, respectively. Thus, despite the above-mentioned local changes in the fluctuations, the overall flexibility of the ACE2 enzyme is not significantly changed as a result of adding drug molecules.
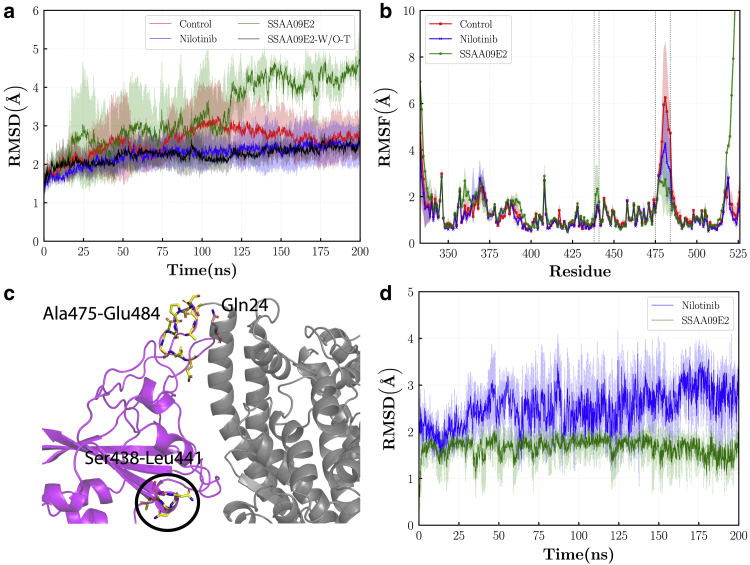
Fig. 4a shows the RMSD of the RBD. For the control and Nilotinib cases, the RMSD fluctuates around ~2.5 Å after ~150 ns of simulation. The final RMSD for the RBD with the Nilotinib is close to the control case. The RBD has the most unstable behavior in the SSAA09E2 case in which the RMSD fluctuates around ~4 Å. The average RMSD values over 200 ns of the simulation in triplicates are 2.580.69, 2.250.45, and 3.311.1 Å for the control, Nilotinib, and SSAA09E2 cases, respectively. However, by analyzing the RMSF plot of the RBD, it can be seen that the higher instabilities in the SSAA09E2 case are mainly stemmed from the large fluctuations of the C-terminal residues. Fig. 4 b shows the RMSF plot of the RBD protein. The most notable increase in the flexibility is at the terminal residues His519-Gly526 in which the RMSF is much larger in the SSAA09E2 case. It should be noted that because of computational limitations, the whole spike protein (PDB: 7AD1 (54)) was not simulated in this study and we just focused on the residues Thr333-Gly526, which represent the RBD. Thus, the spike glycoprotein residues before and after residues Thr333-Gly526 from both termini are not modeled in this work. However, large fluctuations and flexibility of the C-terminal residues, His519-Gly526, in the case of SSAA09E2 may be effective in triggering conformational changes in the rest of the spike protein structure. To analyze the effects of fluctuations of C-terminal residues on the overall RMSD of the SSAA09E2 case, we recalculated the RMSD by neglecting His519-Gly526 residues (Fig. 4 a). The RMSD of the SSAA09E2 case drops to 2.230.34 Å in the absence of C-terminal residues, which is close to the Nilotinib and control cases. Another significant change in the flexibility of the RBD takes place in the loop region Ala475-Glu484 (Fig. 4 c). Both the Nilotinib and SSAA09E2 drugs decrease the flexibility of residues in this region which can be effective on the RBD interactions with the N-terminal domain of the ACE2 enzyme. For instance, the hydrogen bond formed between Asn487 of the RBD and Gln24 of the ACE2 is important in the SARS-CoV-2-binding process (5,55). Moreover, the presence of the SSAA09E2 molecule can affect the fluctuations of three other regions of the RBD protein. For instance, the SSAA09E2 increases the flexibility of residues Ser438-Leu441 whereas the Nilotinib molecule does not affect the fluctuations of this region. Less significant increases in the RMSF can be observed for the SSAA09E2 case at residues Ser359-Tyr369 and Leu390-Phe400, which are close to the C-terminal and may be affected by its large fluctuations. The average of RMSF over all residues is 1.711.84 Å for the SSAA09E2 case, which is significantly larger than 1.391.04 and 1.260.79 Å for the control and Nilotinib cases, respectively, mainly because of huge fluctuations at the terminal residues. Neglecting the C-terminal residues leads to significant drops in both mean and standard deviation of the averaged RMSF to 1.410.87 Å.
Figure 4.
(a) RMSD of the RBD, SSAA09E2-W/O-T represents the SSAA09E2 case in which the RBD terminal residues of His519-Gly526 are not considered in the RMSD calculation. (b) RMSF of the RBD, residues Ala475-Glu484 and Ser438-Leu441 are shown by regions between two vertical dashed lines. (c) The snapshot of ACE2-RBD complex with Ala475-Glu484, Ser438-Leu441, and Gln24 that are showed by licorice-stick representation. (d) RMSD of ligand molecules. In the parts (a), (b), and (d), means and standard deviations are shown by solid lines and filled regions, respectively. To see this figure in color, go online.
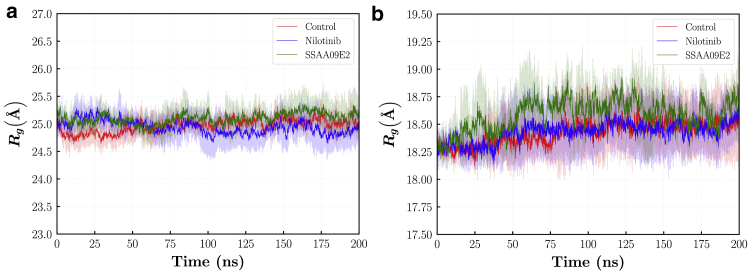
Fig. 4d illustrates the RMSD of the ligand molecules at the binding pocket. Both SSAA09E2 and Nilotinib are stable at the binding pocket during the 200 ns simulation with the average RMSD of 1.660.32 and 2.510.61 Å, respectively. It should be noted that the Nilotinib molecule is larger in comparison with the SSAA09E2. Although at the best docking pose almost half of the Nilotinib molecule is in the druggable pocket, another half of the molecule is freer to fluctuate out of the binding pocket (see Video S1). This leads to higher total deviations from the reference structure. Moreover, higher fluctuations of the Nilotinib molecule may help this drug to interfere with the hydrogen bonds between ACE2 and RBD. Despite higher RMSD, visual analysis of trajectory shows that the Nilotinib molecule is fixed at the pocket during the 200 ns of simulation and does not escape from its initial location. Fig. 5 shows the radius of gyration of the ACE2 and RBD proteins in all simulations. is a measure for the compactness of proteins and shows the stability of protein folding. It can be seen that for the ACE2 protein, in all cases the proteins are stably folded and the differences in the compactness are negligible. For the RBD, the is slightly higher for the SSAA09E2 case. The average is 18.420.23, 18.430.22, 18.580.24 Å for the control, Nilotinib, and SSAA09E2 cases, respectively. However, this small increase of less than ~ may be stemmed from the higher fluctuations of terminal residues and cannot be considered as a meaningful change in the compactness of the RBD structure. Thus, drugs do not influence the consistent shape and size of the ACE2-RBD complex during the 200 ns of simulation.
Figure 5.
of the (a) ACE2, and (b) RBD proteins. Means and standard deviations are shown by solid lines and filled regions, respectively. To see this figure in color, go online.
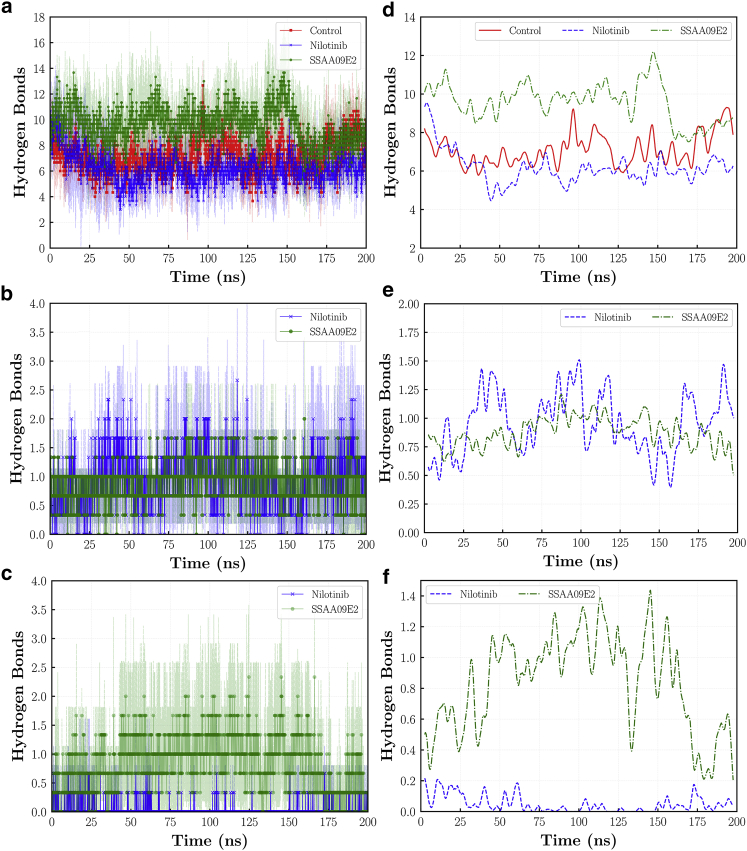
Hydrogen bonds have a significant role in the strength of protein-protein interactions (56). A recent study shows that the average number of hydrogen bonds formed between the RBD and cell receptors can be almost two times more for the novel SARS-CoV-2 in comparison with the SARS-CoV (57). This can be a reason for the greater binding energy of ACE2-RBD and more infectivity of the SARS-CoV-2 virus. We studied the hydrogen bonds between the ACE2 and RBD proteins in various cases. To make plots more clear, in addition to the average number of hydrogen bonds at each frame, running averages over every 50 frames are plotted. As illustrated in Fig. 6 a, the SSAA09E2 and Nilotinib molecules have different effects on the number of hydrogen bonds between the ACE2 and RBD proteins. In the case of SSAA09E2, more hydrogen bonds form between the two proteins. On the other hand, Nilotinib weakens the structural integrity by reducing the number of hydrogen bonds. The average numbers of hydrogen bonds during the 200 ns of simulation in triplicates are 7.131.96, 6.202.05, and 9.572.50 for the control, Nilotinib, and SSAA09E2 cases, respectively. Thus, on average, the Nilotionib case has ~1 fewer hydrogen bonds in comparison with the control system. On the other hand, the SSAA09E2 drug can result in a tighter binding of the ACE2 and RBD proteins, because on average, ~2 more hydrogen bonds are formed in this case. Moreover, we analyzed the number of hydrogen bonds between the ligand molecules and the ACE2 or RBD proteins. Fig. 6, b, c, e, and f illustrates the number of hydrogen bonds formed between the SSAA09E2 and Nilotinib molecules with the ACE2 and RBD proteins. Both ligands can form hydrogen bonds with the ACE2 protein. On average, Nilotinib has 0.950.84 hydrogen bonds during the 200 ns of simulation with the ACE2 protein, which is slightly greater than 0.880.54 bonds made between SSAA09E2 and ACE2. On the other hand, the average number of hydrogen bonds of SSAA09E2 with the RBD is 0.820.93, which is significantly larger than 0.050.23 for the Nilotinib case. Thus, we can expect the formation of one or two hydrogen bonds between the SSAA09E2 and RBD during the simulation time, whereas the formation of such a hydrogen bond between the Nilotinib and RBD is much less expected.
Figure 6.
Number of hydrogen bonds formed between (a) the ACE2 and RBD proteins, (b) drug molecules and ACE2, and (c) drug molecules and RBD. Rolling average of average hydrogen bonds are also plotted for (d) ACE2-RBD, (e) ligand-ACE2, and (f) ligand-RBD. In parts (a)–(c), means and standard deviations are shown by solid lines and filled regions, respectively. To see this figure in color, go online.
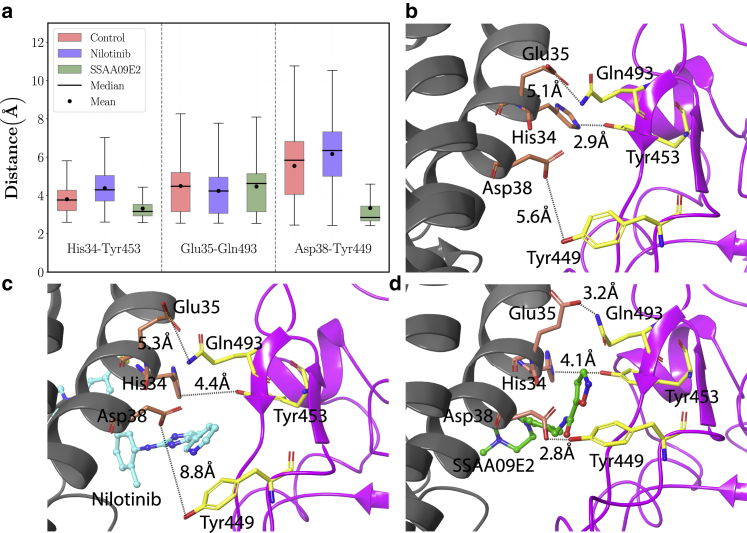
Moreover, we analyzed the distance between three pairs of key residues at the interface of the ACE2-RBD complex. Fig. 7 a shows the box plots of His34/NE2-Tyr453/OH, Glu35/OE2-Gln493/NE2, and Asp38/OD2-Tyr449/OH distances calculated by using all the frames of the concatenated simulation trajectories (made by pooling together three trajectory replicates for each case). Each box plot shows the minimum, first quartile, median, average, third quartile, and maximum of distance distributions. Fig. 7, b–d show the snapshots of the residues and their distances at the last frame of simulation (200 ns in the first trajectory replicate) for various cases. The box plots of Fig. 7 a illustrate that the average and median distances of Asp38-Tyr449 are significantly reduced in the case of SSAA09E2. The Asp38-Tyr449 distance ranges between ~2.4 and 4.6 Å in the SSAA09E2 case, in which 50% of data lie in the range of ~2.7–3.4 Å. On the other hand, for the control case, the distance ranges between ~2.4 and 10.8 Å, and half of the data lie between 4.0 and 6.8 Å. Moreover, the median and average of data are 2.8 and 3.3 Å for the SSAA09E2 case, respectively. In the control case, the median of data is 5.8 Å and the average is 5.5 Å. Therefore, the average distance in the control case is ~1.6 times greater than the SSAA09E2 case. Moreover, the Asp38-Tyr449 distance fluctuates in a significantly wider range in the control case. For instance, ~50 of data in the control case lies between ~5.8 and 10.8 Å, which is larger than the maximal distance of 4.6 Å in the SSAA09E2 case. Thus, SSAA09E2 can significantly reduce the average and lower the fluctuations of the distance between Asp38 and Tyr449. The same but less significant effect can be seen for the distance between His34 and Tyr453 in the SSAA09E2 case. Whereas in the control case the distance ranges between 2.6 and 5.8 Å, the upper bound is reduced to 4.4 Å for the SSAA09E2 case. Moreover, the average and median distances for His34-Tyr453 and Asp38-Tyr449 are increased in the Nilotinib case. However, the ligand effects are less significant on the distance between Glu35 and Gln493. The distances between key residues are consistent with the hydrogen bond analysis results that show a tighter ACE2-RBD binding in the case of SSAA09E2.
Figure 7.
(a) Box plots of His34-Tyr453, Glu35-Gln493, Asp38-Tyr449 distances, and snapshots of residues and their distances at the last frame of simulation (200 ns) for the (b) control, (c) Nilotinib, and (d) SSAA09E2 cases. Distances are shown by dashed black lines. To see this figure in color, go online.
Analysis of hydrogen bonds and key residues distances is essential in determining two different mechanisms of action for the Nilotinib and SSAA09E2 molecules. Although the Nilotinib molecule is weakening ACE2-RBD interactions by reducing the average number of hydrogen bonds, SSAA09E2 increases the strength of protein-protein binding at the ACE2-RBD interface. On the other hand, although both drugs can form at least one hydrogen bond with the ACE2 protein during the majority of simulation trajectory, the formation of hydrogen bonds is less probable between the RBD and Nilotinib. Thus, the Nilotinib molecule may have more freedom to fluctuate at the binding pocket, whereas the SSAA09E2 is bound with a greater number of hydrogen bonds especially with the RBD. Therefore, Nilotinib may work by a disruption mechanism in which the binding between the ACE2 and RBD proteins is targeted, whereas SSAA09E2 has another mechanism of action that involves more powerful binding at the interface. The tighter binding at the interface may trigger higher fluctuations at other regions such as C-terminal residues and induce structural changes in the rest of the spike protein.
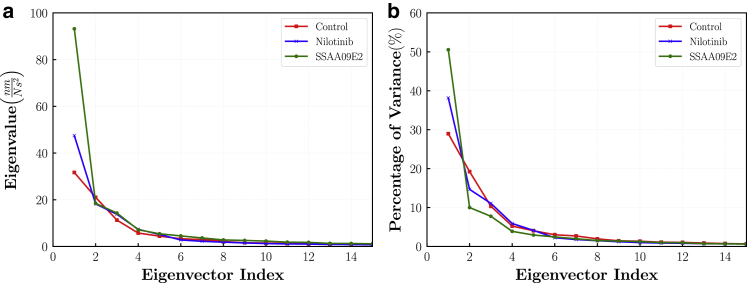
We employed the PCA or essential dynamics analysis to identify dominant motion modes of the ACE2 and RBD in various cases. For each case, the concatenated trajectory made by pooling together all three trajectory replicates was used for the PCA analysis. The covariance matrix of fluctuations was calculated for backbone atoms and diagonalized to find the eigenvalues and eigenvectors. The conformational area that is sampled by each system was then analyzed by projecting the trajectory into the essential phase space along the calculated eigenvectors. Usually, a few numbers of low-frequency eigenvectors are responsible for the majority of overall fluctuations. Fig. 8 shows the eigenvalues and their percentage of variance that is defined as the contribution of each individual eigenvalue to the sum of eigenvalues. The traces of covariance matrices are 109.37, 124.37, 184.39 for the control, Nilotinib, and SSAA09E2 cases suggesting that the tertiary structural conformations are more dynamic in the cases with the ligand molecules. As shown in Fig. 8 b, our results indicate that the first three eigenvectors account for 58.5, 63.9, 68.3% of overall fluctuations in the control, Nilotinib, and SSAA09E2 cases, respectively. Thus, the first three eigenvectors contribute more to the overall essential dynamics of proteins in the cases with ligand molecules. Fig. 8 b also illustrates that the first eigenvector has a greater percentage of variance for the cases with the Nilotinib and SSAA09E2 drug molecules, which shows the importance of the first eigenvector in the dynamics of systems with ligand molecules.
Figure 8.
(a) Eigenvectors and corresponding eigenvalues for various systems. (b) Percentage of variance of eigenvectors. To see this figure in color, go online.
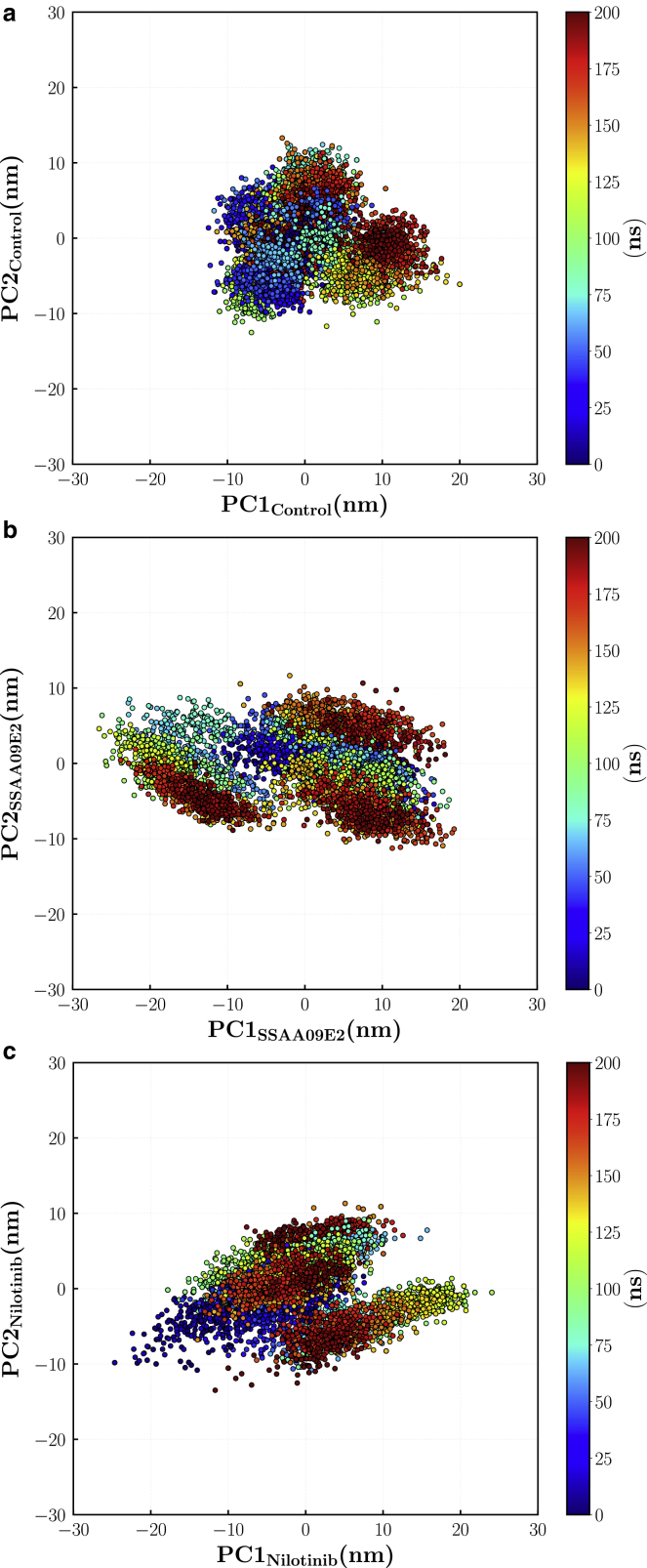
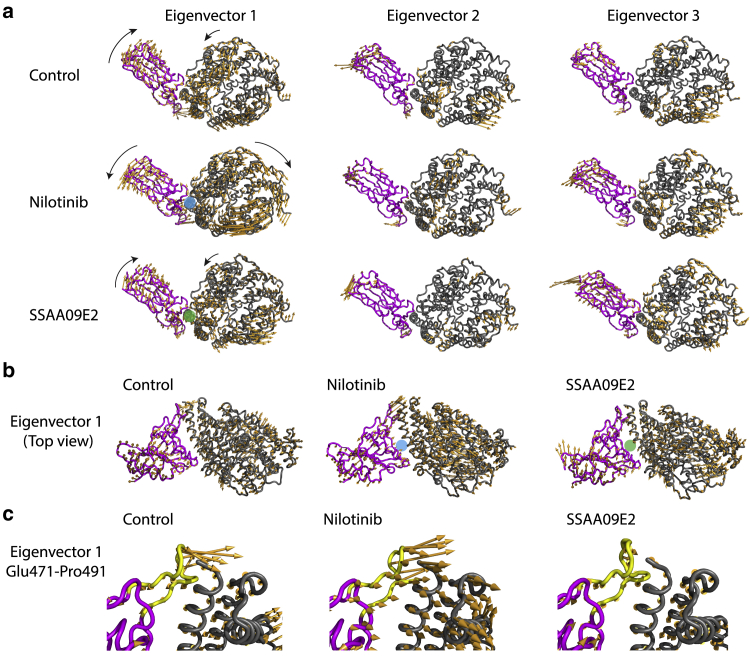
Fig. 9 shows the conformational space sampled by the ACE2-RBD complex in various cases. Each point shows a frame of 200 ns trajectory projected onto a two-dimensional space along the first two eigenvectors. Principal components are calculated for the concatenated trajectory made by pooling together all three trajectory replicates. Fig. 9 shows the effect of ligand molecules on intensifying dynamics of the ACE2-RBD complex. The largest covered area is for the SSAA09E2 case, which shows that a larger area of the conformational space is sampled especially along the first principal component. Therefore, in this case proteins have more dynamic tertiary structural conformations. Moreover, the area covered by the ACE2-RBD complex bound with Nilotinib illustrates the higher flexibility of this case in comparison with the control case. For a better analysis of the dominant dynamics, porcupine plots (58) are employed to show characteristics of dynamic fluctuations for the first three eigenvectors. In Fig. 10, vectors indicate the motion of each backbone atom along the eigenvector direction. The length of each arrow represents the magnitude of the corresponding motion. As expected, the largest motions happen at the loop regions. For the first eigenvector, the overall rotation direction (indicated by black arrows in Fig. 10 a) of the ACE2-RBD complex bound with the SSAA09E2 molecule is similar to the control case. However, unlike the control case, residues Glu471-Pro491 of the RBD (Fig. 10 c, some of the residues in this loop region are shown in Fig. 4 c) does not move toward the ACE2 protein in the SSAA09E2 case. As mentioned before, the Glu471-Pro491 loop region and its large-scale motions can affect the interactions of the RBD with the N-terminal residues of the ACE2 enzyme and formation of key hydrogen bonds such as Asn487 (RBD)-Gln24 (ACE2). The difference between the dominant motions is more significant in the Nilotinib case. In contrast to the control case, the ACE2 and RBD proteins bound with Nilotinib move away from each other by a counterclockwise rotation in the RBD and a clockwise rotation in the ACE2. Results of essential dynamics analysis clarify the difference in the mechanisms of action of SSAA09E2 and Nilotinib. Although SSAA09E2 changes the motion dynamics locally, especially at the terminal residues and loops close to the ACE2 interface, Nilotinib induces completely different large-scale motions in both proteins.
Figure 9.
Dynamic conformations projected onto the two first principal vectors for (a) control, (b) SSAA09E2, (c) and Nilotinib cases. To see this figure in color, go online.
Figure 10.
(a) Front-view snapshots of porcupine plots of essential dynamics for the three lowest-frequency principal components, binding pockets are annotated with blue (Nilotinib) and green (SSAA09E2) circles for the first eigenvectors. (b) Top-view snapshots of the porcupine plots for the first eigenvector of the contol, Nilotinib, and SSAA09E2 cases, (c) large-scale motions of residues Glu471-Pro491 of the RBD, shown by yellow cartoons (backbone atoms only), for control, Nilotinib, and SSAA09E2 cases. To see this figure in color, go online.
Conclusions
Molecular docking and MD methods were employed to study the intervention and destabilization mechanisms of two small therapeutics. Results suggest that the SSAA09E2 and Nilotinib molecules may have different mechanisms of action to intervene with the ACE2-RBD binding process or destabilize the protein complex after initial binding. Our ligand-protein interactions study at the best docking position shows that both drugs are able to form hydrogen bonds and other forms of attractive interactions with some key residues at the interface of ACE2 and RBD. Moreover, MD simulations were performed to study the structural changes induced by drug molecules. The stable binding of SSAA09E2 at the pocket between ACE2 and RBD leads to higher flexibility in the residues Ser438-Ser443 and C-terminal residues of the RBD and less fluctuations at the residues Ser477-Gly484 of the RBD. In the case of SSAA09E2, more hydrogen bonds are formed between the ACE2 and RBD structures and the ligand can form hydrogen bonds with both proteins during the simulation trajectory. In addition to hydrogen bonds, the distance between the Asp38 (ACE2) and Tyr449 (RBD) is significantly lower for the SSAA09E2 case. Thus, the SSAA09E2 molecule may result in a tighter binding of ACE2 and RBD proteins. Moreover, the tighter binding at the interface and significant increase in the fluctuations of terminal residues may trigger large-scale structural changes in the whole spike glycoprotein, which are beyond the scope of this study. The PCA analysis shows that in the first dominant motion of the SSAA09E2 case, both ACE2 and RBD proteins have similar large-scale rotation directions to the control case. However, some large-scale motions such as dynamics of residues Glu471-Pro491 are different in the case of SSAA09E2 that may affect the ACE2-RBD binding process. On the other hand, the Nilotinib is less stable at the binding pocket and half of its structure, which is posed parallel to the ACE2 interface with the RBD can fluctuate out of the druggable pocket of the ACE2-RBD complex. However, Nilotinib is able to weaken the interactions between the ACE2 and RBD proteins by reducing the average number of hydrogen bonds. Our essential dynamics analysis illustrates that both drugs are destabilizing the ACE2-RBD complex by triggering large-scale motions that lead to increase in the conformational area that is sampled by the ACE2-RBD complex. In the case of Nilotinib, filtering the trajectory by the first three eigenvectors shows that the most dominant motions of ACE2 and RBD proteins are completely different from the control system. In contrast to the control system, Nilotinib induces a divergent large-scale motion that rotates ACE2 and RBD in opposite directions. Our structural analysis broadens the understanding of small molecules destabilization and intervention mechanisms that would be an effective strategy to prevent infection at its early stages. However, considering the intrinsic limitations of all-atomistic MD simulations, such as small size scales, short timescales, and sampling issues, further experimental and numerical studies should be conducted to fully understand the mechanisms of action of SSAA09E2, Nilotinib, and other similar chemical compounds. More specifically, one of the future works would be analyzing drug effects on the whole spike protein at longer timescales and potential changes in the enzymatic activity of ACE2 proteins as a result of these two drugs.
Author contributions
M.R. designed research, performed research, analyzed data, and wrote the article. M.N. performed research, analyzed data, and wrote the article. Y.L. designed and directed research, performed research, analyzed data, and wrote the article.
Acknowledgments
The authors acknowledge the support of this work from the National Institutes of Health grant of R01HL131750, and National Science Foundation grant of CBET 2039310. This work used the Extreme Science and Engineering Discovery Environment through allocation MCB180032.
Editor: Alexandr Kornev.
References
- 1.Tai W., He L., Du L. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell. Mol. Immunol. 2020;17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nguyen H.L., Lan P.D., Li M.S. Does SARS-CoV-2 bind to human ACE2 more strongly than does SARS-CoV? J. Phys. Chem. B. 2020;124:7336–7347. doi: 10.1021/acs.jpcb.0c04511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xie Y., Karki C.B., Li L. Spike proteins of SARS-CoV and SARS-CoV-2 utilize different mechanisms to bind with human ACE2. Front. Mol. Biosci. 2020;7:591873. doi: 10.3389/fmolb.2020.591873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shang J., Ye G., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lan J., Ge J., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 6.Yan R., Zhang Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang C., Li W., Bosch B.-J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020;11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jahanshahlu L., Rezaei N. Monoclonal antibody as a potential anti-COVID-19. Biomed. Pharmacother. 2020;129:110337. doi: 10.1016/j.biopha.2020.110337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shanmugaraj B., Siriwattananon K., Phoolcharoen W. Perspectives on monoclonal antibody therapy as potential therapeutic intervention for Coronavirus disease-19 (COVID-19) Asian Pac. J. Allergy Immunol. 2020;38:10–18. doi: 10.12932/AP-200220-0773. [DOI] [PubMed] [Google Scholar]
- 10.Chaturvedi P., Han Y., Vuković L. Adaptive evolution of peptide inhibitors for mutating SARS-CoV-2. Adv. Theory Simul. 2020 doi: 10.1002/adts.202000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cao L., Goreshnik I., Baker D. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370:426–431. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adedeji A.O., Severson W., Sarafianos S.G. Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J. Virol. 2013;87:8017–8028. doi: 10.1128/JVI.00998-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei T.Z., Wang H., Li H.y. In silico screening of potential spike glycoprotein inhibitors of SARS-CoV-2 with drug repurposing strategy. Chin. J. Integr. Med. 2020;26:663–669. doi: 10.1007/s11655-020-3427-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wishart D.S., Feunang Y.D., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao H., Caflisch A. Molecular dynamics in drug design. Eur. J. Med. Chem. 2015;91:4–14. doi: 10.1016/j.ejmech.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 17.Lin X., Li X., Lin X. A review on applications of computational methods in drug screening and design. Molecules. 2020;25:1375. doi: 10.3390/molecules25061375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagan M.F., Zandi R. Recent advances in coarse-grained modeling of virus assembly. Curr. Opin. Virol. 2016;18:36–43. doi: 10.1016/j.coviro.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Razizadeh M., Nikfar M., Liu Y. Coarse-grained modeling of pore dynamics on the red blood cell membrane under large deformations. Biophys. J. 2020;119:471–482. doi: 10.1016/j.bpj.2020.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou Y., Sohrabi S., Liu Y. Mechanical properties of nanoworm assembled by DNA and nanoparticle conjugates. J. Nanosci. Nanotechnol. 2016;16:5447–5456. doi: 10.1166/jnn.2016.12068. [DOI] [PubMed] [Google Scholar]
- 21.Prieto-Martínez F.D., López-López E., Medina-Franco J.L. In: In Silico Drug Design. Roy Kunal., editor. Elsevier; 2019. Computational drug design methods–current and future perspectives; pp. 19–44. [Google Scholar]
- 22.Deganutti G., Prischi F., Reynolds C.A. Supervised molecular dynamics for exploring the druggability of the SARS-CoV-2 spike protein. J. Comput. Aided Mol. Des. 2021;35:195–207. doi: 10.1007/s10822-020-00356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sisk J.M., Frieman M.B., Machamer C.E. Coronavirus S protein-induced fusion is blocked prior to hemifusion by Abl kinase inhibitors. J. Gen. Virol. 2018;99:619–630. doi: 10.1099/jgv.0.001047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coleman C.M., Sisk J.M., Frieman M.B. Abelson kinase inhibitors are potent inhibitors of severe acute respiratory syndrome coronavirus and Middle East respiratory syndrome coronavirus fusion. J. Virol. 2016;90:8924–8933. doi: 10.1128/JVI.01429-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foà R., Bonifacio M., Ferrara F. Philadelphia-positive acute lymphoblastic leukaemia (ALL) in Italy during the COVID-19 pandemic: a Campus ALL study. Br. J. Haematol. 2020;190:e3–e5. doi: 10.1111/bjh.16758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Breccia M., Abruzzese E., Saglio G., Campus CML Working Group Chronic myeloid leukemia management at the time of the COVID-19 pandemic in Italy. A campus CML survey. Leukemia. 2020;34:2260–2261. doi: 10.1038/s41375-020-0904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gordon J.C., Myers J.B., Onufriev A. H++: a server for estimating pKas and adding missing hydrogens to macromolecules. Nucleic Acids Res. 2005;33:W368–W371. doi: 10.1093/nar/gki464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou J., Yin J., Zhang P. Computational prediction of mutational effects on SARS-CoV-2 binding by relative free energy calculations. J. Chem. Inf. Model. 2020;60:5794–5802. doi: 10.1021/acs.jcim.0c00679. [DOI] [PubMed] [Google Scholar]
- 29.Kim S., Chen J., Bolton E.E. PubChem 2019 update: improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hanwell M.D., Curtis D.E., Hutchison G.R. Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012;4:17. doi: 10.1186/1758-2946-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Volkamer A., Kuhn D., Rarey M. Combining global and local measures for structure-based druggability predictions. J. Chem. Inf. Model. 2012;52:360–372. doi: 10.1021/ci200454v. [DOI] [PubMed] [Google Scholar]
- 32.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 33.Salentin S., Schreiber S., Schroeder M. PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015;43:W443–W447. doi: 10.1093/nar/gkv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris G.M., Huey R., Olson A.J. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gasteiger J., Marsili M. Iterative partial equalization of orbital electronegativity-a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 36.Forli S., Huey R., Olson A.J. Computational protein-ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016;11:905–919. doi: 10.1038/nprot.2016.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morris G.M., Huey R., Olson A.J. Using AutoDock for ligand-receptor docking. Curr. Protoc. Bioinformatics. 2008;Chapter 8:Unit 8.14. doi: 10.1002/0471250953.bi0814s24. [DOI] [PubMed] [Google Scholar]
- 38.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris G.M., Goodsell D.S., Olson A.J. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
- 40.Huang J., Rauscher S., MacKerell A.D., Jr. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatcher E., Guvench O., Mackerell A.D. CHARMM additive all-atom force field for aldopentofuranoses, methyl-aldopentofuranosides, and fructofuranose. J. Phys. Chem. B. 2009;113:12466–12476. doi: 10.1021/jp905496e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jorgensen W.L., Chandrasekhar J., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 43.Kim S., Lee J., Im W. CHARMM-GUI ligand reader and modeler for CHARMM force field generation of small molecules. J. Comput. Chem. 2017;38:1879–1886. doi: 10.1002/jcc.24829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanommeslaeghe K., Hatcher E., Mackerell A.D. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vanommeslaeghe K., Raman E.P., MacKerell A.D., Jr. Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012;52:3155–3168. doi: 10.1021/ci3003649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hess B., Bekker H., Fraaije J.G. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 47.Martyna G.J., Klein M.L., Tuckerman M. Nosé-Hoover chains: the canonical ensemble via continuous dynamics. J. Chem. Phys. 1992;97:2635–2643. [Google Scholar]
- 48.Berendsen H.J., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 49.DeLano W.L. The PyMOL User’s Manual. DeLano Scientific; San Carlos, CA, USA: 2002. [Google Scholar]
- 50.Hussain M., Jabeen N., Aziz B. Structural variations in human ACE2 may influence its binding with SARS-CoV-2 spike protein. J. Med. Virol. 2020;92:1580–1586. doi: 10.1002/jmv.25832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ali A., Vijayan R. Dynamics of the ACE2-SARS-CoV-2/SARS-CoV spike protein interface reveal unique mechanisms. Sci. Rep. 2020;10:14214. doi: 10.1038/s41598-020-71188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei Y., Aris P., Xia X. Predicting mammalian species at risk of being infected by SARS-CoV-2 from an ACE2 perspective. Sci. Rep. 2021;11:1702. doi: 10.1038/s41598-020-80573-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He J., Tao H., Xiao Y. Molecular mechanism of evolution and human infection with SARS-CoV-2. Viruses. 2020;12:428. doi: 10.3390/v12040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Juraszek J., Rutten L., Langedijk J.P.M. Stabilizing the closed SARS-CoV-2 spike trimer. Nat. Commun. 2021;12:244. doi: 10.1038/s41467-020-20321-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Andrade J., Gonçalves P.F.B., Netz P.A. Why does the novel coronavirus spike protein interact so strongly with the human ACE2? A thermodynamic answer. ChemBioChem. 2021;22:865–875. doi: 10.1002/cbic.202000455. [DOI] [PubMed] [Google Scholar]
- 56.Xu D., Tsai C.J., Nussinov R. Hydrogen bonds and salt bridges across protein-protein interfaces. Protein Eng. 1997;10:999–1012. doi: 10.1093/protein/10.9.999. [DOI] [PubMed] [Google Scholar]
- 57.Veeramachaneni G.K., Thunuguntla V.B., Bondili J.S. Structural and simulation analysis of hotspot residues interactions of SARS-CoV 2 with human ACE2 receptor. J. Biomol. Struct. Dyn. 2021;39:4015–4025. doi: 10.1080/07391102.2020.1773318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tai K., Shen T., McCammon J.A. Analysis of a 10-ns molecular dynamics simulation of mouse acetylcholinesterase. Biophys. J. 2001;81:715–724. doi: 10.1016/S0006-3495(01)75736-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.