Abstract
The entry of the severe acute respiratory syndrome coronavirus 2 virus in human cells is mediated by the binding of its surface spike protein to the human angiotensin-converting enzyme 2 (ACE2) receptor. A 23-residue long helical segment (SBP1) at the binding interface of human ACE2 interacts with viral spike protein and therefore has generated considerable interest as a recognition element for virus detection. Unfortunately, emerging reports indicate that the affinity of SBP1 to the receptor-binding domain of the spike protein is much lower than that of the ACE2 receptor itself. Here, we examine the biophysical properties of SBP1 to reveal factors leading to its low affinity for the spike protein. Whereas SBP1 shows good solubility (solubility > 0.8 mM), circular dichroism spectroscopy shows that it is mostly disordered with some antiparallel β-sheet content and no helicity. The helicity is substantial (>20%) only upon adding high concentrations (≥20% v/v) of 2,2,2-trifluoroethanol, a helix promoter. Fluorescence correlation spectroscopy and single-molecule photobleaching studies show that the peptide oligomerizes at concentrations >50 nM. We hypothesized that mutating the hydrophobic residues (F28, F32, and F40) of SBP1, which do not directly interact with the spike protein, to alanine would reduce peptide oligomerization without affecting its spike binding affinity. Whereas the mutant peptide (SBP1mod) shows substantially reduced oligomerization propensity, it does not show improved helicity. Our study shows that the failure of efforts, so far, to produce a short SBP1 mimic with a high affinity for the spike protein is not only due to the lack of helicity but is also due to the heretofore unrecognized problem of oligomerization.
Significance
A short peptide that mimics the binding interface of the human ACE2 receptor to the severe acute respiratory syndrome coronavirus 2 spike protein would be valuable for both diagnosis and treatment of the coronavirus disease 2019. Yet, peptide mimics of the helical binding motif of the receptor have not succeeded in replicating the high affinity of the latter to the spike protein. Here, we identify dual causes for this failure: a lack of helical structure for the peptides and an unexpected tendency for them to oligomerize. Efforts to replicate the natural spike protein:ACE2 interface need to focus on both these properties to succeed.
Introduction
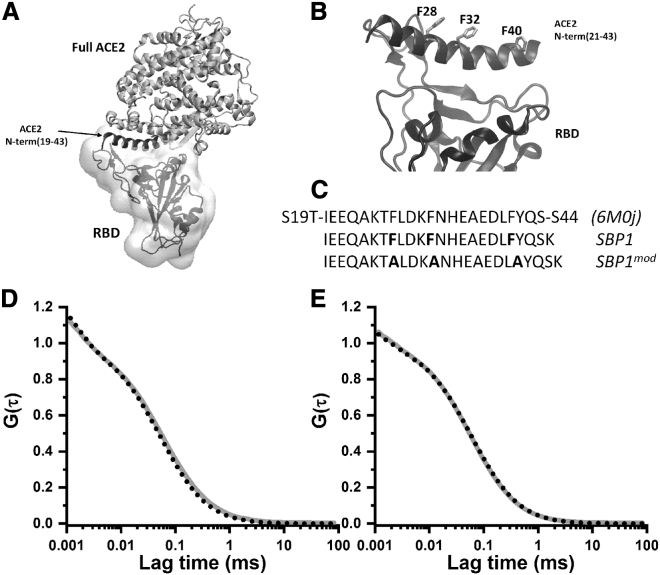
The primary entry pathway of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus into the human cell is through the interactions of one of its envelope proteins (the “spike” protein), and the human ACE2 membrane receptor (1, 2, 3). In high-resolution structures obtained using cryo-electron microscopy (4, 5, 6) and crystallography (7,8), it is apparent (Fig. 1 A) that these two proteins predominantly interact via the receptor-binding domain (RBD) of the spike protein and the residues 21–43 from the N-terminal α-helix of ACE2. This helical segment (called the spike binding peptide (SBP1) henceforth) has been of considerable interest to the scientific community because, hypothetically, the peptide segment in its isolated form may competitively bind to the spike RBD and neutralize the virus to protect against the coronavirus disease infection (9,10). Additionally, it may be possible to use SBP1 as a recognition element for detection purposes. Indeed, artificial proteins that have been designed based on this helix have displayed affinities (100 pM to 10 nM) (11) that are much greater than that of the original ACE2 protein (15 nM) (6). Similarly, large immunoadhesins composed of the entire ACE2 (12) have been successful in binding to RBD and are considered to be promising candidates for the coronavirus disease 2019 treatment. A dissociation constant of 9 nM was measured for ACE2-Fc with RBD and of 0.03 nM for a modified ACE2-Fc (12). However, the goal of directly applying the SBP1 peptide (or simple variants) has remained elusive. Whereas initial reports suggested that the SBP1 peptide had 47 nM binding affinity to the RBD (9), later revisions have suggested that the dissociation constant is much higher (>1 μM) (13). Moreover, the binding data have been inconsistent because as the affinity showed dependence on the source of the spike protein RBD (9,13).
Figure 1.
(A) The ACE2:RBD complex according to crystal structure PDB: 6M0J (7). The N-terminal helical segment (19–43) serves as the primary binding motif for ACE2 with RBD (surface and secondary structure representation). (B) The broad binding interface formed between residues 21 and 43 of the ACE2 N-terminal helix and the mostly disordered binding segment of the RBD. The helix contains three hydrophobic residues which do not interact with the RBD but stabilize the secondary structure by participating in coiled-coil interactions with other helices within ACE2. (C) The primary sequence of the segment (S19–S44) crystallized in PDB: 6M0J (7) and that for the derived peptides SBP1 (the helix in Fig. 1B, except for S44) and SBP1mod used in our biophysical studies. SBP1mod differs from SBP1 in terms of replacing the three hydrophobic phenylalanine residues with alanine (indicated in bold). (D and E) Normalized autocorrelation data obtained from Rh110-labeled peptides (SBP1 and SBP1mod) with 1 μM RBD (black dotted line) and without RBD (gray solid line). (D) 30 nM SBP1 and (E) 35 nM SBP1mod.
Here, we examine the biophysical properties of SBP1 to understand its low affinity to the spike RBD and whether small modifications in either the peptide or its solution conditions can increase its binding affinity to the spike RBD. We hypothesize that the helical secondary structure of SBP1 within ACE2 may be altered in solution form. Additionally, given the considerable solution exposure to the hydrophobic residues of the isolated SBP1, self-interactions may bury the RBD binding surface of the peptide. We test these hypotheses using far-ultraviolet (UV) circular dichroism (CD) spectroscopy to determine the secondary structure and fluorescence correlation spectroscopy (FCS) to determine the tendency of aggregation. We find that, indeed, the peptide has very little helicity to start with and has a tendency to form stable oligomers even at submicromolar concentrations. We employ the single-molecule photobleaching (smPB) technique to investigate the stoichiometry of the oligomers. We also ask if changing the solution conditions by including helix-promoting agents can stabilize the original helical structure. To reduce the tendency of aggregation, we mutated the hydrophobic phenylalanine residues (F28, F32, and F40) of SBP1 (Fig. 1, B and C) to alanine and studied the properties of the resultant peptide, which we term SBP1mod. As seen in Fig. 1 B, the three phenylalanine residues point away from the RBD binding interface. Within the ACE2 structure, F28, F32, and F40 contribute toward helix stability by participating in hydrophobic coiled-coil interactions (Fig. 1 A). However, in the isolated SBP1 they would be solvent exposed and may contribute to the self-aggregation propensity of the peptide. Furthermore, amino acid helix-propensity scale estimates (14) indicate that alanines have better helix forming propensities relative to phenylalanines.
Materials and methods
Peptide synthesis
The peptides were synthesized on Rink Amide 4-Methylbenzhydrylamine (MBHA) resin (100–200 mess size and 0.35 mmol/g loading capacity) using an automated solid-phase peptide synthesizer in the laboratory (PS3; Protein Technologies, Tucson, AZ) using the standard 9-fluorenylmethoxycarbonyl (Fmoc) chemistry. This method of synthesis on Rink Amide resin amidates the C-terminal carboxylate end of the peptide. The sequence of the wild-type version of ACE2 N-terminal fragment (residues 21–43 in Fig. 1, B and C), is H2N-IEEQAKTFLDKFNHEAEDLFYQS-CONH2. We added an extra lysine to the C-terminal, with an orthogonal protection using the 4-Methyltrityl group (N-α-Fmoc-N-ε-4-methyltrityl-L-lysine), so that the C-terminus of the peptide could be modified (e.g., with dye labeling) later on, if needed. This synthesized peptide with the sequence H2N-IEEQAKTFLDKFNHEAEDLFYQSK-CONH2 is termed SBP1 (Fig. 1 C). We further synthesized a modified version of SBP1, termed SBP1mod, wherein the three phenylalanine residues (F28, F32, and F40) of the former have been mutated (Fig. 1 C) to alanine to yield a peptide with the following sequence: H2N-IEEQAKTALDKANHEAEDLAYQSK-CONH2.
All the Fmoc amino acids, solvents, and reagents used during the synthesis were purchased from Merck Schuchardt (Hohenbrunn, Germany). The crude peptides were subsequently characterized using a matrix-assisted laser desorption ionization-time-of-flight mass spectrometer (UltrafleXtreme; Bruker, Billerica, MA) in the laboratory and were lyophilized thereafter. The lyophilized peptides were stored at 4°C.
Rhodamine 110 (Rh110) labeling (Rh110-SBP1 and Rh110-SBP1mod) was performed by covalently attaching the 5-(and-6)-carboxyrhodamine 110, succinimidyl ester (mixed isomers, 5(6)-CR110, SE) dye to the N-terminal free amine of SBP1 and SBP1mod peptides. The labeling was done on the peptides attached to the resins, and the excess dye was thoroughly washed with N,N-dimethylformamide (DMF) afterwards. The 5(6)-CR110, SE dye was purchased from Thermo Fisher Scientific (Waltham, MA). The Cyanine 3 (Cy3)-labeled SBP1 (Cy3-SBP1) peptide was similarly synthesized by attaching Cy3 N-hydroxysuccinimide (NHS) ester dye (Lumiprobe, Hunt Valley, MD) to the N-terminal of the peptide. Matrix-assisted laser desorption ionization-time-of-flight and electrospray ionization mass spectrometry mass spectra of the unlabeled and N-terminal Cy3-labeled peptides are shown in the Figs. S1–S4. It is clear from the mass spectra that the dye labeling was nearly complete (∼100%).
RBD preparation and purification
The RBD of the SARS-CoV-2 spike protein (residues 331–532 (15)) was cloned in a pTRIP lentiviral vector (16) under a cytomegalovirus (CMV) enhancer and chicken β-actin promoter. The RBD gene is flanked with a tPA sequence (MDAMKRGLCCVLLLCGAVFVSPSEI) at the N-terminus for its efficient secretion and an HRV-3C precision protease site linked to 10× Histidine followed by a Twin-Strep tag at the C-terminus for recombinant purification. This construct was a kind gift from Dr. Minhaj Sirajuddin, Institute of Stem Cell Science and Regenerative Medicine (Bangalore, India). Human embryonic kidney (HEK) 293T cells that were adapted to grow in suspension were maintained in Freestyle 293 media with 1% FBS at 37°C in the presence of 10% CO2 and were used for expression of the RBD. 100 mL of cells at a density of 1 million/mL were transfected with 100 μg of the RBD plasmid complexed with 300 μg of linear polyethylenimine and grown further. The cells were harvested 48 h post-transfection by centrifugation at 4000 × g for 15 min.
The supernatant containing the secreted RBD protein was concentrated to 30 mL using a 10-kDa Vivaflow concentrator (Sartorius, Gottingen, Germany). The concentrate was diluted with 200 mL of wash buffer containing 35 mM Tris (pH 8), 100 mM NaCl, 5% glycerol, 1 mM PMSF, 1 mM DTT, and concentrated again to 50 mL. The buffer-exchanged supernatant was then added to 500 μL of pre-equilibrated Strep-Tactin beads (GE Healthcare, Chicago, IL) and allowed to bind overnight at 4°C. The beads were washed with 50 mL of wash buffer on a gravity flow column, followed by elution with wash buffer containing 10 mM desthiobiotin (Sigma-Aldrich, St. Louis, MO). 500 μL elution fractions were collected and further concentrated using an Amicon 10-kDa concentrator (Millipore, Burlington, MA). The concentrated protein was aliquoted and flash-frozen in liquid nitrogen and stored at −80°C until further use.
FCS measurements
We used FCS for measuring the binding affinity of the peptides with the RBD of the SARS-CoV-2 virus as well as for measuring the hydrodynamic radii (Rh) of the peptides. These measurements were performed using a home-built FCS instrument (17,18). Briefly, a 488-nm laser beam was expanded and collimated using a 1:4 telescope set up before focusing into the sample using an apochromatic 60× water immersion objective with the numerical aperture of 1.2 (Olympus, Center Valley, PA). The fluorescence was collected using the same objective and focused onto a 15-μm core-diameter optical fiber after filtering through a suitable emission filter (Chroma Technology, Rockingham, VT). The fiber was used as a confocal pinhole to reject the out-of-focus fluorescence. The fluorescence was detected by a single-photon avalanche photodiode (PerkinElmer, Waltham, MA), and the data were collected and processed using a hardware correlator (PicoHarp 300; PicoQuant, Berlin, Germany). FCS data were fitted with Eq. 1 in Origin 6 software (OriginLab, Northampton, MA).
To measure RBD binding, the stock concentration of both the peptides, Rh110-labeled SBP1 (3.64 μM) and SBP1mod (13 μM), were prepared at pH 7.5 and were diluted appropriately during the experiment using 20 mM phosphate-buffered saline (PBS) (pH 7.5), 146 mM NaCl, 5.4 mM KCl, 20 mM Na2HPO4, 0.4 mM KH2PO4, and 2 mM NaN3). A 41-μM stock of RBD solution was prepared in solution with the same buffer composition. All stocks were flash-frozen and stored at −80°C. They were immediately used for the experiment after thawing.
A two-component, three-dimensional diffusion model with a triplet component (19) was used to fit the FCS data (Eq. 1):
| (1) |
Here, G(t) is the correlation function at time t, f is the fraction of the triplet component, and τ is the corresponding triplet lifetime. Also, g1 and g2 are the amplitudes of the correlation function corresponding to the two diffusing components (free peptide and peptide bound to RBD), τD1 and τD2 are the corresponding diffusion times, a is the structure parameter for the optical probe volume (assumed to be a Gaussian ellipsoid), and bl denotes the background signal). Free Rh110 was used as a standard to calibrate the instrument (diffusion coefficient of 4.4 × 10−6 cm2 s−1 in water at 25°C and the corresponding Rh of ∼0.56 nm (20)). The diffusion times of the Rh110-labeled peptides were converted to Rh by comparing their diffusion times with that of free Rh110 in solution.
The average Rh of the peptides were also measured using FCS. Rh110-labeled SBP1 and SBP1mod stocks were prepared in pH 7.5 PBS, as stated in the previous section. The concentrations of the stocks (CStock = 2.7 μM) were determined by monitoring the absorbance at λmax = 499 nm (using Rh110 ε (499 nm) = 80,000 M−1 cm−1). These stocks were then diluted in PBS to a final concentration of 140 nM. The data were also fitted with Eq. 1 using a two-species approximation. Here, the two components correspond to the monomeric peptide and the free dye in solution, respectively. The same solutions were diluted to low nanomolar concentrations to follow time-dependent changes in the Rh of the peptides.
Far-UV CD spectroscopy
UV CD measures the secondary structure content of a protein or peptide. Dry lyophilized powders of the unlabeled SBP1 and SBP1mod peptides were dissolved in pH 7.5, 20 mM PBS (composition: 146 mM NaCl, 5.4 mM KCl, 0.4 mM KH2PO4, 20 mM Na2HPO4 with 2 mM NaN3) at concentrations of 0.8 mM. These stocks were then centrifuged at 2000 × g for 10 min to discard large aggregates, if any. The concentration of the solution before and after centrifugation was measured spectrophotometrically, as described below. The concentration did not change after centrifugation, indicating that the peptide is soluble at 0.8 mM concentration. The supernatants were then divided into small aliquots, flash-frozen in liquid nitrogen, and finally stored at −80°C. They were thawed quickly and used for the experiments straightaway. The concentrations of the stocks were estimated by diluting them (at 20-fold dilution) in the same buffer (described above) and measuring the absorbance at 280 nm on an Analytik Jena (Jena, Germany) SPECORD 205 UV–Vis Spectrophotometer (using tyrosine ε (280 nm) = 1280 M−1 cm−1). For the experiments, these stock solutions were diluted 20-fold in pH 7.5, 20 mM phosphate buffer with sodium fluoride (composition: 150 mM NaF, 0.4 mM KH2PO4, 20 mM Na2HPO4), yielding a final peptide concentration of 50 μM. Here, NaCl was replaced by NaF to avoid unwanted saturation of the detector at shorter wavelengths (deep UV) without altering the ionic strength of the buffer.
Steady-state far-UV CD spectra were recorded on a JASCO (Easton, MD) J-1500 Circular Dichroism Spectrophotometer. Spectra are recorded by monitoring the CD signal (after baseline subtraction using only phosphate buffer with sodium fluoride) at room temperature (25°C) from 300 to 190 nm with a data interval of 0.1 nm, CD scale of 200 mdeg/1.0 dOD, data integration time of 4 s, and bandwidth of 1.00 nm, at a scanning speed of 50 nm/min and average of five successive accumulations. The secondary structural analysis to calculate the secondary structure content was performed using the Beta Structure Selection (BeStSel) software (21,22) (http://bestsel.elte.hu/), and the parameters obtained from the deconvolution were plotted using OriginPro 2019.
Sample preparation and measurements for smPB experiments
smPB (23, 24, 25, 26) can detect the stoichiometry of protein oligomers, down to the level of individual oligomers (27, 28, 29, 30). The coverslips were precleansed using alkali and piranha solutions, as described earlier (26), and then finally plasma cleaned before starting any smPB measurements. Peptide stocks (CStock = 100 μM) were prepared in pH 7.5 PBS, as stated earlier. The concentrations of the stocks were determined by diluting them (25-fold) in PBS and monitoring the absorbance at 545 nm (using Cy3 ε (545 nm) = 1.3 × 105 M−1 cm−1). These stocks were then diluted in PBS to a final concentration of 1 nM. Concentrations were reconfirmed from the FCS count rates using a standard solution of known concentration. The resultant solution was thoroughly mixed with a 0.25% polyvinyl alcohol (PVA) solution and spin coated (for 30 s) on the top of a clean glass coverslip with a spinning speed of 3000 rpm at room temperature until a thin homogeneous film was created and the coverslip became completely dry. During this process, PVA polymerizes into a solid matrix, leaving each monomer or aggregate as an individual particle (i.e., they are spatially well separated from each other), and these molecules, therefore, do not exhibit any diffusive movements when they are subjected to the smPB measurements.
smPB images were acquired using a total internal reflection fluorescence microscope that was assembled earlier in the laboratory (26). An objective lens with a high numerical aperture (NA = 1.49, 100×; Nikon, Tokyo, Japan) was used for both illuminating (through the evanescent field) and simultaneously collecting the emission. Cy3-labeled peptides were excited using a 543-nm He-Ne laser (Melles Griot, Rochester, NY). Fluorescence emission was separated from the excitation light with the help of a dichroic mirror (565 nm) and a band-pass filter (605/55 nm, BA577-633; Nikon, Tokyo, Japan) in the emission path. The fluorescence photons were finally focused into an electron-multiplying charge-coupled device camera (iXON DV887ECS-UVB; Andor, Belfast, United Kingdom). The Nikon Perfect Focus System (Nikon, Tokyo, Japan) was utilized (in a dark environment) to focus on the plane of the coverslip containing the fluorescently labeled peptides, then the acquisition was started. The power at back aperture of the objective was 1.3 mW, and the images were recorded at 90 ms/frame.
smPB data analysis
We used Fiji (Windows 64 bit; ImageJ, National Institutes of Health, Bethesda, MD) (31) software to obtain the stoichiometry of the particles from the smPB data. The TrackMate plugin (32) was used to track the particles. A minimal threshold was applied to the images to suppress the background (100 in the images shown here). For a single spot, the diameter was restricted to 3 × 3 pixels (pixel size, 156 nm). A subsequent z-axis profile of the individual particles with time reported the stoichiometry. The statistics of oligomers were background subtracted (minor fluorescence impurities) using a 0.25% PVA solution without the oligomers. Finally, the distribution was corrected for “prebleaching”. Although the “B-value” (prebleaching probability) for carboxytetramethylrhodamine (TAMRA) is known previously, we used the relative bleaching time of Cy3 versus TAMRA to correct for “prebleaching” in the final distribution.
Results
Measurement of the binding of SBP1 and SBP1mod to the RBD
We measured the binding of both the peptides with the RBD of the spike protein by FCS. The following concentrations of the constituents were used for the experiments: 30 nM of SBP1, 35 nM of SBP1mod, and 1 μM of (unlabeled) RBD. Each sample was incubated for 30 min. Fig. 1, D and E shows the autocorrelation traces of SBP1 and SBP1mod with and without RBD, respectively. The results were fitted with Eq. 1 using two components, where one component corresponds to the free peptide and the other component corresponds to the peptide-RBD complex. SBP1 (Fig. 1 D) shows a τD1 of 78 ± 1.6 μs, which becomes 62 ± 1 μs in the presence of RBD. SBP1mod (Fig. 1 E) shows a τD1 of 63 ± 1 μs, that becomes 68 ± 1 μs in the presence of RBD. In each experiment, τD2 was fixed to the free dye diffusion time of 21.5 μs. The slight reduction of the diffusion time in the SBP1 experiment is ascribed to a somewhat higher dissociation of the peptide oligomers. Overall, our FCS results show negligible binding of SBP1 or SBP1mod to the RBD at these concentrations.
Measurement of the secondary structure content of the peptides
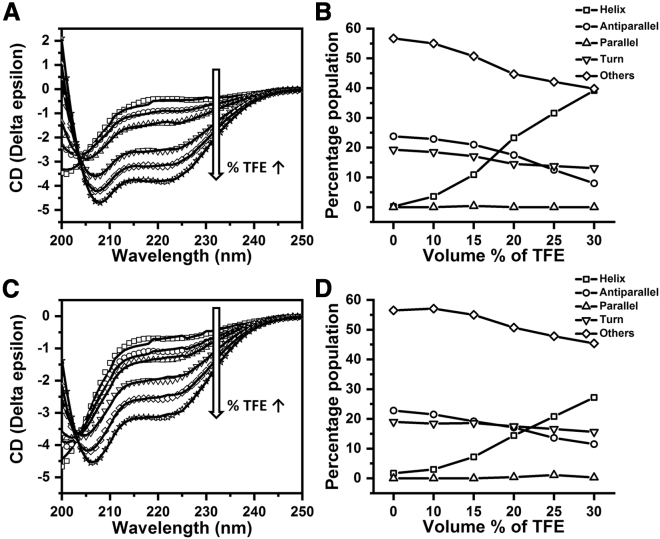
We measured the secondary structure content of the SBP1 peptide by performing a far-UV CD measurement (Fig. 2 A). The CD spectra were recorded using 50-μM peptide, diluted from 0.8-mM stock (the peptide is soluble at least up to 0.8 mM, see Materials and methods). The CD spectra were analyzed in terms of the underlying secondary structure using the BeStSel software (Fig. 2 B) (21,22). It is apparent from the data that the SBP1 peptide does not have significant helical content (Fig. 2 A, square). Even the modified peptide (SBP1mod) has negligible helical content (Fig. 2 C, square). SBP1mod has the same sequence as SBP1, except for three phenylalanine residues (F28, F32, and F40) in the latter, which have been mutated to alanine. We also monitored the increase of helicity with the addition of 2,2,2-trifluoroethanol (TFE), at different TFE concentrations (Fig. 2, A and C) to promote helical conformations (33, 34, 35, 36). The helicity increases with TFE, but the peptides become considerably helical only when the TFE concentration is 20% (v/v) or higher (Fig. 2, B and D). It is interesting to note that, despite the greater helix forming propensities expected for alanine relative to phenylalanine (14), SBP1 has higher helical content than SBP1mod in the presence of TFE.
Figure 2.
CD spectra of (A) SBP1 and (C) SBP1mod incubated in buffer (square) and with 10% (v/v, circle), 15% (up-triangle), 20% (down-triangle), 25% (rhombus), and 30% (star) 2,2,2-trifluoroethanol (TFE), respectively. 50 μM of each of these two peptides were incubated in 20 mM phosphate buffer with 150 mM NaF (pH 7.5) at 25°C. Spectra (in symbols) shown are the averages of five consecutive accumulations; they were fitted (solid lines) using the online version of the BeStSel software (21,22) for secondary structure determination. Variation of different secondary structures for SBP1 (B) and SBP1mod (D) as a function of TFE concentration, showing helix (square), antiparallel (circle), parallel (up-triangle), turn (down-triangle), and others (rhombus). Solid lines are guide to the eye.
Measurement of Rh of SBP1 and SBP1mod
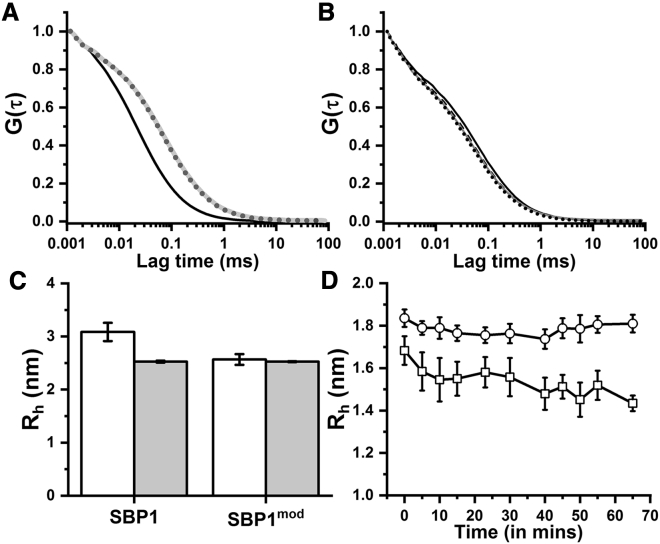
The Rh of the peptides were measured using FCS. For this purpose, SBP1 and SBP1mod were labeled with the Rh110 dye at the N-terminal free amine. Peptide solutions were prepared in 20 mM PBS from a stock solution of 2.7 μM. At a concentration of 140 nM, both the peptides had (Rh) > 2.5 nm (Figs. 3, A and C and S5). Interestingly, SBP1 had an Rh of 3.1 ± 0.2 nm, which is larger than that of the SBP1mod peptide (2.6 ± 0.1 nm). After 24 h of incubation, both the peptides had a very similar Rh of ∼2.5 nm (Fig. 3 C). These values are too large for a monomeric 24-residue peptide. We hypothesized that this indicates oligomerization of both peptides, with SBP1 forming a larger oligomer at 140 nM than SBP1mod. If the large size is indeed due to noncovalent oligomerization, then the oligomers should dissociate at sufficiently low concentrations. When we incubate the peptides at a concentration of 3 nM for 70 min, we observed a decrease in the Rh for SBP1 to 1.68 ± 0.06 nm immediately after dilution, then to a value of 1.43 ± 0.04 nm in 65 min (Fig. 3, B and D, square). Rh of SBP1mod decreases to ∼1.8 nm but remains similar thereafter. This suggests that the major fraction of the peptide population for SBP1 was in an oligomeric state at 140 nM and became nearly monomeric at low nanomolar concentrations. Overall, our FCS results show that both SBP1 and SBP1mod form oligomers at higher concentrations but dissociate to a monomeric or near-monomeric population at lower concentrations.
Figure 3.
(A) Normalized fluorescence autocorrelation data obtained from free Rh110 in solution (black solid line), 140 nM Rh110-labeled SBP1 after 30 min (gray solid line) and 24 h (dark gray dotted line) of incubation. (B) Autocorrelation traces obtained from 3 nM Rh110-labeled SBP1 at time t = 0 (black solid line) and after 20 min (black dashed line), 40 min (gray, solid line), and 1 h (black, dotted line) of incubation. (C) Rh-values determined from the FCS experiments for Rh110-labeled SBP1 and SBP1mod at 140 nM after vortexing for 30 min (white) and after 24 h (gray). The error bars are plotted as SEs of the mean (n = 5). (D) Rh of Rh110-labeled SBP1 (square) and SBP1mod (circle) as a function of time at a final peptide concentration of 3 nM. The error bars are plotted as SEs of the mean (n = 10 for SBP1, and n = 8 for SBP1mod). Solid lines are guide to the eye.
We note that the same data collected for the FCS experiment can be analyzed with the photon-counting histogram (PCH) technique (37). In principle, PCH can yield the stoichiometry of the oligomers. However, PCH fits including more than two components, given the quality of the data, do not provide unique parameter values. Nevertheless, PCH analysis confirms that the solution must contain at least one multimeric component in addition to monomers (see Fig. S6).
We also investigated whether the fluorescence label has a substantial role in the oligomerization process. We carried out dynamic light scattering measurements using DynaPro (Protein Solutions, Lakewood, NJ) of both SBP1 and SBP1mod (unlabeled). Fig. S7 shows the correlation traces of SBP1 and SBP1mod at 1 μM (Fig. S7, A and B) and at 10 μM (Fig. S7, C and D, respectively) concentrations. We observe multiple traces that show the presence of larger aggregates in the solution. We conclude that the peptide has a tendency to aggregate, even without any fluorescent labels. We note that we cannot access concentration below this for such a small peptide because of the lack of sensitivity of dynamic light scattering (compared with FCS).
Investigating the nature of the oligomer with smPB
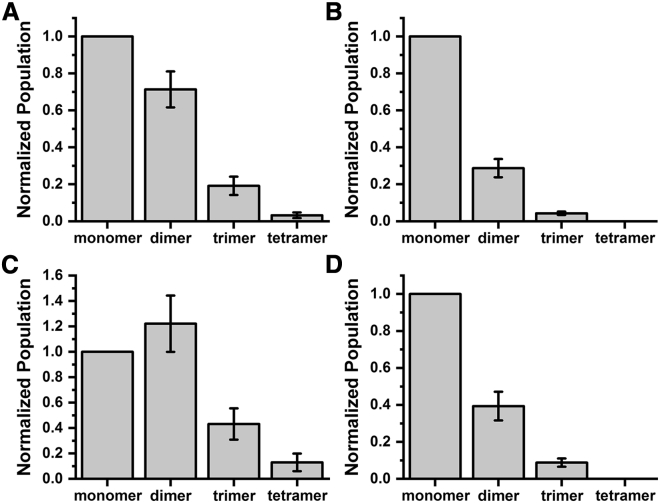
The stoichiometry of the initial oligomers and the presumed monomers after dissociation can be directly observed using smPB measurement (28,30,38). We performed smPB on two different samples: 1) a freshly diluted one and 2) a sample that is incubated for 24 h at a concentration of 1 nM. Samples of Cy3-labeled SBP1 are subjected to smPB measurements using a total internal reflection fluorescence microscope (Figs. S8 and S9), as described in the Materials and methods. We observe a distribution of sizes in both the cases (Fig. 4, A and B). However, the population at the initial time has a mean stoichiometry of 1.6 ± 0.2, whereas after 24 h of incubation, it becomes 1.3 ± 0.1. We verified that the overall concentration of the peptide remains relatively unchanged (within 30%) during the 1-day incubation period (Fig. S10). This confirms the oligomeric nature of the peptide as observed by FCS and also implies that there is a heterogeneous mixture of oligomers in the population. These measurements overestimate the monomers because some of the oligomers may be “prebleached” (38), despite using a Perfect Focus System (Nikon, Tokyo, Japan) along with the shutter. This can be corrected for, as shown in Fig. 4, C and D. After the prebleaching correction, the stoichiometry at the initial time has a mean of 1.9 ± 0.3, whereas after the 24 h of incubation, it has a mean of 1.4 ± 0.1. We note that we have not corrected for dissociation that may occur within a short time (∼5 min) of sample preparation. We also note that the Rh observed in FCS for the SBP1mod is slightly higher than that of SBP1, whereas the single-molecule data show a similar or lower oligomer stoichiometry (Fig. S10). This is possibly due to a difference in shape between the two. We also cannot rule out small differences caused by the difference in the fluorescence labels used in the two experiments.
Figure 4.
A smPB study shows the population distribution of Cy3-labeled SBP1 oligomers (A) at time 0 and (B) after 24-h incubation. (C and D) are the same as (A) and (B), respectively, but after prebleaching correction (38). The errors are plotted as the SE of means. For fresh SBP1, N = 323 points (three sets); for SBP1 after 24 h, N = 319 points (three sets). Each set consists of six regions of interest.
Discussion
A short 23-residue peptide mimicking the binding interface of the spike protein with ACE2 would be a prime candidate for recognition of the spike protein and for reducing its infectious abilities by competitive inhibition of its binding to ACE2. However, initial efforts with the SBP1 have failed to achieve the binding constant observed for ACE2 with the spike RBD (13), and our own experiments using FCS show an absence of binding (Fig. 1, D and E). The real dissociation constant is actually several micromoles (13), much higher than the initially reported value of 47 nM (9). So the concentration of the peptides needs to be >1 μM to have substantial binding. However, if the concentration of the viral protein in the pathological sample is small, this would mean that almost all of that peptide would be free, providing an unwanted, large background. This is the reason why the artificial peptides have failed as diagnostics so far. A systematic understanding of this failure can lead to strategies for tuning the binding affinity of SBP1 to the spike. Here, we have studied the biophysical properties of the SBP1 peptide that should indicate where the problem lies.
First and foremost, SBP1 does not retain the secondary structure observed in the ACE2 protein. This is perhaps not surprising because the ACE2 environment provides stabilizing interactions to keep the SBP1 sequence helical. However, what is perhaps notable is how little helicity is observed for this isolated peptide. Analyses of our CD data (Fig. 2, A–D) show that the helicity is less than 1%. There is considerable turn and some β-sheet structure, which suggests that this peptide takes an alternative form when isolated. However, such a secondary structure is rather unlikely to form a binding interface with the spike protein. A simplistic docking and molecular dynamics study (Supporting materials and methods, Section S2) shows that nonhelical conformations of the SBP1 peptide tend to produce lower binding energy scores. They form fewer hydrogen bonds and salt bridges at the RBD interface relative to the stable helical peptide conformation (Fig. S11; Tables S2–S4) presented by the ACE2 receptor. What is also notable is that the helix promoter TFE is only moderately effective in promoting helicity within SBP1 because the peptide becomes strongly helical only at TFE concentrations exceeding 20%.
A peptide that does not fold into a well-defined secondary structure can be aggregation prone. However, the solubility of SBP1 is reasonably high, which suggests that the peptide has the means to stabilize its structure, although a nonnative one. Probing the size (Rh) of the peptide using FCS yields a Rh of 2.5 nm (Fig. 3, A and C). We note that if the helix retained its structure, the expected Rh would be ∼1.06 nm (see Supporting materials and methods). On the other hand, if the peptide is unfolded and behaves like a random coil, the expected Rh would be 1.32 nm (see Supporting materials and methods). So the observed radius of 2.5 nm indeed suggests that the peptide is oligomeric at concentrations above 140 nM. A PCH analysis of the data also suggests oligomerization (Fig. S6). However, the peptide dissociates into a monomeric and/or low oligomeric state at low nM concentrations (Fig. 3 D). A part of this dissociation happens immediately upon dilution, whereas for SBP1, a further dissociation happens over time, resulting in a final Rh of ∼1.5 nm. SBP1mod remains somewhat larger (∼1.8 nm), possibly because it retains some dimeric population.
The FCS results, of course, cannot determine what the oligomeric state is and whether the final size obtained at low concentrations is that of a true monomer. This required us to probe the solution with smPB. Our results show that the SBP1 solution at low concentrations, immediately after dilution from higher concentrations, is a mixture of monomer, dimer, trimer, and even higher oligomers (Fig. 4 C). The solution after a 24-h incubation period is still not monomeric, but it is considerably enriched with the monomeric species (Fig. 4 D). We note that oligomerization is a concentration-driven process. The relative fraction of monomers is substantial only at low nM concentrations and would be progressively smaller at higher concentrations. So at μM levels, which will be appropriate for a diagnostic sample, there would be a negligible fraction of monomers available for binding to the RBD. This is what is shown by our RBD binding experiments (Fig. 1, D and E).
We reasoned that oligomerization can be a factor in stabilizing a random conformation. An inspection of the peptide bound to the RBD (Fig. 1, A and B) shows that the surface of the helix that is on the opposite side from the binding interface has three phenylalanine residues that can be expected to make it prone to form oligomers. We mutated these residues with alanine, which is known to promote helix formation (14) and is not very hydrophobic, to create the variant peptide SBP1mod (Fig. 1 C). However, the results remain qualitatively similar for the new peptide. The helicity of SBP1mod is only marginally higher than that of SBP1 (in buffer). Further, SBP1mod still forms oligomers, although of a smaller size at the higher concentrations and of a somewhat higher size at lower concentrations (Figs. 3, C and D and S5).
Overall, our results indicate that designs of high-affinity spike RBD binders based on the SBP1 peptide must overcome at least two major hurdles. These are the lack of helicity of the SBP1 peptide and its tendency to oligomerize in solution. Also, simple alterations of the sequence are unlikely to make this peptide change these fundamental attributes. Indeed, in recent reports (39) yet to be peer-reviewed, the lack of helicity of SBP1 has been tackled using chemical staples. Nevertheless, the binding affinity of such modified peptides to the spike remains low. Our results suggest that this may be due to the oligomerization of the peptide, which has not been accounted for in all such approaches so far. If the goal is to design a short SBP1-based peptide as a spike protein binder, one must stabilize both the helical secondary structure and the monomeric state of the peptide.
Author contributions
S.M. designed the research with help from R.V. A. Das synthesized the peptides and performed the circular dichroism and dynamic light scattering experiments. V.V. performed the fluorescence correlation spectroscopy experiments. A. Dey, S.D., and A.G. performed the single-molecule photobleaching measurements. A. Dey analyzed the single-molecule photobleaching data. D.S.R. performed the photon-counting histogram analysis of the FCS data. S.K. made the clone, and S.Y. prepared the protein (receptor-binding domain). M.D. and K.K.V. carried out computational studies and analysis. All the authors contributed to writing and editing the manuscript.
Acknowledgments
We thank Gitanjali A. Dhotre, Ankona Datta, and Rajasree Kundu for their help in characterizing the peptides. We thank U. S. Sandra, Ullas Kolthur-Seetharam, and Vinothkumar Kutti Ragunath for helping us at various stages of this project.
S.Y. is supported by National Centre for Biological Sciences-Institute of Stem Cell Science and Regenerative Medicine graduate program fellowship and S.K. is supported by inStem postdoctoral fellowship. S.M. acknowledges support from the Department of Atomic Energy, Government of India, provided under project number RTI4003. This work was supported by the Department of Atomic Energy, Government of India, provided under project number RTI4003.
Editor: Jochen Mueller.
Footnotes
Anirban Das and Vicky Vishvakarma contributed equally to this work.
Supporting material can be found online at https://doi.org/10.1016/j.bpj.2021.06.017.
Supporting citations
References 40, 41, 42, 43, 44, 45, 46, 47 appear in the Supporting materials and methods
Supporting material
References
- 1.Zhou P., Yang X.-L., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., Kleine-Weber H., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H., Penninger J.M., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46:586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan R., Zhang Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Walls A.C., Park Y.-J., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wrapp D., Wang N., McLellan J.S. Cryo-EM structure of the 2019-NCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lan J., Ge J., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 8.Shang J., Ye G., Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang G., Pomplun S., Pentelute B.L. The first-in-class peptide binder to the SARS-CoV-2 spike protein. bioRxiv. 2020 doi: 10.1101/2020.03.19.999318. [DOI] [Google Scholar]
- 10.Whisenant J., Burgess K. Blocking coronavirus 19 infection via the SARS-CoV-2 spike protein: initial steps. ACS Med. Chem. Lett. 2020;11:1076–1078. doi: 10.1021/acsmedchemlett.0c00233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao L., Goreshnik I., Baker D. De novo design of picomolar SARS-CoV-2 miniprotein inhibitors. Science. 2020;370:426–431. doi: 10.1126/science.abd9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen-Dvashi H., Weinstein J., Diskin R. Coronacept – a potent immunoadhesin against SARS-CoV-2. bioRxiv. 2020 doi: 10.1101/2020.08.12.247940. [DOI] [Google Scholar]
- 13.Zhang G., Pomplun S., Pentelute B.L. Investigation of ACE2 N-terminal fragments binding to SARS-CoV-2 spike RBD. bioRxiv. 2020 doi: 10.1101/2020.03.19.999318. [DOI] [Google Scholar]
- 14.Pace C.N., Scholtz J.M. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 1998;75:422–427. doi: 10.1016/s0006-3495(98)77529-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Malladi S.K., Singh R., Varadarajan R. Design of a highly thermotolerant, immunogenic SARS-CoV-2 spike fragment. J. Biol. Chem. 2020;296 doi: 10.1074/jbc.RA120.016284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentili M., Kowal J., Manel N. Transmission of innate immune signaling by packaging of CGAMP in viral particles. Science. 2015;349:1232–1236. doi: 10.1126/science.aab3628. [DOI] [PubMed] [Google Scholar]
- 17.Sengupta P., Balaji J., Maiti S. Measuring diffusion in cell membranes by fluorescence correlation spectroscopy. Methods. 2002;27:374–387. doi: 10.1016/s1046-2023(02)00096-8. [DOI] [PubMed] [Google Scholar]
- 18.Abhyankar R., Sahoo B., Maiti S. Amyloid diagnostics: probing protein aggregation and conformation with ultrasensitive fluorescence detection. Proc. SPIE. 2012;8233 [Google Scholar]
- 19.Wohland T., Maiti S., Macháň R. IOP Publishing; Bristol, United Kingdom: 2020. An Introduction to Fluorescence Correlation Spectroscopy. [Google Scholar]
- 20.Gendron P.-O., Avaltroni F., Wilkinson K.J. Diffusion coefficients of several rhodamine derivatives as determined by pulsed field gradient-nuclear magnetic resonance and fluorescence correlation spectroscopy. J. Fluoresc. 2008;18:1093–1101. doi: 10.1007/s10895-008-0357-7. [DOI] [PubMed] [Google Scholar]
- 21.Micsonai A., Wien F., Kardos J. Accurate secondary structure prediction and fold recognition for circular dichroism spectroscopy. Proc. Natl. Acad. Sci. U S A. 2015;112:E3095–E3103. doi: 10.1073/pnas.1500851112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Micsonai A., Wien F., Kardos J. BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res. 2018;46:W315–W322. doi: 10.1093/nar/gky497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ying L., Xie X.S. Fluorescence spectroscopy, exciton dynamics, and photochemistry of single allophycocyanin trimers. J. Phys. Chem. B. 1998;102:10399–10409. [Google Scholar]
- 24.Gordon M.P., Ha T., Selvin P.R. Single-molecule high-resolution imaging with photobleaching. Proc. Natl. Acad. Sci. U S A. 2004;101:6462–6465. doi: 10.1073/pnas.0401638101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H., Guo P. Single molecule photobleaching (SMPB) technology for counting of RNA, DNA, protein and other molecules in nanoparticles and biological complexes by TIRF instrumentation. Methods. 2014;67:169–176. doi: 10.1016/j.ymeth.2014.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dey S., Maiti S. Single-molecule photobleaching: instrumentation and applications. J. Biosci. 2018;43:447–454. [PubMed] [Google Scholar]
- 27.Shu D., Zhang H., Guo P. Counting of six pRNAs of phi29 DNA-packaging motor with customized single-molecule dual-view system. EMBO J. 2007;26:527–537. doi: 10.1038/sj.emboj.7601506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding H., Wong P.T., Steel D.G. Determination of the oligomer size of amyloidogenic protein beta-amyloid(1-40) by single-molecule spectroscopy. Biophys. J. 2009;97:912–921. doi: 10.1016/j.bpj.2009.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakajo K., Ulbrich M.H., Isacoff E.Y. Stoichiometry of the KCNQ1 - KCNE1 ion channel complex. Proc. Natl. Acad. Sci. U S A. 2010;107:18862–18867. doi: 10.1073/pnas.1010354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zijlstra N., Blum C., Subramaniam V. Molecular composition of sub-stoichiometrically labeled α-synuclein oligomers determined by single-molecule photobleaching. Angew. Chem. Int. Ed. Engl. 2012;51:8821–8824. doi: 10.1002/anie.201200813. [DOI] [PubMed] [Google Scholar]
- 31.Schindelin J., Arganda-Carreras I., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tinevez J.-Y., Perry N., Eliceiri K.W. TrackMate: an open and extensible platform for single-particle tracking. Methods. 2017;115:80–90. doi: 10.1016/j.ymeth.2016.09.016. [DOI] [PubMed] [Google Scholar]
- 33.Shiraki K., Nishikawa K., Goto Y. Trifluoroethanol-induced stabilization of the α-helical structure of β-lactoglobulin: implication for non-hierarchical protein folding. J. Mol. Biol. 1995;245:180–194. doi: 10.1006/jmbi.1994.0015. [DOI] [PubMed] [Google Scholar]
- 34.Cammers-Goodwin A., Allen T.J., Kemp D.S. Mechanism of stabilization of helical conformations of polypeptides by water containing trifluoroethanol. J. Am. Chem. Soc. 1996;118:3082–3090. [Google Scholar]
- 35.Luo P., Baldwin R.L. Mechanism of helix induction by trifluoroethanol: a framework for extrapolating the helix-forming properties of peptides from trifluoroethanol/water mixtures back to water. Biochemistry. 1997;36:8413–8421. doi: 10.1021/bi9707133. [DOI] [PubMed] [Google Scholar]
- 36.Roccatano D., Colombo G., Mark A.E. Mechanism by which 2,2,2-Trifluoroethanol/water mixtures stabilize secondary-structure formation in peptides: a molecular dynamics study. Proc. Natl. Acad. Sci. U S A. 2002;99:12179–12184. doi: 10.1073/pnas.182199699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen Y., Müller J.D., Gratton E. The photon counting histogram in fluorescence fluctuation spectroscopy. Biophys. J. 1999;77:553–567. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dey S., Das A., Maiti S. Correction of systematic bias in single molecule photobleaching measurements. Biophys. J. 2020;118:1101–1108. doi: 10.1016/j.bpj.2019.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curreli F., Victor S.M.B., Debnath A.K. Stapled peptides based on human angiotensin-converting enzyme 2 (ACE2) potently inhibit SARS-CoV-2 infection in vitro. bioRxiv. 2020 doi: 10.1101/2020.08.25.266437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cantor C.R., Schimmel P.R. W. H. Freeman; New York: 1980. Biophysical Chemistry Part III: The behavior of biological macromolecules, [Google Scholar]
- 41.Wilkins D.K., Grimshaw S.B., Smith L.J. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- 42.Dominguez C., Boelens R., Bonvin A.M.J.J. HADDOCK: A protein−protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 43.van Zundert G.C.P., Rodrigues J.P., Bonvin A.M.J.J. The HADDOCK2.2 Web Server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 44.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 45.Jo S., Kim T., Im W. CHARMM-GUI: A Web-Based Graphical User Interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 46.Phillips J.C., Hardy D.J., Tajkhorshid E. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020;153:44130. doi: 10.1063/5.0014475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J., MacKerell Jr A.D. CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.