Abstract
Adenium obesum (Forssk.) Roem. & Schult. belonging to the family Apocynaceae, is remarkable for its horticultural and ornamental values, poisonous nature, and medicinal uses. In order to have understanding of cp genome characterization of highly valued medicinal plant, and the evolutionary and systematic relationships, the complete plastome / chloroplast (cp) genome of A. obesum was sequenced. The assembled cp genome of A. obesum was found to be 154,437 bp, with an overall GC content of 38.1%. A total of 127 unique coding genes were annotated including 96 protein-coding genes, 28 tRNA genes, and 3 rRNA genes. The repeat structures were found to comprise of only mononucleotide repeats. The SSR loci are compososed of only A/T bases. The phylogenetic analysis of cp genomes revealed its proximity with Nerium oleander.
Keywords: Chloroplast genome, Plastome, Poisonous plant, Medicinal plant, Adenium obesum, Apocynaceae
1. Introduction
Adenium obesum (Forssk.) Roem. & Schult. (family Apocynaceae), the ‘Desert Rose’ is a poisonous, medicinal plant, distributed from Africa to Arabia, and is used traditoinally in the treatment of various ailments e.g. skin diseases, wounds, muscle pain, joint pain, venereal diseases, tooth decay, septic wounds, and nasal infections (Dimmitt and Hanson, 2002, Mouza and Hossain, 2015, Hossain et al., 2017). It is also used as a pesticide (Versiani et al., 2014), arrow poison for hunting in Africa (Oyen, 2008) and fish toxin (Wiseman, 2009). The A. obesum plant extract reported to possess cytotoxic (Almehdar et al., 2012), antimicrobial (Hossain et al., 2017) and anti-influenza (Kiyohara et al., 2012) activities. The phytochemical compounds identified from A. obesum include cardiac glycosides (cardenolides), pregnanes, triterpenes, flavonoids, and acetyldigitoxigenin (Versiani et al., 2014). The molecular docking of acetyldigitoxigenin elucidates the plausible mechanisms underlying the anticancer properties (Gurung et al., 2020).
The recent development in plastome or chloroplast (cp) genomics due to massive progress in the next-generation sequencing (NGS) platforms (Eid et al., 2009, Rothberg et al., 2011, Pattnaik et al., 2014, Jain et al., 2016, Shendure et al., 2017) and bioinformatics tools (Mavromatis et al., 2007, Knudsen et al., 2010, Huang et al., 2012, McElroy et al., 2012, Shendure and Aiden, 2012, Yang and Rannala, 2012, Caboche et al., 2014, Shcherbina, 2014, Kwon et al., 2015, Langmead and Nellore, 2018) have greatly impact on biotechnology application (Spök et al., 2008, Zhang et al., 2015, Daniell et al., 2016). We herein for the first time report the cp genome characterization of highly valued medicinal plant A. obesum, and discuss its structure including gene content, repeat organization, and phylogeny.
2. Materials and methods
2.1. DNA sequencing, assembly and annotation
The fresh leaves of A. obesum were collected from the wild condition of desert habitat near to Riyadh, Saudi Arabia. The total genomic DNA was isolated using QIAGEN DNeasy DNA extraction kit. The de novo sequencing base calling was performed using the Illumina Pipeline 1.3.2 (Nie et al., 2012). The raw reads were filtered using FastQC to obtain the high-quality clean data by removing adaptor sequences using trimmomatic and low-quality reads with Q-value ≤ 20. The filtered reads were assembled using Spades (Bankevich et al., 2012), and annotated using GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Tillich et al., 2017, Hansen et al., 2007). Further downstream analysis from the assembled cp genome included the repeat structure (Benson, 1999, Timme et al., 2007) and small inversion (Nagano et al., 1991, Yang et al., 2010, Doorduin et al., 2011, Castro et al., 2013, Beier et al., 2017).
2.2. Comparison of cp genome and phylogenetic analysis
The cp genome of A. obesum were plotted using the mVISTA (http://genome.lbl.gov/vista/mvista/submit.shtml) program with a total number of nine complete cp genomes of Apocynaceae [i.e. (1) Asclepias nivea Forssk., (2) Carissa macrocarpa (Eckl.) A. DC., (3) Catharanthus roseus (L.) G. Don, (4) Cynanchum auriculatum Buch.-Ham. ex Wight, (5) Echites umbellatus Jacq., (6) Nerium oleander L., (7) Oncinotis tenuiloba Stapf, (8) Pentalinon luteum (L.) B.F. Hansen & Wunderlin, and (9) Rhazya stricta Decne.] (Table 1).
Table 1.
The ingroup and outgroup taxon with their classification and GenBank accession number included in the phylogenetic analyses. The GenBank accession number marked with * were included in the mVISTA alignment.
| Sl. No. | Taxon | Order | Family | Subfamily | Tribe | Subtribe | GenBank |
|---|---|---|---|---|---|---|---|
| Ingroup | |||||||
| 1. | Adenium obesum (Forssk.) Roem. & Schult. | Gentianales | Apocynaceae | Apocynoideae | Nerieae | Neriinae | MN765097* |
| 2. | Asclepias nivea Forssk. | Gentianales | Apocynaceae | Asclepiadoideae | Asclepiadeae | Asclepiadinae | NC_022431.1* |
| 3. | Cynanchum auriculatum Buch.-Ham. ex Wight | Gentianales | Apocynaceae | Asclepiadoideae | Asclepiadeae | Cynanchinae | NC_029460.1* |
| 4. | Carissa macrocarpa (Eckl.) A. DC. | Gentianales | Apocynaceae | Rauvolfioideae | Carisseae | NC_033354.1* | |
| 5. | Catharanthus roseus (L.) G. Don | Gentianales | Apocynaceae | Rauvolfioideae | Vinceae | Catharanthinae | NC_021423.1* |
| 6. | Rhazya stricta Decne. | Gentianales | Apocynaceae | Rauvolfioideae | Amsonieae | NC_024292.1* | |
| 7. | Echites umbellatus Jacq. | Gentianales | Apocynaceae | Apocynoideae | Echiteae | Echitinae | NC_025655.1* |
| 8. | Pentalinon luteum (L.) B.F. Hansen & Wunderlin | Gentianales | Apocynaceae | Apocynoideae | Echiteae | Pentalinoninae | NC_025658.1* |
| 9. | Nerium oleander L. | Gentianales | Apocynaceae | Apocynoideae | Nerieae | Neriinae | NC_025656.1* |
| 10. | Oncinotis tenuiloba Stapf | Gentianales | Apocynaceae | Apocynoideae | Baisseeae | NC_025657.1* | |
| 11. | Anethum graveolens L. | Apiales | Apiaceae | NC_029470.1 | |||
| 12. | Ilex delavayi Franch. | Aquifoliales | Aquifoliaceae | KX426470.1 | |||
| 13. | Helianthus annuus L. | Asterales | Apocynaceae | NC_007977.1 | |||
| 14. | Viburnum betulifolium Batalin | Dipsacales | Adoxaceae | NC_037951.1 | |||
| 15. | Eucommia ulmoides Oliv. | Garryales | Eucommiaceae | NC_037948.1 | |||
| 16. | Gentiana tibetica King ex Hook. f. | Gentianales | Gentianaceae | NC_025319.1 | |||
| 17. | Iodes cirrhosa Turcz. | Icacinales | Icacinaceae | NC_036254.1 | |||
| 18. | Premna microphylla Turcz. | Lamiales | Lamiaceae | NC_026291.1 | |||
| 19. | Iochroma australe Griseb. | Solanales | Solanaceae | NC_029833.1 | |||
| Outgroup | |||||||
| 20. | Cornus controversa Hemsl. | Cornales | Cornaceae | MG525004.1 | |||
The cp sequences of 48 genes [e.g. ATP synthase genes (atpA, atpB, atpE, atpF, atpH, and atpI), c-type cytochrome synthesis gene (ccsA), envelope membrane protein gene (cemA), Maturase gene (matK), cytochrome b6/f genes (petA, petB, petD, petG, and petN), Photosystem I genes (psaA, psaB, psaC, and psaJ), Photosystem II genes (psbA, psbC, psbE, psbH, psbI, psbJ, psbK, psbN, and psbT), Rubisco gene (rbcL), Large-subunit ribosomal protein genes (rpl14, rpl2, rpl20, rpl32, rpl33, and rpl36), RNA polymerase subunit genes (rpoB, rpoC1, and rpoC2), Small-subunit ribosomal protein genes (rps14, rps15, rps18, rps19, rps2, rps3, rps4, rps7, and rps8), Genes of unknown function (ycf3, and ycf4)] were retrieved from 19 ingroup taxon comprising 10 species of the family Apocynaceae, the representative of the family Apiaceae, Aquifoliaceae, Apocynaceae, Adoxaceae, Eucommiaceae, Gentianaceae, Icacinaceae, Lamiaceae, Solanaceae, and the outgroup from the family Cornaceae (Table 1), and aligned using Clustal X (Thompson et al., 1994), and the molecular phylogenetic analysis was performed by Maximum Evolution method (Rzhetsky and Nei, 1992) using in MEGA X (Kumar et al., 2018).
3. Results and discussion
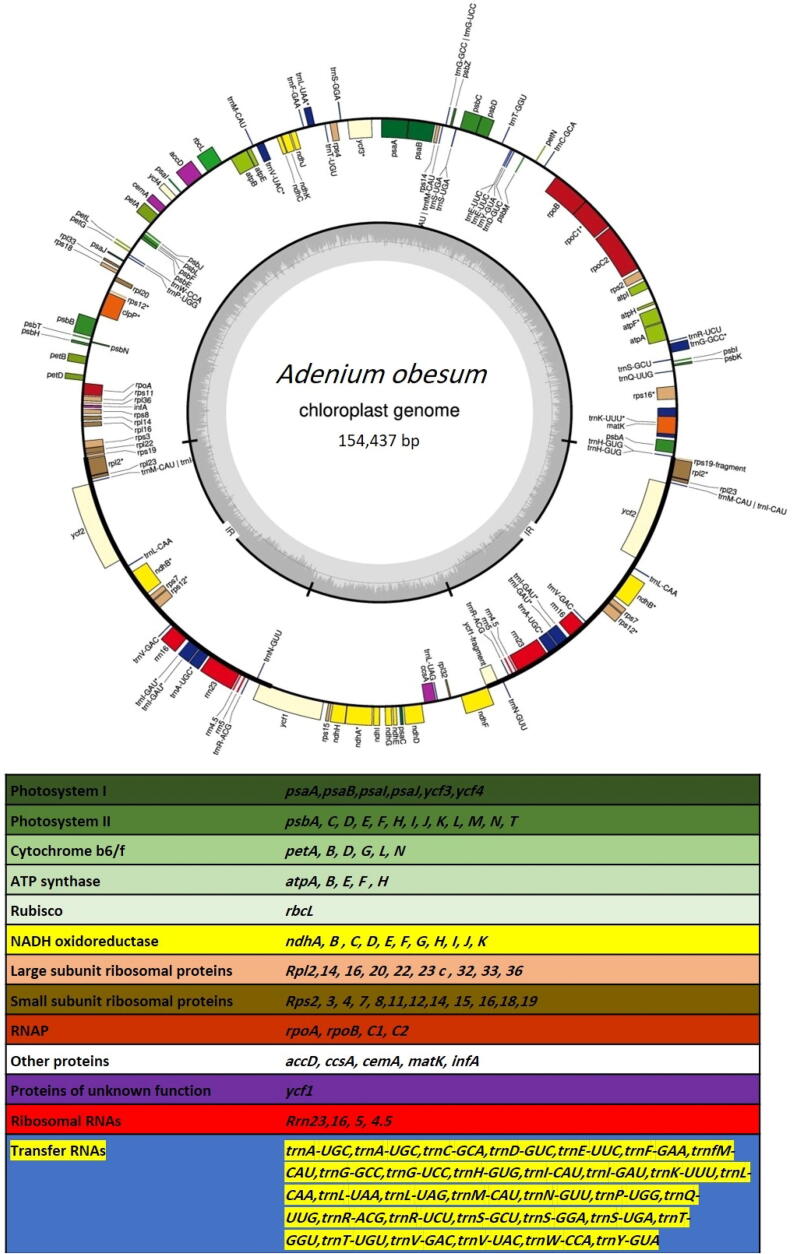
The present study reports assembly of the complete cp genome map as a conserved circular structure comprising a total length of 154,437 bp (including LSC, SSC, IRa, and IRb), with an overall GC content of 38.1% (Fig. 1). The results revealed the gene contents, orientation, and the conservation as well as polymorphisms were found in the chloroplast genome as similar to those of other cp genome of angiosperms (Daniell et al., 2016). A total numner of 127 genes were annotated including 96 protein-coding genes, 28 tRNA genes, and 3 rRNA genes (NCBI GenBank accession number: MN765097).
Fig. 1.
The gene map and genes contained in the cp genome of A. obesum.
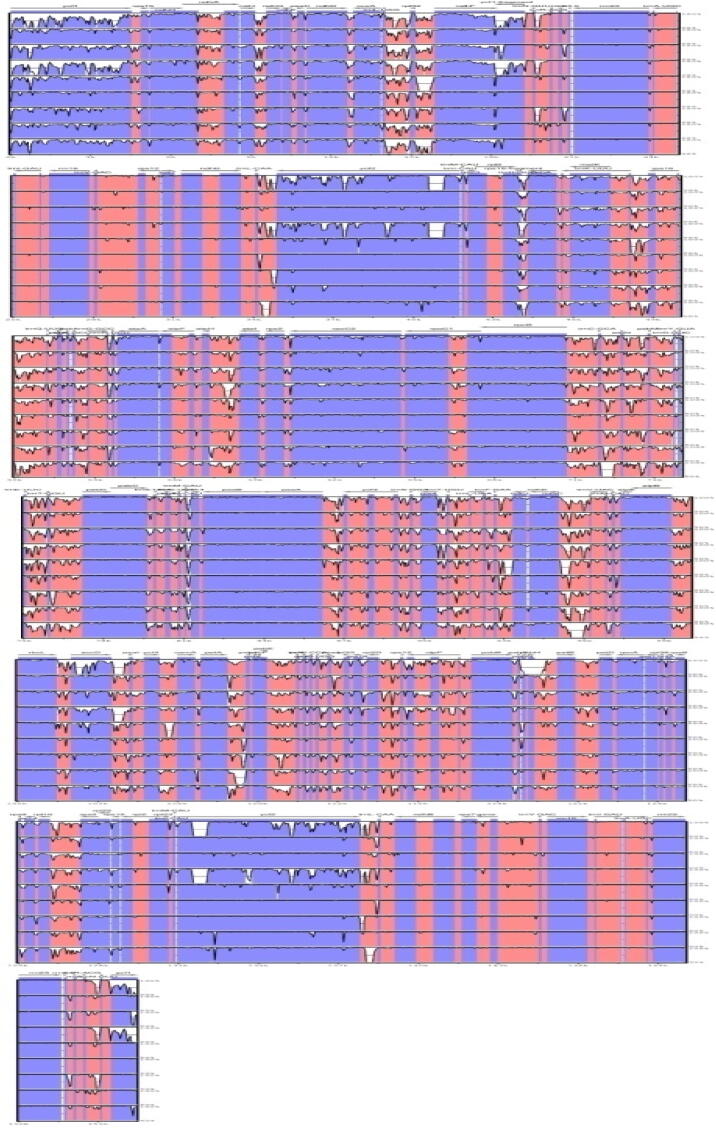
The sequence identity of A. obesum plotted with the nine different complete cp genomes from the family Apocynaceae e.g. A. nivea, C. macrocarpa, C. roseus, C. auriculatum, E. umbellatus, N. oleander, O. tenuiloba, P. luteum and R. stricta using the mVISTA revealed high similarities amongst them with few regions where the identities was below 90% (Fig. 2).
Fig. 2.
The percent identity plot for comparison of cp genome of A. obesum with the other Apocynaceae genomes. Lane from up to down: A. nivea, C. macrocarpa, C. roseus, C. auriculatum, E. umbellatus, N. oleander, O. tenuiloba, P. luteum, and R. stricta.
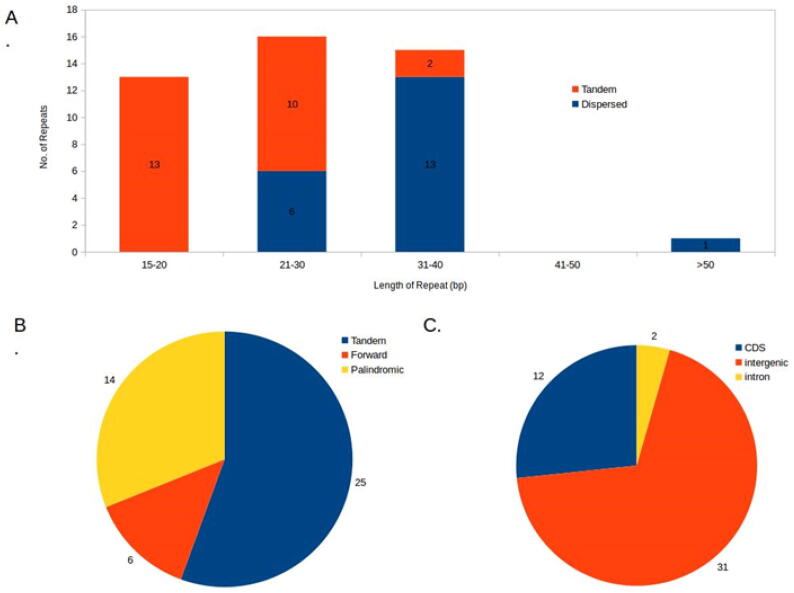
Moreover, the present study depicted the distribution and location of repeated structures and microsatellites in the cp genome. The microsatellites or simple sequence repeats (SSRs) are tandem repeats which ranges from 1 to 6 bp and are present commonly in cp genomes (Meng et al., 2018). SSRs have been served as an important marker for molecular characterization of plant species. A total of 40 SSRs were predicted in A. obesum (Table 2) which were composed of a length of at least 10 bp, all of which were found to be homopolymers containing multiple A or T nucleotides at each locus. These reveal that SSR loci are rich in A–T content in the A. obesum cp genome which supports previous chloroplast SSRs reports (Li et al., 2017). Among these SSRs, four SSRs were situated in coding regions and 31 were located in the intergenic regions (Table 2). A total number of 19 genes including 11 protein-coding genes and 8 tRNA genes contained one or two introns (Table 3). Furthermore, five SSRs were found in intronic regions. Thus, most of the repeats were situated in the intergenic region. Tandem and dispersed repeats were analyzed for A. obesum cp genomes and a total of 25 tandem and 19 dispersed repeats were observed (Fig. 3).
Table 2.
The SSR loci in the cp genome of Adenium obesum.
| Start | End | Repeat | Repeat length of consensus | Locus | Region | |
|---|---|---|---|---|---|---|
| 2109 | 2189 | (A)10 (A)12 |
81 | ycf1 | CDS | |
| 2914 | 2925 | (A)12 | 12 | ycf1 | CDS | |
| 9557 | 9566 | (T)10 | 10 | ndhI-ndhG | intergenic | |
| 13,831 | 13,840 | (A)10 | 10 | ccsA-trnL-UAG | intergenic | |
| 15,378 | 15,388 | (T)11 | 11 | rpl32-ndhF | intergenic | |
| 15,614 | 15,624 | (A)11 | 11 | rpl32-ndhF | intergenic | |
| 23,933 | 23,950 | (T)18 | 18 | rrn23-trnA-UGC | intergenic | |
| 43,878 | 43,887 | (A)10 | 10 | rpl2-rps19-fragment | intergenic | |
| 49,254 | 49,266 | (T)13 | 13 | rps16 | intron | |
| 52,132 | 52,141 | (A)10 | 10 | psbI-trnS-GCU | intergenic | |
| 53,347 | 53,356 | (T)10 | 10 | trnG-GCC | intron | |
| 53,607 | 53,620 | (T)14 | 14 | trnG-GCC-trnR-UCU | intergenic | |
| 53,763 | 53,775 | (T)13 | 13 | trnR-UCU-atpA | intergenic | |
| 55,426 | 55,435 | (A)10 | 10 | atpA-atpF | intergenic | |
| 56,129 | 56,139 | (T)11 | 11 | atpF | intron | |
| 57,865 | 57,874 | (T)10 | 10 | atpH-atpI | intergenic | |
| 58,082 | 58,093 | (T)12 | 12 | atpH-atpI | intergenic | |
| 60,138 | 60,147 | (A)10 | 10 | rps2-rpoC2 | intergenic | |
| 62,367 | 62,377 | (T)11 | 11 | rpoC2 | CDS | |
| 72,576 | 72,585 | (T)10 | 10 | trnC-GCA-petN | intergenic | |
| 79,736 | 79,747 | (T)12 | 12 | psbC-trnS-UGA | intergenic | |
| 88,261 | 88,272 | (T)12 | 12 | ycf3 | intron | |
| 95,456 | 95,465 | (T)10 | 10 | ndhC-trnV-UAC | intergenic | |
| 96,134 | 96,143 | (A)10 | 10 | ndhC-trnV-UAC | intergenic | |
| 97,206 | 97,257 | (T)12, (T)13 | 52 | trnM-CAU-atpE | intergenic | |
| 104,329 | 104,341 | (T)13 | 13 | psaI-ycf4 | intergenic | |
| 105,304 | 105,315 | (A)12 | 12 | ycf4-cemA | intergenic | |
| 105,629 | 105,642 | (T)14 | 14 | ycf4-cemA | intergenic | |
| 109,707 | 109,720 | (T)14 | 14 | psbE-petL | intergenic | |
| 109,940 | 109,949 | (A)10 | 10 | psbE-petL | intergenic | |
| 111,353 | 111,366 | (T)14 | 14 | trnP-UGG-psaJ | intergenic | |
| 113,000 | 113,009 | (A)10 | 10 | rps18-rpl20 | intergenic | |
| 115,212 | 115,222 | (T)11 | 11 | clpP | intron | |
| 120,988 | 120,997 | (A)10 | 10 | petB-petD | intergenic | |
| 122,446 | 122,455 | (A)10 | 10 | petD-rpoA | intergenic | |
| 125,125 | 125,136 | (T)12 | 12 | rps8-rpl14 | intergenic | |
| 125,645 | 125,710 | (A)10, (T)10 | 66 | rpl14-rpl16 | intergenic | |
| 128,414 | 128,424 | (T)11 | 11 | rpl22 | CDS | |
| 128,795 | 128,804 | (T)10 | 10 | rps19-rpl2 | intergenic | |
| 148,732 | 148,749 | (A)18 | 18 | trnA-UGC-rrn23 | intergenic |
Table 3.
The intron containing genes in the cp genome of Adenium obesum.
| Gene | Location | Exon I bp | Intron I bp | Exon II bp | Intron II bp | Exon III bp |
|---|---|---|---|---|---|---|
| trnA-UGC | IR | 35 | 818 | 38 | ||
| trnI-GAU | IR | 35 | 943 | 42 | ||
| rps12* | LSC-IR | 234 | 536 | 25 | 114 | |
| ndhB | IR | 777 | 684 | 756 | ||
| rpl2 | IR | 391 | 649 | 434 | ||
| trnK-UUU | LSC | 35 | 2476 | 37 | ||
| rps16 | LSC | 226 | 843 | 41 | ||
| trnG-GCC | LSC | 23 | 691 | 37 | ||
| atpF | LSC | 411 | 706 | 144 | ||
| rpoC1 | LSC | 1613 | 749 | 451 | ||
| ycf3 | LSC | 155 | 773 | 228 | 738 | 124 |
| trnL-UAA | LSC | 37 | 491 | 50 | ||
| trnV-UAC | LSC | 37 | 586 | 38 | ||
| clpP | LSC | 229 | 657 | 291 | 746 | 71 |
| rpl2 | IR | 434 | 649 | 391 | ||
| ndhB | IR | 756 | 684 | 777 | ||
| rps12 | IR | 25 | 536 | 234 | ||
| trnI-GAU | IR | 42 | 943 | 35 | ||
| trnA-UGC | IR | 38 | 818 | 35 |
*rps12 is trans-spliced gene with 5′ end exon located in the LSC region and the duplicated 3′ end exon located in IR regions.
Fig. 3.
(A-C). The repeat structure analysis in the cp genome of A. obesum. The cutoff value for tandem repeat is 15 bp and 30 bp for dispersed repeat. A. Frequency of repeats by length; B. Repeat type; C. The location distribution of all the repeats.
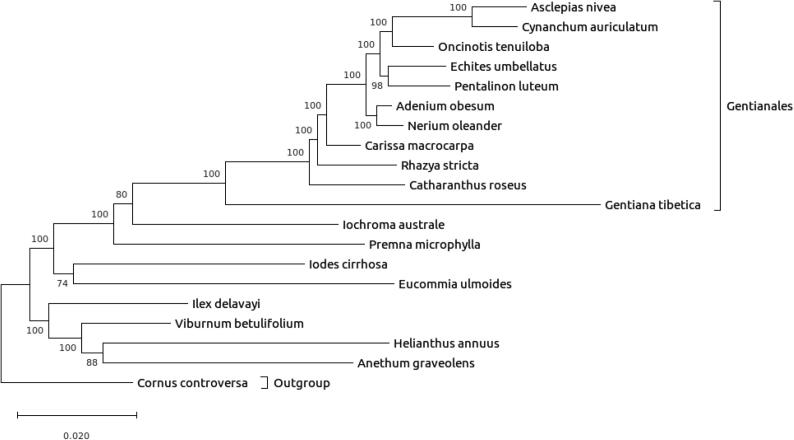
The phylogenetic relationships of a total number of 48 cp genes from the 19 cp genomes including the family Apocynaceae and the representative members of the family Apiaceae (Apiales), Aquifoliaceae (Aquifoliales), Adoxaceae (Dipsacales), Eucommiaceae (Garryales), Gentianaceae (Gentianales), Icacinaceae (Icacinales), Lamiaceae (Lamiales), Solanaceae (Solanales), and the outgroup at the family Cornaceae (Cornales) revealed the proximity of A. obesum (Subfamily Apocynoideae, Tribe Nerieae, Subtribe Neriinae) with Nerium oleander (Subfamily Apocynoideae, Tribe Nerieae, Subtribe Neriinae) (Fig. 4). The family Apocynaceae is one of the 10 largest angiosperm families with c. 4,500 species under c. 370 genera globally with the greatest diversity in the tropics and subtropics (Stevens, 2001, Endress et al., 2014, APG, 2016). Apart from the large number of molecular phylogenetic studies on the family Apocynaceae (e.g. Liede and Täuber, 2000, Liede and Täuber, 2002, Liede, 2001, Liede and Meve, 2001, Liede and Meve, 2002, Meve and Liede, 2001, Meve and Liede, 2002, Meve and Liede, 2004a, Meve and Liede, 2004b, Potgieter and Albert, 2001, Liede and Kunze, 2002, Liede et al., 2002a, Liede et al., 2002b, Verhoeven et al., 2003, Rapini et al., 2003, Rapini et al., 2004, Rapini et al., 2006, Rapini et al., 2007, Simões et al., 2004, Simões et al., 2006, Simões et al., 2007, Liede-Schumann et al., 2005, Venter et al., 2006, Endress et al., 2007, Goyder et al., 2007, Ionta and Judd, 2007, Lahaye et al., 2007, Livshultz et al., 2007, Meve and Liede-Schumann, 2007, Wanntorp and Forster, 2007), the family has also been intensely studied for their pollination biology, plant–herbivore interactions, and secondary chemistry (Wyatt and Broyles, 1994, Góngora Castillo et al., 2012, Courdavault et al., 2014, Agrawal et al., 2015). The phylogenetic nesting of the family Asclepiadaceae in Apocynaceae s.s. has been demonstrated repeatedly (Wanntorp, 1988, Judd et al., 1994, Sennblad and Bremer, 1996, Potgieter and Albert, 2001). The most recent classification of Apocynaceae (Endress et al., 2014) segregated the family into five subfamilies, two paraphyletic which correspond to the former Apocynaceae s.s. (Rauvolfioideae and Apocynoideae) and three monophyletic that relates to the former Asclepiadaceae (Periplocoideae, Secamonoideae, and Asclepiadoideae).
Fig. 4.
The maximum likelihood tree inferred from the cp genome of A. obesum analyzed together with the members of the family Apocynaceae and Aquifoliaceae, Adoxaceae, Eucommiaceae, Gentianaceae, Icacinaceae, Lamiaceae, and Solanaceae.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to extend their sincere appreciation to the Deanship of Scientific Research at King Saud University for its funding of the research through the research group project #RG-1438-015. This study was supported by the KRIBB Initiative Program of the Republic of Korea. JL thanks the support from the Chungnam National University, Daejeon, Republic of Korea.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Mohammad Ajmal Ali, Email: ajmalpdrc@gmail.com.
Arun Bahadur Gurung, Email: arunbgurung@gmail.com.
References
- Agrawal A.A., Ali J.G., Rasmann S., Fishbein M. Macroevolutionary trends in the defense of milkweeds against monarchs: latex, cardenolides, and tolerance of herbivory. In: Oberhauser K.S., Altizer S., Nail K., editors. Monarchs in a changing world: Biology and conservation of an iconic insect. Cornell University Press; Ithaca, NY, USA: 2015. pp. 47–59. [Google Scholar]
- Almehdar H., Abdallah H.M., Osman A.M., Abdel-Sattar E.A. In vitro cytotoxic screening of selected Saudi medicinal plants. J. Nat. Med. 2012;66:406–412. doi: 10.1007/s11418-011-0589-8. [DOI] [PubMed] [Google Scholar]
- APG, IV., 2016. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20.
- Bankevich A., Nurk S., Antipov D., Gurevich A., Dvorkin M., Kulikov A.S., Lesin V., Nikolenko S., Pham S., Prjibelski A., Pyshkin A., Sirotkin A., Vyahhi N., Tesler G., Alekseyev M.A., Pevzner P.A. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19(5):455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier S., Thiel T., Münch T., Scholz U., Mascher M. MISA-web: A web server for microsatellite prediction. Bioinform. 2017;33(16):2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caboche S., Audebert C., Lemoine Y., Hot D. Comparison of mapping algorithms used in high-throughput sequencing: application to Ion Torrent data. BMC Genom. 2014;15:264. doi: 10.1186/1471-2164-15-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro I., Pinto-Carnide O., Ortiz J.M., Martin J.P. Chloroplast genome diversity in Portuguese grapevine (Vitis vinifera L.) cultivars. Mol. Biotechnol. 2013;54(2):528–540. doi: 10.1007/s12033-012-9593-9. [DOI] [PubMed] [Google Scholar]
- Courdavault, V.N., Papon, M., Clastre, N., Giglioli Guivarc'h, B., St Pierre, V., Burlat., 2014. A look inside an alkaloid multisite plant: the Catharanthus logistics. Current Opinion in Plant Biol. 19, 43–50. [DOI] [PubMed]
- Daniell H., Lin C.S., Yu M., Chang W.J. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 2016;17(1):134. doi: 10.1186/s13059-016-1004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimmitt M.A., Hanson C. The genus Adenium in cultivation. Diplorhynchus Welw Fic & Hiern (Apocynaceae) Mededelingen. Cactus Success J. 2002;63:223–225. [Google Scholar]
- Doorduin L., Gravendeel B., Lammers Y., Ariyurek Y., Chin A.W.T., Vrieling K. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 2011;18:93–105. doi: 10.1093/dnares/dsr002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J., Fehr A., Gray J., Luong K., Lyle J., Otto G., Peluso P., Rank D., Baybayan P., Bettman B., Bibillo A., Bjornson K., Chaudhuri B., Christians F., Cicero R., Clark S., Dalal R., Dewinter A., Dixon J., Foquet M., Gaertner A., Hardenbol P., Heiner C., Hester K., Holden D., Kearns G., Kong X., Kuse R., Lacroix Y., Lin S., Lundquist P., Ma C., Marks P., Maxham M., Murphy D., Park I., Pham T., Phillips M., Roy J., Sebra R., Shen G., Sorenson J., Tomaney A., Travers K., Trulson M., Vieceli J., Wegener J., Wu D., Yang A., Zaccarin D., Zhao P., Zhong F., Korlach J., Turner S. Real-time DNA sequencing from single polymerase molecules. Science. 2009;323(5910):133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- Endress M.E., Liede-Schumann S., Meve U. An updated classification for Apocynaceae. Phytotaxa. 2014;159:175–194. [Google Scholar]
- Endress, M.E., van der Ham, R.W.J.M., Nilsson, S., Civeyrel, L., Chase, M.W., Sennblad, B., Potgieter, K., Joseph, J., Powell, M., Lorence, D., Zimmerman, Y.-M., Albert, V.A., 2007. A phylogenetic analysis of Alyxieae (Apocynaceae) based on rbcL, matK, trnL intron, trnL-F spacer sequences, and morphological characters. Ann. Miss. Bot. Gard. 94, 1–35.
- Góngora Castillo E., Childs K.L., Fedewa G., Hamilton J.P., Liscombe D.K., Magallanes Lundback M., Mandadi K.K., Nims E., Runguphan W., Vaillancourt B., Varbanova-Herde M., DellaPenna D., McKnight T.D., O’Connor S., Robin Buell C. Development of transcriptomic resources for interrogating the biosynthesis of monoterpene indole alkaloids in medicinal plant species. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0052506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyder, D.J., Nicholas, A., Liede-Schumann, S., 2007. Phylogenetic relationships in subtribe Asclepiadinae (Apocynaceae: Asclepiadoideae). Ann. Miss. Bot. Gard. 94, 423–434.
- Gurung A.B., Ali M.A., Lee J., Al-Hemaid F., Farah M.A., Al-Anazi K.M. Molecular docking elucidates the plausible mechanisms underlying the anticancer properties of acetyldigitoxigenin from Adenium obesum. Saudi J. Biological Sciences. 2020;27(7):1907–1911. doi: 10.1016/j.sjbs.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen D.R., Dastidar S.G., Cai Z., Penaflor C., Kuehl J.V., Boore J.L., Jansen R.K. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early-diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae) Mol. Phylogen. Evol. 2007;45:547–563. doi: 10.1016/j.ympev.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Hossain M.A., Akhtar S.M., Sadri S.A. Two new flavonoids from Adenium obesum grown in Oman. J. King Saud Univ. Sci. 2017;29:62–69. [Google Scholar]
- Huang W., Li L., Myers J.R., Marth G.T. ART: a next-generation sequencing read simulator. Bioinformatics. 2012;28:593–594. doi: 10.1093/bioinformatics/btr708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionta, G.M., Judd, W.S., 2007. Phylogenetic relationships in Periplocoideae (Apocynaceae s. l.) and insights into the origin of pollinia. Ann. Miss. Bot. Gard. 94, 360–375.
- Jain M., Olsen H.E., Paten B., Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17:239. doi: 10.1186/s13059-016-1103-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd W.S., Sanders R.W., Donoghue M.J. Angiosperm family pairs: preliminary phylogenetic analyses. Harvard Papers Bot. 1994;5:1–51. [Google Scholar]
- Kiyohara H., Ichino C., Kawamura Y., Nagai T., Sato N., Yamada H., Salama M.M., Abdel-Sattar E. In vitro anti-influenza virus activity of a cardiotonic glycoside from Adenium obesum (Forssk.) Phytomed. 2012;19:111–114. doi: 10.1016/j.phymed.2011.07.004. [DOI] [PubMed] [Google Scholar]
- Knudsen B., Forsberg R., Miyamoto M.M. A computer simulator for assessing different challenges and strategies of de novo sequence assembly. Genes. 2010;1(2):263–282. doi: 10.3390/genes1020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35(6):1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon T., Yoo W.G., Lee W.-J., Kim W., Kim D.-W. Next-generation sequencing data analysis on cloud computing. Genes Genom. 2015;37:489–501. [Google Scholar]
- Lahaye R., Klackenberg J., Källersjö M., van Campo E., Civeyrel L. Phylogenetic relationships between derived Apocynaceae s.l. and within Secamonoideae based on four chloroplast sequences. Ann. Miss. Bot. Gard. 2007;94:376–391. [Google Scholar]
- Langmead B., Nellore A. Cloud computing for genomic data analysis and collaboration. Nat. Rev. Genet. 2018;19(4):208–219. doi: 10.1038/nrg.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Lin F., Huang P., Guo W., Zheng Y. Complete chloroplast genome sequence of Decaisnea insignis: Genome organization, genomic resources and comparative analysis. Scientific Rep. 2017;7:10073. doi: 10.1038/s41598-017-10409-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liede S. Subtribe Astephaninae (Apocynaceae–Asclepiadoideae) reconsidered: new evidence based on cpDNA spacers. Ann. Miss. Bot. Gard. 2001;88:657–668. [Google Scholar]
- Liede S., Kunze H. Cynanchum and the Cynanchinae (Apocynaceae – Asclepiadoideae) – a molecular, anatomical and latex triterpenoid study. Organisms Diversity & Evol. 2002;2:239–269. [Google Scholar]
- Liede S., Meve U. New combinations and new names in Malagasy Asclepiadoideae (Apocynaceae) Adansonia, sér.3. 2001;23:347–351. [Google Scholar]
- Liede S., Meve U. Dissolution of Cynanchum sect. Macbridea (Apocynaceae – Asclepiadoideae) Nordic J. Bot. 2002;22:1–13. [Google Scholar]
- Liede S., Täuber A. Sarcostemma R. Br. (Apocynaceae–Asclepiadoideae) – a controversial generic circumscription reconsidered: evidence from trnL-F spacers. Plant Syst. Evol. 2000;225:133–140. [Google Scholar]
- Liede S., Täuber A. Circumscription of the genus Cynanchum (Apocynaceae Asclepiadoideae) Syst. Bot. 2002;27:789–800. [Google Scholar]
- Liede S., Meve U., Täuber A. What is the subtribe Glossonematinae (Apocynaceae: Asclepiadoideae)? A phylogenetic study based on cpDNA spacer. Bot. J. Linn. Soc. 2002;139:145–158. [Google Scholar]
- Liede S., Täuber A., Schneidt J. Molecular considerations on the Tylophorinae K. Schum. (Apocynaceae-Asclepiadoideae) Edinburgh J. Bot. 2002;59:377–403. [Google Scholar]
- Liede-Schumann S., Rapini A., Goyder D.J., Chase M.W. Phylogenetics of the New World subtribes of Asclepiadeae (Apocynaceae–Asclepiadoideae): Metastelmatinae, Oxypetalinae, and Gonolobinae. Syst. Bot. 2005;30:184–195. [Google Scholar]
- Livshultz T., Middleton D.J., Endress M.E., Williams. J.K. Phylogeny of Apocynoideae and the APSA clade (Apocynaceae s.l.) Ann. Miss. Bot. Gard. 2007;94:324–359. [Google Scholar]
- Mavromatis K., Ivanova N., Barry K., Shapiro H., Goltsman E., McHardy A.C., Rigoutsos I., Salamov A., Korzeniewski F., Land M., Lapidus A., Grigoriev I., Hugenholtz P., Kyrpides N.C. Use of simulated data sets to evaluate the fidelity of metagenomic processing methods. Natural Methods. 2007;4:495–500. doi: 10.1038/nmeth1043. [DOI] [PubMed] [Google Scholar]
- McElroy K.E., Luciani F., Thomas T. GemSIM: general, error-model based simulator of next-generation sequencing data. BMC Genom. 2012;13:74. doi: 10.1186/1471-2164-13-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J., Li X., Li H., Yang J., Wang H., He J. Comparative analysis of the complete chloroplast genomes of four Aconitum medicinal species. Molecules. 2018;23:1015. doi: 10.3390/molecules23051015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meve U., Liede S. Reconsideration of the status of Lavrania, Larryleachia and Notechidnopsis (Asclepiadoideae-Ceropegieae) S. African J. Bot. 2001;67:161–168. [Google Scholar]
- Meve U., Liede S. Floristic exchange between mainland Africa and Madagascar: A case study of Apocynaceae-Asclepiadoideae. J. Biogeo. 2002;29:865–873. [Google Scholar]
- Meve U., Liede S. Generic delimitations in tuberous Periplocoideae (Apocynaceae) from Africa and Madagascar. Annals Bot. 2004;93:407–414. doi: 10.1093/aob/mch057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meve U., Liede S. Subtribal division of Ceropegieae (Apocynaceae—Asclepiadoideae) Taxon. 2004;53:61–72. [Google Scholar]
- Meve U., Liede-Schumann S. Ceropegia (Apocynaceae, Ceropegieae, Stapeliinae): paraphyletic, but still taxonomically sound. Ann. Miss. Bot. Gard. 2007;94:392–406. [Google Scholar]
- Mouza K.G., Hossain M.A. Determination of total phenolics, flavonoids and antioxidant activity of root crude extracts of Adenium obesum traditionally used for the treatment of bone dislocations and rheumatism. Asian Pac. J. Trop. Dis. 2015;5(Suppl. 1):S155–S158. [Google Scholar]
- Nagano Y., Ishikawa H., Matsuno R., Sasaki Y. Nucleotide sequence and expression of the ribosomal protein L2 gene in pea chloroplasts. Plant Mol. Biol. 1991;17:541–545. doi: 10.1007/BF00040653. [DOI] [PubMed] [Google Scholar]
- Nie, X., Lv, S., Zhang, Y., Du, X., Wang, L., Biradar, S.S., Tan, X., Wan, F., Weining, S., 2012. Complete chloroplast genome sequence of a major invasive species, crofton weed (Ageratina adenophora). PLoS One 7: e36869. [DOI] [PMC free article] [PubMed]
- Oyen, L.P.A., 2008. Adenium obesum (Forssk.) Roem. & Schult. In: Schmelzer, G.H., Gurib-Fakim, A., (Eds.), Plant Resources of Tropical Africa, Backhuys. Wageningen. 11(1), Medicinal Plants 1.
- Pattnaik S., Gupta S., Rao A.A., Panda B. SInC: an accurate and fast error-model based simulator for SNPs, Indels and CNVs coupled with a read generator for short-read sequence data. BMC Bioinf. 2014;15:1–9. doi: 10.1186/1471-2105-15-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potgieter K., Albert V.A. Phylogenetic relationships within Apocynaceae s.l. based on trnL intron and trnL-F spacer sequences and propagule characters. Ann. Miss. Bot. Gard. 2001;88:523–549. [Google Scholar]
- Rapini A., Chase M.W., Goyder D.J., Griffiths J. Asclepiadeae classification: evaluating the phylogenetic relationships of New World Asclepiadoideae (Apocynaceae) Taxon. 2003;52:33–50. [Google Scholar]
- Rapini A., Fontella-Pereira J., Lamare E., Liede-Schumann S. Taxonomy of Peplonia (including Gonioanthela) and a reinterpretation of Orthosieae (Asclepiadoideae, Apocynaceae) Kew Bull. 2004;59:531–539. [Google Scholar]
- Rapini A., Chase M.W., Konno T.U.P. Phylogenetics of South American Asclepiadeae (Apocynaceae) Taxon. 2006;55:119–124. [Google Scholar]
- Rapini A., van den Berg C., Liede-Schumann S. Diversification of Asclepiadoideae (Apocynaceae) in the New World. Ann. Miss. Bot. Gard. 2007;94:407–422. [Google Scholar]
- Rothberg J.M., Hinz W., Rearick T.M., Schultz J., Mileski W., Davey M., Leamon J.H., Johnson K., Milgrew M.J., Edwards M., Hoon J., Simons J.F., Marran D., Myers J.W., Davidson J.F., Branting A., Nobile J.R., Puc B.P., Light D., Clark T.A., Huber M., Branciforte J.T., Stoner I.B., Cawley S.E., Lyons M., Fu Y., Homer N., Sedova M., Miao X., Reed B., Sabina J., Feierstein E., Schorn M., Alanjary M., Dimalanta E., Dressman D., Kasinskas R., Sokolsky T., Fidanza J.A., Namsaraev E., McKernan K.J., Williams A., Roth G.T., Bustillo J. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475:348–352. doi: 10.1038/nature10242. [DOI] [PubMed] [Google Scholar]
- Rzhetsky A., Nei M. A simple method for estimating and testing minimum evolution trees. Mol. Biol. Evol. 1992;9:945–967. [Google Scholar]
- Sennblad B., Bremer B. The familial and subfamilial relationships of Apocynaceae and Asclepiadaceae evaluated with rbcL data. Plant Syst. Evol. 1996;202:153–175. [Google Scholar]
- Shcherbina A. FASTQSim: platform-independent data characterization and in silico read generation for NGS datasets. BMC Res. Notes. 2014;7:533. doi: 10.1186/1756-0500-7-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., Aiden E.L. The expanding scope of DNA sequencing. Nature Biotech. 2012;30:1084–1994. doi: 10.1038/nbt.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shendure J., Balasubramanian S., Church G.M., Gilbert W., Rogers J., Schloss J.A., Waterston R.H. DNA sequencing at 40: Past, present and future. Nature. 2017;550:345–353. doi: 10.1038/nature24286. [DOI] [PubMed] [Google Scholar]
- Simões A.O., Endress M.E., van der Niet T., Conti E., Kinoshita L.S. Tribal and intergeneric relationships of Mesechiteae (Apocynoideae, Apocynaceae): evidence from three noncoding plastid DNA regions and morphology. American J. Bot. 2004;91:1409–1418. doi: 10.3732/ajb.91.9.1409. [DOI] [PubMed] [Google Scholar]
- Simões A.O., Endress M.E., van der Niet T., Kinoshita L.S., Conti E. Is Mandevilla (Apocynaceae, Mesechiteae) monophyletic? Evidence from five plastid DNA loci and morphology. American J. Bot. 2006;94:1409–1418. doi: 10.3732/ajb.91.9.1409. [DOI] [PubMed] [Google Scholar]
- Simões A.O., Livshultz T., Conti E., Endress M.E. Phylogeny and systematics of the Rauvolfioideae (Apocynaceae) based on molecular and morphological evidence. Ann. Miss. Bot. Gard. 2007;94:268–297. [Google Scholar]
- Spök A., Karner S., Stein A.J., Rodríguez C.E. Plant molecular farming; opportunities and challenges. JRC Scientific Technical Reports. 2008 [Google Scholar]
- Stevens, P.F., 2001 onwards. Angiosperm phylogeny website, version 12. http://www.mobot.org/MOBOT/research/APweb/.
- Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillich M., Lehwark P., Pellizzer T., Ulbricht-Jones E.S., Fischer A., Bock R., Greiner S. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme R.E., Kuehl J.V., Boore J.L., Jansen R.K. A comparative analysis of the Lactuca and Helianthus (Asteraceae) plastid genomes: Identification of divergent regions and categorization of shared repeats. American J. Bot. 2007;94:302–312. doi: 10.3732/ajb.94.3.302. [DOI] [PubMed] [Google Scholar]
- Venter H.J.T., Dold A.P., Verhoeven R.L., Ionta G. Kappia lobulata (Apocynaceae, Periplocoideae), a new genus from South Africa. South African J. Bot. 2006;72:529–533. [Google Scholar]
- Verhoeven R.L., Liede S., Endress M.E. The tribal position of Fockea and Cibirhiza (Apocynaceae: Asclepiadoideae): evidence from pollinium structure and cpDNA sequence data. Grana. 2003;42:70–81. [Google Scholar]
- Versiani M.A., Ahmed S.K., Ikram A., Ali S.T., Yasmeen K., Faizi S. Chemical constituents and biological activities of Adenium obesum (Forsk.) Roem. et Schult. Chem Biodivers. 2014;11(2):171–180. doi: 10.1002/cbdv.201200254. [DOI] [PubMed] [Google Scholar]
- Wanntorp H.E. The genus Microloma (Asclepiadaceae) Opera Botanica. 1988;98:1–69. [Google Scholar]
- Wanntorp L., Forster P.I. Phylogenetic relationships between Hoya and the monotypic genera Madangia, Absolmsia, and Micholitzia (Apocynaceae, Marsdenieae): Insights from flower morphology. Ann. Miss. Bot. Gard. 2007;94:36–55. [Google Scholar]
- Wiseman, J., 2009. SAS Survival Handbook (Revised Edition). William Morrow Paperbacks, pp. 240.
- Wyatt, R., Broyles., S.B., 1994. Ecology and evolution of reproduction in milkweeds. Ann. Rev. Ecol. & Syst. 25, 423– 441.
- Yang, M., Zhang, X., Liu, G., Yin, Y., Chen, K., Yun, Q., Zhao, D., Al-Mssallem, I.S., Yu, J., 2010. The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS One 5: e12762. [DOI] [PMC free article] [PubMed]
- Yang Z., Rannala B. Molecular phylogenetics: Principles and practice. Nat. Rev. Genet. 2012;13(5):303–314. doi: 10.1038/nrg3186. [DOI] [PubMed] [Google Scholar]
- Zhang J., Khan S.A., Hasse C., Ruf S., Heckel D.G., Bock R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science. 2015;347:991–994. doi: 10.1126/science.1261680. [DOI] [PubMed] [Google Scholar]