Abstract
Bacillus anthracis is a gram positive, deadly spore forming bacteria causing anthrax and these bacteria having the complex mechanism in the cell wall envelope, which can adopt the changes in environmental conditions. In this, the membrane bound cell wall proteins are said to progressive drug target for the inhibition of Bacillus anthracis. Among the cell wall proteins, the SrtA is one of the important mechanistic protein, which mediate the ligation with LPXTG motif by forming the amide bonds. The SrtA plays the vital role in cell signalling, cell wall formation, and biofilm formations. Inhibition of SrtA leads to rupture of the cell wall and biofilm formation, and that leads to inhibition of Bacillus anthracis and thus, SrtA is core important enzyme to study the inhibition mechanism. In this study, we have examined 28 compounds, which have the inhibitory activity against the Bacillus anthracis SrtA for developing the 3D-QSAR and also, compounds binding selectivity with both open and closed SrtA conformations, obtained from 100 ns of MD simulations. The binding site loop deviate in forming the open and closed gate mechanism is investigated to understand the inhibitory profile of reported compounds, and results show the closed state active site conformations are required for ligand binding specificity. Overall, the present study may offer an opportunity for better understanding of the mechanism of action and can be aided to further designing of a novel and highly potent SrtA inhibitors.
Keywords: Bacillus anthracis, Cell adhesion, Molecular dynamics, Pharmacophore modelling, Sortase, QSAR
1. Introduction
Bacillus anthracis (B. anthracis) is a spore forming bacteria and it is a Gram-positive pathogenic microbe causing the highly resilient and deadly disease called anthrax (Spencer, 2003). It causes the destructive infection in common cattle’s and sometimes the infections are seen in humans and due to its destructive infection feature, it has been used for bioweapons (Hadler, 2007). The main advantages of this bacterium are, adjusting the environmental conditions, in both favourable or unfavourable conditions and survive (Haruta and Kanno, 2015). In unfavourable conditions, the bacterium forms the dormant spore and these spores are living long last in the contaminated soil, and on favourable condition, the bacterium survives back (Swick et al., 2016). In this connection, the cell surface proteins are imperial in this mechanistic function, as they play the role in spore formations, biofilm formations, and also in the host-pathogen interactions (Blanchette and Orihuela, 2012). So that, most of the pathogenic microbes are targeted for cell surface proteins, and here gram-negative bacteria holds the surface protein in the outer layer, and gram-positive bacteria use the thick cell membrane to anchor the cell surface proteins (Navarre and Schneewind, 1999, Silhavy et al., 2010). This surface proteins are arranged in the ornamental arrangement display through the mode of covalent or non-covalent interactions with peptidoglycan layer or lipid membrane (teichoic acid or lipoteichoic acid) (Brown et al., 2013).
Among this cell wall anchoring proteins, the sortase proteins are well known in sorting signals and also anchors the protein in to the peptidoglycan layer (Marraffini et al., 2006). These sortase are classified into A-F and each proteins function to play the mechanistic function and among that, Sortase A (SrtA) is the core important protein in anchoring the cell membrane (Selvaraj et al., 2018a). SrtA are also called as housekeeping genes, that mainly function to link the surface proteins in to the bacterial cell wall through amide bond mediated transpeptidation reaction (Spirig et al., 2011). Recent evidences suggest that, inhibition of SrtA gene leads to inhibition of microbes by the lack of pathogenicity and cell wall formations and called universal drug target of all Gram-positive pathogens (Marraffini et al., 2006, Spirig et al., 2011). Thus, most of the researchers are focused on targeting the SrtA for the inhibition of the microbes, and here the SrtA in B. anthracis have the special feature of active site loop (β7/β8), plays the anchoring role that undergoes the transformation of disorder to order or conformational changes upon sorting signal results in recognition of lipid II (Bentley et al., 2007, Frankel et al., 2005). This β7/β8 is a flexible loop function as the active site and holds the multiple conformations in recognition of lipid II (Chan et al., 2015, Jacobitz et al., 2017). In this region, the hydrophobic residues Val168 and Lue169 are positioned in β6-β7 loop for initiating the hydrophobic interactions with LPXTG peptides N-terminal domain (Weiner et al., 2010). In the β6-β7 loop, the Val168 and Lue169 are the hydrophobic residues and the Pro168, Asp169, Lys170 and Trp171 are function as catalytic residues (Chan et al., 2015).
In this study, we have examined the β7/β8 active site loop, structural and conformational changes for its recognition of SrtA inhibitors. For understanding and reporting the binding selectivity of SrtA inhibitors, we have considered the reported rhodanine, pyridazinone and pyrazolethione derivatives from Suree et al., 2009 having the anti-bacterial activity against the B. anthracis (Suree et al., 2009). Suree et al., 2009 has performed the SAR studies in providing the anti-bacterial activity against the B. anthracis and reported the activity in micromolar (µM) range. Among these inhibitors reported by Suree et al., 2009, 28 synthetic compounds are having the potential activity against the B. anthracis and said to have strong activity than the available inhibitors (Suree et al., 2009). We have taken the activity of the reported 28 molecules and executed the 3D-QSAR model for understanding the statistical model in both active and inactive ligands. In addition, we have also performed the 100 ns molecular dynamics study to understand the conformational changes in the β7/β8 active site loop and its binding specificity of rhodanine, pyridazinone and pyrazolethione derivatives in different conformations. Overall, this study will provide the detail insights on analysing the binding specificity of β7/β8 loop conformation of SrtA with respect to the rhodanine, pyridazinone and pyrazolethione derivatives, and this study will provide the base for discovering of novel lead molecules for further development.
2. Materials and methods
2.1. Data set preparation
The dataset for this work is taken from Suree et al. (2009) and the compounds having the activity against the Staphylococcus aureus is eliminated and only the compounds having the activity against the B. anthracis is considered for this work (Suree et al., 2009). Based on these criteria, only 28 compounds are taken for the study and these rhodanine, pyridazinone and pyrazolethione derivatives are prepared by using LigPrep/Confgen with OPLS-AA force field (Chen and Foloppe, 2010, Watts et al., 2010). The bioactivity in form of IC50 values are converted into pIC50 using the formula (pIC50 = -log IC50), and for accuracy in prediction of micromolar and nanomolar conversion, we have used the pIC50 calculator (https://www.sanjeevslab.org/tools.html). From the 28 molecules, 21 training set and 7 test set compounds in order to maintain the 3:1 ratio (Selvaraj, 2011). For activity threshold, the active molecules are set to have the pIC50 of 5.9 and inactive molecules are set to have the pIC50 of 4.3 (Raskevicius and Kairys, 2017). Based on this, the eight active molecules, five inactive molecules, and other in middle state molecules are subject to pharmacophore modelling (Chaudhari and Pahelkar, 2019). The protein considered for this study is the solution structure of NMR derived Ba-SrtA (PDB ID: 2KW8) prepared with the protein preparation wizard (Sastry et al., 2013, Weiner et al., 2010).
2.2. Pharmacophore modelling and 3D QSAR
Based on the active molecules, and up to seven common pharmacophores is generated using the PHASE module from Schrodinger and based on the fitness value, best pharmacophore is selected and reported (Agrawal et al., 2013, Suryanarayanan et al., 2013, Vijayalakshmi et al., 2014). Further, the common pharmacophore is evaluated, visualized and 3D-QSAR model is initiated with PHASE module from Schrodinger (Reddy et al., 2012). For the 3D-QSAR, the 28 molecules are sub-categorized in to 7 test molecules and 21 training molecules for correlating the actual vs. predicted activity with the PLS regression factor of 3 (Reddy et al., 2013). Till getting the best statistical values for the model based on R2 and Q2, the test set and training set of the molecules are shuffled randomly and values are noted for the best 3D-QSAR model (Dessalew and Singh, 2008, Singh et al., 2006).
2.3. External validation
For the obtained 3D-QSAR, the external validation is performed for the execution of predictive abilities of generated model in robustness confirmation (Lorca et al., 2018). For this the r2, K values and are the parameters considered and the “r” is calculated by
Here the and are the actual and predicted activity, and are the average values of the actual and predicted pIC50 values of test set (Alexander et al., 2015, Majumdar and Basak, 2018). For the reliable 3D-QSAR, the statistical value of r2 should be close to 1 and regression of y against from origin: should be considered by k (slope) close to 1 and it is predicted by
The cross validated co-efficient , is predicted by
Also, the external server SIMCA-P 13.0 demo version is utilized for prediction of applicability domain (AD), to analyse the direct properties of distance matrix, multivariate descriptor, that are related to training set based on the PCA (Principal Component Analysis) (Kar et al., 2018). Through this applicability domain (AD), the prediction errors in the data set from the source compound will be avoided (Weaver and Gleeson, 2008).
2.4. Molecular dynamics based conformation analysis
For the prepared Ba-SrtA (PDB ID: 2KW8), the long scale Molecular Dynamics (MD) simulations is performed using the GROMACS (http://www.gromacs.org) MD simulation package (Pronk et al., 2013, Van Der Spoel et al., 2005). For solvent SPC water model is placed inside the dodecahedron box, and minimized along with protein using steepest descent method with maximum force of 100 kJ mol−1 nm−1 by the inclusion of GROMAS96 force field (Yagasaki et al., 2020). Energy minimization is performed to eliminate bad contacts and optimize the structure in the least energy minimized conformation state (Chopra et al., 2008). The room temperature of 300 K is provided for the system for comparing the room temperature future experiments, and controlled Nosé–Hoover thermostat dynamics with a damping coefficient of 2 ps−1 (Selvaraj et al., 2018b). NPT and NVT are performed initially, and the MD simulation step is processed for the timescale of 100 ns. For the ligand bound complex, the inclusion of topology from PRODRG server and similar protocol is followed, and the ligand complex MD is simulated for 20 ns (Selvaraj et al., 2020b).
2.5. Molecular docking simulations
From the MD simulations, the open and closed state conformations are taken and pre-processed with maestro tools (Vijayalakshmi et al., 2013). The GRID for docking is generated using the grid generation method, considering the β7/β8 active site loop with the 2.0 Å vdW radii of receptor atoms and with a partial charge cut off 0.25 (Tripathi et al., 2012). Mainly, the ligand conformations are set to rigid, as the QSAR based bioactive conformations derives the activity of the ligand molecules (Vilar and Costanzi, 2012). Docking is performed with Quantum Polarized Ligand Docking (QPLD) method for its inclusion of QM based partial charges that generate high accuracy by DFT method (6-31G*/B3LYP) (Selvaraj et al., 2020a, Selvaraj et al., 2014a, Selvaraj et al., 2014b). Final molecules are ranked based on Gscore, Energy and interactions, that are visualized on the Maestro visualizer (Park et al., 2009, Smak et al., 2021).
2.6. Conformation based activity prediction
The linear interaction approximation (LIA) model for the predicted binding affinities is calculated by Liaison program in Schrodinger 2014 (Selvaraj et al., 2015). It generates the fit between the known and predicted binding energy and this approach is utilized for the complex generated from the QPLD complex in both open and closed state conformation docking results (Evenseth et al., 2019). The calculation of binding affinities is based on the following equation
Herein, Uelec, Uvdw, and Ucav are representative of electrostatic, van der Waals, as well as cavity energy terms in the SGB (Surface Generalized Born) continuum solvent model (Matossian et al., 2014). Here the open and closed conformation-based docking results and their respective pIC50 (Experimental Value) is subjected to STRIKE to build a linear equation representing binding affinity.
3. Results
3.1. Pharmacophore modelling
In this study, the bioactive compounds from rhodanine, pyridazinone and pyrazolethione derivatives of 28 molecules are subject to Pharmacophore modelling and 3D-QSAR. Among the 28 molecules (Supplementary Fig. 1–2D structure information), 21 are categorized as training set molecules and 7 molecules are categorized as test set molecules, as shown in the Table 1. Phase pharmacophore modelling applies tree-based partition algorithm, that provides the common pharmacophore available between the active ligands and from this, we have got AAAHR (3 hydrogen bond acceptor, 1 hydrophobic and 1 aromatic ring), and AAADR (3 hydrogen bond acceptor, 1 hydrophobic and 1 aromatic ring) pharmacophore for the further analysis (Selvaraj and Singh, 2014). The mathematical data summary for both obtained pharmacophore AAAHR and AAADR listed in Table 2. The angles and distances of pharmacophore hypothesis AAAHR are given in supplementary material Table S1.
Table 1.
Structures and actual versus predicted pIC50 of compounds (* Test Set Compounds).
| Compounds | X | R1 | R2 | R3 | R4 | Actual | Predicted | Residual |
|---|---|---|---|---|---|---|---|---|
| IC50 | IC50 | |||||||
| Rhodanine | ||||||||
| 1–1 | -Ph | 4-NO2 | 3-Br, 2-OH, 5-NO2 |
4.69 | 4.52 | 0.179 | ||
| 1–2 | -Me | 3-Br, 2-OH, 5-NO2 |
4.88 | 5 | −0.11 | |||
| 1–3 | -Pr | -H | 4.27 | 4.29 | −0.01 | |||
| 1–4 | -Et | 2-NO2 | 4.13 | 4.31 | −0.17 | |||
| 1–5 | -Allyl | -H | 4.56 | 4.52 | 0.04 | |||
| Pyridazinone | ||||||||
| 2–1* | -SH | -OEt | -Ph | -H | 5.85 | 5.88 | −0.02 | |
| 2–5 | -OMe | -SH | -Ph | -H | 5.74 | 5.5 | 0.24 | |
| 2–6 | –OH | OCH2Ph | -Ph | -H | 4.30 | 3.9 | 0.40 | |
| 2–7 | –OH | -OMe | -Ph | -H | 4.30 | 4.24 | 0.06 | |
| 2–8 | –OH | -SEt | -Ph | -H | 4.30 | 4.59 | −0.28 | |
| 2–9* | -SH | -SEt | -Ph | -H | 6.52 | 6.11 | 0.41 | |
| 2–10 | -SH | -OEt | -Ph | –H | 5.49 | 5.87 | −0.37 | |
| 2–11 | -SH | -OEt | -Ph | 4-NO2 | 5.17 | 5.41 | −0.23 | |
| 2–13 | -SH | -OEt | -Ph | 3-F | 5.74 | 5.7 | 0.04 | |
| 2–14* | -SH | -OEt | -Ph | 3-Me | 5.77 | 5.68 | 0.09 | |
| 2–15 | -SH | -OEt | -Ph | 3,5-Cl2 | 4.85 | 5.5 | −0.64 | |
| 2–16 | -SH | -OEt | Cyclohexyl | 5.85 | 5.96 | −0.10 | ||
| 2–17* | -SH | -OEt | -Ph | -H | 5.92 | 5.94 | −0.01 | |
| 2–18 | -OEt | -SH | -Ph | -H | 5.92 | 5.66 | 0.26 | |
| 2–19* | -OEt | -SH | -Ph | 3-F | 6.04 | 6.16 | −0.11 | |
| 2–20 | -OEt | -SH | -Ph | 3-Me | 6.39 | 6.25 | 0.15 | |
| 2–21 | -OEt | -SH | -Ph | 3,5-Cl2 | 5.28 | 5.48 | −0.19 | |
| 2–35* | -OEt | -Cl | -Ph | -H | 6.52 | 5.26 | 1.26 | |
| 2–36 | -OEt | -Cl | -Ph | 4-NO2 | 3.60 | 4.04 | −0.43 | |
| 2–47 | -Cl | -Cl | -Ph | 3,5-Cl2 | 4.85 | 4.71 | 0.14 | |
| Pyrazolethione | ||||||||
| 3–9* | S | 4-N = N-Ph | H | 5.85 | 5.85 | 0.004 | ||
| 3–17* | S | 6.52 | 6.72 | −0.19 | ||||
| 3–12 | S | 2,4,6-Br3 | H | 5.85 | 5.9 | −0.05 | ||
Table 2.
Quantitative structure activity relationship (QSAR) results for the two best common pharmacophore hypotheses.
| AAAHR | AAADH | |
|---|---|---|
| SD | 0.34 | 0.41 |
| R2 | 0.87 | 0.82 |
| F | 40.3 | 25.4 |
| P | 6.079e-08 | 2.555e-06 |
| RMSE | 0.2637 | 0.4111 |
| Q2 | 0.74 | 0.6806 |
| Pearson R | 0.88 | 0.96 |
3.2. 3D QSAR and validation
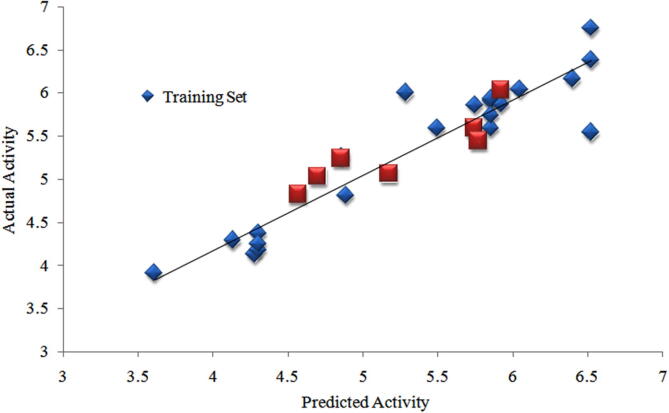
The obtained pharmacophore is subject to the development of 3D-QSAR prediction and for this the structurally different compounds were distributed uniformly with wide range of pIC50 value in both training and test set. Alignment was done using the phase, and pharmacophore of active molecules AAAHR and AAADR is considered for the 3D-QSAR model development with three PLS factors and re-evaluated by test set compounds. For assessing the good QSAR model, the statistical parameters like R2, Q2, RMSE, SD, and F are assessed. Based on this, two 3D QSAR models are generated showing good and consistent R2 greater than 0.82, standard deviation (SD) lesser than 0.4 and moderate F-test values. From the R2, the two models interpreting structure activity relationship with the training set is satisfactory. We also consider the Q2, as tropsha said high value of R2 is essential but not appropriate to make sense of QSAR model (Golbraikh and Tropsha, 2002, Tropsha, 2012). The AAAHR based QSAR model shows good external predictive ability with the Q2 value 0.74 and this AAAHR hypothesis has highest Q2 value and it is the best model among the other generated models. In addition, the AAAHR (AAAHR pharmacophore hypothesis in Fig. S2) based QSAR model shows the low RMSE value which also favours the best model. Results of data points generated from the 3D-QSAR are plotted with respected to X and Y axis showing the actual vs predicted pIC50 represented in the Fig. 1, by difference in test and training set. This generated model is externally evaluated with 7 test set compounds and internally by, leave-n-out technique. The generated best model base by cross validated coefficient with 0.56 value, high k slope values of regression lines with 1.002 and non-cross validated correlation coefficient (r2) with 0.51 value. This obtained values are possible to predict new future derivatives and the summary of statistics are provided in the Table S2.
Fig. 1.
QSAR based statistical plot representing the actual vs predicted pIC50 values for AAAHR hypothesis (Scatter Plot).
3.3. 3D QSAR visualization
In the 3D QSAR, the favourable and unfavourable regions could be visualized in 3D space, by the contour cubes generated. This could be applied to alteration of atoms in the ligands as the enrichment factor, and thus, 3D QSAR visualization is important along with the statistical values. The contours for the most active and least active molecule are provided in Fig. 2 and here blue colour contours represent the sterically favoured spatial regions to enhance the activity and red contours represent the sterically unfavoured regions. In addition, it can state more properties like positive/negative ionic, hydrophobic, hydrogen bond donor and bond acceptor (electron withdrawing) hypothesis that links non-covalent interactions with receptor excluded volumes. Receptor excluded volumes are the topology of receptor site, that provides the clarity of hydrophobic space assessed by the contour’s positions. Fig. 2a provide the most active compound (pyridazinone derivative (2–9)) with available of more favoured contour regions, and 2b provides the least active compound (pyridazinone derivatives (2–36)) with available of more unfavoured contour regions, predicted from the hydrophobic property using QSAR model. Most active compound show higher favourable regions around the aromatic ring, and Electron withdrawing group (N), while the inactive compound shows high red colour contours indicating the visualization of difference among both compounds.
Fig. 2.
QSR visualization for the most active compound (a) and for the least active compound (b) representing the respective favorable regions in blue contours and unfavorable regions in red color contour.
3.4. Validation of applicability domain
The applicability domain is validated for representing the test set molecules are reliable and for that DmodX value of 3.4 with 7 test set ligands are executed. Residual values with both predicted and experimental standard deviation of X-residuals (DmodX) are provided in Fig. S3. This values and figure represent the consolidation for the test set is reliable for the developed QSAR model. It also confirms that, based on the obtained statistical values, future set of compounds could also predict with high reliability. While the QSAR visualization provides the atom-based contours, this statistical data provides the conformations for the dataset, especially for the test set.
3.5. Molecular dynamics simulation for conformation analysis
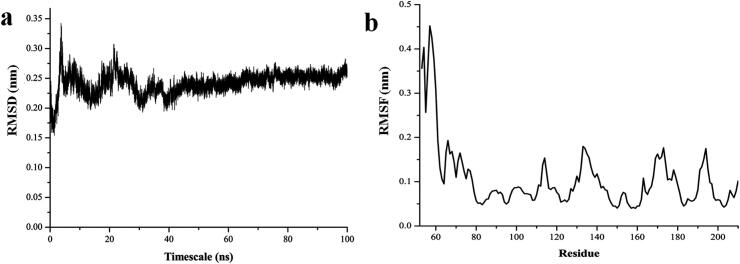
The solution structure of Ba-SrtA (PDB ID: 2KW8) having multiple conformations, and the average conformation is showing the open state conformation β7/β8 active site loop. This active site of β7/β8 loop is analysed for its conformational changes using the MD simulations for the timescale of 100 ns. The event of simulation, in respect to each conformational change are noted for each ns intervals. Those snapshots are calculated for the Root Mean Square Deviation (RMSD) from its initial reference structure and plotted in the Fig. 3a shows the Cα atoms vs. timescale (ns). The results of RMSD are interesting by showing the initial fluctuation up to 30th ns, from the 30th to 40th ns, the RMSD plot is getting consolidated and finally from 40th ns, the simulation event is stable till the end of the 100th ns. This cause for fluctuation is checked with residue wise fluctuations using the Root Mean Square fluctuation (RMSF) and the points are plotted in the Fig. 3b. Results of RMSF shows clear evidence of fluctuations in the β7/β8 loop with amino acids between the Gly53-Asp64 residues. This loop causes the deviations in higher level, and that leads to deviations that occur between 0 ns and 40th ns.
Fig. 3.
Molecular dynamics results showing the (a) RMSD graph and (b) RMSF graph for the timescale of 100 ns for the Ba-SrtA.
3.6. Open and closed lid conformations
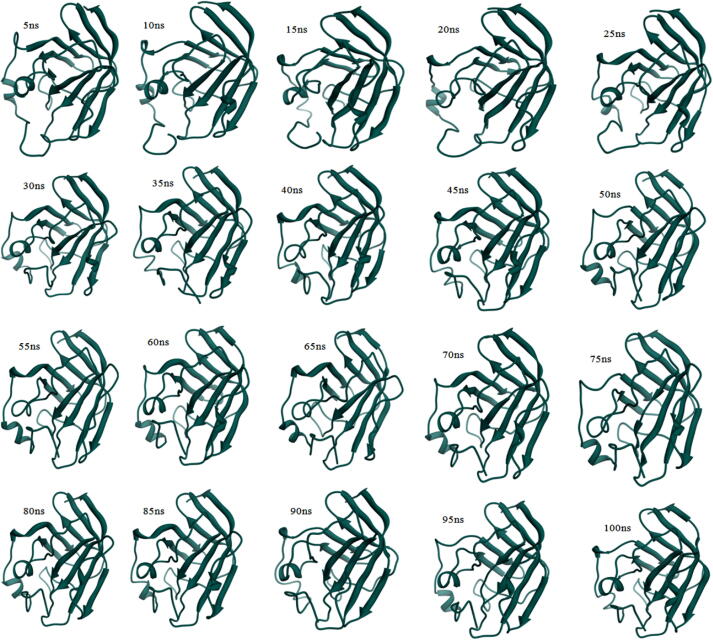
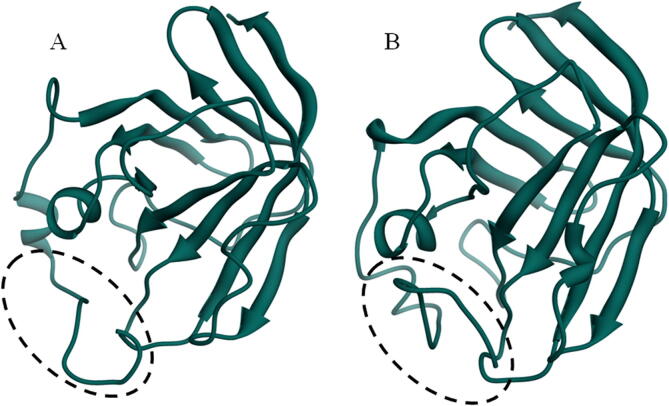
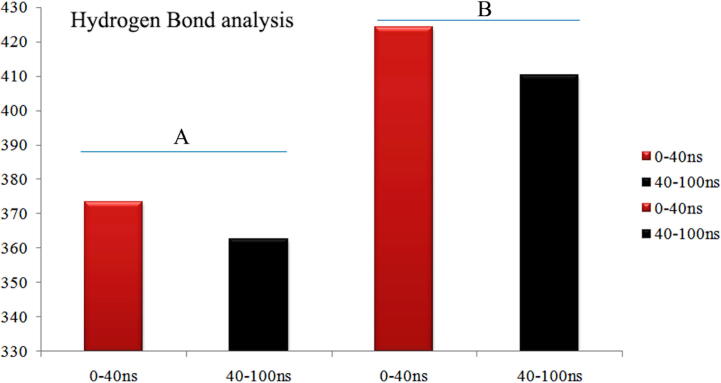
From the MD simulations, the β7/β8 loop deviation results are interesting and from that, the trajectories are subjected to dissection. The trajectory visualization of N-terminal loop from the Gly53-Asp64 residues show wide open active site in the 0-30th ns, from that 30th ns to 40th ns consolidation of β7/β8 loop by closing the active site. For evidence, the snapshot of Ba-SrtA at each 5th ns interval till 100th ns is provided in the Fig. 4 and merged conformations are provided in Fig. S4. This shows clearly that, the Ba-SrtA undergoes conformational changes, by open to closed lid conformations and after 40th ns, the closed lid conformations are stable till the 100th ns. Fig. 5 shows the both feature of open and closed lid conformations of Ba-SrtA, which clearly shows the transformation of open β7/β8 loop turns closed lid conformations. For understanding the role of water molecules in open and closed lid conformations, the hydrogen bond analysis with protein and water is accounted and provided in the Fig. 6. Here, the plot is considered for initial 0-40th ns (open lid conformations) and other plot is considered for 40th-100th ns (Closed lid conformations) from the pair above 0.35 nm (A) & within 0.35 nm (B). Results shows that average hydrogen of both above 0.35 nm and within 0.35 nm is higher in 0-40th ns, but lower in 40th to 100th ns, which is due to closure of active site results in protection of hydrophobicity in β7/β8 loop active site to interact with water.
Fig. 4.
Trajectory isolation of SrtA from different timescale with the interval of 5 ns showing the open to close active site loop transition occurs in between 30th ns to 40th ns.
Fig. 5.
Focused active site loop of (a) open form in the 0th ns and the (b) closed form taken from the 40th ns of MD simulations.
Fig. 6.
Hydrogen bond analysis of SrtA structure with water contact shows that pair above 0.35 nm (A) –and pair within 0.35 nm (B) - are showing high contacts in 0–40 ns and after that the hydrogen bonds has been decreased.
3.7. Binding selectivity of SrtA inhibitors
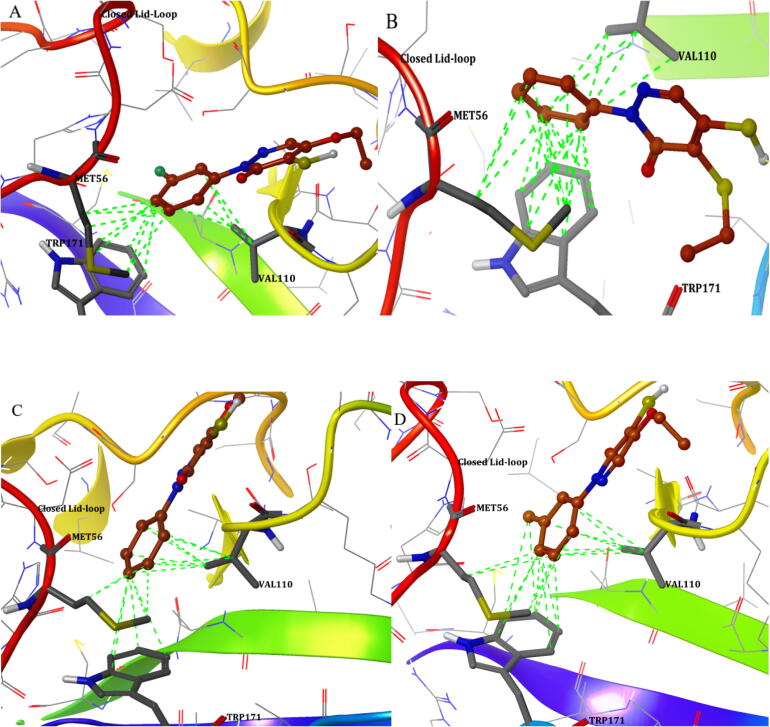
For understanding the binding selectivity of rhodanine, pyridazinone and pyrazolethione derivatives of 28 molecules obtained from the 3D-QSAR conformations, both open and closed lid conformations are subject to QM based molecular docking. Binding selectivity amino acids are fixed in the β7/β8 loop active site and both grids hold the conformational changes by open and closed lid state. Docking pattern shows the closed binding pocket favours strong interactions with three amino acids, namely Met56, Val110 and Trp171 and among the active compounds (Scoring values provided in Table 3), 92% molecules bind strong with these amino acids through the mode of hydrophobic interactions. One of the active compounds pyridazinone-2–17 shows similar binding mode along with another amino acid Ala58. Among the active compounds, the aromatic ring function in the pyridazinone and pyrazolethione are core important for the activity and shows direct interactions with the hydrophobic binding pocket, which is shown in the Fig. 7. In the Fig. 7, the hydrophobic interactions are visualized in green colour, especially around the aromatic ring bound to Met56, Val110 and Trp171.
Table 3.
scoring parameters of active compounds in QSAR towards closed lid structure of SrtA.
| pIC50 | IFD score | Glide score | Glide Energy | Binding Energy | Hydrophilic interactions | |
|---|---|---|---|---|---|---|
| Pyridazinone-2–19 | 6.046 | 489.21 | −6.591 | −42.62 | −38.61 | Met56, Val110 and Trp171 |
| Pyridazinone-2–9 | 6.523 | 480.67 | −6.486 | −43.19 | −40.51 | Met56, Val110 and Trp171 |
| Pyrazolethione-3–12 | 6.523 | 477.99 | −6.209 | −39.57 | −38.67 | Met56, Val110 and Trp171 |
| Pyridazinone-2–5 | 5.745 | 481.21 | −5.916 | −36.24 | −39.24 | Met56, Val110 and Trp171 |
| Pyridazinone-2–20 | 6.398 | 467.26 | −5.858 | −40.29 | −39.98 | Met56 and Val110 |
| Pyridazinone-2–16 | 5.854 | 459.38 | −5.614 | −36.21 | −38.21 | Met56, Val110 and Trp171 |
| Pyridazinone-2–35 | 6.523 | 461.50 | −5.512 | −38.29 | −38.20 | Met56, Val110 and Trp171 |
| Pyridazinone-2–17 | 5.854 | 446.55 | −5.367 | −39.18 | −36.19 | Met56, Ala58, Val110 and Trp171 |
Fig. 7.
Active compounds showing the aromatic ring structure showing hydrophobic interactions with Met56, Val110 and Trp171.
3.8. Molecular Dynamics: Ligand stability
For the ligand bound MD simulations, top five active compounds based on activity and scoring parameters are selected. As the closed state binding is favour the active compounds binding through the hydrophobic interactions, that active ligand stability is accessed through MD simulations for the timescale of 20 ns. Trajectory analysis shows that, the ligand bound inside the closed pocket through hydrophobic interactions doesn’t allow the ligand to unbound in the dynamic conditions. The active molecules are stable inside the pocket throughout the 20 ns of MD simulations and also shows strong binding within the active site. Especially, the amino acids, Met56, Val110 and Trp171 are strongly holding the ligand aromatic ring feature throughout the 20 ns. Ligand bound MD simulation is calculated for RMSD mean values for pyridazinone-2–19, pyridazinone-2–9, pyrazolethione-3–12, pyridazinone-2–5 and pyridazinone-2–20 active compounds and all the compounds are showing in between the range of 0.2 nm to 0.4 nm as shown in the Fig. S5. Only for the Pyridazinone-2–5 compound, the RMSD values are showing the higher range of 0.4 nm, but interestingly, all the compounds show strong and similar mode of binding.
3.9. Binding mode analysis with open/closed active site
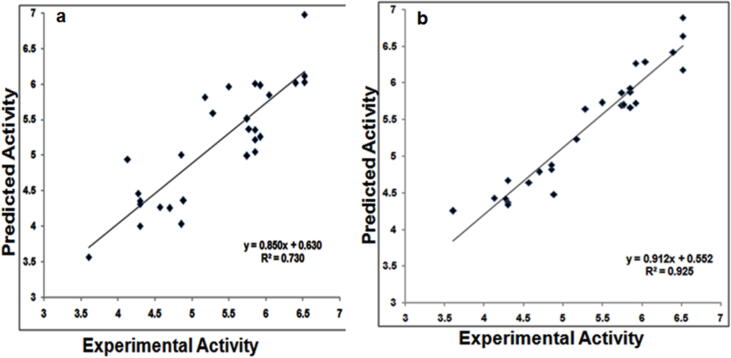
While we know the experimental activity of the 28 compounds, we want to know about the ligand activity is for closed state conformation or else with open state conformations. For that, docking results of 28 compounds with both open and closed lid is analysed for its binding affinity. The Uele, Uvdw and Ucav values obtained from liaison, along with experimental pIC50 values are plotted with strike to get the data points. For these data points, the LIA model is created for understanding its R2 values. Here the obtained values are based on the reference activity and theoretical activity for both open and closed lid. The correlation analysis shows in the Fig. 8, predicts that, the open lid protein conformations show r 2 of 0.730 and closed lid conformations shows R2 of 0.925. This confirms that, the protein conformation obtained from the 40th ns of molecular dynamics simulation is more active and suitable for molecular modelling calculations.
Fig. 8.
R2 cross validation of theoretical and experimental activity with respect to open lid (a) and closed lid (b) conformation.
4. Discussion
This research provides the detail insights of SrtA inhibition with respect to structural conformation of Ba-SrtA and mode of binding selectivity of ligand molecules. For that, 28 molecules from rhodanine, pyridazinone and pyrazolethione derivatives with experimental activity against the Bacillus anthracis are considered for this work. The initial part of the work is carried out using the ligand-based drug designing methods, which includes pharmacophore modelling and 3D-QSAR and the results shows AAAHR pharmacophore with the R2 of 0.82 and Q2 of 0.74. According to tropsha et al, this obtained 3D-QSAR is the best predicted statistical model is a better model for validating future compounds. The obtained statistical model is satisfactory and that is validated externally by leave-n-out technique. On Visualization, the most active ligand and least active ligand shows the favourable and unfavourable regions, that can be adjusted for improving the activity. Most active compound show less unfavoured regions by displaying the red contours and high favourable regions by displaying the blue colour contours. For least active compound, results show the high unfavourable regions in displaying high red colour contours. The ligand conformations obtained from the 3D-QSAR are analysed for its actual binding ability with both open and closed lid conformations obtained from the 100 ns MD simulations. This β7/β8 loop feature is previously reported in other similar species like Bacillus subtilis, Enterococcus faecalis, Bacillus cereus, Bacillus anthracis, Staphylococcus aureus for its importance with the drug-protein interactions. In Ba SrtA β7/β8 loop active site is widely open in the initial position is shown with evidence of closing inside due to the hydrophobic effect. This happens in between the 30th to 40th ns and remains closed throughout the MD simulations. From this long scale MD simulations, we have provided the detail conformational change that shows the switching mechanism of transition of open to closed lid conformations. This molecular level mechanism of switching is core important to understand in the process of developing new lead compounds. In the 40th ns, the β7/β8 loop active site function as protector of hydrophobic environment and so, it is tough to design the suitable compounds in considering the closed lid and if the compounds design based on open lids, that may not work as strong inhibitors. But the evidence of open and close lid conformation switch with respect to inhibitor binding is lack and thus, the molecular docking with QPLD is performed with both conformations. Interaction studies and energy binding with both open and close lid conformations shows strong binding with closed active site than the open active site, through the hydrophobic interactions with Met56, Val110 and Trp171. In the ligand molecules from rhodanine, pyridazinone and pyrazolethione derivatives aromatic ring is showing high binding interactions and these interactions are stable even in the dynamic conditions. Regression analysis on comparing the experimental activity with the docked complex shows close similarity with the closed conformations. This statistical value demonstrates the closed site is required for showing the realistic interactions and if the interaction studies are made with open state conformations, the interactions may not be accurate. Overall, this study may be one of the clear reports for understanding the importance of closed state conformations, which is also closed to the reported experimental binding of the reported compounds.
5. Conclusion
Overall, this study concludes the ligand-based features of rhodanine, pyridazinone and pyrazolethione with the pharmacophore modelling and 3D-QSAR by predicting the common pharmacophore of AAAHR and the 3D-QSAR shows the statistical values valid QSAR model. Contour shows higher allocation of favourable regions in the most active compound and unfavourable regions in the least active compounds. These results suggest the placement of atoms that can be modified in augmenting the activity of those compounds. The ligand conformation subject to open and closed lid conformations from the 100 ns MD simulations. Docking and binding interactions shows an open lid conformation of protein represents the R2 with 0.730 whereas closed lid conformation produces R2 with 0.925. This confirms that, the conformation obtained from the 40th ns of molecular dynamics simulation is more active and suitable for molecular modelling calculation. Finally, the 3D-QSAR model, molecular dynamics and docking studies performed here will provide the novel insights for the development of new and potent lead compounds for the inhibition of SrtA. Overall, this study will provide the information for considering the closed lid conformations for designing future lead compounds for the inhibition of SrtA in Bacillus anthracis.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgements
The Author CS and SKS, thankfully acknowledge the Tamil Nadu State Council for Higher Education (TANSCHE) for the research grant (Au/S.o. (P&D): TANSCHE Projects: 117/2021). The author AB extend their appreciation to Taif University for funding current work by Taif University Researchers Supporting Project number (TURSP-2020/295), Taif University, Taif, Saudi Arabia.
Funding support
The current work was funded by Tamil Nadu State Council for Higher Education (TANSCHE) for the research grant (Au/S.o. (P&D): TANSCHE Projects: 117/2021) and Taif University Researchers Supporting Project number (TURSP-2020/295), Taif university, Taif, Saudi Arabia
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2021.05.009.
Contributor Information
Chandrabose Selvaraj, Email: selnikraj@bioclues.org.
Sanjeev Kumar Singh, Email: sksingh@alagappauniversity.ac.in.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- Agrawal R., Jain P., Dikshit S.N., Bahare R.S. 3D QSAR and docking study of gliptin derivatives as DPP-IV inhibitors. Comb. Chem. High Throughput Screen. 2013;16:249–273. doi: 10.2174/1386207311316040001. [DOI] [PubMed] [Google Scholar]
- Alexander D.L., Tropsha A., Winkler D.A. Beware of R (2): simple, unambiguous assessment of the prediction accuracy of QSAR and QSPR models. J. Chem. Inf. Model. 2015;55:1316–1322. doi: 10.1021/acs.jcim.5b00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley M.L., Gaweska H., Kielec J.M., McCafferty D.G. Engineering the substrate specificity of Staphylococcus aureus Sortase A. The beta6/beta7 loop from SrtB confers NPQTN recognition to SrtA. J. Biol. Chem. 2007;282:6571–6581. doi: 10.1074/jbc.M610519200. [DOI] [PubMed] [Google Scholar]
- Blanchette K.A., Orihuela C.J. Future perspective on host-pathogen interactions during bacterial biofilm formation within the nasopharynx. Future Microbiol. 2012;7:227–239. doi: 10.2217/fmb.11.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, S., Santa Maria, J.P., Jr., Walker, S., 2013. Wall teichoic acids of gram-positive bacteria. Annu. Rev. Microbiol. 67, 313-336. [DOI] [PMC free article] [PubMed]
- Chan A.H., Yi S.W., Terwilliger A.L., Maresso A.W., Jung M.E., Clubb R.T. Structure of the Bacillus anthracis Sortase A enzyme bound to its sorting signal: A flexible amino-terminal appendage modulates substrate access. J. Biol. Chem. 2015;290:25461–25474. doi: 10.1074/jbc.M115.670984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhari H.K., Pahelkar A. 3D QSAR, docking, molecular dynamics simulations and MM-GBSA studies of extended side chain of the antitubercular drug (6S) 2-nitro-6- {[4- (trifluoromethoxy) benzyl] oxy}-6,7-dihydro-5H-imidazo[2,1-b] [1,3] oxazine. Infect. Disord. Drug Targets. 2019;19:145–166. doi: 10.2174/1871526518666181015145545. [DOI] [PubMed] [Google Scholar]
- Chen I.J., Foloppe N. Drug-like bioactive structures and conformational coverage with the LigPrep/ConfGen suite: comparison to programs MOE and catalyst. J. Chem. Inf. Model. 2010;50:822–839. doi: 10.1021/ci100026x. [DOI] [PubMed] [Google Scholar]
- Chopra G., Summa C.M., Levitt M. Solvent dramatically affects protein structure refinement. Proc. Natl. Acad. Sci. U. S. A. 2008;105:20239–20244. doi: 10.1073/pnas.0810818105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dessalew N., Singh S.K. 3D-QSAR CoMFA and CoMSIA study on benzodipyrazoles as cyclin dependent kinase 2 inhibitors. Med. Chem. 2008;4:313–321. doi: 10.2174/157340608784872244. [DOI] [PubMed] [Google Scholar]
- Evenseth L.M., Warszycki D., Bojarski A.J., Gabrielsen M., Sylte I. In silico methods for the discovery of orthosteric GABAB receptor compounds. Molecules. 2019;24 doi: 10.3390/molecules24050935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel B.A., Kruger R.G., Robinson D.E., Kelleher N.L., McCafferty D.G. Staphylococcus aureus sortase transpeptidase SrtA: insight into the kinetic mechanism and evidence for a reverse protonation catalytic mechanism. Biochemistry. 2005;44:11188–11200. doi: 10.1021/bi050141j. [DOI] [PubMed] [Google Scholar]
- Golbraikh A., Tropsha A. Beware of q2! J. Mol. Graph. Model. 2002;20:269–276. doi: 10.1016/s1093-3263(01)00123-1. [DOI] [PubMed] [Google Scholar]
- Hadler J.L. Learning from the 2001 anthrax attacks: immunological characteristics. J. Infect. Dis. 2007;195:163–164. doi: 10.1086/510317. [DOI] [PubMed] [Google Scholar]
- Haruta S., Kanno N. Survivability of microbes in natural environments and their ecological impacts. Microbes Environ. 2015;30:123–125. doi: 10.1264/jsme2.ME3002rh. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobitz A.W., Kattke M.D., Wereszczynski J., Clubb R.T. Sortase transpeptidases: structural biology and catalytic mechanism. Adv. Protein Chem. Struct. Biol. 2017;109:223–264. doi: 10.1016/bs.apcsb.2017.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar S., Roy K., Leszczynski J. Applicability domain: a step toward confident predictions and decidability for QSAR modeling. MethodsMol. Biol. 2018;1800:141–169. doi: 10.1007/978-1-4939-7899-1_6. [DOI] [PubMed] [Google Scholar]
- Lorca M., Morales-Verdejo C., Vasquez-Velasquez D., Andrades-Lagos J., Campanini-Salinas J., Soto-Delgado J., Recabarren-Gajardo G., Mella J. Structure-activity relationships based on 3D-QSAR CoMFA/CoMSIA and design of aryloxypropanol-amine agonists with selectivity for the human beta3-adrenergic receptor and anti-obesity and anti-diabetic profiles. Molecules. 2018;23 doi: 10.3390/molecules23051191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S., Basak S.C. Beware of external validation! - a comparative study of several validation techniques used in QSAR modelling. Curr. Comput. Aided Drug Des. 2018;14:284–291. doi: 10.2174/1573409914666180426144304. [DOI] [PubMed] [Google Scholar]
- Marraffini L.A., Dedent A.C., Schneewind O. Sortases and the art of anchoring proteins to the envelopes of gram-positive bacteria. Microbiol. Mol. Biol. Rev. 2006;70:192–221. doi: 10.1128/MMBR.70.1.192-221.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matossian M., Vangelderen C., Papagerakis P., Zheng L., Wolf G.T., Papagerakis S. In silico modeling of the molecular interactions of antacid medication with the endothelium: novel therapeutic implications in head and neck carcinomas. Int.. J. Immunopathol. Pharmacol. 2014;27:573–583. doi: 10.1177/039463201402700413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarre W.W., Schneewind O. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 1999;63:174–229. doi: 10.1128/mmbr.63.1.174-229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.S., Dessal A.L., Smrcka A.V., Stern H.A. Evaluating docking methods for prediction of binding affinities of small molecules to the G protein betagamma subunits. J. Chem. Inf. Model. 2009;49:437–443. doi: 10.1021/ci800384q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pronk S., Pall S., Schulz R., Larsson P., Bjelkmar P., Apostolov R., Shirts M.R., Smith J.C., Kasson P.M., van der Spoel D., Hess B., Lindahl E. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskevicius V., Kairys V. Predicting Isoform-specific binding selectivities of benzensulfonamides using QSAR and 3D-QSAR. Curr. Comput. Aided Drug Des. 2017;13:75–83. doi: 10.2174/1573409912666161129153820. [DOI] [PubMed] [Google Scholar]
- Reddy K.K., Singh S.K., Dessalew N., Tripathi S.K., Selvaraj C. Pharmacophore modelling and atom-based 3D-QSAR studies on N-methyl pyrimidones as HIV-1 integrase inhibitors. J. Enzyme Inhib. Med. Chem. 2012;27:339–347. doi: 10.3109/14756366.2011.590803. [DOI] [PubMed] [Google Scholar]
- Reddy K.K., Singh S.K., Tripathi S.K., Selvaraj C. Identification of potential HIV-1 integrase strand transfer inhibitors: in silico virtual screening and QM/MM docking studies. SAR QSAR Environ. Res. 2013;24:581–595. doi: 10.1080/1062936X.2013.772919. [DOI] [PubMed] [Google Scholar]
- Sastry G.M., Adzhigirey M., Day T., Annabhimoju R., Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J. Comput. Aided Mol. Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Selvaraj, C., Dinesh, D.C., Panwar, U., Boura, E., Singh, S.K., 2020a. High-Throughput Screening and Quantum Mechanics for Identifying Potent Inhibitors against Mac1 Domain of SARS-CoV-2 Nsp3. IEEE/ACM Trans Comput Biol Bioinform PP. [DOI] [PMC free article] [PubMed]
- Selvaraj C., Omer A., Singh P., Singh S.K. Molecular insights of protein contour recognition with ligand pharmacophoric sites through combinatorial library design and MD simulation in validating HTLV-1 PR inhibitors. Mol. BioSyst. 2015;11:178–189. doi: 10.1039/c4mb00486h. [DOI] [PubMed] [Google Scholar]
- Selvaraj C., Panwar U., Dinesh D.C., Boura E., Singh P., Dubey V.K., Singh S.K. Microsecond MD Simulation and Multiple-Conformation Virtual Screening to Identify Potential Anti-COVID-19 Inhibitors Against SARS-CoV-2 Main Protease. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.595273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj C., Priya R.B., Singh S.K. Exploring the biology and structural architecture of sortase role on biofilm formation in gram positive pathogens. Curr. Top. Med. Chem. 2018;18:2462–2480. doi: 10.2174/1568026619666181130133916. [DOI] [PubMed] [Google Scholar]
- Selvaraj C., Sakkiah S., Tong W., Hong H. Molecular dynamics simulations and applications in computational toxicology and nanotoxicology. Food Chem. Toxicol. 2018;112:495–506. doi: 10.1016/j.fct.2017.08.028. [DOI] [PubMed] [Google Scholar]
- Selvaraj C., Singh P., Singh S.K. Molecular insights on analogs of HIV PR inhibitors toward HTLV-1 PR through QM/MM interactions and molecular dynamics studies: comparative structure analysis of wild and mutant HTLV-1 PR. J. Mol. Recognit. 2014;27:696–706. doi: 10.1002/jmr.2395. [DOI] [PubMed] [Google Scholar]
- Selvaraj C., Singh P., Singh S.K. Molecular modeling studies and comparative analysis on structurally similar HTLV and HIV protease using HIV-PR inhibitors. J. Recept. Signal Transduct. Res. 2014;34:361–371. doi: 10.3109/10799893.2014.898659. [DOI] [PubMed] [Google Scholar]
- Selvaraj C., Singh S.K. Validation of potential inhibitors for SrtA against Bacillus anthracis by combined approach of ligand-based and molecular dynamics simulation. J. Biomol. Struct. Dyn. 2014;32:1333–1349. doi: 10.1080/07391102.2013.818577. [DOI] [PubMed] [Google Scholar]
- Selvaraj, C.T., SK. Reddy, KK. Singh, SK, 2011. Tool development for Prediction of pIC 50 values from the IC 50 values-A pIC 50 value calculator. Curr. Trends in Biotechnol. Pharmacy 1, 2.
- Silhavy T.J., Kahne D., Walker S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010;2 doi: 10.1101/cshperspect.a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.K., Dessalew N., Bharatam P.V. 3D-QSAR CoMFA study on indenopyrazole derivatives as cyclin dependent kinase 4 (CDK4) and cyclin dependent kinase 2 (CDK2) inhibitors. Eur. J. Med. Chem. 2006;41:1310–1319. doi: 10.1016/j.ejmech.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Smak P., Chandrabose S., Tvaroska I., Koca J. Pan-selectin inhibitors as potential therapeutics for COVID-19 treatment: in silico screening study. Glycobiology. 2021 doi: 10.1093/glycob/cwab021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer R.C. Bacillus anthracis. J. Clin. Pathol. 2003;56:182–187. doi: 10.1136/jcp.56.3.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirig T., Weiner E.M., Clubb R.T. Sortase enzymes in Gram-positive bacteria. Mol. Microbiol. 2011;82:1044–1059. doi: 10.1111/j.1365-2958.2011.07887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suree N., Yi S.W., Thieu W., Marohn M., Damoiseaux R., Chan A., Jung M.E., Clubb R.T. Discovery and structure-activity relationship analysis of Staphylococcus aureus sortase A inhibitors. Bioorg. Med. Chem. 2009;17:7174–7185. doi: 10.1016/j.bmc.2009.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryanarayanan V., Singh S.K., Tripathi S.K., Selvaraj C., Reddy K.K., Karthiga A. A three-dimensional chemical phase pharmacophore mapping, QSAR modelling and electronic feature analysis of benzofuran salicylic acid derivatives as LYP inhibitors. SAR QSAR Environ. Res. 2013;24:1025–1040. doi: 10.1080/1062936X.2013.821421. [DOI] [PubMed] [Google Scholar]
- Swick, M.C., Koehler, T.M., Driks, A., 2016. Surviving Between Hosts: Sporulation and Transmission. Microbiol Spectr 4. [DOI] [PMC free article] [PubMed]
- Tripathi, S.K., Singh, S.K., Singh, P., Chellaperumal, P., Reddy, K.K., Selvaraj, C., 2012. Exploring the selectivity of a ligand complex with CDK2/CDK1: a molecular dynamics simulation approach. J Mol Recognit 25, 504-512. [DOI] [PubMed]
- Tropsha A. Recent trends in statistical QSAR modeling of environmental chemical toxicity. Exp. Suppl. 2012;101:381–411. doi: 10.1007/978-3-7643-8340-4_13. [DOI] [PubMed] [Google Scholar]
- Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J. GROMACS: fast, flexible, and free. J. Comput. Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Vijayalakshmi P., Selvaraj C., Shafreen R.M., Singh S.K., Pandian S.K., Daisy P. Ligand-based pharmacophore modelling and screening of DNA minor groove binders targeting Staphylococcus aureus. J. Mol. Recognit. 2014;27:429–437. doi: 10.1002/jmr.2363. [DOI] [PubMed] [Google Scholar]
- Vijayalakshmi P., Selvaraj C., Singh S.K., Nisha J., Saipriya K., Daisy P. Exploration of the binding of DNA binding ligands to Staphylococcal DNA through QM/MM docking and molecular dynamics simulation. J. Biomol. Struct. Dyn. 2013;31:561–571. doi: 10.1080/07391102.2012.706080. [DOI] [PubMed] [Google Scholar]
- Vilar S., Costanzi S. Predicting the biological activities through QSAR analysis and docking-based scoring. MethodsMol. Biol. 2012;914:271–284. doi: 10.1007/978-1-62703-023-6_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts K.S., Dalal P., Murphy R.B., Sherman W., Friesner R.A., Shelley J.C. ConfGen: a conformational search method for efficient generation of bioactive conformers. J. Chem. Inf. Model. 2010;50:534–546. doi: 10.1021/ci100015j. [DOI] [PubMed] [Google Scholar]
- Weaver S., Gleeson M.P. The importance of the domain of applicability in QSAR modeling. J. Mol. Graph. Model. 2008;26:1315–1326. doi: 10.1016/j.jmgm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Weiner E.M., Robson S., Marohn M., Clubb R.T. The Sortase A enzyme that attaches proteins to the cell wall of Bacillus anthracis contains an unusual active site architecture. J. Biol. Chem. 2010;285:23433–23443. doi: 10.1074/jbc.M110.135434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagasaki T., Matsumoto M., Tanaka H. Lennard-Jones parameters determined to reproduce the solubility of NaCl and KCl in SPC/E, TIP3P, and TIP4P/2005 water. J. Chem. Theory Comput. 2020;16:2460–2473. doi: 10.1021/acs.jctc.9b00941. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.