Abstract
Countries in the Middle-East (ME) are tackling two corona virus outbreaks simultaneously, Middle-Eastern Respiratory Syndrome Coronavirus (MERS-CoV) and the current Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Both viruses infect the same host (humans) and the same cell (type-II alveolar cells) causing lower respiratory illnesses such as pneumonia. Molecularly, MERS-CoV and SARS-CoV-2 enter alveolar cells via spike proteins recognizing dipeptidyl peptidase-4 and angiotensin converting enzyme-II, respectively. Intracellularly, both viruses hide in organelles to generate negative RNA strands and initiate replication using very similar mechanisms. At the transcription level, both viruses utilise identical Transcription Regulatory Sequences (TRSs), which are known recombination cross-over points during replication, to transcribe genes. Using whole genome alignments of both viruses, we identify clusters of high sequence homology at ORF1a and ORF1b. Given the high recombination rates detected in SARS-CoV-2, we speculate that in co-infections recombination is feasible via TRS and/or clusters of homologies. Accordingly, here we recommend mitigation measure and testing for both MERS-CoV and SARS-CoV-2 in ME countries.
Keywords: SARS-CoV-2, MERS-CoV, COVID-19, humans, dromedary camels, recombination
1. PROSPECTIVE
The world is facing the worst public health crisis since the Spanish flu in the 1918s. Different countries are battling an outbreak caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). SARS-CoV-2 was first detected among patients with pneumonia in Wuhan, China in December 2019, then spread worldwide mainly by air-travel [1]. The virus causes a respiratory illness called Coronavirus Disease 2019 (COVID-19) and has varied presentation from asymptomatic to mild and in some cases leads to severe disease with multi-organ failure and death. To date the World Health Organization (WHO) has reported 54,050 cases and 329 deaths in UAE and 226,486 cases and 2151 deaths in KSA. In the Middle East (ME) specifically UAE and KSA, SARS-CoV-2 has caused significant outbreak, however the swift governmental scaled responses have strongly limited its spread [2]. Both countries, among others in the Arabian Peninsula, are particularly unique in that they are simultaneously battling another coronavirus called Middle Eastern Respiratory Syndrome Coronavirus (MERS-CoV). According to the WHO, the UAE reported its first case of MERS-CoV in July, 2013. Since then it has confirmed at least 91 as of September 2020 [3]. Similarly, Saudi Arabia, where MERS-CoV was first reported in September 2012, has reported 2156 cases as of June, 2020 [4,5].
At the onset of MERS-CoV outbreak, dromedary camels were recognized as reservoirs with the primary mode of transmission to humans being from direct or indirect contact with infected animals [6,7]. More importantly secondary mode of transmission has been confirmed from human to human after close contact [6]. Infected dromedary camels can shed viral-RNA from oral, nasal and gastrointestinal secretions [6,7]. While for SARS-CoV-2 no definite animal reservoir has been identified to date, however it spreads primarily through droplets and occasionally through airborne routes with secondary transmission from direct contact or through fomites. Given the co-circulation of MERS-CoV and SARS-CoV-2 in the ME, it raises concerns about the possibility of genomes recombining if present simultaneously in a reservoir (camels) or a host (humans). Given the high mortality rate of MERS-CoV (35%) and contagiousness of SARS-CoV-2, one could only imagine the worst.
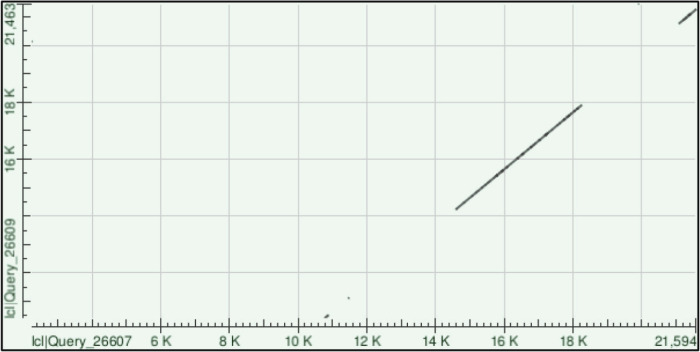
Viral genome recombination is a fundamental evolutionary process with some coronaviruses such as avian CoVs (the infectious bronchitis virus) repairing genomes through recombination [8,9]. The process of RNA recombination occurs when viral replicases synthesize RNA templates from donor RNA viruses during viral replication, where the nascent RNA is used as a template for RNA synthesis [10]. Several factors such as homology, genome size, secondary structures, replication rates and replicase complexes often modulated RNA recombination [11]. Accordingly, we aligned both SARS-CoV-2 and MERS-CoV genomes using Blast-2 and found four clusters of high homology sequences (Figure 1).
Figure 1.
Homology clusters between SARS-CoV-2 and MERS-CoV. Severe Acute Respiratory Syndrome Coronavirus 2 genome sequence (GenBank number MN996527.1) was aligned with MERS.CoV genome sequence (GenBank number MG987421.1) using Blast 2 sequences from NCBI [12]. Clusters of genome homologies along the whole genome was presented in a Dot plot graph [13].
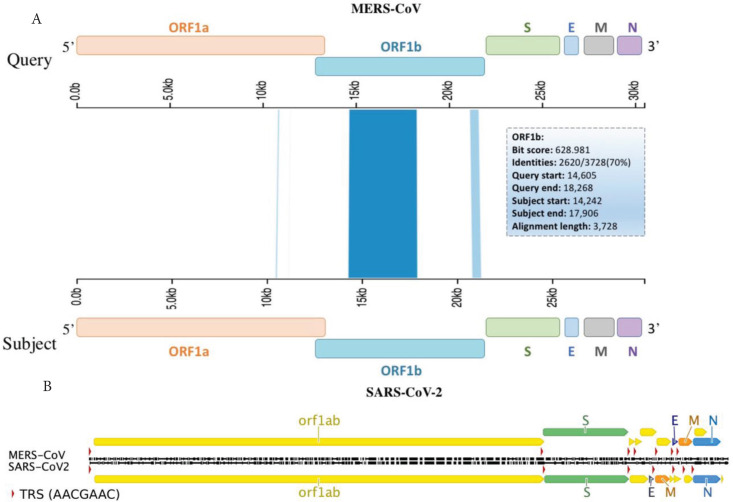
Interestingly, two clusters mapped to ORF1a and ORF1b with some clusters showing up to 100% sequence homology (Table 1). Such findings highly suggest recombination could take place with-in ORF1a and/or b and crossover at the next homology cluster (Figure 2A), at any downstream Transcription Regulatory Sequences (TRS) or span the rest of the genome, which includes S, E, M and N genes (Figure 2B).
Table 1.
Sequence homology information between MERS-CoV and SARS-CoV-2 aligned clusters
| Cluster No. | Query | Subject | Score | Identities/Matches | Alignment length (bps) | Gene |
|---|---|---|---|---|---|---|
| MERS-CoV | SARS-CoV-2 | |||||
| 1 | 14605–18268 | 14242–17906 | 628 bits (340) | 2620 (70%) | 3728 | ORF1b |
| 2 | 21081–21594 | 20749–21263 | 102 bits (55) | 372 (71%) | 525 | ORF1b |
| 3 | 10778–10895 | 10414–10530 | 56.5 bits (30) | 90 (76%) | 119 | ORF1a |
| 4 | 11464–11486 | 11108–11130 | 38.1 bits (20) | 22 (96%) | 23 | ORF1a |
| 5 | 2246–2261 | 20138–20123 | 30.7 bits (16) | 16 (100%) | 16 | ORF1a/ORF1b |
| 6 | 19910–19925 | 21463–21448 | 30.7 bits (16) | 16 (100%) | 16 | ORF1b |
All of the data was extracted from Blast 2 sequences using NCBI after aligning MERS-CoV genome with SARS-CoV-2 genome.
Figure 2.
Viral genes aligned with TRSs and sequence homology clusters. Schematic diagram of MERS-CoV and SARS-CoV-2 viral genomes (A) aligned with sequence homology clusters highlighted in blue and (B) identical TRSs (red triangles). These hotspots should enable recombination prediction points based on homology clusters and TRS sites within viral genes.
Once recombined, the successful inheritance of the chimeric genomes is determined by the relative fitness of the recombinant viral progeny [14–16]. Such recombination observations have been reported in several CoVs such as mouse hepatitis virus and SARS-CoV, where both viruses share 69% homology within their 3′ UTRs [17]. This homology was found to have mutual recognition of cis-acting genomic structures in both CoV strains that resulted in a competent viral replication [17]. In addition, the discontinuous transcription criteria of CoVs, which is defined by the common dissociation and re-joining of the polymerase complex and nascent RNA during transcription leads to a template-switching, a known phenomenon in RNA viruses recombination [18,19].
This model of recombination “copy choice” is mediated by conserved nucleotide sequences called TRSs, which are present at the 5′ of each ORF and 5′ of the genome [20]. TRSs allow for leader-body joining due to the presence of a conserved Core Sequence (CS) that is identical to the genome leader (CS-L) and all mRNA coding sequences (mCS-B), which facilitate the base pairing of CS-L with the complementary nascent negative-strand CS-B (cCS-B) [20,21]. Furthermore, secondary structure analysis of the TRS-L region from the transmissible Bovine Coronavirus and transmissible gastroenteritis virus show that the exposed CS-L in the loop of a structured hairpin is relevant for transcription providing evidence for the TRS-L selection during the template switch [20,21]. In reference to SARS-CoV-2, seven TRSs conserved motifs, 5′AACGAAC3′, were found upstream of ORF1ab, S, ORF3a, M, ORF7a, ORF8 and N genes, details in Table 2. These motifs are completely identical to TRS-L core sequences in MERS-CoV [22]. Hence, providing evidence of a highly similar transcription mechanism between MERS-CoV and SARS-CoV-2 suggesting high recombination probabilities during co-infections.
Table 2.
Identical TRSs between MERS-CoV and SARS-CoV-2 viral genomes
| Gene | Transcription regulatory sequences (TRS) | Nucleotide position |
|---|---|---|
| ORF 1ab | 5′ TCTAAACGAACTTTAA 3′ | 00032–00047 |
| S | 5′ ACTAAACGAACAATGT 3′ | 21,518–21,533 |
| ORF3a | 5′ CATAAACGAACTTATG 3′ | 25,347–25,362 |
| M | 5′ TCTAAACGAACTAAAT 3′ | 26,435–26,450 |
| ORF7a | 5′ ATTAAACGAACATGAA 3′ | 27,349–27,364 |
| ORF8 | 5′ CCTAAACGAACATGAA 3′ | 27,850–27,865 |
| N | 5′ TCTAAACGAACAAACT 3′ | 28,222–28,237 |
It is not clear whether MERS-CoV and SARS-CoV-2 have recombined, in spite of preliminary evidence supporting our hypothesis. Both MERS-CoV and SARS-CoV-2 are thought to have originated from bat CoV recombining with other CoVs [1,7,23]. This explains the 50% sequence homology between MERS-CoV and SARS-CoV-2 genomes [23,24]. Sequence homology no doubt increases the chances of genomes undergoing recombination regardless of the group boundaries. In fact, several studies have shown recombination taking place in RNA-dependent RNA polymerase (RdRp) of N and M genes between SARS-CoV and other CoVs, which resulted in host-shifting [25,26]. The co-exitance of CoVs with MERS-CoV in single reservoirs or hosts is not undocumented in the ME. Indeed, studies have reported up to three CoVs [MERS-CoV, betacoronavirus 1 (β1-HKU23-CoV), and human CoV 229E (HCoV-229E)] co-infecting dromedary camels, during the 2014-2015 MERS-CoV outbreak [27]. Phylogenetic analysis of isolates demonstrated five distinct recombination lineages, all of which contained human and camel MERS-CoV sequence [27]. As such for both MERS-CoV and SARS-CoV-2 genomes to merge they must simultaneously infect the same reservoir or host. The latter is highly plausible given that humans contract both infections and both viruses attack the lungs causing similar lower respiratory illnesses such as pneumonia [6,23].
Molecularly, MERS-CoV enters cells by recognizing Dipeptidyl Peptidase-4 (DPP4) receptors using spike proteins [28]. In humans, cells expressing DPP4 are at lower respiratory tract type-II alveolar cells [28]. Cell membranes anchor DPP4 along with Transmembrane Protease Serine Protease-2 (TMPRSS2) via CD-9 [28]. MERS-CoV therefore uses the protease activity of TMPRSS2, among other proteases, to induce conformational changes on its spike proteins required for cell-entry [28]. Interestingly, TMPRSS2 is also necessary for SARS-CoV-2 cell entry [29,30]. SARS-CoV-2 similarly uses spike proteins to enter cells, but they recognize a receptor called Angiotensin Converting Enzyme-II (ACE2) [23]. In humans, the expression of ACE2 is widely spread throughout the respiratory tract [31,32]. This explains why COVID-19 is a disease affecting both the upper and lower respiratory tract. It is also worth noting that type-II alveolar cells do express membrane ACE2 [31,32]. Thus, strongly supporting the possibility of both MERS-CoV and SARS-CoV-2 infecting the same cell in the lungs.
The genomes of MERS-CoV and SARS-CoV-2 could also recombine in a reservoir, here dromedary camels. It is currently unclear whether SARS-CoV-2 could infect dromedary camels. However, SARS-CoV-2 does seem to infect domesticated animals [33,34]. In one study, ferrets and cats inoculated with SARS-CoV-2 showed efficient viral replication and shedding as early as 3-days post-inoculation [33]. It is SARS-CoV-2’s inherent ability to recognize diverse ACE2 receptors across different species that makes its host spectrum unpredictable [35]. In the ME, the peak of MERS-CoV human infections runs from March until May, which coincides with dromedary camels shedding the highest viral RNA from November till January [7,36].
Proactive measures should be implemented to mitigate the chances of both viral genomes recombining. These measures must include separating dromedary camels and their carers from the general public. Additionally, studies should focus on understanding the susceptibility of dromedary camels to SARS-CoV-2 by regularly screening for viral RNA and antisera. Finally, in cities still suffering from MERS-CoV infections, real time quantitative PCR must be used to screen for MERS-CoV in pneumonia suspected patients as well as in COVID-19 diagnosed patients. Whole viral genome sequencing should also be used to monitor any recombination events between SARS-CoV-2 and MERS-CoV in patients and dromedary camels testing positive for MERS-CoV and SARS-CoV-2.
Here we present our hypothesis supporting the possibility of MERS-CoV and SARS-CoV-2 recombining in a single host or reservoir. In type II alveolar cells both viral genomes could recombine at TRSs and/or homology clusters as shown in Figure 2. It is therefore, that we strongly recommend extra-precautionary measures to avoid MERS-CoV and SARS-CoV-2 merging into a novel coronavirus.
Footnotes
Data availability statement: Datasets analyzed are available in NCBI under the accession number: SARS-CoV-2 (MN996527.1) and MERS.CoV (MG987421.1). These datasets were derived from the following public domain resources: https://www.ncbi.nlm.nih.gov/nuccore.
CONFLICTS OF INTEREST
The authors declare they have no conflicts of interest.
FUNDING
This perspective was funded by Khalifa University of Science and Technology Competitive Internal Research Award CIRA-ADEK and by Abu Dhabi Award for Research Excellence (AARE) 2019.
SUPPLEMENTARY MATERIALS
Supplementary data related to this article can be found at
.
REFERENCES
- [1].Ji W, Wang W, Zhao X, Zai J, Li X. Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol. 2020;92:433–40. doi: 10.1002/jmv.25682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Algaissi AA, Alharbi NK, Hassanain M, Hashem AM. Preparedness and response to COVID-19 in Saudi Arabia: building on MERS experience. J Infect Pub Health. 2020;13:834–8. doi: 10.1016/j.jiph.2020.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].World Health Organization . Middle East respiratory syndrome coronavirus (MERS-CoV) – United Arab Emirates. Geneva, Switzerland: World Health Organization; 2020. [Google Scholar]
- [4].AlRuthia Y, Somily AM, Alkhamali AS, Bahari OH, AlJuhani RJ, Alsenaidy M, et al. Estimation of direct medical costs of middle east respiratory syndrome coronavirus infection: a single-center retrospective chart review study. Infect. Drug Resist. 2019;12:3463–73. doi: 10.2147/IDR.S231087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].World Health Organization . Middle East respiratory syndrome coronavirus (MERS-CoV) – The Kingdom of Saudi Arabia. Geneva, Switzerland: World Health Organization; 2020. [Google Scholar]
- [6].Memish ZA, Perlman S, Van Kerkhove MD, Zumla A. Middle East respiratory syndrome. Lancet. 2020;395:1063–77. doi: 10.1016/S0140-6736(19)33221-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Omrani AS, Al-Tawfiq JA, Memish ZA. Middle East respiratory syndrome coronavirus (Mers-coV): animal to human interaction. Pathog Glob Health. 2015;109:354–62. doi: 10.1080/20477724.2015.1122852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Smati R, Silim A, Guertin C, Henrichon M, Marandi M, Arella M, et al. Molecular characterization of three new avian Infectious Bronchitis Virus (IBV) strains isolated in Quebec. Virus Genes. 2002;25:85–93. doi: 10.1023/a:1020178326531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Wang L, Junker D, Collisson EW. Evidence of natural recombination within the S1 gene of infectious bronchitis virus. Virology. 1993;192:710–16. doi: 10.1006/viro.1993.1093. [DOI] [PubMed] [Google Scholar]
- [10].Cheng CP, Nagy PD. Mechanism of RNA recombination in carmo- and tombusviruses: evidence for template switching by the RNA-dependent RNA polymerase in vitro. J Virol. 2003;77:12033–47. doi: 10.1128/jvi.77.22.12033-12047.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Domingo E. Chapter 2 - Molecular basis of genetic variation of viruses: error-prone replication. In: Domingo E, editor. Virus as Populations: Composition, Complexity, Dynamics, and Biological Implications. Academic Press; 2016. pp. 35–71. [DOI] [Google Scholar]
- [12].Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–14. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- [13].Wintersinger JA, Wasmuth JD. Kablammo: an interactive, web-based BLAST results visualizer. Bioinformatics. 2015;31:1305–6. doi: 10.1093/bioinformatics/btu808. [DOI] [PubMed] [Google Scholar]
- [14].Nagy PD, Pogany J, Simon AE. RNA elements required for RNA recombination function as replication enhancers in vitro and in vivo in a plus-strand RNA virus. EMBO J. 1999;18:5653–65. doi: 10.1093/emboj/18.20.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Figlerowicz M, Nagy PD, Bujarski JJ. A mutation in the putative RNA polymerase gene inhibits nonhomologous, but not homologous, genetic recombination in an RNA virus. Proc Natl Acad Sci U S A. 1997;94:2073–8. doi: 10.1073/pnas.94.5.2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cheng CP, Serviene E, Nagy PD. Suppression of viral RNA recombination by a host exoribonuclease. J Virol. 2006;80:2631–40. doi: 10.1128/JVI.80.6.2631-2640.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Goebel SJ, Taylor J, Masters PS. The 3′ cis-acting genomic replication element of the severe acute respiratory syndrome coronavirus can function in the murine coronavirus genome. J Virol. 2004;78:7846–51. doi: 10.1128/JVI.78.14.7846-7851.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mateos-Gomez PA, Morales L, Zuñiga S, Enjuanes L, Sola I. Long-distance RNA-RNA interactions in the coronavirus genome form high-order structures promoting discontinuous RNA synthesis during transcription. J Virol. 2013;87:177–86. doi: 10.1128/JVI.01782-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lai MM. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. https://pubmed.ncbi.nlm.nih.gov/1579113/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Alonso S, Izeta A, Sola I, Enjuanes L. Transcription regulatory sequences and mRNA expression levels in the coronavirus transmissible gastroenteritis virus. J Virol. 2002;76:1293–308. doi: 10.1128/jvi.76.3.1293-1308.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Dufour D, Mateos-Gomez PA, Enjuanes L, Gallego J, Sola I. Structure and functional relevance of a transcription-regulating sequence involved in coronavirus discontinuous RNA synthesis. J Virol. 2011;85:4963–73. doi: 10.1128/JVI.02317-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].van Boheemen S, de Graaf M, Lauber C, Bestebroer TM, Raj VS, Zaki AM, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3:e00473–12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–3. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914.e10–921.e10. doi: 10.1016/j.cell.2020.04.011. https://pubmed.ncbi.nlm.nih.gov/32330414/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Stavrinides J, Guttman DS. Mosaic evolution of the severe acute respiratory syndrome coronavirus. J Virol. 2004;78:76–82. doi: 10.1128/jvi.78.1.76-82.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rest JS, Mindell DP. SARS associated coronavirus has a recombinant polymerase and coronaviruses have a history of host-shifting. Infect Genet Evol. 2003;3:219–25. doi: 10.1016/j.meegid.2003.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sabir JSM, Lam TTY, Ahmed MMM, Li L, Shen Y, Abo-Aba SE, et al. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–4. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- [28].Widagdo W, Sooksawasdi Na Ayudhya S, Hundie GB, Haagmans BL. Host determinants of MERS-CoV Transmission and Pathogenesis. Viruses. 2019;11:280. doi: 10.3390/v11030280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Matsuyama S, Nao N, Shirato K, Kawase M, Saito S, Takayama I, et al. Enhanced isolation of SARS-CoV-2 by TMPRSS2-expressing cells. Proc Natl Acad Sci U S A. 2020;117:7001–3. doi: 10.1073/pnas.2002589117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hoffmann M, Kleine-Weber H, Schroeder S, Krüger N, Herrler T, Erichsen S, et al. SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271.e8–80.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Xu H, Zhong L, Deng J, Peng J, Dan H, Zeng X, et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int J Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Hamming I, Timens W, Bulthuis ML, Lely AT, Navis G, van Goor H. Tissue distribution of ACE2 protein, the functional receptor for SARS coronavirus. A first step in understanding SARS pathogenesis. J Pathol. 2004;203:631–7. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Shi J, Wen Z, Zhong G, Yang H, Wang C, Huang B, et al. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. 2020;368:1016–20. doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wan Y, Shang J, Graham R, Baric RS, Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol. 2020;94:e00127–20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yuan Z. The diverse of ACE2 receptor made the inference of host Difficult copy.pdf. figshare. 2020 [Google Scholar]
- [36].Khalafalla AI, Lu X, Al-Mubarak AI, Dalab AH, Al-Busadah KA, Erdman DD. MERS-CoV in upper respiratory tract and lungs of dromedary camels, Saudi Arabia, 2013-2014. Emerg Infect Dis. 2015;21:1153–8. doi: 10.3201/eid2107.150070. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.