Abstract
Introduction: Influenza infection poses a significant public health threat. The core for disease prevention and control relies on strengthened surveillance activities, particularly in Saudi Arabia, the country that hosts the largest annual mass gathering event worldwide. This study aimed to assess the molecular and seasonal pattern of influenza virus subtypes in western Saudi Arabia to inform policy decisions on influenza vaccine.
Methods: This cross-sectional study was conducted at King Abdulaziz Medical City, western Saudi Arabia. Medical records and surveillance database of laboratory-confirmed influenza cases were reviewed from October 2015 to 2019. A panel of real-time polymerase chain reactions was performed to detect influenza A and B. Extracted RNA from a subset of positive samples was used to determine influenza A subtypes and influenza B lineages.
Results: This study included a total of 1928 patients with laboratory-confirmed influenza infections. Influenza peaks were observed in October each season, with variant predominant strains. Influenza virus subtypes co-circulate with no reports of co-infection. Influenza A(H3N2) was reported in 42% of the cases, then influenza B (30.7%) and influenza A(H1N1)pdm09 (27.3%). Healthcare workers represented 9.4% of the cases. One-third of the cases (30.4%) were admitted to the hospital with a median admission duration of 4 days. The influenza B viruses were subtyped in 218 cases. Victoria lineage was predominant (64.1%) in 2015 and 2016; however, Yamagata was predominant in the next two consecutive seasons (94.4% and 85.4%, respectively).
Conclusion: The burden due to influenza B may be underestimated with an observed vaccine mismatch. A quadrivalent influenza vaccine is recommended to reduce the health impact associated with influenza B infections. Molecular surveillance of the influenza viruses should be enhanced continuously for a better understanding of the influenza activity and assessment of vaccine effectiveness.
Keywords: Influenza, molecular, epidemiological, Saudi Arabia
1. INTRODUCTION
Influenza viruses are recognized globally as persistent threats to human health. They are the primary respiratory pathogens that cause annual epidemics with high economic loss and approximately 250,000–500,000 deaths annually worldwide [1,2].
Influenza viruses are characterized by two mechanisms that direct the evolution of new variants; antigenic drift and genetic shift [3,4]. Genetic shift is a sudden, substantial change in the influenza A virus and related to the unexpected emergence of new subtypes, and occurrence of pandemics; however, earlier reports suggest that they do not play a role in the genetic variation of influenza B viruses [5,6]. Nevertheless, the antigenic drift through several mechanisms that include insertions, deletions, and substitutions are frequent among influenza A and different lineages of influenza B viruses [7–10].
Studies that aimed at comparing the epidemiological and clinical characteristics of influenza virus subtypes are limited and showed variant results [11,12].
In Saudi Arabia, reports on influenza viruses have focused on estimating prevalence and clinical outcomes among other causes of acute respiratory illnesses in young children [13,14] or among pilgrims [15]. Data are limited on the isolation and partial identification of influenza subtypes, especially influenza B viruses [16].
Continued changes in Haemagglutinin (HA) and Neuraminidase (NA) genes of influenza A and B strains necessitate enhanced surveillance and monitoring of emerging strains, particularly in Saudi Arabia, the country that hosts the largest religious mass gathering event throughout the year (Umrah and Hajj). This study was conducted to identify influenza A subtypes and B lineages and their association with the clinical characteristics and outcomes among patients with laboratory-confirmed influenza infection in a tertiary care hospital located in western Saudi Arabia.
2. MATERIALS AND METHODS
This cross-sectional study was conducted at King Abdulaziz Medical City (KAMC), Jeddah, Saudi Arabia. KAMC-Jeddah is a referral tertiary care hospital located in western Saudi Arabia. This medical city and the three affiliated primary care centers in Jeddah provide care to the Saudi National Guard population and their families, in addition to the healthcare workers and their dependents. Data were collected as part of the routine influenza surveillance program.
Medical records and surveillance database of all laboratory-confirmed influenza cases diagnosed at KAMC-Jeddah were reviewed. Data were collected throughout the four consecutive influenza seasons from October 2015 to 2019. No specific sampling technique was applied because all the positive samples were included. The studied medical files represented patients who had flu-like illnesses and visited the hospital outpatient department, primary health care centers, emergency department, or inpatient during the study period. Cases were managed according to the hospital’s influenza management protocol. The decision on hospital admission or emergency department referral was based on the case clinical assessment.
Studied variables included: patient demographic data (age, gender, nationality and occupation being health care worker), clinical data (date of diagnosis, symptoms and signs, hospital admission and discharge, and associated comorbidities or complications), and influenza virus subtypes.
2.1. Clinical Samples
Nasopharyngeal Swabs (NPS) from patients with suspected influenza infections were collected in viral transport media (Copan UTM 321c, USA) as part of the routine ongoing influenza diagnosis/surveillance program. These samples were initially screened for influenza A(H1N1)pdm09, influenza A/non-H1N1, and influenza B using Xpert Flu kits (Cepheid, USA) at the molecular virology laboratory in KAMC as per the manufacturer’s instructions. All patients with laboratory-confirmed samples during the study period from 2015 to 2019 were included. Samples were stored at −80°C for further nucleic acid extraction.
2.2. Influenza A/2009 H1N1 and Influenza B Detection
A panel of Real-Time Polymerase Chain Reactions (RT-PCR) was performed to detect influenza A/2009 H1N1 and influenza B using Cepheid Xpert Flu Assay (Cat. No. GXFLU-10A) according to the manufacturer’s instructions (Cepheid) (Supplementary Material).
2.3. Subtyping and Lineage Determination of Influenza A/non-H1N1 and Influenza B Viruses by RT-PCR
Nasopharyngeal samples are not usually stored at the main laboratory for a longer duration due to limited storage capacity; however, at the time of analysis, a subset of the stored laboratory-confirmed samples was available from three consecutive seasons (2015–2018). No samples were available for the 2018 and 2019 season. These samples were transported in dry ice containers for further virus subtyping and lineage determination at the Special Infectious Agents Unit at King Fahad Medical Research Center, King Abdulaziz University, Jeddah, Saudi Arabia.
Extracted RNA from positive samples was used to determine influenza A/non-H1N1 subtype using primers and probes targeting haemagglutinin gene (HA) in H3, H5, and H7 subtypes in this sequential order (NIID-H3 TMPrimer-F1, NIID-H3 TMPrimer-R1 and NIID-H3 Probe1, H5-1201F, H5-1387R and H5-1285P, NIID-H7 TMPrimer-F1, NIID-H7 TMPrimer-R1 and NIID-H7 Probe1) according to WHO protocol [17].
Similarly, RNA from specimens that were positive for influenza B was used to determine influenza B lineages using influenza B primers (BHA-188F and BHA-270R) and probes specific for Victoria (Probe-VIC2) and Yamagata (Probe-YAM2) lineages according to WHO protocol [17] (Supplementary Material).
Stringent precautions were taken to avoid cross-contamination of samples during all steps, and all positive and some negative samples were re-extracted and retested independently to confirm the results.
2.4. Ethical Approval
This study was approved by the Institutional Review Board at King Abdullah International Medical Research Center. No informed consent was required for influenza detection, subtyping, and molecular characterization. The patient data were kept confidential and analyzed anonymously.
2.5. Statistical Analysis
Data were collected and analyzed using IBM SPSS software version 26 (IBM, Armonk, NY, USA). Descriptive statistics, including frequency, percentage, mean, standard deviation, median, and Interquartile Range (IQR) were used. The Chi-squared test was applied to compare the categorical variables of influenza subtypes and other demographic and clinical data. Epidemiological week and year were used to show the influenza peaks and seasonal variations. The level of significance was determined at p < 0.05.
3. RESULTS
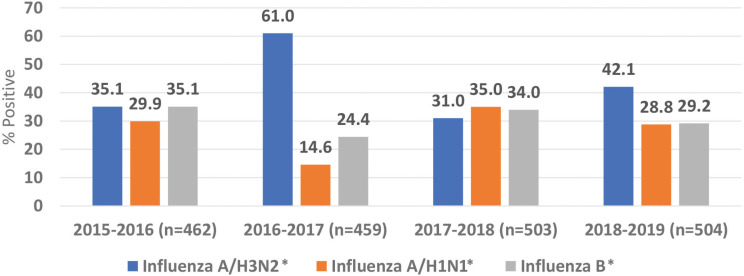
This study included a total of 1928 patients with laboratory-confirmed influenza infections over four seasons from October 2015 to 2019. The average number of laboratory-confirmed cases per season was 482 (range 459–504) (Figure 1).
Figure 1.
Distribution of influenza A and B virus subtypes. *p < 0.05.
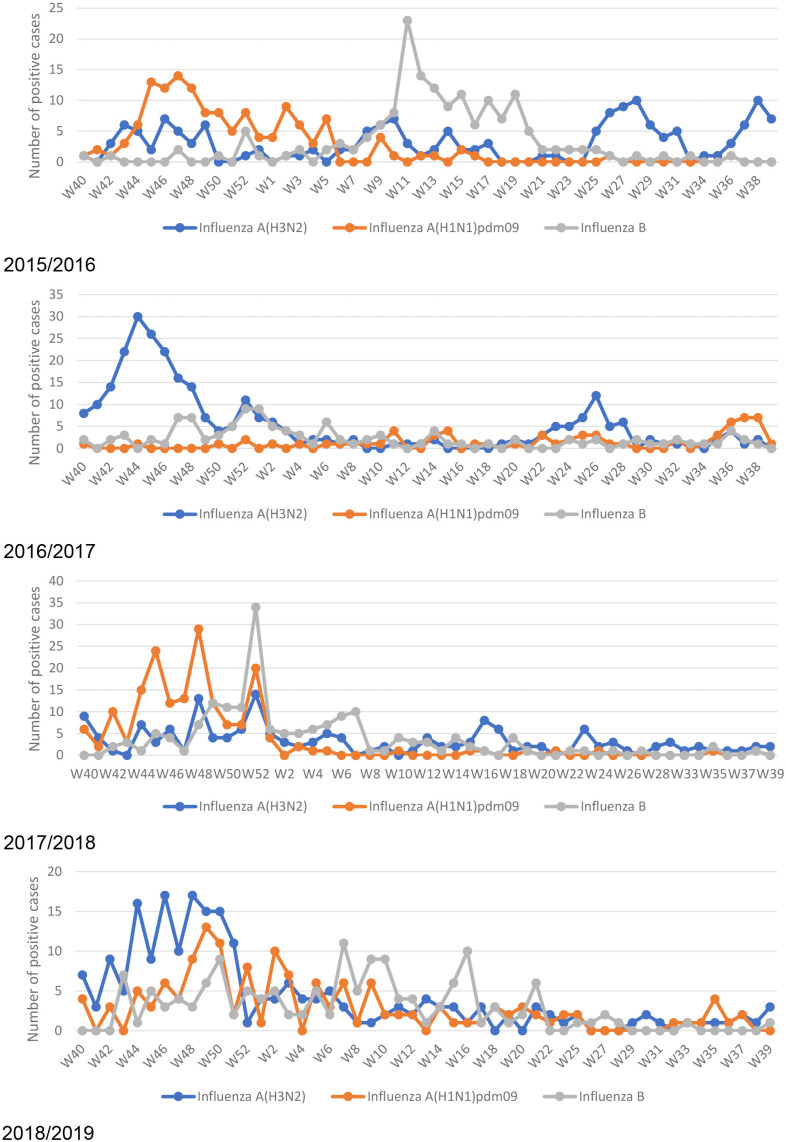
Influenza peaks were observed in October each season, with variant predominant strains. Except for a high peak of the influenza B subtype in March 2016, a mild peak also appeared during summer months (June and July) coincided with Umrah/Hajj season. Throughout the studied seasons, influenza virus subtypes co-circulate with no reports of co-infection (Figure 2).
Figure 2.
Seasonal pattern of influenza A and B subtypes.
Influenza A(H3N2) was reported in 42% of the cases, then influenza B (30.7%) and influenza A(H1N1)pdm09 (27.3%). Half of the patients were females (51.0%), and their median age was 31. Most of the cases (50.2%) were reported among adult patients (19–60 years old). The majority of the cases were Saudi (93.2%), and healthcare workers represented 9.4% of the cases with no predominance of one subtype (Table 1).
Table 1.
Demographic characteristics of patients with laboratory-confirmed cases
| Variables | Influenza subtypes | Total (n = 1928) | ||
|---|---|---|---|---|
| Influenza A(H3N2) (n = 810, 42.0%) | Influenza A(H1N1)pdm09 (n = 526, 27.3%) | Influenza B (n = 592, 30.7%) | ||
| Gender | ||||
| Male | 404 (49.9) | 250 (47.5) | 291 (49.2) | 945 (49.0) |
| Female | 406 (50.1) | 276 (52.5) | 301 (50.8) | 983 (51.0) |
| Age† (years) | ||||
| ≤5 | 167 (20.6)* | 53 (10.1)* | 105 (17.7)* | 325 (16.9) |
| 6–18 | 104 (12.8) | 37 (7.0) | 125 (21.1) | 266 (13.8) |
| 19–60 | 366 (45.2) | 331 (62.9) | 271 (45.8) | 968 (50.2) |
| >60 | 173 (21.4) | 105 (20.0) | 91 (15.4) | 369 (19.1) |
| Nationality | ||||
| Saudi | 755 (93.2) | 485 (92.2) | 557 (94.1) | 1797 (93.2) |
| Non-Saudi | 55 (6.8) | 41 (7.8) | 35 (5.9) | 131 (6.8) |
| Job category | ||||
| Health care worker | 73 (9.0) | 57 (10.8) | 52 (8.8) | 182 (9.4) |
| Non-health care worker | 737 (91.0) | 469 (89.2) | 540 (91.2) | 1746 (90.6) |
Median age of all patients = 31 years, IQR = (11.0–55.0 years), median age of the admitted patients was 40.2 years (IQR = 6–66 years).
Significant association comparing each subtype to the others (p < 0.05).
Column % was used to show distribution within each influenza subtype.
The most common symptoms and signs were fever (65.4%), cough (61.2%) and sore throat (41.6%), and the most common comorbidities were diabetes mellitus (10.4%), cardiovascular (10.6%) and chronic respiratory illnesses (8.0%) (Table 2).
Table 2.
Clinical characteristics and co-morbidities among the laboratory-confirmed cases
| Clinical manifestations and co-morbidities | Influenza subtypes | Total (n = 1928) | ||
|---|---|---|---|---|
| Influenza A(H3N2) (n = 810, 42.0%) | Influenza A(H1N1)pdm09 (n = 526, 27.3%) | Influenza B (n = 592, 30.7%) | ||
| Clinical symptoms and signs | ||||
| Fever | 564 (69.6)* | 281 (53.4)* | 415 (70.1)* | 1260 (65.4) |
| Cough | 520 (64.2)* | 268 (51.0)* | 391 (66.0)* | 1179 (61.2) |
| Sore throat | 350 (43.2) | 190 (36.1)* | 263 (44.4) | 803 (41.6) |
| Runny nose | 342 (42.2)* | 158 (30.0)* | 227 (38.3) | 727 (37.7) |
| Shortness of breath | 177 (21.9) | 110 (20.9) | 113 (19.1) | 400 (20.7) |
| Body aches | 144 (17.8) | 109 (20.7) | 110 (18.6) | 363 (18.8) |
| Headache | 87 (10.7) | 77 (14.6) | 76 (12.8) | 240 (12.4) |
| Vomiting | 74 (9.1) | 44 (8.4) | 62 (10.5) | 180 (9.3) |
| Sneezing | 67 (8.3) | 48 (9.1) | 41 (6.9) | 156 (8.1) |
| Diarrhea | 45 (5.6) | 21 (4.0) | 42 (7.1) | 108 (5.6) |
| Nausea | 37 (4.6) | 30 (5.7) | 25 (4.2) | 92 (4.8) |
| Comorbidities | ||||
| Diabetes mellitus | 86 (10.6) | 59 (11.2) | 56 (9.5) | 210 (10.4) |
| Hypertension | 94 (11.6) | 57 (10.8) | 54 (9.1) | 205 (10.6) |
| Chronic respiratory disease | 79 (9.8)* | 31 (5.9)* | 45 (7.6) | 155 (8.0) |
| Chronic kidney disease | 26 (3.2) | 13 (2.5) | 13 (2.2) | 52 (2.7) |
| Immunosuppression | 32 (4.0) | 12 (2.3) | 24 (4.1) | 68 (3.5) |
Significant association comparing each subtype to the others (p < 0.05).
Column % was used to show distribution of the clinical symptoms and comorbidities within each subtype.
One-third of the influenza cases (30.4%) were admitted to the hospital with a median admission duration of 4 days. The median age of the admitted patients was 40.5 years (IQR = 6–66.2 years). Among the influenza A(H3N2) patients, 34.7% were admitted, compared to lesser proportions among patients with influenza A(H1N1)pdm09 and Influenza B (27.9% and 26.7%, respectively). More than 40% of influenza A(H1N1)pdm09 patients were admitted for more than 5 days, compared to 37.3% among influenza B and 28.9% among influenza A(H3N2) patients. Pneumonia, as a complication of influenza, was observed in 1.8%, and deaths occurred in 1.1% of the cases. Almost similar proportions of pneumonia and mortality were observed among patients with different virus subtypes (Table 3).
Table 3.
Hospital admission and outcomes among the studied cases
| Variables | Influenza subtypes | Total | ||
|---|---|---|---|---|
| Influenza A(H3N2) (n = 810, 42.0%) | Influenza A(H1N1)pdm09 (n = 526, 27.3%) | Influenza B (n = 592, 30.7%) | ||
| Hospital admission | ||||
| Yes (n = 586, 30.4%) | 281 (34.7)* | 147 (27.9) | 158 (26.7)* | 586 (30.4) |
| No (n = 1342, 69.6%) | 529 (65.3) | 379 (72.1) | 434 (73.3) | 1342 (69.6) |
| Duration of admission (days) | ||||
| 1–2 | 99 (35.2)* | 41 (27.9) | 41 (25.9) | 181 (30.9) |
| 3–5 | 101 (35.9) | 47 (32.0) | 58 (36.7) | 206 (35.2) |
| 6–7 | 35 (12.5) | 16 (10.9) | 18 (11.4) | 69 (11.8) |
| >7 | 46 (16.4) | 43 (29.3) | 41 (25.9) | 130 (22.2) |
| Pneumonia as complication | ||||
| Yes | 15 (1.9) | 11 (2.1) | 8 (1.4) | 34 (1.8) |
| No | 795 (98.1) | 515 (97.9) | 584 (98.6) | 1894 (98.2) |
| Outcome | ||||
| Discharge | 802 (99.0) | 518 (98.5) | 586 (99.0) | 1906 (98.9) |
| Death | 8 (1.0) | 8 (1.5) | 6 (1.0) | 22 (1.1) |
Significant association comparing each subtype to the others (p < 0.05).
Column % was used to show distribution of hospital admission and outcomes within each subtype.
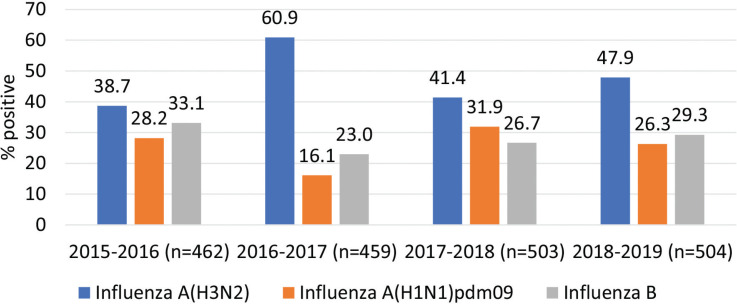
Figure 3 depicts hospital admission per influenza season. Admission due to influenza B infection was almost similar from 2015–2016 to 2018–2019 (range between 23% and 33.1%); however, a variation was observed in the admission proportions due to influenza A(H3N2) (range between 38.7% and 60.9%) and influenza A(H1N1)pdm09 (range between 16.1% and 31.9%) (Figure 3).
Figure 3.
Hospital admission distributed per influenza season.
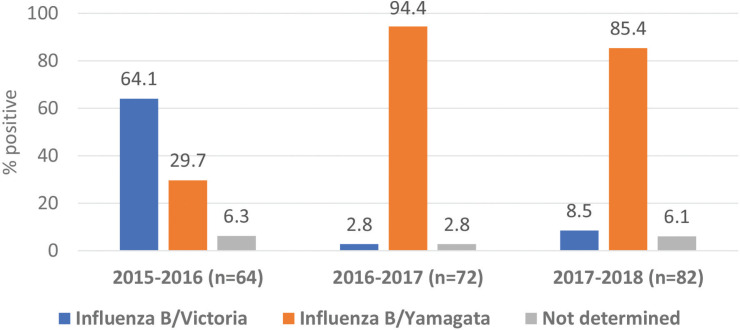
The influenza B viruses were subtyped in 218 cases only. Victoria lineage was predominant (64.1%) in 2015 and 2016; however, Yamagata was predominant in the next two consecutive seasons (94.4% and 85.4%, respectively). Also, co-circulation of both B lineages was observed throughout the seasons. No samples were stored for further analysis of influenza A/non-H1N1 and influenza B during the season 2018 and 2019 (Figure 4).
Figure 4.
Influenza B lineages from 2015 to 2018.
4. DISCUSSION
This study investigated seasonality, subtypes, lineages, and clinical pattern of a relatively large number of laboratory-confirmed influenza cases among Saudi and non-Saudi population with eligibility for medical service at KAMC in western Saudi Arabia, over four consecutive seasons. It added more evidence on the continuous co-circulation of influenza viruses throughout the years and vaccine mismatch for the influenza B lineages [18–20].
During the study period, influenza A(H3N2) was the most frequent circulating subtype (42.0%), followed by influenza B (30.7%) and influenza A(H1N1)pdm09 (27.3%). This finding was different from a previous study from 15 countries in the Eastern Mediterranean Region of confirmed influenza among patients with suspected severe acute respiratory infection, where influenza A(H1N1)pdm09 was the frequent circulating subtype (41.7%); however, influenza B infections were the second (28.7%) [18]. Another study in the Middle East and North Africa (MENA) region (16 countries) from April 2009 to December 2017 (no data was reported from Saudi Arabia), showed that influenza A(H1N1)pdm09 was the most frequently circulating subtype (50.8%) followed by influenza B (17.2%) and influenza A(H3N2) (15.9%) [21]. These data should be interpreted, taking into consideration the inclusion of data during the 2009 influenza A(H1N1)pdm09 pandemic.
Concerning the predominant influenza subtype per season, influenza A(H3N2) and influenza B were equally common in influenza season 2015 and 2016, influenza A(H3N2) was predominant in two influenza seasons (2016–2019), and during the season 2017 and 2018, influenza B was the predominant subtype. These findings were different than those reported in the Eastern Mediterranean Region, where influenza B was the predominant circulating subtype in the influenza season 2016 and 2017, and influenza A(H1N1)pdm09 was predominant in influenza season 2017 and 2018 [18,22]. Influenza A(H3N2) showed faster and more diverse antigenic drift than influenza A(H1N1)pdm09 [21]. This drift and the trend for more severe infection may be the reason for the predominance of influenza A(H3N2). Meanwhile, influenza B represented one-third of the cases in each season in this study. This observation was higher than the previously reported case, where nearly one-fourth of all cases were due to influenza B virus in the MENA region from 2011 to 2016 [23].
A primary high amplitude influenza peak was observed in October and November each year, earlier than most northern hemisphere countries in the region [19,21,23], and continued till March to April. However, a smaller secondary peak was found during March and April and/or June and July, which has also been reported by another study in the MENA region [23]. The secondary peak during June and July coincided with Umrah/Hajj season during the studied years, although these peaks and the most common influenza subtypes are usually unexpected [23]. For instance, a secondary high amplitude peak due to the influenza B virus was observed during March and April in the season 2015 and 2016. Hajj represents the largest mass gathering event that occurs annually. Pilgrims visit the holy city of Makkah in the western region of Saudi Arabia throughout the year; however, number of pilgrims significantly increase during the Umrah season, and they reach the peak during the Hajj time, where almost 2 million pilgrims from over 180 countries gather over 5–7 days [24]. Hajj time follows the lunar calendar, thus it circulates over different times of the Gregorian calendar. During the study period from 2015 to 2019, the Umrah peak season and Hajj were during the summertime (mostly from June to August). Pilgrims from the southern hemisphere are a potential source for spreading the influenza virus during their stay and travel from Makkah. According to this potential risk, the Saudi Ministry of Health requires all internal pilgrims (from inside the country), healthcare staff, security, and food services personnel to receive the southern hemisphere influenza vaccine. So, both the Southern Hemisphere (SH) and the Northern Hemisphere (NH) influenza vaccines are distributed in Saudi Arabia. Yet, the currently observed influenza seasonality pattern supports to start the annual influenza vaccination campaign in Saudi Arabia at an earlier time to reduce the number of cases that start to increase as early as October [25].
This study analyzed subsets of nasopharyngeal samples from three seasons (2015–2018). Both B/Victoria-lineage and B/Yamagata-lineage were co-circulating. However, B/Victoria-lineage was predominant during influenza season 2015 and 2016, and B/Yamagata-lineage was predominant in the seasons 2016–2018. Nasopharyngeal samples were stored for the season 2018 and 2019. Data on influenza B sequences are very scarce from the region. A literature review of published influenza sequences in the Eastern Mediterranean Region since 2012 showed only 49 influenza B sequences, compared to 1036 influenza A sequences [19]. The trivalent influenza vaccine, the only vaccine used in this region over consecutive seasons, contained B/Victoria-lineage with an increased probability of vaccine mismatch. Simultaneously, the current data did not show an increased admission during the seasons with vaccine mismatch, which may be associated with the severity of the infections. Despite its relatively small sample size, this is the first study, to the best of our knowledge, to identify influenza B lineages in Saudi Arabia. It provides the need for regular investigation of Influenza B lineages on yearly basis. By contrast, our findings highlight the need for a shift from the trivalent into quadrivalent influenza vaccine, especially in consideration of the difficulty of predicting the dominant influenza B lineage each season [6,23]. Cross-protection studies of trivalent vaccine containing B/Victoria-lineage or B/Yamagata-lineage strain among middle-aged and elderly persons [26] or immunologically naïve children [27] did not show consistent findings.
Limitations of the current study included being a single-center experience and the relatively small number of influenza B positive samples analyzed for specific lineages. This also includes lack of sequence data on influenza A strains. This surveillance information is essential in studying virus evolution and assessing the effectiveness of both influenza vaccines and neuraminidase inhibitors that are used for the treatment of influenza infections (e.g. Oseltamivir and Zanamivir). Potential for selection bias is possible related to the inclusion of patients with moderate to severe symptoms who require physician consultation, making the study findings limited to a similar population. Accordingly, a community-based influenza surveillance program all year-round at the national level with a standard molecular detection assay for influenza viruses’ subtypes and lineages will provide in-depth information about the circulating influenza strains nationwide. This information will significantly contribute to the influenza vaccine decision and preparation in collaboration with the WHO. Another potential selection bias is related to the molecular subtype analysis of a subset of samples for influenza B lineages. However, the current findings may pave the way for further studies, especially with the lack of data on influenza B subtypes. The potential of selection bias in the current study makes it difficult to compare with other studies and more evidence is required to build up conclusions based on the current findings.
The similarity of the study sample with the general population and the longitudinal study over four-season period are considered among the study strengths.
In conclusion, few studies have been conducted in this region related to the influenza seasonality pattern and circulating influenza viruses, especially the influenza B lineages. Yet, understanding the impact of climatic factors such as absolute humidity and temperature and variations linked to Umrah and Hajj seasons requires further investigation. The current study monitored the activity of circulating influenza viruses in a tertiary care hospital in Jeddah city the main destination of the international pilgrims and the majority of the local pilgrims from inside Saudi Arabia). Data showed the co-circulation of influenza virus subtypes throughout the study years. The burden due to influenza B needs to be monitored regularly as it used to be underestimated compared to the influenza A virus. Besides, a quadrivalent influenza vaccine is recommended to reduce the burden of the disease associated with influenza B infections. The unique situation of Saudi Arabia related to the Hajj and Umrah is a trigger for continuous strengthened influenza surveillance, which will impact the global surveillance and public health preparedness against influenza virus spread.
ACKNOWLEDGMENTS
The authors would like to thank Sanofi Pasteur who funded this investigator sponsored study. Sanofi Pasteur was provided the opportunity to review a preliminary version of this manuscript for factual accuracy, but the authors are solely responsible for final content and interpretation. Also, thanks to Sulafah Hakami and Omaima Hassan from the Infection Prevention and Control Department, KAMC, Jeddah for their assistance in the data collection. Also, thanks to Noha Jawi and Rotana Magadmi from Molecular Biology laboratory for RNA extraction.
Footnotes
Data availability statement: The authors confirm that the data supporting the findings of this study are available within the article.
CONFLICTS OF INTEREST
The authors declare they have no conflicts of interest.
AUTHORS’ CONTRIBUTION
ABA and FF conceived the study. FF performed the statistical analysis. MSA, ALA, EIA and SAE performed the laboratory analysis. ABA and FF interpreted the results and wrote the first draft of the manuscript. All authors reviewed and approved the final version of the manuscript.
SUPPLEMENTARY MATERIALS
Supplementary data related to this article can be found at
.
REFERENCES
- [1].Paules C, Subbarao K. Influenza. Lancet. 2017;390:697–708. doi: 10.1016/S0140-6736(17)30129-0. [DOI] [PubMed] [Google Scholar]
- [2].Thompson CI, Barclay WS, Zambon MC. Changes in in vitro susceptibility of influenza A H3N2 viruses to a neuraminidase inhibitor drug during evolution in the human host. J Antimicrob Chemother. 2004;53:759–65. doi: 10.1093/jac/dkh155. [DOI] [PubMed] [Google Scholar]
- [3].Rota PA, Hemphill ML, Whistler T, Regnery HL, Kendal AP. Antigenic and genetic characterization of the haemagglutinins of recent cocirculating strains of influenza B virus. J Gen Virol. 1992;73:2737–42. doi: 10.1099/0022-1317-73-10-2737. [DOI] [PubMed] [Google Scholar]
- [4].Yoon SW, Webby RJ, Webster RG. Evolution and ecology of influenza A viruses. In: Compans R, Oldstone M, editors. Influenza Pathogenesis and Control - Volume I. Current Topics in Microbiology and Immunology. Switzerland: Springer, Cham; 2014. pp. 359–75. [DOI] [PubMed] [Google Scholar]
- [5].Chen R, Holmes EC. The evolutionary dynamics of human influenza B virus. J Mol Evol. 2008;66:655–63. doi: 10.1007/s00239-008-9119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ali G, Amer HM, Almajhdi FN. Hemagglutinin and neuraminidase genes of influenza B viruses circulating in Riyadh, Saudi Arabia during 2010-2011: evolution and sequence analysis. J Med Virol. 2014;86:1003–16. doi: 10.1002/jmv.23819. [DOI] [PubMed] [Google Scholar]
- [7].Nerome R, Hiromoto Y, Sugita S, Tanabe N, Ishida M, Matsumoto M, et al. Evolutionary characteristics of influenza B virus since its first isolation in 1940: dynamic circulation of deletion and insertion mechanism. Arch Virol. 1998;143:1569–83. doi: 10.1007/s007050050399. [DOI] [PubMed] [Google Scholar]
- [8].McCullers JA, Wang GC, He S, Webster RG. Reassortment and insertion-deletion are strategies for the evolution of influenza B viruses in nature. J Virol. 1999;73:7343–8. doi: 10.1128/JVI.73.9.7343-7348.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Shaw M, Cooper L, Xu X, Thompson W, Krauss S, Guan Y, et al. Molecular changes associated with the transmission of avian influenza A H5N1 and H9N2 viruses to humans. J Med Virol. 2002;66:107–14. doi: 10.1002/jmv.2118. [DOI] [PubMed] [Google Scholar]
- [10].Chi XS, Bolar TV, Zhao P, Tam JS, Rappaport R, Cheng SM. Molecular evolution of human influenza A/H3N2 virus in Asia and Europe from 2001 to 2003. J Clin Microbiol. 2005;43:6130–2. doi: 10.1128/JCM.43.12.6130-6132.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Wie SH, So BH, Song JY, Cheong HJ, Seo YB, Choi SH, et al. A comparison of the clinical and epidemiological characteristics of adult patients with laboratory-confirmed influenza A or B during the 2011-2012 influenza season in Korea: a multi-center study. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062685. e62685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dawood FS, Fiore A, Kamimoto L, Bramley A, Reingold A, Gershman K, et al. Burden of seasonal influenza hospitalization in children, United States, 2003 to 2008. J Pediatr. 2010;157:808–14. doi: 10.1016/j.jpeds.2010.05.012. [DOI] [PubMed] [Google Scholar]
- [13].Bakir TMF, Halawani M, Ramia S. Viral aetiology and epidemiology of acute respiratory infections in hospitalized Saudi children. J Trop Pediatr. 1998;44:100–3. doi: 10.1093/tropej/44.2.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Al-Shehri MA, Sadeq A, Quli K. Bronchiolitis in Abha, Southwest Saudi Arabia: viral etiology and predictors for hospital admission. West Afr J Med. 2005;24:299–304. doi: 10.4314/wajm.v24i4.28193. [DOI] [PubMed] [Google Scholar]
- [15].Rashid H, Shafi S, Haworth E, Memish ZA, El Bashir H, Ali KA, et al. Influenza vaccine in Hajj pilgrims: policy issues from field studies. Vaccine. 2008;26:4809–12. doi: 10.1016/j.vaccine.2008.07.001. [DOI] [PubMed] [Google Scholar]
- [16].Almajhdi FN, Ali G. Report on influenza A and B viruses: their coinfection in a saudi leukemia patient. Biomed Res Int. 2013;2013 doi: 10.1155/2013/290609. 290609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].World Health Organization . WHO information for the molecular detection of influenza viruses. Geneva, Switzerland: WHO; 2017. Available from: https://www.who.int/influenza/gisrs_laboratory/WHO_information_for_the_molecular_detection_of_influenza_viruses_20171023_Final.pdf. [Google Scholar]
- [18].Elhakim M, Hafiz Rasooly M, Fahim M, Sheikh Ali S, Haddad N, Cherkaoui I, et al. Epidemiology of severe cases of influenza and other acute respiratory infections in the Eastern Mediterranean Region, July 2016 to June 2018. J Infect Public Health. 2020;13:423–9. doi: 10.1016/j.jiph.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Khan W, El Rifay AS, Malik M, Kayali G. Influenza research in the eastern mediterranean region: a review. Oman Med J. 2017;32:359–64. doi: 10.5001/omj.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Malik M, Mahjour J, Khan W, Alwan A. Influenza in the Eastern Mediterranean region: identifying the unknowns for detection and control of epidemic and pandemic threats. East Mediterr Health J. 2016;22:428–9. doi: 10.26719/2016.22.7.428. [DOI] [PubMed] [Google Scholar]
- [21].Al Khatib HA, Al Thani AA, Gallouzi I, Yassine HM. Epidemiological and genetic characterization of pH1N1 and H3N2 influenza viruses circulated in MENA region during 2009-2017. BMC Infect Dis. 2019;19:314. doi: 10.1186/s12879-019-3930-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Horton KC, Dueger EL, Kandeel A, Abdallat M, El-Kholy A, Al-Awaidy S, et al. Viral etiology, seasonality and severity of hospitalized patients with severe acute respiratory infections in the Eastern Mediterranean Region, 2007-2014. PLoS One. 2017;12 doi: 10.1371/journal.pone.0180954. e0180954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Caini S, El-Guerche Séblain C, Ciblak MA, Paget J. Epidemiology of seasonal influenza in the Middle East and North Africa regions, 2010-2016: circulating influenza A and B viruses and spatial timing of epidemics. Influenza Other Respir Viruses. 2018;12:344–52. doi: 10.1111/irv.12544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Yezli S, Assiri AM, Alhakeem RF, Turkistani AM, Alotaibi B. Meningococcal disease during the Hajj and Umrah mass gatherings. Int J Infect Dis. 2016;47:60–4. doi: 10.1016/j.ijid.2016.04.007. [DOI] [PubMed] [Google Scholar]
- [25].World Health Organization Influenza. Vaccination timing. 2017 Available from: http://www.who.int/influenza/vaccines/tropics/vaccination_timing/en/ (accessed February 25, 2020).
- [26].Camilloni B, Neri M, Lepri E, Iorio AM. Cross-reactive antibodies in middle-aged and elderly volunteers after MF59-adjuvanted subunit trivalent influenza vaccine against B viruses of the B/Victoria or B/Yamagata lineages. Vaccine. 2009;27:4099–103. doi: 10.1016/j.vaccine.2009.04.078. [DOI] [PubMed] [Google Scholar]
- [27].Levandowski RA, Regnery HL, Staton E, Burgess BG, Williams MS, Groothuis JR. Antibody responses to influenza B viruses in immunologically unprimed children. Pediatrics. 1991;88:1031–6. https://pubmed.ncbi.nlm.nih.gov/1945607/ [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.