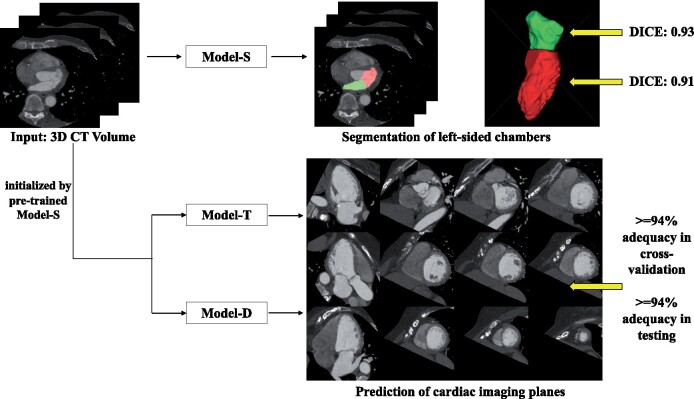
Abstract
Aims
To develop an automated method for bloodpool segmentation and imaging plane re-slicing of cardiac computed tomography (CT) via deep learning (DL) for clinical use in coronary artery disease (CAD) wall motion assessment and reproducible longitudinal imaging.
Methods and results
One hundred patients who underwent clinically indicated cardiac CT scans with manually segmented left ventricle (LV) and left atrial (LA) chambers were used for training. For each patient, long-axis (LAX) and short-axis planes were manually defined by an imaging expert. A DL model was trained to predict bloodpool segmentations and imaging planes. Deep learning bloodpool segmentations showed close agreement with manual LV [median Dice: 0.91, Hausdorff distance (HD): 6.18 mm] and LA (Dice: 0.93, HD: 7.35 mm) segmentations and a strong correlation with manual ejection fraction (Pearson r: 0.95 LV, 0.92 LA). Predicted planes had low median location (6.96 mm) and angular orientation (7.96) errors which were comparable to inter-reader differences (P > 0.71). 84–97% of DL-prescribed LAX planes correctly intersected American Heart Association segments, which was comparable (P > 0.05) to manual slicing. In a test cohort of 144 patients, we evaluated the ability of the DL approach to provide diagnostic imaging planes. Visual scoring by two blinded experts determined ≥94% of DL-predicted planes to be diagnostically adequate. Further, DL-enabled visualization of LV wall motion abnormalities due to CAD and provided reproducible planes upon repeat imaging.
Conclusion
A volumetric, DL approach provides multiple chamber segmentations and can re-slice the imaging volume along standardized cardiac imaging planes for reproducible wall motion abnormality and functional assessment.
Keywords: Deep learning, Computed tomography, Left ventricle, Left atrium, Wall motion abnormality
Graphical Abstract
Introduction
Accurate and reproducible morphofunctional assessment of the left ventricle (LV) is crucial as LV morphology, volumes, ejection fraction (EF), and regional function are critical parameters used in the diagnosis,1 clinical management, prognostication, and follow-up of numerous cardiovascular and systemic diseases.2,3 The assessment of LV parameters is included in clinical guidelines1–3 and is used for both inclusion criteria and endpoints in clinical trials.4 In addition, regional LV wall motion abnormalities for 17 American Heart Association (AHA) LV segments are assessed using standardized views and are important for the evaluation of cardiac pathology, including coronary artery disease (CAD).5,6 Beyond the LV, the assessment of the left atrium (LA) provides additional insight into cardiovascular disease and function and is particularly important in evaluating patients with atrial fibrillation, valvular disease, and diastolic heart failure.7
Cardiac computed tomography (CT) is a safe and cost-effective non-invasive imaging modality for the evaluation of suspected CAD8 and acute chest pain.9 Cardiac CT is an important prognostic tool in CAD and can be used to follow-up patients with atherosclerosis who are at increased risk of worsening stenosis severity and number of coronary vessels involved.10,11 While qualitative morphofunctional assessment is possible by reviewing phases of the cardiac cycle in a cine loop, quantitative assessment requires accurate segmentation, often requiring manual annotation of the images. Further, as images are acquired volumetrically, visualization of wall motion abnormalities requires generating standard imaging planes such as multiple long-axis (LAX) planes and one short-axis (SAX) stack.12 Currently, this requires specialized viewing software13 and manual processing which may lead to inter-reader variability, limiting clinical use.
Several deep learning (DL) algorithms have been developed for cardiac image segmentation14,15 but an approach that can perform segmentation and determine standard imaging planes has not been described. Previously, regions of interest16,17 and three orthogonal projections (axial, coronal, and sagittal)18,19 have been used as inputs for neural networks designed to segment cardiac chambers. More recently, fully convolutional networks (FCN) such as the ‘U-Net’20 have been shown to provide high pixel-wise segmentation accuracy and capture more global context. For example, a 2D U-Net with cross-entropy loss has shown the highest LV segmentation accuracy (Dice = 0.968) in the ‘ACDC’ dataset which is the largest publicly available cardiac MRI dataset;21 Baskaran et al. 22 successfully applied a 2D U-Net to segment four-chamber images in cardiac CT in a 2D slice-by-slice fashion with high accuracy (Dice > 0.91); and Vigneault et al.23 modified a conventional U-Net to also predict scaling and rotation of 2D MRI images. However, predicting SAX and LAX imaging planes requires significant global context. Unfortunately, 3D approaches significantly increase the memory demands. As a result, input image volumes are typically cropped or downsampled and networks have less features. For example, recent efforts first utilize a localization network for coarse detection of heart and then applied a 3D U-Net for segmentation of the heart region.24,25
We propose a variant of a 3D U-Net that performs pixel-wise segmentation and simultaneously predicts vectors which define the SAX and LAX imaging planes. We hypothesized that down-sampled 3D volumes will maintain sufficient network complexity and image information for accurate blood-pool segmentation and plane slicing and the shared network architecture will leverage similarities between the tasks. A fast, automatic, and reproducible method to both assess left-sided heart chamber volumes and generate standard cardiac imaging planes from volumetric cardiac CT images would increase clinical utility and reproducibility by avoiding the need for manual interaction. In this article, we test the ability of this DL framework to perform automated multi-chamber segmentation and LAX and SAX plane re-slicing of cardiac CT image volumes.
Materials and methods
Training population
Studies for training were identified from an existing cohort of patients with available expert segmentations. Under institutional review board approval (Study #191797), 100 electrocardiogram (ECG)-gated contrast enhanced, cardiac CT studies between June 2012 and June 2018 were retrospectively identified as meeting the inclusion criteria defined below.
Strict inclusion criteria were utilized to maximize training quality: each study had (i) reconstructed images across the entire cardiac cycle at 10% intervals and (ii) sufficient quality for bloodpool segmentation as determined by image analysis expert (author D.M.V.) and slice planning as determined by a cardiovascular imaging expert (author M.R.). Manual segmentations of LV and LA bloodpool were generated using a standardized processing pipeline (described below). For each patient, two frames [end diastole (ED) and end systole (ES)] served as training samples for the DL model.
Training images were collected at two institutions (67 UCSD, 33 NIH) with three CT systems. Two were long z-axis scanners with 256 detector rows (GE Revolution, n = 41 studies) and 320 detector rows (Toshiba AquilionONE, n = 47) allowing for a single heartbeat axial 16 cm acquisition throughout the cardiac cycle. Retrospective gating using a conventional low-pitch (0.18 0.02, range 0.16–0.22) helical acquisition over several heart beats was used with the third scanner (Siemens SOMATOM Force, n = 12). The training studies were performed for clinical cardiac indications: pre-operative assessment of patients undergoing transcatheter aortic valve replacement (TAVR, n = 39), suspected coronary artery disease (CAD, n = 38), and pre-operative assessment of pulmonary vein ablation (PVA, n = 23).
Manual segmentation and volumetric assessment
Pixel-wise manual segmentations of the LV and LA blood volumes, LVm and LAm respectively, (see Figure 1) were confirmed by an image analysis expert (author D.M.V.) with 7 years of experience in cardiac image segmentation using ITK-SNAP (Philadelphia, PA, USA).26 From each segmentation, blood chamber volumes were obtained, and the function of each chamber was measured via EF (LVEFm and LAEFm, respectively).
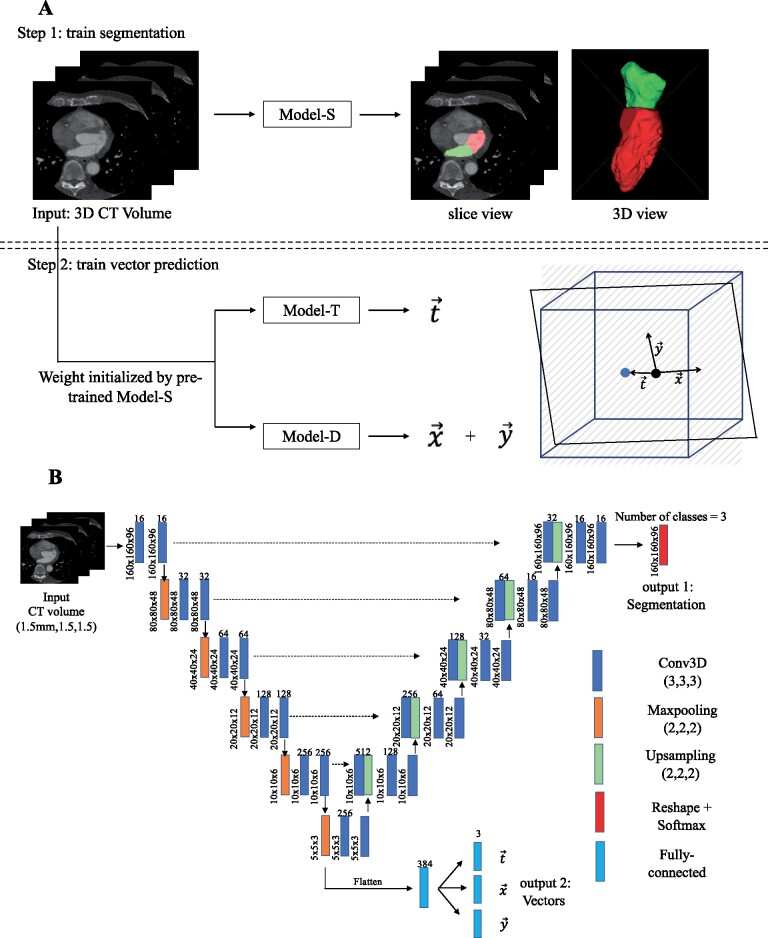
Figure 1.
Deep learning model training approach and model architecture. (A) 3D computed tomography volumes were first resampled to uniform spatial resolution (1.5 mm isotopically) and uniform dimension (160 × 160 × 96) and then served as an input to all models. Step 1: Model-S was trained to predicted LVDL (red) and LADL (green). Step 2: Model-T and Model-D were initialized by Model-S and then trained to predict imaging plane vectors DL, DL, and DL. A graphic illustration of these three vectors in relationship to the image volume is shown. The blue cube represents the computed tomography volume with a re-sliced plane in black. The blue dot is the centre of volume and black dot is the centre of plane. is the displacement between the blue and black dot and and are directional vectors of the 2D plane in the volume’s coordinate system. (B) U-Net architecture with added branch consisting of four fully connected layers after the last max-pooling layer in the down-sampling path was used. Conv3D, 3D convolution layer.
Manual imaging planes and plane vectors
Manual cardiac imaging planes were defined from the volumetric scans according to standardized guidelines27 by a fellowship-trained cardiovascular imaging expert (author M.R.). A SAX plane at the level of the mitral valve (MVm), a two-chamber plane (2CHm), a three-chamber plane (3CHm), and a four-chamber plane (4CHm) were identified for each patient. The short-axis stack (SAXm) of images was defined to span from the MVm to the LV apex using 8 mm slice spacing. Each plane was described by three vectors: xm and ym (defining the plane’s orientation) and tm (defining the plane’s centre). These vectors are illustrated in Figure 1A.
Model architecture
The DL model is an adaptation of the standard U-Net architecture20 with two modifications: (i) 3D convolution, maxpooling, and upsampling layers were utilized to accommodate 3D CT image volumes as the input and (ii) a fully connected layer was added after the last max-pooling layer in the down-sampling path to regress the plane vectors (Figure 1B). 3D CT images at ED and ES were resampled to 1.5 mm isotropic voxels for all patients. The DL model labels each voxel as one of three classes (LV, LA, or background). The three vectors (DL, DL, and DL) were predicted using three individual fully connected layers. The total loss of the model was defined to be:
with wi (i = seg, t, x, y) the weight assigned to each loss: the categorical cross-entropy loss of the segmentation, the mean square error of the predicted translation vector DL, the cosine proximity error for DL, and the cosine proximity error for DL.
Model training
Training was performed in stages. The first stage (‘Model-S’) was trained to perform LV and LA segmentations (LVDL and LADL, respectively) by assigning wseg = 1, wt = wx= wy = 0. ‘Model-S’ served as the initialization for the training of subsequent cardiac plane models. For each plane, two models were trained: one predicted the translation vector DL (‘Model-T’: wseg = wt = 1, wx= wy = 0) while another predicted directional vectors DL and DL (‘Model-D’: wseg = wx = wy =1, wt = 0). Each plane (2CH, 3CH, 4CH, and MV) was trained individually which led to four translation-vector models and four direction-vector models, and independent prediction of planes. Models were trained on ED and ES image volumes.
Training and validation were performed using five-fold cross-validation with random shuffling for robust unbiased evaluation. As a result, each model was trained on 80 studies (160 volumes) and evaluated on 20 validation studies (40 volumes). We report model performance across all folds.
Model evaluation and statistics
Chamber segmentation and assessment of function
Deep learning segmentation accuracy was evaluated using the Dice coefficient (a volumetric metric) and Hausdorff distance (HD, a surface-based metric). Dice coefficient is defined as 2(|VmanualVDL|)/(|VmanualVDL) and measures the overlap between manual and DL segmentation. The HD measures the local maximum distance between two surfaces Smanual and SDL. Differences in segmentation accuracy between CT vendors and between different clinical indications were evaluated using one-way analysis of variance (ANOVA) for Dice scores.
The accuracy of the segmentation-derived functional assessment was evaluated by comparing EFDL to manually derived EFm.
Quantitative evaluation of deep learning-predicted imaging plane vectors
2CHDL, 3CHDL, 4CHDL, and MVDL planes were derived from three predicted vectors output by ‘Model-T’ and ‘Model-D’. SAXDL was defined to be parallel to MVDL and span the LV as defined by the DL segmentation. For each plane, the differences between DL prediction PlaneDL and the manual plane Planem was assessed via the displacement error between centre of PlaneDL and that of Planem and angulation error of PlaneDL:
where normal vectors were calculated as the cross-product of and and is the vector length.
Inter- and intra-reader differences in imaging plane vectors
Ten studies were randomly selected to assess inter- and intra-reader differences in the manual delineation of cardiac planes. Reader 1 was the cardiac imaging expert mentioned above (author M.R.) who generated the plane annotations for DL training and Reader 2 was a fellowship-trained cardiothoracic radiologist with 12 years of experience (author S.K.). Inter-reader differences were assessed by comparison of this 10-study subset of Planem1 defined by Reader 1 to Planem2 defined by Reader 2. Intra-reader differences were assessed via repeat delineation by Reader 1 six months after the initial plane definition.
The DL algorithm was trained on annotations made by Reader 1. Therefore, two tests were used to evaluate DL predicted planes. First, the difference between the DL and Reader 1 on the study subset described above was compared to intra-reader differences. Second, the difference between DL planes and those defined by Reader 2 was compared to inter-reader differences.
Visual assessment of plane quality
For unbiased assessment of the DL performance (Supplementary material online, Figure S1A and B), Reader 2 assessed, in a randomized, blinded fashion, both the manual planes used for training (Planem, defined by Reader 1) and resulting DL-predicted planes (PlaneDL). Each plane was scored as one of four classes: perfect, excellent, good, and inadequate with the first three classes considered as diagnostically adequate. The criteria for each class can be found in the Supplementary material online.
Quantitative assessment of slice position: AHA wall assessment
The AHA 17-segment model was used to assess whether manual and DL-predicted LAX planes intersected the corresponding AHA segment at the mid-ventricular slice (Supplementary material online, Figure S1C).
Run-time for deep learning-based approach
We performed all DL trainings by using Keras (https://keras.io/) with TensorFlow (https://www.tensorflow.org/) on an 8-core Ubuntu (version: 18.04.3) workstation with 32 GB RAM equipped with a GeForce GTX 1080 Ti (NVIDIA Corporation, Santa Clara, CA, USA).
The times needed to train the models and to predict the DL chamber segmentations at ED and ES as well as vectors for four planes (three LAX and one MV plane) were recorded for each study.
Testing population
We tested our approach on 144 consecutive ECG-gated cardiac cine CT studies acquired at our institution between January and December 2019 under the same IRB approval. Studies were independent from training data. All studies had complete cardiac cycle reconstructions, radiology reports which assessed cardiac function, and a field-of-view which captured the entire LA and LV chambers. Studies were not included if the patients had congenital heart disease, images were taken for lead extraction planning, or studies showed metal implants in the LA or LV (e.g. mitral valve or leads in the LV chamber). Studies with coronary stents were included. The testing studies were performed for the following clinical cardiac indications: suspected CAD (n = 74), pre-operative assessment of PVA (n = 48), TAVR (n = 10), and others (n = 12). For each case, the diagnostic utility of DL-predicted frames was scored independently by Reader 2 and Reader 3 (author L.H., a fellowship-trained cardiovascular radiologist) using the criteria defined above, see Supplementary material online. In addition, each reader made a visual prediction of LVEF (to the nearest 5%) which was compared to the automated segmentation value.
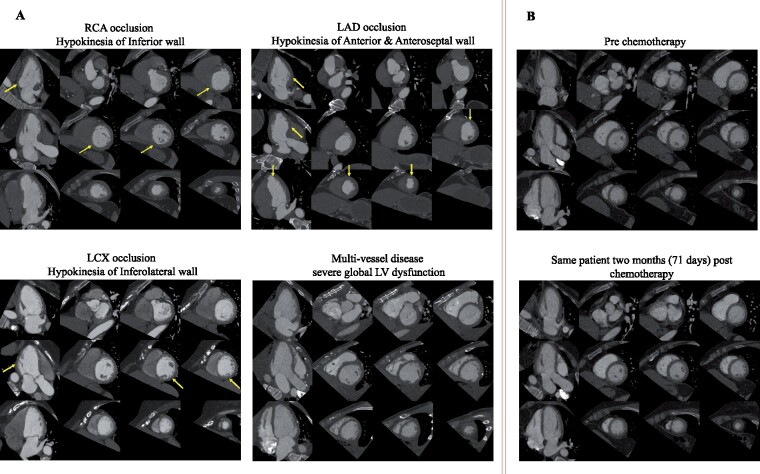
To highlight the utility in the evaluation of segmental LV wall motion abnormality (WMA), we show four studies with a radiologically confirmed diagnosis of coronary artery disease: (i) right coronary artery stenosis with inferior wall WMA, (ii) left anterior descending stenosis with anterior, anteroseptal wall, and apical cap WMA, (iii) left circumflex stenosis with mild WMA in the inferolateral wall, and (iv) three-vessel stenosis with globally reduced cardiac function. All stenoses and corresponding WMA were identified on clinical radiologic assessment. To assess the utility of PlaneDL in longitudinal CT imaging, we show PlanesDL generated for two CT studies obtained 71 days apart in the same patient (before and after chemotherapy) as part of the cardio-oncology evaluation.
Statistical evaluation
Unless otherwise indicated above, data are reported as median (with interquartile range) given non-normality on Shapiro–Wilk testing. Pearson correlation and two-tailed paired Student’s t-test were performed to test agreement between the DL- and manually derived EF. Two-tailed categorical z-test was used to evaluate differences in the proportion of successful intersecting between PlaneDL and Planem. Spearman correlation was used to assess visual and DL-derived EF in the validation cohort. Statistical significance was set at a P 0.05. All analyses were performed in Python version 3.6 with scipy (version 1.1.0).
Results
Of the 100 patients used for training, 60 (60%) were male (age: 67 ± 16) and 40 (40%) were female (age 69 ± 18). The bloodpool in the training data had a median intensity of 495 HU (IQR: 401–607, range: 277–885 HU) for the LV and 541 HU (IQR: 429–664, range: 257–1014 HU) for the LA.
Of the 144 patients used for testing, 89 (61.8%) were male (age: 59 ± 16) and 55 (38.2%) were female (age: 63 ± 15). The bloodpool in the testing data had a median intensity of 525 HU (IQR: 433–616, range: 320–1022 HU) for the LV and 544 HU (IQR: 451–632, range: 295–1207 HU) for the LA.
Chamber segmentation and assessment of function
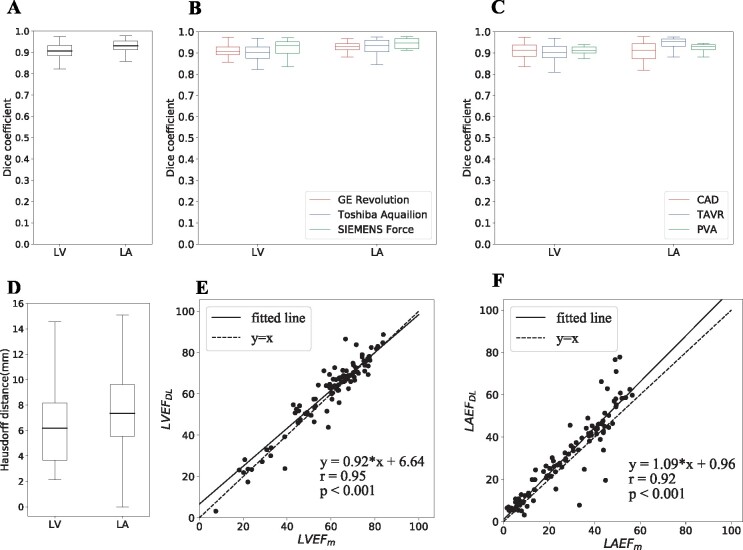
DL-predicted segmentations had median Dice coefficients of 0.907 (IQR: 0.884–0.932) and 0.931 (IQR: 0.913–0.954) (Figure 2A), and median HDs of 6.2 mm (IQR: 3.7–8.2) and 7.3 mm (IQR: 5.6–9.6) for the LV and LA, respectively (Figure 2D). There were no statistically significant differences (P > 0.05) in Dice across vendors but Dice in the LA did vary with respect to clinical indications (P = 0.001) on ANOVA testing (Figure 2B and C).
Figure 2.
Close agreement between deep learning and manual chamber segmentation and function assessment. (A) Dice coefficient for two chambers of interest, the left ventricle and left atrial was high. (B) Dice coefficient for three computed tomography scanners. (C) Dice coefficient for three types of clinical indications. (D) Hausdorff distance for left ventricle and left atrial. (E) Correlation of left ventricular ejection fraction derived using manual and deep-learning segmentation was close to identity (dashed) line with a fit (solid) of left ventricle EFDL = 0.92EFm + 6.64 and Pearson correlation r = 0.95 with P < 0.001. (F) Left atrial ejection fraction correlation was close to identity (dashed) line with fit (solid) of left atrial EFDL = 1.09EFm + 0.96, and Pearson correlation r = 0.92 with P < 0.001.
There was close agreement in LVEF (Pearson correlation r = 0.95, P = 0.49) as well as between LA EF (r = 0.92, P = 0.29) (Figure 2E and F).
Quantitative evaluation of deep learning-predicted imaging plane vectors
The median displacement error between Planem and PlaneDL was 7.0 mm (IQR: 5.0–9.5) across all planes, 6.3 mm (IQR: 4.3–8.5) for 2CH, 6.2 mm (IQR: 4.3–8.8) for 3CH, 7.2 mm (IQR: 5.5–9.6) for 4CH and 7.6 mm (IQR: 5.3–11.0) for MV. The median orientation error between planes was 8.0 (IQR: 5.0–11.7) across all planes, 9.5 (IQR: 6.0–13.9) for 2CH, 8.3 (IQR: 4.8–13.0) for 3CH, 7.2 (IQR: 4.6–11.2) for 4CH and 7.5 (IQR: 4.8–9.4) for MV.
Intra and inter-reader differences in imaging plane vectors
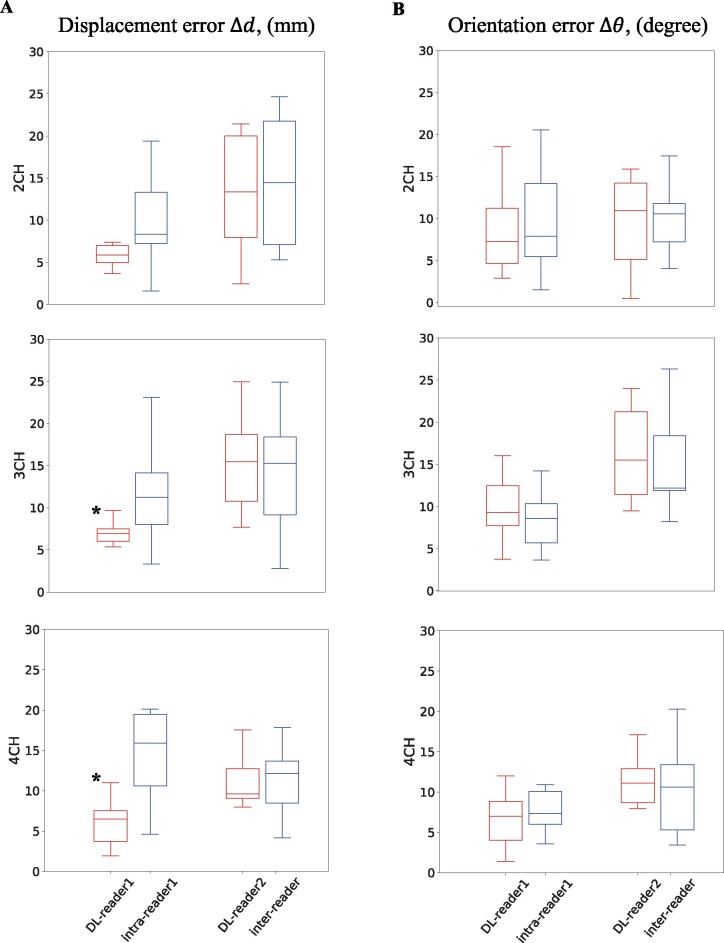
Deep learning-reader orientation differences were not significantly different (P > 0.05) compared with the corresponding inter- and intra-reader difference (Figure 3, Table 1). In terms of displacement differences the 3CH and 4CH DL-reader1 difference was significantly smaller (P < 0.05) than the intra-reader difference (Figure 4A).
Figure 3.
Comparison of long-axis plane location and angulation differences between readers and deep learning. Plane displacement (A) and orientation (B) differences between deep learning and Reader 1 (first red boxplot) were compared to intra-Reader 1 differences (first blue boxplot) and differences between deep learning and Reader 2 (second red boxplot) were compared to inter-Reader differences (second blue boxplot) for each long-axis plane. The asterisk (*) indicates significant differences. CH, chamber.
Table 1.
Comparison of LAX plane location and angulation differences between readers and deep learning
| Intra-reader 1 difference | DL-reader 1 difference | P-value | Inter-reader difference | DL-reader 2 difference | P-value | ||
|---|---|---|---|---|---|---|---|
| 2CH | (mm) | 8.3 (7.3,13.3) | 5.9 (5.0,7.0) | 0.20 | 14.4 (7.1,21.8) | 13.4 (7.9,20.0) | 0.91 |
| (degree) | 7.8 (5.4,14.1) | 7.3 (4.7,11.2) | 0.57 | 10.6 (7.2,11.8) | 10.9 (5.1,14.2) | 0.75 | |
| 3CH | (mm) | 11.2 (8.0,14.2) | 6.9 (6.0,7.5)a | 0.04 | 15.3 (9.2,18.4) | 15.5 (10.8,18.7) | 0.76 |
| (degree) | 8.6 (5.7,10.3) | 9.3 (7.7,12.5) | 0.35 | 12.2 (11.9,18.4) | 15.5 (11.4,21.2) | 0.71 | |
| 4CH | (mm) | 15.9 (10.6,19.5) | 6.5 (3.7,7.5)a | 0.003 | 12.1 (8.5,13.7) | 9.6 (9.1,12.7) | 0.84 |
| (degree) | 7.3 (6.0,10.1) | 7.0 (4.0,8.8) | 0.35 | 10.6 (5.3,13.4) | 11.1 (8.7,12.9) | 0.82 |
Intra-reader 1 differences represent variation in planes planned by the same reader 6 months apart. Given that the DL approach was trained on slice planning by reader 1, DL-reader 1 differences were compared to intra-reader 1 differences. Inter-reader variation captures variation in slice planning by two different readers. DL-reader 2 differences were compared to inter-reader values. Differences were reported as median (IQR).
Indicates a significant difference (P < 0.05).
Figure 4.
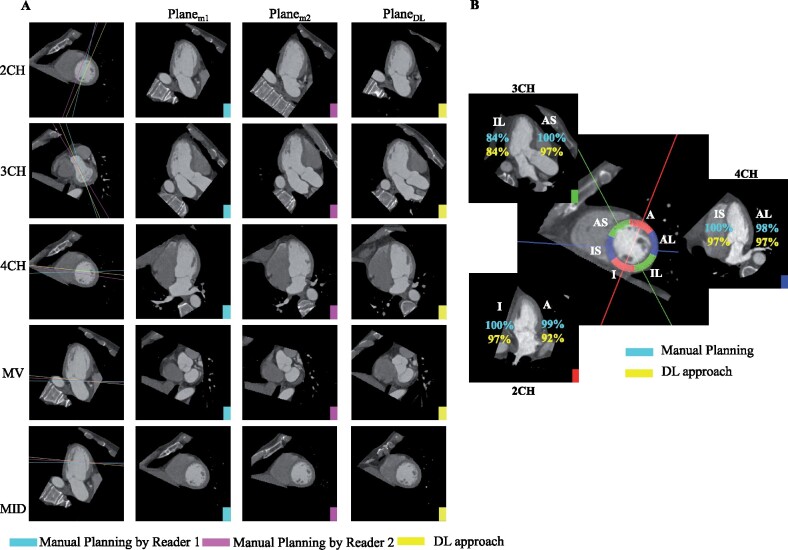
Deep learning agrees with manual slice planning and correctly visualizes corresponding AHA segments. (A) Rows 1–3: For each long-axis imaging plane, Column 1 shows the slice location on the short-axis slice (mid-ventricular slice for 2CH and 4CH and basal slice for 3CH). Columns 2–4 depict the corresponding images. Rows 4–5: For the short-axis mitral valve and mid-ventricular plane, Column 1 shows the slice position of the plane on the 2CH long-axis. Columns 2–4 depict the short-axis images: Planem1, (plane resliced by Reader 1), Planem2, (plane resliced by Reader 2) and PlaneDL (deep learning-derived slice). (B) Six mid-ventricular AHA segments are shown as arcs on a mid-ventricular short-axis slice. The percentage of cases in which a wall was correctly intersected by the associated long-axis plane are shown for the 2CH (red solid line, lower left), 3CH (green solid line, top left), and 4CH (blue solid line, right) planes. There is close agreement between the manual planes (reported in cyan) and deep learning (yellow). A, anterior; AL, anterolateral; AS, anteroseptal; I, inferior; IL, inferolateral; IS, inferoseptal.
Visual assessment of plane quality
The deep-learning approach yielded diagnostically adequate imaging planes for a large percentage (≥94%) of cases across all slice plane locations (Table 2, representative patient shown in Figure 4A, distribution of scores in Table S1).
Table 2.
Diagnostic adequacy of manual and deep-learning imaging planes as scored by cardiothoracic imaging expert
| Planem (%) | PlaneDL (%) | |
|---|---|---|
| 2CH | 100 | 100 |
| 3CH | 100 | 94 |
| 4CH | 100 | 98 |
| SAX | 100 | 100 |
Planem, plane manually resliced; PlaneDL, plane predicted by DL model.
Quantitative assessment of slice position via AHA wall assessment
The proportion of cases with correct AHA segment inclusion was not significantly different between Planem and PlaneDL for all AHA walls (two-sided z test, P > 0.05) except the anterior wall which had a lower likelihood of being visualized by the 2CH plane (P = 0.02) (Figure 4B, Table 3).
Table 3.
Assessment of AHA wall visualization for manual and DL-based cardiac planes
| Planem (%) | PlaneDL (%) | P-value | ||
|---|---|---|---|---|
| 2CH | Inferior | 100 | 97 | 0.08 |
| Anterior | 99 | 92 | 0.02* | |
| 3CH | Inferolateral | 84 | 84 | 1 |
| Anteroseptal | 100 | 97 | 0.08 | |
| 4CH | Inferoseptal | 100 | 97 | 0.08 |
| Anterolateral | 98 | 97 | 0.65 |
Percentage of cases in which the LAX plane correctly intersects corresponding AHA wall was shown.
Significant P-values are shown by asterisk.
Run-time for deep learning-based approach
Each model was trained for 50 epochs with each epoch using approximately 170 s in our workstation. The time needed to predict two segmentations (ED and ES) as well as four imaging planes (2CHDL, 3CHDL, 4CHDL, and MVDL) was 29.7 ± 4.0 s.
Assessment of the utility of deep learning-predicted planes on test cases
DL yielded diagnostically adequate imaging planes for a large percentage (≥99% by Reader 2 and ≥94% by Reader 3) of cases across all slice plane locations (Table 4, Table S2).
Table 4.
Diagnostic adequacy of deep-learning imaging planes in the testing group as scored by imaging experts
| Reader 2 (%) | Reader 3 (%) | |
|---|---|---|
| 2CH | 99 | 99 |
| 3CH | 100 | 94 |
| 4CH | 100 | 95 |
| SAX | 100 | 100 |
There was close agreement between the visual estimation of ejection fraction by readers and quantification via automated segmentation. Specifically, linear regression demonstrated a strong correlation (Spearman =0.93 and 0.95 for Reader 2 and 3, respectively). In addition, classification of EF < 40%, 40–50%, and >50% with the DL approach agreed with visual prediction in 88.9% and 80.5% of cases for Reader 2 and 3, respectively (Table 5).
Table 5.
The close agreement of classification of ejection fraction between visual estimation by expert readers and automated quantification via deep learning left ventricle segmentation
| Reader 2 |
Reader 3 |
||||||
|---|---|---|---|---|---|---|---|
| <40% | 40–50% | >50% | <40% | 40–50% | >50% | ||
| DL predict | <40% | 30 | 3 | 0 | 31 | 1 | 1 |
| 40–50% | 0 | 7 | 4 | 8 | 2 | 1 | |
| >50% | 0 | 9 | 91 | 2 | 15 | 83 | |
The classification of EF into <40%, 40–50%, and >50% with the DL approach agreed with visual prediction in 88.9% and 80.5% of cases for Reader 2 and 3, respectively.
Deep learning-predicted planes of four test CT studies with CAD provided visualization of both regional and global LV wall motion abnormalities recorded in the radiology report (Figure 5A, Supplementary material online, Video S1). The DL model generates similar planes in two CT studies of the same patient acquired as part of a longitudinal study (Figure 5B, Supplementary material online, Video S2).
Figure 5.
Utility of deep-learning slice planning in evaluation of regional wall motion abnormalities and longitudinal assessment of patients. (A) Four exemplar cases were selected based on radiologically observed with either global (bottom right) or regional (the rest three) left ventricle wall motion abnormality due to coronary artery disease. Yellow arrows point out the walls with regional dysfunction where the readers can refer to in Supplementary material online, Video S1. (B) Deep learning-predicted planes for the same patient imaged before chemotherapy and 2 months after the therapy which shows high reproducibility. The videos for these planes showing the cardiac function in one cardiac cycle are submitted as Supplementary material online, Videos. Window level = 500 HU, width = 900 HU.
Discussion
In this study, we developed and evaluated a DL network that automatically predicts cardiac chamber volumes as well as standardized cardiac imaging planes from volumetric CT data using a shared model architecture. The DL approach generated high-quality segmentations (median Dice = 0.907 and 0.931 for LV and LA, respectively) and had a strong correlation (Pearson r > 0.9) with manually derived EF. Furthermore, DL-predicted planes had low errors in spatial displacement and angulation and intersected the relevant mid-ventricular myocardial segment in a high proportion of cases. Testing the DL network in a series of 144 consecutive cine CT cases demonstrated our approach generates diagnostically useful imaging planes and automated segmentation leads to EF estimation that is in agreement with visual interpretation. We highlight the utility of our DL-predicted planes in the evaluation of LV wall motion abnormality and for reproducible longitudinal assessment in a sample of test cases with known pathological findings.
In our approach, a 3D image volume (at 1.5 mm isotropic voxel size) was utilized as the input for blood pool segmentation and image plane re-slicing. While a 3D approach significantly increases the memory utilization, the choice was intended to preserve volumetric context to enable accurate slicing of the LAX and SAX imaging planes. Other DL approaches for CT chamber segmentation have utilized portions of the volume as the image input; Dormer et al.16 and Zreik et al. 17 input patches and performed patch-wise image classification while Mortazi et al. 18 and Wang et al. 19 used three orthogonal projections (axial, coronal, and sagittal) as inputs. Despite the feature vector depth being limited due to memory constraints, our 3D approach led to segmentation accuracy comparable to recent work by Baskaran et al.22 which applied a 2D U-Net to segment CT in a 2D slice-by-slice fashion and achieved Dice >0.91 for all four chambers.
The feasibility of predicting imaging planes was previously shown in 2D by Vigneault et al.23 in which a conventional U-Net was modified to predict a scaling and rotation of MRI images. Other efforts to use DL for plane prediction have focused on cardiac MRI. Le et al.28 and Blansit et al.29 recently presented DL-based cardiac MRI plane prescription by using DL to localize the anatomical landmarks that mimic how an expert performs slice planning on 2D MR. However, this is not required during CT acquisitions as they are volumetric.
Compared with manual annotation, our approach automatically and quickly analyzed the series of 3D CT image volumes (∼30 s for volumes at ED and ES). This represents a significant improvement as it usually takes a trained expert around 20 min to obtain volumes from two 3D image volumes at ED and ES30 and it leads to interobserver variability. Furthermore, the processing time of our approach can be readily improved through parallelization, as we currently predict imaging planes sequentially. While optimization of prediction time was not the focus of this study, the fast computation time increases the likelihood of clinical translation.
Our model was trained using annotations provided by one imaging expert and scored by a second independent blinded reader. The use of a second, independent reader confirms the utility of our imaging planes as it demonstrates that the DL approach leads to imaging planes that were not significantly different that the observer inter- and intra-reader variations in slice planning. Further, the inter- and intra-reader variations illustrate the extent of uncertainty in planning imaging planes as a range of plane angles and position can provide correct visualization. While this leads to variability between observers, our approach provides robust results that are comparable to a trained expert.
Robust and automated prediction of cardiac volumes and imaging planes could be used to measure multiple important clinical parameters. Apart from EF, the availability of LAX planes can enable the assessment of additional measures such as contour-based global longitudinal and circumferential strain. Furthermore, labelling of other cardiac chambers or the myocardium in the training data would enable measurements such as RV volumes or myocardial masses.
Our fully automated approach may enable rapid and reproducible assessment of global function as well as regional wall motion abnormalities in patients, such as those with CAD and other cardiomyopathies who are frequently evaluated with cardiac CT. In addition, automatic slicing of standardized cardiac planes can be used for reproducible longitudinal assessment of patients undergoing serial cardiac exams and in clinical trials.
Our approach has limitations. First, we observed lower performance for 3CH plane predictions relative to 2CH and 4CH planes. However, the success rate for the inferolateral wall was decreased in both 3CHm and 3CHDL. This could be explained by the difficulty in planning 3CH views as there is a balance between correct visualization of the left ventricular outflow tract and the intersection of the inferolateral wall. This suggests that anatomical variability may be limiting the performance of the DL approach. Second, the time and effort needed to derive both cardiac planes and blood chamber segmentations limited our training size. However, the achieved accuracy suggests clinical utility is possible even with the limited training data that was available. Lastly, the accuracy of the algorithm in patients with a wider range of phenotypes including implanted medical devices, significant changes in iodine contrast timing/intensity, and decreases in overall image quality were not studied but are planned for future work.
In conclusion, DL can automatically perform multi-chamber volumetric assessments and generate standardized cardiac imaging planes from CT images. This approach has the promise for regional cardiac visualization and reproducible assessment of cardiac function.
Supplementary material
Supplementary material is available at European Heart Journal – Digital Health.
Funding
National Institutes of Health (NIH) (NHLBI HL145817 and HL143113 to F.C.).
Conflict of interest: none declared.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author. The source code is available in https://github.com/ucsd-fcrl/AI_chamber_segmentation_plane_re-slicing.git.
Supplementary Material
References
- 1. Yancy CW, Januzzi JL, Allen AL, et al. 2017 ACC/AHA/HFSA Focused Update of the 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Failure Society of Amer. Circulation 2017;136:e137–e161. [DOI] [PubMed] [Google Scholar]
- 2. Al-Khatib SM, Stevenson WG, Ackerman MJ, et al. 2017 AHA/ACC/HRS guideline for management of patients with ventricular arrhythmias and the prevention of sudden cardiac death: executive summary. Circulation 2018;138:e210–e271. [DOI] [PubMed] [Google Scholar]
- 3. Nishimura RA, Otto CM, Bonow RO, et al. 2017 AHA/ACC focused update of the 2014 AHA/ACC guideline for the management of patients with valvular heart disease: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol 2017;70:252–289. [DOI] [PubMed] [Google Scholar]
- 4. Moss AJ, Zareba W, Hall WJ, et al. Prophylactic implantation of a defibrillator in patients with myocardial infarction and reduced ejection fraction. N Engl J Med 2002;346:877–883. [DOI] [PubMed] [Google Scholar]
- 5. McVeigh ER, Pourmorteza A, Guttman M, et al. Regional myocardial strain measurements from 4DCT in patients with normal LV function. J Cardiovasc Comput Tomogr 2018; doi:10.1016/j.jcct.2018.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Contijoch FJ, Groves DW, Chen Z, Chen MY, McVeigh ER.. A novel method for evaluating regional RV function in the adult congenital heart with low-dose CT and SQUEEZ processing. Int J Cardiol. 2017;249:461–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goette A, Kalman JM, Aguinaga L, et al. EHRA/HRS/APHRS/SOLAECE expert consensus on atrial cardiomyopathies: definition, characterization, and clinical implication. Europace 2016;18:1455–1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Neglia D, Rovai D, Caselli C, et al. Detection of significant coronary artery disease by noninvasive anatomical and functional imaging. Circ Cardiovasc Imaging 2015;8. [DOI] [PubMed] [Google Scholar]
- 9. Hoffmann U, Truong QA, Schoenfeld DA, et al. Coronary CT angiography versus standard evaluation in acute chest pain. N Engl J Med 2012;367:299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hulten EA, Carbonaro S, Petrillo SP, Mitchell JD, Villines TC.. Prognostic value of cardiac computed tomography angiography. J Am Coll Cardiol 2011;57:1237–1247. [DOI] [PubMed] [Google Scholar]
- 11. Cho I, Chang H-J, Sung JM, et al. Coronary computed tomographic angiography and risk of all-cause mortality and nonfatal myocardial infarction in subjects without chest pain syndrome from the CONFIRM registry (Coronary CT Angiography Evaluation for Clinical Outcomes: an International Multicenter registry). Circulation 2012;126:304–313. [DOI] [PubMed] [Google Scholar]
- 12. Lu MT, Ersoy H, Whitmore AG, Lipton MJ, Rybicki FJ.. Reformatted four-chamber and short-axis views of the heart using thin section (≤2 mm) MDCT images. Acad Radiol 2007;14:1108–1112. doi:10.1016/j.acra.2007.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ecabert O, Peters J, Walker MJ, et al. Segmentation of the heart and great vessels in CT images using a model-based adaptation framework. Med Image Anal 2011;15:863–876. [DOI] [PubMed] [Google Scholar]
- 14. Chen C, Qin C, Qiu H, et al. Deep learning for cardiac image segmentation: a review. Front Cardiovasc Med 2020;7:25. doi: 10.3389/fcvm.2020.00025. [DOI] [PMC free article] [PubMed]
- 15. Litjens G, Ciompi F, Wolterink JM, et al. State-of-the-art deep learning in cardiovascular image analysis. JACC Cardiovasc Imaging 2019;12:1549–1565. [DOI] [PubMed] [Google Scholar]
- 16. Dormer JD, Fei B, Halicek M, Ma L, Reilly CM, Schreibmann E. Heart chamber segmentation from CT using convolutional neural networks. In: Gimi B, Krol A, eds. Medical Imaging 2018: Biomedical Applications in Molecular, Structural, and Functional Imaging, vol. 10578. USA: SPIE; 2018. p100. [DOI] [PMC free article] [PubMed]
- 17. Zreik M, Leiner T, de Vos BD, van Hamersvelt RW, Viergever MA, Isgum I. Automatic segmentation of the left ventricle in cardiac CT angiography using convolutional neural networks. In: 2016 IEEE 13th International Symposium on Biomedical Imaging (ISBI). IEEE; 2016. p40-43. https://ieeexplore.ieee.org/document/7493206.
- 18. Mortazi A, Burt J, Bagci U. Multi-planar deep segmentation networks for cardiac substructures from MRI and CT. 2017. http://arxiv.org/abs/1708.00983.
- 19. Wang C, Smedby Ö. Automatic whole heart segmentation using deep learning and shape context. In: Pop M, Sermesant M, Jodoin P-M, et al. , eds. Statistical Atlases and Computational Models of the Heart. ACDC and MMWHS Challenges. Cham: Springer International Publishing; 2018.. p242–249. [Google Scholar]
- 20. Ronneberger O, Fischer P, Brox T.. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells WM, Frangi AF, eds. Medical Image Computing and Computer-Assisted Intervention – MICCAI 2015. Cham: Springer International Publishing; 2015. p234–241. [Google Scholar]
- 21. Bernard O, Lalande A, Zotti C, et al. Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: is the problem solved? IEEE Trans Med Imaging 2018;37:2514–2525. [DOI] [PubMed] [Google Scholar]
- 22. Baskaran L, Maliakal G, Al’Aref SJ, et al. Identification and quantification of cardiovascular structures from CCTA. JACC Cardiovasc Imaging 2020;13:1163–1171. [DOI] [PubMed] [Google Scholar]
- 23. Vigneault DM, Xie W, Ho CY, Bluemke DA, Noble JA.. Ω-Net (Omega-Net): fully automatic, multi-view cardiac MR detection, orientation, and segmentation with deep neural networks. Med Image Anal 2018;48:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu Z, Wu Z, Feng J.. CFUN: Combining Faster R-CNN and U-net Network for Efficient Whole Heart Segmentation. 2018.
- 25. Wang C, MacGillivray T, Macnaught G, Yang G, Newby D. A two-stage 3D Unet framework for multi-class segmentation on full resolution image. arXiv. April 2018. http://arxiv.org/abs/1804.04341.
- 26. Yushkevich PA, Piven J, Hazlett HC, et al. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage 2006;31:1116–1128. [DOI] [PubMed] [Google Scholar]
- 27. Kramer CM, Barkhausen J, Bucciarelli-Ducci C, Flamm SD, Kim RJ, Nagel E.. Standardized cardiovascular magnetic resonance imaging (CMR) protocols: 2020 update. J Cardiovasc Magn Reson. 2020;22:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Le M, Lieman-Sifry J, Lau F, Sall S, Hsiao A, Golden D.. Computationally efficient cardiac views projection using 3D convolutional neural networks. In: Cardoso MJ, Arbel T, Carneiro G, et al. , eds. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support. Cham: Springer International Publishing; 2017. p109–116. [Google Scholar]
- 29. Blansit K, Retson T, Masutani E, Bahrami N, Hsiao A.. Deep learning-based prescription of cardiac MRI planes. Radiol Artif Intell 2019;1:e180069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bai W, Sinclair M, Tarroni G, et al. Automated cardiovascular magnetic resonance image analysis with fully convolutional networks. J Cardiovasc Magn Reson 2018;20:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author. The source code is available in https://github.com/ucsd-fcrl/AI_chamber_segmentation_plane_re-slicing.git.