Abstract
The novel coronavirus disease 2019 (COVID‐19) is considered to be a significant health challenge worldwide because of its rapid human‐to‐human transmission, leading to a rise in the number of infected people and deaths. The detection of COVID‐19 at the earliest stage is therefore of paramount importance for controlling the pandemic spread and reducing the mortality rate. The real‐time reverse transcription‐polymerase chain reaction, the primary method of diagnosis for coronavirus infection, has a relatively high false negative rate while detecting early stage disease. Meanwhile, the manifestations of COVID‐19, as seen through medical imaging methods such as computed tomography (CT), radiograph (X‐ray), and ultrasound imaging, show individual characteristics that differ from those of healthy cases or other types of pneumonia. Machine learning (ML) applications for COVID‐19 diagnosis, detection, and the assessment of disease severity based on medical imaging have gained considerable attention. Herein, we review the recent progress of ML in COVID‐19 detection with a particular focus on ML models using CT and X‐ray images published in high‐ranking journals, including a discussion of the predominant features of medical imaging in patients with COVID‐19. Deep Learning algorithms, particularly convolutional neural networks, have been utilized widely for image segmentation and classification to identify patients with COVID‐19 and many ML modules have achieved remarkable predictive results using datasets with limited sample sizes.
Keywords: COVID‐19 diagnosis, CT, deep learning, machine learning, X‐ray
1. INTRODUCTION
Since the end of 2019, the world is experiencing a pandemic caused by the novel highly transmissible coronavirus called severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2).1, 2 The newly discovered viral‐induced lung disease was termed COVID‐19, which has rapidly spread to 222 countries and territories, causing over 102 million confirmed cases, and 2.2 million deaths by January 31, 2021. 3
COVID‐19 patients present with cough, fever, dyspnoea, fatigue, and myalgia. However, hemoptysis, chest pain, sputum production, rhinorrhea, headache, sore throat, and gastrointestinal manifestations are uncommon symptoms in COVID‐19.4, 5, 6 Although infected people predominantly suffer mild symptoms or are asymptomatic, a significant number of patients rapidly progress to severe acute respiratory failure with a higher risk of death, specifically in elderly, as well as in people who have underlying comorbidities, for instance, chronic respiratory or cardiovascular disease, diabetes, and cancer.7, 8
The early and accurate COVID‐19 diagnosis has the potential to control the epidemic spread and reduce mortality. The reverse‐transcription polymerase chain reaction (RT‐PCR) of the nose and nasopharyngeal swabs is considered an essential method for the clinical detection of SARS‐CoV‐2 infection. 9 Nevertheless, there are some shortcomings, including relative low sensitivity in the early stage of the disease, a time‐consuming procedure, and a shortage of RT‐PCR kits.10, 11, 12, 13 Meanwhile, medical imaging showed a high positive rate in the detection of the disease, in particular by computed tomography (CT) that may be considered as a primary and a reliable diagnostic tool for COVID‐19 detection and follow‐up.13, 14
In recent years, numerous approaches of machine learning (ML) have been successfully applied in the healthcare and medical fields to cope with challenges such as the accurate diagnosis and prediction of disease outcomes. Therefore, researchers have undertaken efforts to apply ML techniques to assist in mitigating the COVID‐19 pandemic. This review aims to discuss various ML algorithms developed to construct automated COVID‐19 diagnostic and prognostic systems based on medical imaging. Our contributions in this review include:
This review provides a list of publicly available data sets for training and testing of ML models. The data sets encompass collections of medical chest images of COVID‐19, other common pneumonia, and healthy controls.
We explore CT manifestations at different stages of the disease, and report on the main findings for COVID‐19 using X‐ray images.
We focus on analysing developed ML models for COVID‐19 diagnosis using image modalities (CT scan, X‐ray, and ultrasound imaging), and we briefly describe the general workflow of an image‐based COVID‐19 diagnostic system.
We illustrate the current ML techniques that are applied in COVID‐19 research for image segmentation and classification tasks, and we discuss the common limitations of ML methods in detecting COVID‐19 infections.
The rest of our review manuscript is organized as follows: In Section 2, we list the popular and current open‐source data sets of medical imaging data from healthy samples, COVID‐19, and other pneumonia patients. We then present the predominant features of medical imaging during SARS‐CoV‐2 infection, and highlight recent research which employed ML algorithms for classification and diagnosis of COVID‐19. Areas that used CT‐based, X‐ray‐based, and ultrasound‐based diagnosis are emphasized in Section 3 while the subsequent Section 4 introduces studies for the assessment of COVID‐19 severity and the prediction of mortality using medical imaging. In Section 5, the manuscript provides a brief discussion that summarizes the limitations of ML methods for COVID‐19 imaging‐based diagnosis. Finally, we draw conclusion in Section 6.
2. THE MEDICAL IMAGING DATA SETS
In the event of a novel disease, one of the challenges facing scientists is data insufficiency and its variation over different geographic regions. As data is an essential component of ML techniques, 15 the quality of data representation plays a critical role in the performance of ML models; thus the availability of large quantities of data with balanced representation provides opportunities to achieve high performing detection and prediction models. 16 Although various datasets of CT scan and radiologic (X‐ray) images are publicly available, the data sizes, specifically of early‐stage COVID‐19, are limited. Therefore, collaboration and exchange of resources among researchers are required to develop diagnostic ML systems and to better combat the COVID‐19 pandemic.
Table 1 lists publicly accessible data sets of CT, X‐ray, and ultrasound images of healthy, COVID‐19, and other common forms of pneumonia.
Table 1.
Chest medical imaging datasets of COVID‐19, pulmonary diseases, and healthy samples
| Data set | Modality | Description |
|---|---|---|
| Zhang et al. 17 | Chest CT |
The data set encompasses collections of CT images of COVID‐19, other common PNA patients, and normal from the China Consortium of Chest CT Image Investigation (CC‐CCII). It also includes lesion segmentation data set of 750 slices from 150 patients with COVID‐19. The images of lesion segmentation data set were manually segmented into the background, lung field, GGO, and CL. The data set is publicly available at http://ncov-ai.big.ac.cn/download?lang=en |
| Cohen et al. 18 | Chest CT and X‐ray |
The data set is composed of 20 COVID‐19 CT scans, and 761 X‐ray (679 frontal and 82 lateral views) images from COVID‐19 and other common PNA patients, includes metadata. The authors extracted the images from websites and online publications. The data set is publicly available at https://github.com/ieee8023/covid-chestxray-data_set |
| COVID19‐CT 19 | Chest CT |
The data set is comprised of 349 images from 216 patients with COVID‐19 and 397 non COVID‐19 images. The images extracted from online publications. The data set is publicly available at https://github.com/UCSD-AI4H/COVID-CT |
| COVID‐19 and common PNA chest CT data set 20 | Chest CT |
The COVID‐19 and common PNA chest CT data set is comprised of 416 confirmed COVID‐19 scans, and 412 other common PNA scans. The data set is publicly available at https://data.mendeley.com/datasets/3y55vgckg6/1 |
| COVID‐19 Radiography Database 21 | Chest X‐ray |
The database is composed of 219 images of COVID‐19, 1345 images of viral PNA, and 1341 normal images. The data set was created from different articles and public datasets. The data set is publicly available at https://drive.google.com/file/d/1xt7g5LkZuX09e1a8rK9sRXIrGFN6rjzl/view?usp=sharing |
| SIRM COVID‐19 data set 22 | Chest CT and X‐ray |
Italian Society of Medical and Interventional Radiology (SIRM) COVID‐19 data set contains 94 COVID‐19 X‐rays and 290 COVID‐19 CTs. The data set is publicly available at https://www.sirm.org/category/senza-categoria/covid-19/ |
| Feng et al. 23 | Laboratory data and CT characteristics | The data set contains CT features, clinical characteristics, and laboratory data from 141 COVID‐19 patients. CT characteristics include GGO, CL, crazy‐paving, and air bronchogram. The data is available in Excel worksheet within the supplementary information of the paper. |
| POCUS data set 24 | ultrasound | The data set contains 654 COVID‐19 images, 172 normal, and 277 bacterial PNA images. The images are sampled from ultrasound videos from different online sources. The data set is publicly available at GitHub-jannisborn/covid19_pocus_ultrasound: Open source ultrasound (POCUS) data collection initiative for COVID‐19 |
| ICLUS‐DB 25 | ultrasound | The Italian COVID‐19 Lung Ultrasound DataBase (ICLUS‐DB) comprises 277 lung ultrasound videos from 35 COVID‐19 patients. Data could be requested through https://iclus-web.bluetensor.ai/login/?next=/ |
| ChestX‐Ray8 26 | Chest X‐ray | The database was constructed from 108,948 labeled frontal images from 32,717 other common patients and normal cases collected between 1992 and 2015. The images have one or multi labels, including PNA, pneumothorax, atelectasis, nodule, mass, cardiomegaly, infiltration, effusion, and normal. The data set is publicly available at https://nihcc.app.box.com/v/ChestXray-NIHCC |
| CheXpert 27 | Chest X‐ray | The data set encompasses 224,316 labeled chest radiographs from 65,240 normal and other common patients collected from studies between the 2002 and 2017 at the Stanford Hospital. The images have one or multi labels, including PNA, pneumothorax, CL, lung lesion, lung opacity, atelectasis, nodule, fracture, enlarged cardiom, cardiomegaly, infiltration, edema, effusion, and no finding. The data set is publicly available at https://stanfordmlgroup.github.io/competitions/chexpert/ |
Abbreviations: CL, consolidation; CT, computed tomography; GGO, ground‐glass opacity; PNA, pneumonia.
It is worth noting that the majority of published papers reviewed here developed ML models for COVID‐19 diagnosis and detection using privatly‐sourced data sets.
3. ML FOR IMAGE‐BASED COVID‐19 DIAGNOSIS AND CLASSIFICATION
Several studies have recently appeared in the area of chest imaging segmentation and classification using ML to identify patients with COVID‐19. 28 The general workflow to build an image‐based COVID‐19 diagnostic system using ML algorithms is described in Figure 1. In brief, it requires:
Figure 1.
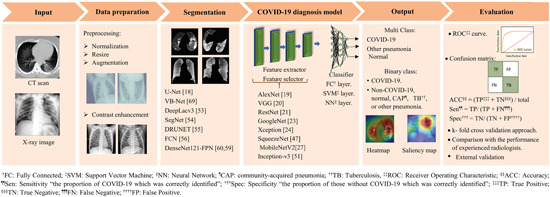
Illustration of the general workflow of using machine learning techniques and medical imaging for COVID‐19 diagnosis. This pipeline presents the various steps for building an image‐based COVID‐19 diagnostic system and commonly used machine learning techniques [Color figure can be viewed at wileyonlinelibrary.com]
(1) Medical imaging data set and corresponding accurate output labels; (2) data preparation and preprocessing: a curation process for medical imaging data is required for optimal training, validation, and testing of ML models; (3) a suitable segmentation technique. The U‐net 29 is the most popular convolutional network used in COVID‐19 applications to extract both lung region and lesions (region of interest [ROI]) from the background of medical images. Of note, this step was performed more often in the studies based on CT scans than in those based on X‐ray images, making CT segmentation an essential process of the COVID‐19 diagnostic model; (4) an appropriate ML technique for elaboration of the diagnostic model. The studies discussed below have been almost entirely based on deep learning (DL) network architectures, mainly convolutional neural networks (CNN). A few establish their ML models from scratch, while the majority used a variety of pre‐trained CNN networks in COVID‐19 research. The CNN architectures are constructed of a stack of convolutional and pooling layers performing feature extraction, followed by fully connected layers as a classifier. The following are some well‐known pretrained CNN architectures:
AlexNet is a basic CNN network introduced in the ImageNet Large Scale Visual Recognition Challenge (ILSVRC‐2012). 30 Its architecture has a depth of eight and its architecture comprises five convolutional layers, a limited number of max‐pooling layers, and three fully connected layers. This model uses large filters in the first and second convolutional layers.
VGG 31 contains 16 or more convolutional and fully connected layers. Raising the depth to 16 in VGG‐16 and 19 in VGG‐19 improves the general performance of the model. Each convolutional layer utilizes a small filter (3 × 3).
ResNet 32 is a novel convolutional network because of its architecture (depth and the residual blocks). This model was evaluated with a depth of 152 layers on the ImageNet data set. 33
GoogleNet 34 has devised a new module termed inception. This network is 27 layers deep including pooling layers.
Xception 35 consists of 36 convolutional layers.
Besides the network's architecture, its receptive field and activation function are important concepts in the design of a successful ML model. The receptive field, a small portion of the input that generates a feature in a convolutional layer, has increased as the above networks evolved from AlexNet to other models. Accordingly, the growth of the receptive field may be related to improvements in the model's accuracy. The following equation can be used to calculate the maximum receptive field:
| (1) |
where sn is stride size of layer n, kn is kernel size of layer n, Sn is cumulative stride from layer n, and RF n is the receptive field of a unit from layer n.
From this Equation (1), the receptive field is affected by several factors, including the filter size, and the stride of all the previous layers. Therefore, the receptive field could be modified by changing the filter size and adding more max‐pooling layers to increase Sn . Besides, an increase in the dilation rate leads to an increase in the receptive field.
However, the importance of the receptive field in CNNs has not been described in the studies reported here. Nevertheless, a limited receptive field impedes a convolutional layer to capture larger patchy‐like lung lesions, whereas the high‐level layers with large receptive fields were able to detect the diffuse and patchy‐like lung lesions, such as consolidations. 36
There are various activation functions available but rectified linear unit (ReLU) is the one commonly utilized for DL. The function returns zero for all negative inputs, but it returns the raw value for any positive input. Due to the simplicity of the ReLU function, the model can be trained in a short time, and the function is relatively cheap to compute. Also, the function is an optimal solution to avoid the vanishing gradient problem. Consequently, the ReLU function is preferred when the deeper network and the computational load is a major concern. Moreover, the Leaky Rectified Linear unit (Leaky ReLu) can be used as an activation function to deal with the vanishing gradient problem, but this requires more computation than using ReLU. Sigmoid and softmax are other activation functions that are similar in structure and have common problems, including the slow convergence rate and gradient diffusion. The sigmoid function is used mostly for binary classification such as the two‐class logistic regression, whereas softmax is used for multiclass classification.
While most studies analysed in this review used ReLU as an activation function for their network, Yan et al. 37 used a sigmoid function for a fully connected layer with one output to generate the probability for COVID‐19, and Wang et al. 38 used the softmax function for a progressive classifier consisting of three convolutional layers within a fully connected layer.
Ozturk et al. 39 used two different activation functions, the Leaky ReLU activation function used for their model which ends with softmax layers to produce outputs which achieved a high performance of 98.08%. On the other hand, Toğaçar et al. 32 proposed a diagnostic system based on MobileNet‐V2 40 which utilizes uses ReLU function between layers, and support vector machines (SVM) using sigmoid activation function for the classification task and achieved 99.27% accuracy. Therfore, different activation functions used in different models can achieve high performance.
The main power of convolutional networks compared to traditional ML methods lies in its depth and compositionally.41, 42 This allows the extraction of complex and discriminating features from the input medical image at multiple abstraction levels. Therefore, an architecture of sufficient depth can produce a compact representation which is beneficial for the prediction accuracy. 31 However, a CNN network with fewer convolutional and fully connected layers might perform similarly to deeper networks. 43 Therefore, not only the network depth but also the structure of the cohort data, balanced representation of samples in a data set, and the data quality have a significant impact on the model's performance. Data representation is one of the keys to successful ML algorithms. Table 2 illustrates the representation of imaging data from different ML algorithms used by the studies discussed in this review. (5) validation and evaluation of the model's performance. Validation strategies include internal, temporal, and external validation. Holdout, cross‐validation, and bootstrapping are the most common types of internal validation. In addition, the accuracy, specificity, sensitivity, curves and area under the curve (AUC), receiver operating characteristic (ROC), and F‐score are standard evaluation measures used to assess the performance of a developed model.
Table 2.
The representation of COVID‐19 imaging data from different ML method
| COVID‐19 imaging data representation | Modalities | ML model | Study |
|---|---|---|---|
| 3D feature maps | CT | Attention‐based deep 3D multiple instance learning | 44 |
| Latent high‐level representation | CT | CPM‐Nets 45 multiview representation learning technique | 46 |
| Adaptive feature selection guided deep forest (AFS‐DF) | 47 | ||
| Image pyramid representation | CT | The multiscale spatial pyramid (MSSP) decomposition 48 | 49 |
| Bag representation | CT | Attention based deep 3D multi‐instance learning (AD3D‐MIL) | 44 |
| One‐hot representation | CT | Adapted 3D ResNet‐18 | 17 |
| 64‐dimensional DL features | CT | DenseNet‐like structure 50 | 51 |
| Activation maps | X‐ray | MobileNetV2, 40 SqueezeNet, 52 Deep CNN architecture | 21, 53, 54, 55 |
Abbreviations: CNN, convolutional neural network; CT, computed tomography.
3.1. Using chest CT
CT scans have become one of the reliable and practical tools for rapid diagnosis of COVID‐19 patients 56 because of its speed, high sensitivity, and ability to detect typical imaging features in COVID‐19 patients.13, 57, 58 Some studies11, 14, 59 investigated the diagnostic value of CT as compared with RT‐PCR for COVID‐19 disease and concluded that CT scanned images had high sensitivity (97%–98%) to correctly diagnose COVID‐19; by comparison, RT‐PCR achieved a sensitivity in the range of 71%–83%.
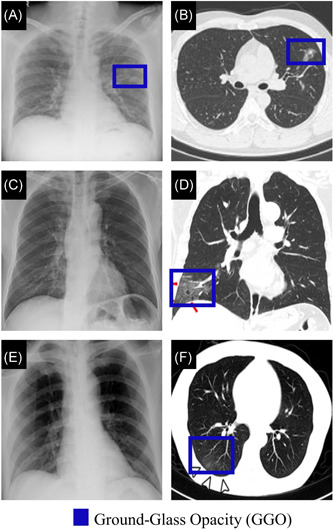
Predominant features of COVID‐19 CT images involve ground‐glass opacity (GGO), consolidation, crazy‐paving pattern, and interlobular septal thickening.49, 60, 61 However, it has been observed that the frequency of these common features in severe COVID‐19 patients are significantly higher than in early‐stage or moderate COVID‐19. 61 Additionally, air bronchogram, pleural effusion, pericardial, nodules, lymphadenopathy, round cystic changes, and bronchial wall thickening are more frequently observed in CT images of severe COVID‐19 patients.49, 60, 61, 62 Figure 2 illustrates manifestations in CT scans at the different COVID‐19 stages using follow‐up CT scans from three COVID‐19 patients.
Figure 2.
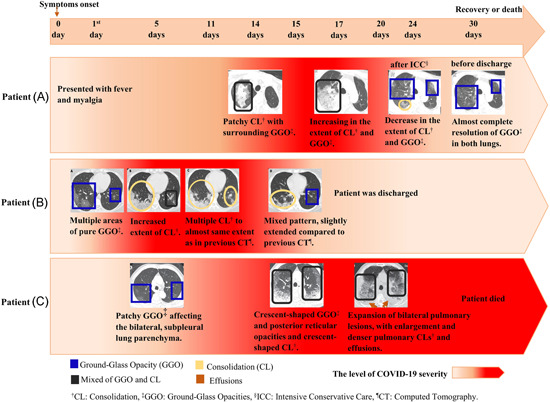
The CT scan findings of COVID‐19 progression from three patients. Patient (A) was a 61‐year‐old male with a history of chronic pleurisy. CT also shows calcified fibrothorax in the left lob due to chronic pleurisy. 63 Patient (B) was a 35‐year‐old female. 64 Patient (C) was a 77‐year‐old male with cerebrovascular disease, cardiovascular disease, and hypertension. 49 CL, consolidation; CT, computed tomography; GGO, ground‐glass opacity; PNA, pneumonia [Color figure can be viewed at wileyonlinelibrary.com]
The lung abnormalities in the COVID‐19 CT scans vary in patients depending on immune status, age group, and disease stage. Wang et al. 64 concluded that ground‐glass opacity was the most common feature in COVID‐19 CT scans, followed by consolidation during the first 11 days following onset of symptoms. While these two patterns constituted approximately 83%–85% of CT manifestations among all COVID‐19 patients in the early stages, pleural effusion was absent. Twelve days from symptom onset, a mixed pattern becomes the predominant CT finding which is consistent with another report. 65 Furthermore, bilateral multiple lobular and subsegmental areas of consolidation were the main findings in intensive care unit (ICU) patients. 4
On the other hand, Li et al. 66 summarized CT findings which were significantly associated with COVID‐19 in comparison to other common viral forms of pneumonia. These included absent of pleural effusion, a lesion range more than 10 cm, multiple affected lobes and peripheral distribution. Moreover, 43.51% of COVID‐19 patients showed mediastinal and hilar lymph node enlargement in CT scans compared to 20.0% in other common viral pneumonia. Figure 3 shows CT images from healthy cases, COVID‐19 and other respiratory diseases.
Figure 3.
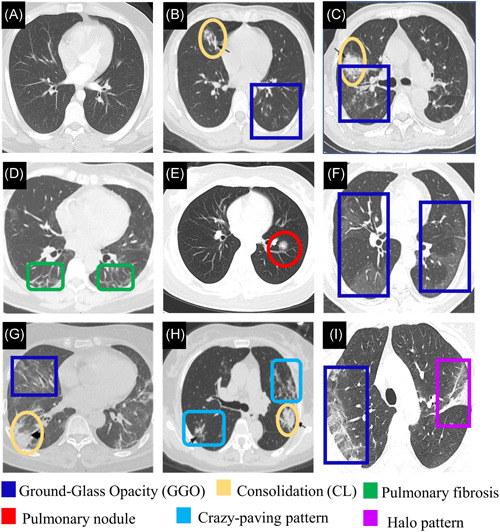
(A) CT image of normal sample. 19 CT images from pneumonia patients (Adapted from Reference [66]. (B) GGO and consolidation are shown in a 23‐year‐old female with influenza infection. (C) a mixed pattern of consolidation and GGO is shown in a 64‐year‐old male with Epstein‐Barr virus infection. CT images from confirmed COVID‐19 patients. (D) Pulmonary fibrosis is shown in both lungs in a CT image of a 56‐year‐old female with moderate COVID‐19 (Adapted from Reference [62]). (E) Pulmonary nodule in a 23‐year‐old female (Adapted from Reference [67]). (F) Confluent pure GGO is presented in COVID‐19 patient (Adapted from Reference [68]). (G) GGO and consolidation with air bronchogram are presented in a CT image of 59‐year‐old female with sever COVID‐19 (adapted from 62 ). (H) Multifocal crazy‐paving pattern and consolidation is shown in a 59‐year‐old female (Adapted from Reference [66]). (I) the right lung with peripheral predominant GGO, and a reversed halo sign in the left in a 45‐year‐old female (Adapted from Reference [69]). CL, consolidation; CT, computed tomography; GGO, ground‐glass opacity [Color figure can be viewed at wileyonlinelibrary.com]
The CT image scans show significant potential for detecting COVID‐19; nevertheless, manual classification of CT features, involving peripheral GGO and consolidation often achieves relatively low sensitivity in distinguishing COVID‐19 patients from other types of pneumonia, especially other viral forms of pneumonia. According to a recent study, 70 seven radiologists were able to differentiate between COVID‐19 and other viral pneumonias with a sensitivity ranging from 70% to 93% using CT images.
Artificial intelligence (AI) could assist radiologists in improving their performance and better cope with such challenges. In this regard, Table 3 summaries selective information from studies employing ML algorithms for COVID‐19 detection using CT images.
Table 3.
Summary of performance of ml models applied for COVID‐19 detection using CT images
| Reference | Number of samples | Segmentation technique | Model's name (ML techniques) | Purpose | ACC/AUC | Sen (%) | Spec (%) |
|---|---|---|---|---|---|---|---|
| 71 | 4352 scans from 3322 patients (1292 COVID‐19, 1735 CAP, and 1325 non‐PNA) | U‐Net | COVNet (3D RestNet50) | COVID‐19 detection | 0.96 | 90 | 96 |
| CAP detection | 0.95 | 87 | 92 | ||||
| Non‐PNA detection | 0.98 | 94 | 96 | ||||
| 72 | 419 cases positive COVID‐19 | CNN | CNN, SVM, random forest and MLP | Prediction of COVID‐19 probability | 0.92 | 84.3 | 82.8 |
| 486 cases negative COVID‐19 | |||||||
| 51 | COVID‐19 data set: 924 COVID‐19, and 342 other PNA | DenseNet121‐FPN | COVID‐19Net (DenseNet‐like structure) | Prediction of COVID‐19 probability | 80.12%/0.88 | 79.35 | 81.16 |
| CT‐EGFR data set: 4106 lung cancer patients | COVID‐19 vs. other PNA | 85%/0.86 | 79.35 | 71.43 | |||
| 73 | 1029 scans of 922 COVID‐19 patients | AH‐Net architecture | Densnet‐121 | COVID‐19 detection from other clinical entities | 89.6%/0.941 | 84.5 | 91.6 |
| 1695 scans of 1695 cancer, CP, and any clinical indication patients | |||||||
| 74 | 1194 CTs of 80 COVID‐19 patients | Not performed | FCONet (pre‐trained DL models) | Multi‐class classifier COVID‐19 vs. other PNA vs. no PNA | 99.87% | 99.58 | 100 |
| 1357 CTs of 100 other PNA patients | |||||||
| 1442 CTs of 126 normal and lung cancer patients | |||||||
| 75 | 150 3D CT scans of COVID‐19 | multi‐view U‐Net | VGG architecture | Binary classification: | |||
| 150 scans CAP | COVID‐19 vs. normal | 96.2%/0.970 | 94.5 | 95.3 | |||
| 150 scans normal | |||||||
| COVID‐19 vs. CAP | 89.1%/0.906 | 87.0 | 86.2 | ||||
| 17 | 835 COVID‐19 patients | DeepLabv3 | 3D ResNet‐18 | Binary classification: | |||
| 888 other PNA patients | COVID‐19 vs. No‐COVID‐19 | 92.49%/0.9797 | 94.93 | 91.13 | |||
| 783 normal cases | Multi‐class classification: | – | – | ||||
| COVID‐19 vs. normal vs. other PNA | 92.49%/0.9813 | ||||||
| 44 | 230 CT scans from 79 patients with COVID‐19 | Not performed | AD3D‐MIL | Binary classification: COVID‐19 vs. normal & CP | 97.9% | – | – |
| 100 CT scans from 100 CP patients | Multi‐class classification: COVID‐19 vs. CP vs. normal | 94.3% | – | – | |||
| 130 CT scans from 130 normal cont | |||||||
| 47 | 1495 COVID‐19 patients | VB‐Net | AFS‐DF | Binary classification: | |||
| 1027 CAP patients | COVID‐19 vs. CAP | 91.79%/0.963 | 93.05 | 89.95 | |||
| 46 | 1495 scans with COVID‐19 | V‐Net | multi‐view ML technique | Binary classification: | |||
| 1027 scans with CAP | COVID‐19 vs. CAP | 95.5% | 96.6 | 93.2 | |||
| 76 | 3389 scans with COVID‐19 from 2565 patients | VB‐Net toolkit | Attention 3D ResNet34 + sampling strategy | Binary classification: | |||
| 1593 scans with CAP from 1080 patients | COVID‐19 vs. CAP | 87.5%/0.944 | 86.9 | 90.1 | |||
| 77 | 132 583 CT slices from 1186 patients: | Manually and three‐dimensional Slicer software | EfficientNet B4 | Binary classification: COVID‐19 vs. other PNA | |||
| 521 COVID‐19 patients | Test data set of 119 patients | 96%/0.95 | 95 | 96 | |||
| 665 non‐COVID‐19 patients | Test data set of 395 patients | 91%/0.95 | 94 | 87 | |||
| 78 | 313 positive COVID‐19 | U‐Net | DeCoVNet (AlexNet, ResNet) | Predicting COVID‐19 probability | 0.959 | 90.7 | 91.1 |
| 229 negative COVID‐19 | |||||||
| 79 | 219 scans from 110 COVID‐19 patients | 3D CNN (V‐Net, IR, RPN) | ResNet‐based | Multi‐class classification: | – | ||
| 224 IAVP patients. 175 healthy | COVID‐19 vs. IAVP vs. healthy | 86.7% | – | – | |||
| 37 | 416 CT scans from 206 COVID‐19 patients | Not performed | MSCNN | Binary classification: | |||
| 412 CP patients | COVID‐19 vs. CP at slice level | 97.7%/0.962 | 99.5 | 95.6 | |||
| COVID‐19 vs. CP at scan level | 87.1%/0.934 | 89.1 | 85.7 |
Abbreviations: ACC, accuracy; AD3D‐MIL, attention‐based deep 3D multiple instance learning; AFS‐DF, adaptive feature selection guided deep forest; AUC, area under the receiver‐operating characteristics curve; CAP, community‐acquired pneumonia; CP, common pneumonia; IAVP, influenza‐A viral pneumonia; MLP, multilayer perceptron; MSCNN, multiscale convolutional neural network; PNA, pneumonia; Sen, sensitivity; Spec, specificity; SVM, support vector machine.
To discriminate COVID‐19 from other forms of pneumonia,37, 73, 80 Wu et al. 80 constructed a multiview fusion model using ResNet50 architecture 32 to assist radiologists in accurately identifying patients with suspected COVID‐19. They trained the model utilizing coronal, axial, and sagittal views of the maximum lung regions from CT scans. The multi‐view module obtained an area under the curve (AUC) of 0.819, and 76% accuracy in the test set which consisted of 50 patients (37 COVID‐19 and 13 other pneumonias). The performance of the multiview fusion model was higher than the single‐view model that used only the axial view and obtained an accuracy of 62% in the testing sets. Yan et al. 37 designed an AI model relying on a multiscale convolutional neural network (MSCNN) using limited training data (164 COVID‐19 patients and 330 patients with other forms of common pneumonias). The authors applied data augmentation, a multiscale spatial pyramid (MSSP) decomposition, 48 and transfer learning for better performance. The AUC values of the proposed system were 0.962 at the 2D slice level, and 0.934 at the 3D scan level.
Also, Ardakani et al. 81 evaluated 10 pretrained CNNs, including ResNet‐101, 32 Xception, 35 AlexNet, 30 MobileNet‐V2, 40 ResNet‐50, 32 SqueezeNet, 52 ResNet‐18, 32 VGG‐16, 31 VGG‐19, 31 and GoogleNet 34 to develop an adjuvant tool that could assist radiologists in differentiating between COVID‐19 infections and other atypical and viral pneumonias. ResNet‐101 32 and Xception 35 appeared with the best diagnostic performance of 0.994 (AUC).
DL models were developed for multi‐class classification tasks to distinguish COVID‐19 infections from other pneumonia and against features irrelevant to infection.44, 74, 79 Xu et al. published the first ML system to screen COVID‐19 patients using CT images 79 and proposed a 3D segmentation CNN model to extract a ROI. They then evaluated two classification models, the traditional residual network (ResNet‐18)‐based 32 model, and the location‐attention model based on ResNet‐18 32 as the backbone, to discriminate COVID‐19 from normal and influenza‐A viral pneumonia (IAVP). The second model obtained better performance with an accuracy of 86.7%. 79 Ko et al. proposed a 2D CNN framework, called the fast‐track COVID‐19 classification network (FCONet), for identifying COVID‐19 in a single CT image. 74 Four pretrained CNN models (ResNet‐50, 32 VGG‐16, 31 Xception, 35 and Inception‐v3 82 ) were evaluated to develop FCONet by transfer learning. ResNet‐50 appeared with the highest diagnostic performance with an accuracy of 99.87% on the testing set of 239 COVID‐19 images, and 96.97% on the external testing set of 264 COVID‐19 low‐quality images, respectively.
COVID‐19 diagnostic systems which outperformed experienced radiologists in distinguishing COVID‐19 patients from other types of pneumonia were also recently proposed. Zhang et al. 17 adopted DeepLacv3 83 as the backbone for lung‐lesion segmentation due to its efficient performance compared to other segmentation tools tested in this study, including SegNet, 84 DRUNET, 85 U‐net, 29 and FCN, 86 and built a 3D classification framework using 3D ResNet‐18. The 2,879 CT slices of COVID‐19 patients and 1816 CT slices of patients with other common pneumonia were manually annotated into seven categories: GGO, consolidation, pleural effusion, and interstitial thickening, lung field, and background for training and evaluation procedures. Their diagnostic classification model obtained 92.49% accuracy in identifying the COVID‐19 group from the No‐COVID‐19 group (normal and other common pneumonia). It also obtained 92.49% accuracy for multiclass classification (COVID‐19 from normal and other common pneumonia). This system yielded a weighted error of 9.29% in a testing set of 18,392 CT slices from 150 subjects compared to an average mean of weighted error of 13.55% when the slices were assessed by eight radiologists. The proposed system by Bai et al. 77 obtained an accuracy of 96% and sensitivity of 89% compared to an average accuracy of 85% and sensitivity of 79% by experts. With the model's assistance, the performance of the radiologists improved to 90% accuracy and 88% sensitivity.
Although significant advancement has been demonstrated by recent studies for COVID‐19 diagnosis using ML techniques on CT images, many of them require manual radiologist‐assisted annotations of the lung lesion. Wang et al. 51 aimed to establish a fully automated diagnostic system, referred to as COVID‐19Net, using deep learning. They used feature pyramid networks (FPN) 87 with a DenseNet121 50 as the backbone for the segmentation model, which pretrained on the ImageNet data set, 33 and fine‐tuned using the VESSEL12 data set, 88 whilst diagnostic and predictive analysis used a DenseNet‐like structure. 50 First, the COVID‐19Net model was trained on CT‐epidermal growth factor receptor (EGFR) data set, 89 consisting of 4106 patients with lung cancer, to predict EGFR mutation status. This allowed the model to learn distinguishing features from CT. Afterwards, the authors transferred the pretrained model to COVID‐19 data set, achieving an accuracy of 85% in identifying COVID‐19 from other pneumonia.
Three‐dimensional deep learning frameworks were developed to distinguish COVID‐19 from community‐acquired pneumonia (CAP).71, 75, 76 Li et al. developed a framework, referred to as COVNet, using CNN models. 71 The COVNet model consisted of U‐Net 29 to extract the lung region as the ROI, RestNet50 32 as the backbone to extract and combine features from all slices, and a fully connected layer to detect COVID‐19 by generating a score of probability for each class of COVID‐19, CAP, and normal. It achieved an AUC of 0.96 for identifying COVID‐19 in the independent testing data set. Ouyang et al. 76 proposed two 3D ResNet34 networks 36 with a novel online attention model. The model was employed on a large testing data set, achieving an AUC of 0.944.
Alongside 3D deep learning models for distinguishing COVID‐19 from CAP, a multiview representation learning technique has been proposed recently. 46 The authors extracted 189‐dimensional features from each image based on the lesion region produced from the segmentation model. Their model achieved 95.5% accuracy, 96.6% sensitivity, and 93.2% specificity. Sun et al. 47 used adaptive feature selection guided deep forest (AFS‐DF) based on extracted location‐specific features from segmented images. For CT segmentation, they applied VB‐Net developed in Reference [90]. The experimental results showed 0.963% and 91.79% in terms of AUC and accuracy, respectively.
Additionally, ML algorithms have been applied for identifying confirmed COVID‐19 patients from negative COVID‐19 cases.72, 78
3.2. Using chest radiographs (X‐ray)
Although CT modality has been utilized as a primary imaging tool for COVID‐19 diagnosis, chest radiology has also been considered as an alternative diagnostic method for this disease. 56 While a recent study reported that the sensitivity of X‐ray (69%) was lower than that of RT‐PCR (91%), X‐ray images showed COVID‐19 signs in six patients who initially tested negative by RT‐PCR. 38
Chest X‐ray imaging is cost‐effective and commonly used for screening purposes. The predominant chest radiographic finding in COVID‐19 patients is consolidation, followed by ground‐glass opacities.38, 91 While lower zone distributions, peripheral distributions, and bilateral involvements presented in X‐rays of COVID‐19 patients, pleural effusions were an uncommon feature. 38 Overall, X‐ray findings in COVID‐19 and other forms of pneumonia are highly similar (Figure 4).
Figure 4.
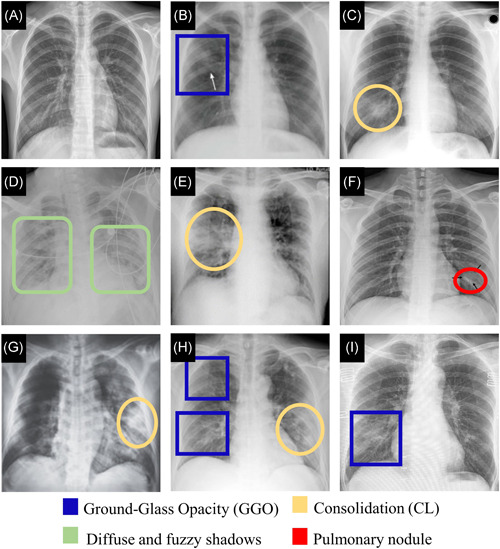
Chest X‐ray images. (A) Normal sample. 92 (B) A female in her 30 s with Mycoplasma pneumoniae pneumonia. X‐ray shows GGO (Adapted from Reference [93]). (C) A 29‐year‐old female with SARS. X‐ray reveals consolidation (Adapted from Reference [94]). (D) A 41‐year‐old male with COVID‐19. X‐ray, obtained 11 days after the symptoms, shows bilateral diffuse patchy and fuzzy shadows (Adapted from Reference [95]). (E) An elderly male with COVID‐19. X‐ray shows consolidative changes in the right lobe (Adapted from Reference [91]). (F) A single nodular consolidation is observed in X‐ray image from a patient with COVID‐19 (Adapted from Reference [68]). (G) A 65‐year‐old male. X‐ray, obtained 9 days after onset of symptoms, shows a progressive infiltrate and consolidation (Adapted from Refernce [96]). (H) X‐ray of patient of COVID‐19 shows patchy ground‐glass opacities and patchy consolidation (Adapted from Reference [68]). (I) A 59‐year‐old female with COVID‐19. X‐ray shows patchy GGO (Adapted from Reference [69]). CL, consolidation; CT, computed tomography; GGO, ground‐glass opacity [Color figure can be viewed at wileyonlinelibrary.com]
Compared to chest CT, the sensitivity of X‐ray is generally low for pulmonary diseases. Therefore, accurate diagnosis of COVID‐19 pneumonia can be more challenging on X‐ray images than on chest CT scans, as shown in Figure 5.
Figure 5.
Comparison of chest X‐ray (left panel) and CT (right panel) of COVID‐19 (A, B) X‐ray and CT of a 34‐year‐old man, the fifth RT‐PCR test was positive after 5 days from onset. X‐ray and CT scan at disease onset show patchy GGO (Adapted from Reference [97]). (C, D) A CT thorax coronal scan and X‐ray performed on the same day of a COVID‐19 patient. GGO was observed on CT but was not visible on X‐ray (Adapted from Reference [91]). (E, F) X‐ray and CT of a 43‐year‐old man. CT scan shows GGOs, and he had negative findings on X‐ray image (Adapted from Reference [38]). CT, computed tomography; GGO, ground‐glass opacity; RT‐PCR, reverse transcription polymerase chain reaction [Color figure can be viewed at wileyonlinelibrary.com]
Nevertheless, radiologic examinations might be an essential diagnostic method for monitoring the progression of lung abnormalities in patients affected by COVID‐19. Numerous studies, presented in Table 4, have developed different ML algorithms for establishing automated systems for COVID‐19 diagnosis using X‐ray images. Two recent studies employed ML models for binary classification tasks to discriminate between COVID‐19 infections and other lung diseases and healthy lungs.53, 98, 99, 103, 106 Certain studies utilized the transfer learning method through pre‐trained CNN models on the ImageNet data set 33 to distinguish COVID‐19 positive patients from healthy controls.53, 103 Also, Brunese et al. 98 proposed a diagnostic model based on pretrained VGG‐16 31 architecture to discriminate COVID‐19 from other pulmonary diseases.
Table 4.
Summary of performance of ML models applied for detection of COVID‐19 using Chest X‐ray
| Reference | Number of samples | Pre‐processing data | Model's name (ML techniques) | Purpose | ACC/AUC | Sen (%) | Spec (%) |
|---|---|---|---|---|---|---|---|
| 39 | 127 COVID‐19 images | Not provided | DarkCovidNet (Darknet‐19) | Binary classification: | |||
| 500 PNA images | COVID‐19 vs. no‐findings | 98.08% | 95.13 | 95.3 | |||
| 500 no‐findings images | Multiclass classification: | ||||||
| COVID‐19 vs. healthy vs. other PNA | 87.02% | 85.35 | 92.18 | ||||
| 98 | 250 COVID‐19 | Resize to 224 × 224 pixels | VGGNet (VGG‐16) | Model 1: healthy vs. COVID‐19 and pulmonary | 96% | 96 | 98 |
| 2753 other pulmonary diseases | Model 2: COVID‐19 vs. other pulmonary | 98% | 87 | 94 | |||
| 3520 healthy controls | |||||||
| 54 | 295 COVID‐19 images | Not provided | MobileNetV2, | Multiclass classification: | |||
| 98 PNA images | SqueeaNet, | COVID‐19 vs. healthy vs. other PNA | 99.27% | 100 | 99.07 | ||
| 65 normal class images | SVM. | ||||||
| 99 | 162 COVID‐19 images | Horizontal flip, width and heigh shift range (0.2), rotation angle | DL‐based decision‐tree | Binary classification: | |||
| 492 TB images | COVID‐19 vs. TB | 100%/1.00 | 100 | 100 | |||
| 585 Normal images | COVID‐19 vs. non‐ TB | 89%/0.89 | 93 | 86 | |||
| 55 | 219 COVID‐19 | Rotate and flip augmentation approaches on training data | CNN, SVM, KNN, Decision Tree. | Multiclass classification: | |||
| 1341 normal | COVID‐19 vs. other viral PNA vs. normal | 98.97% | 89.39 | 99.75 | |||
| 1345 viral PNA. | |||||||
| 100 | 180 COVID‐19; 20 viral PNA; 57 TB; 54 bacterial PNA; 191 normal | Normalization | FC ‐DenseNet103 | Multiclass classification: | |||
| Segmentation | ResNet‐18 | Viral PNA including COVID‐19 vs. bacterial PNA vs. TB vs. normal | 88.9% | 89.5 | 96.4 | ||
| 101 | 231 COVID‐19 images | Resize to 128 × 128 width and height shift, shift range (0.2), and horizontal flip | CapsNet (5 fully convolutional layers) | Binary classification: | |||
| 1050 no‐findings images | COVID‐19 vs. healthy | 97.24% | 97.42 | 97.04 | |||
| 1050 PNA images | Multiclass classification: | ||||||
| COVID‐19 vs. other PNA vs. healthy | 84.22% | 84.22 | 91.79 | ||||
| 102 | 219 COVID‐19; 1345 viral PNA;1341 normal | CVDNet (CNN) | Multiclass classification: | ||||
| COVID‐19 vs. other PNA vs. healthy | 96.69% | 96.84 | – | ||||
| 92 | 284 COVID‐19 images | Resize to 224 × 224 pixels with a resolution of 72 dpi | CoroNet (Xception CNN) | Binary classification: COVID‐19 vs. normal | 99% | 99.3 | 98.6 |
| 330 bacterial PNA images | Multiclass classification: | 89.6% | 89.92 | 96.4 | |||
| 327 viral PNA images | COVID‐19 vs. bacterial PNA vs. viral PNA vs. healthy controls | ||||||
| 310 normal images | COVID‐19 vs. healthy vs. other PNA | 95% | 96.9 | 97.5 | |||
| 103 | 142 COVID‐19 images | Resize to 224 × 224 pixels, horizontal and vertical flipping | nCOVnet (CNN) | Binary classification: | |||
| 142 normal images | COVID‐19 vs. normal | 88.10% | 97.62 | 78.57 | |||
| 104 | 105 samples COVID‐19 | Flipping, translation and rotation, a histogram modification technique | DeTraC (pretrained CNN) | Multiclass classification: | |||
| 11 samples SARS | COVID‐19 vs. normal vs. SARS | 93.1% | 100 | 85.18 | |||
| 80 samples normal | |||||||
| 105 | 364 images COVID‐19 | The image contrast enhancement algorithm (ICEA) | MH‐CovidNet (DL, BPSO and BGWO meta‐heuristic algorithms, SVM) | Multiclass classification: | |||
| 364 images PNA | COVID‐19 vs. healthy vs. other PNA | 99.38% | – | – | |||
| 364 images normal. | |||||||
| 21 | 423 images COVID‐19 | Resize, rotation, and translation | pre‐trained CNN algorithms | Binary classification: COVID‐19 vs. normal | 99.70% | 99.70 | 99.55 |
| 1485 images viral PNA | Multiclass classification: | 97.94% | 97.94 | 98.80 | |||
| 1579 images normal | COVID‐19 vs. normal vs. other viral PNA | ||||||
| 53 | 184 images COVID‐19 | Flipping and rotation | ResNet18, | Binary classification: | |||
| 5000 images Non‐COVID‐19 | ResNet50, SqueezeNet, DenseNet‐161 | COVID‐19 vs. No‐COVID‐19 | 0.992 | 98 | 92.9 |
Abbreviations: ACC, accuracy; AD3D‐MIL, attention‐based deep 3D multiple instance learning; AFS‐DF, adaptive feature selection guided deep forest; AUC, area under the receiver‐operating characteristics curve; CAP, community‐acquired pneumonia; CP, common pneumonia; IAVP, influenza‐A viral pneumonia; MLP, multilayer perceptron; MSCNN, multiscale convolutional neural network; PNA, pneumonia; Sen, sensitivity; Spec, specificity; SVM, support vector machine.
Elaziz et al. 106 utilized novel Fractional Multichannel Exponent Moments (FrMEMs) as features extractor and Manta‐Ray Foraging Optimization (MRFO) modified by Differential Evolution (DE) as features selector. They then trained and evaluated K‐nearest neighbors (KNN) classifier and were able to classify COVID‐19 patients with 98.09% and 96.09% accuracy rates on two different data sets. Yoo et al. 99 proposed a classifier model that comprised three DL‐based decision trees. The first one classified X‐ray images into normal and abnormal images with 98% accuracy. The second decision identified tuberculosis (TB) patients with an accuracy of 80%. The third tree was able to distinguish X‐rays that contained signs of COVID‐19 with 95% accuracy.
Various ML algorithms for multiclass classification to differentiate COVID‐19 infections from other forms of pneumonia and healthy cases have been developed.54, 55, 100, 102, 104, 105 In one study, the authors developed a suitable model for training on a relatively limited data set that comprised 126 cases with COVID‐19, 14 with viral pneumonia, 41 with tuberculosis, 39 with bacterial pneumonia, and 134 normal cases. 100 The authors used fully convolutional‐DenseNet103 107 to extract lung area and heart contour of X‐ray images, followed by a patch‐based CNN based on ResNet‐18 32 as a backbone to identify viral pneumonia including COVID‐19 from other classes. The proposed model showed 88.9% accuracy and 92.5% sensitivity. Abbas et al. 104 adapted a deep CNN architecture, termed Decompose, Transfer, and Compose (DeTraC), that relied on a class decomposition technique. They evaluated DeTraC with various pretrained CNN models to distinguish COVID‐19 from SARS and normal using a limited data set. Experimental results showed VGG‐19 31 with an accuracy of 93.1% and 87.09% sensitivity, and ResNet 108 with 93.1% accuracy and 100% sensitivity. Ouchicha et al. 102 introduced CVDNet, a deep CNN model designed for discriminating COVID‐19 from other pneumonia and healthy controls using a public open‐source X‐ray data set. 21 Their model was able to correctly identify COVID‐19 with an average accuracy of 97.20%. Two different deep CNN models were recently proposed to extract features from X‐rays, followed by optimization algorithms to select the efficient features which were then fed to the SVM classifier.54, 105 While Canayaz 105 applied the image contrast enhancement algorithm on the data set as a preprocessing step, Toğaçar et al. 54 used the Fuzzy Color technique to restructure data classes, and both original and structured images were stacked. These two proposed approaches obtained remarkable classification rates with an accuracy of 99.38% and 99.27%, respectively. Nour et al. 55 proposed a deep CNN model consisting of five convolution layers to extract features from X‐rays collected from an open‐access data set. 21 For the classification task, three different ML algorithms were evaluated to identify COVID‐19, including SVM, KNN, and decision tree. Experimental results showed that the SVM classifier obtained the best performance with 98.97% accuracy.
Four studies have developed ML models for binary and multi‐class classification tasks.21, 39, 92, 101 Chowdhury et al. 21 trained and evaluated eight pretrained CNN models, demonstrating that the performance of all models was quite similar. The classification accuracy rates were 99.70% and 99.41%, respectively, for discriminating COVID‐19 from healthy with and without image augmentation. The proposed approach obtained accuracy rates of 97.94% and 97.74%, respectively, in distinguishing COVID‐19 from normal and other forms of viral pneumonia with and without image augmentation. Ozturk et al. 39 developed a DarkCovidNet model inspired by the architecture of the Darknet‐19 classifier model, 109 obtaining an average accuracy of 87.02% to discriminate COVID‐19 from other pneumonia, and 98.08% to classify COVID‐19 from X‐ray images. Finally, a capsule network model by Toraman et al. 101 comprised of four convolution layers and the primary capsule layer obtained 97.24%, and 84.22% accuracy, respectively, in the binary and multiclass classifier, after training and evaluation using open‐sources data sets.18, 26
3.3. Using lung ultrasonography
Besides to CT and X‐ray modalities, lung ultrasonography is a significant and promising technique for diagnosis and monitoring patients affected by COVID‐19. 110 According to Tung‐Chen et al., 111 ultrasound has a low false‐negative rate in diagnosing patients affected by COVID‐19. They observed that all COVID‐19 abnormalities on CT scans presented on ultrasound as well. Moreover, a limited number of preprint studies, excluded from this review, developed ML models using ultrasound imaging to discriminate COVID‐19 from other pneumonia and healthy.
While existing studies focus on establishing automatic COVID‐19 diagnostic systems based on CT or radiographic images, Horry et al. 112 applied a transfer learning approach to diagnose COVID‐19 infections using three different imaging modes, involving ultrasound, CT scan, and X‐ray. They trained and evaluated eight popular CNN models in a comparative study to identify a suitable model. Experimental results showed the selected VGG‐19 31 model was performing considerably better in distinguishing COVID‐19 from other pneumonia when using ultrasound images. The experimental results showed ultrasound with 100% sensitivity compared to 86% with X‐ray and 83% with CT images.
4. ML FOR IMAGE‐BASED ASSESSMENT OF COVID‐19 SEVERITY AND PROGRESSION
Early assessment of coronavirus disease severity is crucial tool to assist in the prevention of disease progression and in decreasing the fatality rate through early intervention and the delivery of required healthcare to high‐risk patients.
In addition to using medical imaging to develop ML algorithms effective for identifying patients with COVID‐19 amid healthy patients and patients affected by other types of pneumonia, a variety of studies, summarized in Table 5, implemented ML to develop a prognostic algorithm to assess COVID‐19 severity, identify patients who will progress to severe disease, and estimate the mortality risk of a COVID‐19 patient based on CT, radiography (X‐ray), and ultrasound imaging.
Table 5.
Main contributions of studies applied ml approaches for assessment severity of COVID‐19 and prediction of the disease progression and mortality risk using medical imaging
| Reference | Modality | COVID‐19 patient cohort | Main contributions |
|---|---|---|---|
| 113 | Chest X‐ray | 131 images of 84 patients |
• Proposed deep‐learning CNN model with Dense regression layer to produce different severity scores from portable X‐ray for predicting COVID‐19 severity. • Compared traditional and transfer learning that showed better performance. |
| 114 | Chest CT scans |
Severe group:97 scans of 32 patients Non‐severe group: 452 scans of 164 patients |
• Developed a DL model based on U‐Net 29 equipped with the ResNet‐34 115 to segment lung lesions of thick‐section CT scans. • Computed biomarkers (POI and iHU) used as inputs for logistic regression model to classify severe and non‐severe COVID‐19, achieving AUC of 0.97. • Computed changes of lung lesion volume to assess COVID‐19 progressing. |
| 116 | Chest CT scans | 23812 CT images from 408 patients |
• Developed multiple instance learning model using ResNet‐34 115 as backbone to distinguishing severe from non‐severe COVID‐19, achieving AUC of 0.892. • The model was also applied for identifying patients with mild COVID‐19 at hospital admission, who progressed to severe disease. It achieved AUCs of 0.955 and 0.923 in two different subgroups. |
| 117 | Chest CT scans | 72 serial CT of 24 patients | • Developed a DL model based on U‐Net 29 for automated lung segmentation.• Utilized quantification of infected regions to assess COVID‐19 progression.• Created heatmaps to visualize the progression. |
| 118 | Chest X‐ray | 581 patients |
• A convolutional Siamese neural network was built using DenseNet121 50 to provide the pulmonary X‐ray severity score, and assess the disease severity. • To build the model, a training set of X‐ray images was manually annotated using modified version of RALE scoring system, and the model was pretrained with chest X‐ray images from CheXpert 27 for better performance. |
| 23 | Chest CT, and clinical characteristics. |
Stable group: 222 patients Progressive group: 25 patients |
• Applied multivariate logistic regression to identify critical features to construct a nomogram. It was revealed that the features including age, CT severity score, and NLR were the significant, independent risk predictors. • Constructed the nomogram incorporating the predictors to predict the progression risk of patients at admission time. |
| 119 | Ultrasound | 58 lung ultrasound videos from 20 patients |
• An unsupervised and automatic model was proposed to detect and localize the pleural line in ultrasound data using HMM and Viterbi algorithm, achieving 94% and 84% in terms of accuracy for linear and convex probes, respectively. • Depending on pleural line, SVM classifier evaluated the severity of COVID‐19 with accuracy rates of 94% and 88%, respectively, for linear and convex probes. |
| 25 | Ultrasound | 277 lung ultrasound videos from 35 patients |
• Introduce a fully annotated version of the ICLUS‐DB database. • Propose a novel DL network to predict the severity score at frame level, and present uninorms‐based method 120 to estimate the severity score at video level. |
| 121 | Chest CT scan. | 30 patients |
• Utilized a conditional logistic regression model to identify critical predictors of CT scan features to predict the mortality in nonelderly COVID‐19 patients without underlying comorbidities. • It was revealed that the CT severity score was the significant mortality predictor, with the highest specificity and sensitivity of 0.87 and 0.83, respectively. |
Abbreviations: AUC, area under the receiver‐operating characteristics curve; CNN, convolutional neural network; DL, deep learning; HMM, hidden Markov model; ICLUS‐DB, Italian COVID‐19 lung ultrasound database; iHU, the average infection HU; NLR, neutrophil‐to‐lymphocyte ratio; POI, the portion of infection; RALE, radiographic assessment of lung edema; SVM, support vector machine.
Moreover, a number of studies discussed in Section 3.1 have developed systems for prognostic analysis.17, 51 Zhang et al. 17 applied Light Gradient Boosting Machine (LightGBM) and Cox proportional‐hazards (CoxPH) regression models to predict COVID‐19 progression to severe or critical illness based on CT lung lesions and clinical parameters. The DL system developed by Wang et al. 51 succeeded to classify COVID‐19 patients into low‐risk and high‐risk groups.
5. DISCUSSION
With the rapid, global spread of COVID‐19, early and accurate diagnosis of COVID‐19 is essential. While RT‐PCR testing has revealed certain issues during the outbreak of COVID‐19 including low sensitivity in detecting the infection in the early stages, medical imaging can effectively detect the lung abnormalities in suspected cases of COVID‐19. High‐resolution CT scans are able to identify early SARS‐CoV‐2 infection in patients who initially test negative on RT‐PCR test, and ultrasound imaging offers low false‐negative rates for the detection of COVID‐19. Despite the shortcomings of traditional radiography compared with CT, X‐ray imaging may be a fit alternative tool for diagnosing and monitoring disease progression, especially in endemic regions with a high number of infected people, because this technology is widely available, fast, and cost‐effective. Nevertheless, we advocate for the combination of medical imaging, RT‐PCR testing, and laboratory results to identify and diagnose patients suspected of COVID‐19 infection, especially for the identification of asymptomatic patients.
Recent studies have demonstrated that ML is able to assist in the diagnosis of the disease, to improve radiologists' performances in identifying COVID‐19 and in discriminating it from healthy and other forms of pneumonia using various medical imaging tools. Medical imaging‐based supervised ML algorithms and deep learning are the most common methods applied to diagnose COVID‐19 and to assess its severity. The majority of studies discussed above have utilized CNN architectures for building automatic COVID‐19 diagnostic systems. This is due to the remarkable growth in the number of its applications in medical imaging122, 123 and its pivotal role in achieving substantial improvements of performance for multiple medical imaging tasks, including classification, feature extraction, and segmentation.124, 125 On the other hand, logistic regression algorithms have been widely applied for evaluating the relationship between clinical parameters and medical image characteristics to assess the COVID‐19 severity and to predict the mortality risk.
Despite the promising results, the application of ML methods in medical imaging‐based COVID‐19 diagnosis research face some limitations summarized below:
Lack of labeled, annotated training data: Insufficient data of medical images is available for building ML‐based diagnostic systems. Various studies applied augmentation approaches to increase the number of samples for the training phase, and others built systems based on pre‐trained networks to deal with this issue. Nevertheless, the lack of large data sets is a standing issue currently faced by the researchers. Moreover, annotating training data sets requires professional radiologists, not included in a significant number of the studies.
Imbalanced data set: There was a remarkable data imbalance between COVID‐19 samples and non‐COVID‐19 samples (healthy and other lung diseases) in training and validating cohorts. This mainly affects the robustness and performance of the COVID‐19 diagnostic models.
Data quality: The public data sets encompass low‐quality images which were extracted from websites and online publications. Moreover, using medical images from different public data sets may lead to duplication issues. Regarding image resolution, various ML models lower the image resolution to reduce the number of inputs or features. This leads to a decreasing number of parameters to be optimized while diminishing the overfitting risk. However, extensive lowering of the image resolution may result in a loss of significant information needed for correct classification, 126 specifically in a classifier required to discriminate between COVID‐19 and other viral pneumonia which share similar characteristics on CT scans and X‐ray images. The above studies resized the medical images to different resolutions (128 × 128), 101 (224 × 224),75, 77, 92, 98, 103, 113 (256 × 256),74, 80 and (512 × 512). 17 Models featuring those resolutions achieved high rates of accuracy and sensitivity.
Model evaluation: Relatively few models were evaluated using external validation datasets17, 51, 73, 74 to address model generalizability, that is, the ability of a diagnostic and prognostic model to produce accurate predictions based on varied sources of medical imaging data which differ structurally from the datasets used for model development.
Interpretability: To improve the interpretability of COVID‐19 diagnostic models, some studies utilized the gradient‐weighted class activation mapping (Grad‐CAM) 127 heat map approach39, 71, 74, 77, 98, 100, 117 or class activation maps 44 to visualize the region of lesions used by the model for decision making. Other studies provided attention maps37, 73, 76, 78 able to highlight the precise locations of regional lesions. However, these models were not able to visualize unique COVID‐19 features in medical imaging. Interestingly, Zhang et al. 17 visualized prognosis prediction and they provided lesion segmentation with quantitative analysis of all lesion features using CT scans.
Although significant number of models for COVID‐19 detection have been developed, most are unlikely to be validated and implemented in medical practice. Notwithstanding, health experts and radiologists can still benefit from these models by gaining a better understanding of the critical aspects of infected cases. In future studies, large, high‐quality medical images data sets, improvements in interpretability and generalizability will play important roles in determining the robustness and reproducibility of different models.
6. CONCLUSION
Earlier published reviews broadly discussed the use of AI applications in the fight against the COVID‐19 pandemic, including the prediction of pandemic transmission, diagnosis, mortality, and drug discovery. We specifically considered the ML models developed for COVID‐19 detection based on medical images making our review paper valuable for researchers interested in ML and medical imaging. We reviewed and analysed 62 studies that have introduced ML‐based solutions, in particular deep learning algorithms, to cope with the different challenges faced in COVID‐19 detection and in assessing disease severity and progression.
Nevertheless, this review has a certain limitations. One of the them is the lack of studies which compare the performance of ML models between X‐ray and CT scan images. This due to the lack of studies that developed ML models using multiple image modalities. Another limitation is that several studies released recently in high impact journals were not included as they have been published at the time of finalizing this review, and all preprint studies were excluded as well.
Despite the power of ML in COVID‐19 research, the lack of large data sets is a significant issue facing researchers when developing ML algorithms for COVID‐19 detection. While some studies have fine‐tuned or modified the pretrained networks to improve the performance of their diagnostic model on limited data sets, others used data augmentation approaches or capsule networks. However, these models need to be further validated on large data sets. Besides, it is important to evaluate the models using an external validation data set to highlight generalizability to other varied data sources.
While we anticipate rapid development and growth of ML applications in COVID‐19 diagnosis, the emphasis should be on improving the cross‐disciplinary collaborations of ML developers and clinicians to cope with low quality, insufficient data and overfitting risk which negatively impact on the performance of ML classifiers. Moreover, combining findings from medical images with clinical characteristics and laboratory results will further improve the quality and performance of ML models.
Rehouma R, Buchert M, Chen Y‐PP. Machine learning for medical imaging‐based COVID‐19 detection and diagnosis. Int J Intell Syst. 2021;36:5085‐5115. 10.1002/int.22504
REFERENCES
- 1. Gorbalenya AE, Baker SC, Baric RS, et al. The species severe acute respiratory syndrome‐related coronavirus: classifying 2019‐nCoV and naming it SARS‐CoV‐2. Nat Microbiol. 2020;5(4):536‐544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. A novel coronavirus emerging in China—key questions for impact assessment. N Engl J Med. 2020;382(8):692‐694. [DOI] [PubMed] [Google Scholar]
- 3. WHO . Coronavirus disease 2019 (COVID‐19) situation reports. 2021.
- 4. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497‐506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang D, Hu B, Hu C, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061‐1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395(10223):507‐513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Iaccarino G, Grassi G, Borghi C, et al. Age and multimorbidity predict death among COVID‐19 patients: results of the SARS‐RAS study of the Italian society of hypertension. Hypertension. 2020;76(2):366‐372. [DOI] [PubMed] [Google Scholar]
- 8. Curigliano G. Cancer patients and risk of mortality for COVID‐19. Cancer Cell. 2020;38(2):161‐163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tahamtan A, Ardebili A. Real‐time RT‐PCR in COVID‐19 detection: issues affecting the results. Expert Rev Mol Diagn. 2020;20(5):453‐454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Watson J, Whiting PF, Brush JE. Interpreting a covid‐19 test result. BMJ. 2020;369:m1808. [DOI] [PubMed] [Google Scholar]
- 11. Fang Y, Zhang H, Xie J, et al. Sensitivity of chest CT for COVID‐19: comparison to RT‐PCR. Radiology. 2020;296(2):E115‐E117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Woloshin S, Patel N, Kesselheim AS. False negative tests for SARS‐CoV‐2 infection—challenges and implications. N Engl J Med. 2020;383(6):e38. [DOI] [PubMed] [Google Scholar]
- 13. Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J. Chest CT for typical coronavirus disease 2019 (COVID‐19) pneumonia: relationship to negative RT‐PCR testing. Radiology. 2020;296(2):E41‐E45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ai T, Yang Z, Hou H, et al. Correlation of chest CT and RT‐PCR testing for coronavirus disease 2019 (COVID‐19) in China: a report of 1014 cases. Radiology. 2020;296(2):E32‐E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghahramani Z. Probabilistic machine learning and artificial intelligence. Nature. 2015;521(7553):452‐459. [DOI] [PubMed] [Google Scholar]
- 16. Domingos P. A few useful things to know about machine learning. Commun ACM. 2012;55(10):78‐87. [Google Scholar]
- 17. Zhang K, Liu X, Shen J, et al. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID‐19 pneumonia using computed tomography. Cell. 2020;181(6):1423‐1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cohen JP, Morrison P, Dao L, Roth K, Duong TQ, Ghassemi M. COVID‐19 image data collection: prospective predictions are the future. 2020. https://arxiv.org/abs/2006.11988
- 19. He X, Yang X, Zhang S, et al. Sample‐efficient deep learning for COVID‐19 diagnosis based on CT scans. medRxiv. 2020. [Google Scholar]
- 20. Yan T. COVID‐19 and common pneumonia chest CT dataset. In: Data M, trans. 1st ed. Mendeley Data; 2020.
- 21. Chowdhury MEH, Rahman T, Khandakar A, et al. Can AI help in screening viral and COVID‐19 pneumonia? IEEE Access. 2020;8:132665‐132676. [Google Scholar]
- 22. Italian Society of Medical and Interventional Radiology . COVID‐19 database. 2020. https://www.sirm.org/category/senza-categoria/covid-19/. Accessed December 21, 2020.
- 23. Feng Z, Yu Q, Yao S, et al. Early prediction of disease progression in COVID‐19 pneumonia patients with chest CT and clinical characteristics. Nat Commun. 2020;11(1):4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Born J, Brändle G, Cossio M, et al. POCOVID‐Net: automatic detection of COVID‐19 from a new lung ultrasound imaging dataset (POCUS). 2020. https://arxiv.org/abs/2004.12084
- 25. Roy S, Menapace W, Oei S, et al. Deep learning for classification and localization of COVID‐19 markers in point‐of‐care lung ultrasound. IEEE Trans Med Imaging. 2020;39(8):2676‐2687. [DOI] [PubMed] [Google Scholar]
- 26. Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX‐Ray8: hospital‐scale chest X‐ray database and benchmarks on weakly‐supervised classification and localization of common thorax diseases. In: Paper Presented at 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), July 21‐26, 2017.
- 27. Irvin J, Rajpurkar P, Ko M, et al. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. 2019. https://arxiv.org/abs/1901.07031
- 28. Shi F, Wang J, Shi J, et al. Review of artificial intelligence techniques in imaging data acquisition, segmentation and diagnosis for COVID‐19. IEEE Rev Biomed Eng. 2020;14:1‐1. [DOI] [PubMed] [Google Scholar]
- 29. Ronneberger O, Fischer P, Brox T. U‐Net: convolutional networks for biomedical image segmentation. In: Paper Presented at Medical Image Computing and Computer‐Assisted Intervention–MICCAI 2015; 2015; Cham.
- 30. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. In: Proceedings of the 25th International Conference on Neural Information Processing Systems. Vol 1; 2012; Lake Tahoe, Nevada.
- 31. Simonyan K, Zisserman A. Very deep convolutional networks for large‐scale image recognition. 2014. https://arxiv.org/abs/1409.1556v4
- 32. He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Paper Presented at 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June 27–30, 2016.
- 33. Deng J, Dong W, Socher R, Li L, Kai L, Li F‐F. ImageNet: a large‐scale hierarchical image database. In: Paper Presented at 2009 IEEE Conference on Computer Vision and Pattern Recognition, June 20–25, 2009.
- 34. Szegedy C, Wei L, Yangqing J, et al. Going deeper with convolutions. In: Paper Presented at 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June 7–12, 2015.
- 35. Chollet F. Xception: deep learning with depthwise separable convolutions. In: Paper Presented at 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), July 21–26, 2017.
- 36. Hara K, Kataoka H, Satoh Y. Can spatiotemporal 3D CNNs retrace the history of 2D CNNs and ImageNet? In: Paper Presented at 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, June 18–23, 2018.
- 37. Yan T, Wong PK, Ren H, Wang H, Wang J, Li Y. Automatic distinction between COVID‐19 and common pneumonia using multi‐scale convolutional neural network on chest CT scans. Chaos, Solitons Fractals. 2020;140:110153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wong HYF, Lam HYS, Fong AH‐T, et al. Frequency and distribution of chest radiographic findings in patients positive for COVID‐19. Radiology. 2020;296(2):E72‐E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ozturk T, Talo M, Yildirim EA, Baloglu UB, Yildirim O, Rajendra Acharya U. Automated detection of COVID‐19 cases using deep neural networks with X‐ray images. Comput Biol Med. 2020;121:103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L. MobileNetV2: inverted residuals and linear bottlenecks. In: Paper Presented at 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, June 18–23, 2018.
- 41. LeCun Y, Bengio Y, Hinton G. Deep learning. Nature. 2015;521(7553):436‐444. [DOI] [PubMed] [Google Scholar]
- 42. Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw. 2015;61:85‐117. [DOI] [PubMed] [Google Scholar]
- 43. Bressem KK, Adams LC, Erxleben C, Hamm B, Niehues SM, Vahldiek JL. Comparing different deep learning architectures for classification of chest radiographs. Sci Rep. 2020;10(1):13590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Han Z, Wei B, Hong Y, et al. Accurate screening of COVID‐19 using attention‐based deep 3D multiple instance learning. IEEE Trans Med Imaging. 2020;39(8):2584‐2594. [DOI] [PubMed] [Google Scholar]
- 45. Zhang C, Han Z, Cui Y, Fu H, Zhou JT, Hu Q. CPM‐Nets: cross partial multi‐view networks. 2019.
- 46. Kang H, Xia L, Yan F, et al. Diagnosis of coronavirus disease 2019 (COVID‐19) with structured latent multi‐view representation learning. IEEE Trans Med Imaging. 2020;39(8):2606‐2614. [DOI] [PubMed] [Google Scholar]
- 47. Sun L, Mo Z, Yan F, et al. Adaptive feature selection guided deep forest for COVID‐19 classification with chest CT. IEEE J Biomed Health Inform. 2020;24(10):2798‐2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burt P, Adelson E. The laplacian pyramid as a compact image code. IEEE Trans Commun. 1983;31(4):532‐540. [Google Scholar]
- 49. Shi H, Han X, Jiang N, et al. Radiological findings from 81 patients with COVID‐19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis. 2020;20(4):425‐434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Huang G, Liu Z, Maaten LVD, Weinberger KQ. Densely connected convolutional networks. In: Paper Presented at 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), July 21–26, 2017.
- 51. Wang S, Zha Y, Li W, et al. A fully automatic deep learning system for COVID‐19 diagnostic and prognostic analysis. Eur Respir J. 2020;56(2):2000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Iandola FN, Han S, Moskewicz MW, Ashraf K, Dally WJ, Keutzer K. SqueezeNet: AlexNet‐level accuracy with 50x fewer parameters and <0.5MB model size. 2016. https://arxiv.org/abs/1602.07360
- 53. Minaee S, Kafieh R, Sonka M, Yazdani S, Jamalipour Soufi G. Deep‐COVID: predicting COVID‐19 from chest X‐ray images using deep transfer learning. Med Image Anal. 2020;65:101794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Toğaçar M, Ergen B, Cömert Z. COVID‐19 detection using deep learning models to exploit Social Mimic Optimization and structured chest X‐ray images using fuzzy color and stacking approaches. Comput Biol Med. 2020;121:103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nour M, Cömert Z, Polat K. A novel medical diagnosis model for COVID‐19 infection detection based on deep features and bayesian optimization. Appl Soft Comput. 2020;97:106580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zu ZY, Jiang MD, Xu PP, et al. Coronavirus disease 2019 (COVID‐19): a perspective from China. Radiology. 2020;296(2):E15‐E25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chung M, Bernheim A, Mei X, et al. CT imaging features of 2019 novel coronavirus (2019‐nCoV). Radiology. 2020;295(1):202‐207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Herpe G, Lederlin M, Naudin M, et al. Efficacy of chest CT for COVID‐19 pneumonia diagnosis in France. Radiology. 2021;298(2):E81‐E87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Long C, Xu H, Shen Q, et al. Diagnosis of the coronavirus disease (COVID‐19): RT‐PCR or CT? Eur J Radiol. 2020;126:108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ye Z, Zhang Y, Wang Y, Huang Z, Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID‐19): a pictorial review. Eur Radiol. 2020;30(8):4381‐4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li K, Wu J, Wu F, et al. The clinical and chest CT features associated with severe and critical COVID‐19 pneumonia. Invest Radiol. 2020;55(6):327‐331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fu Z, Tang N, Chen Y, et al. CT features of COVID‐19 patients with two consecutive negative RT‐PCR tests after treatment. Sci Rep. 2020;10(1):11548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Koo HJ, Choi S‐H, Sung H, Choe J, Do K‐H. RadioGraphics update: radiographic and CT features of viral pneumonia. Radiographics. 2020;40(4):E8‐E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wang Y, Dong C, Hu Y, et al. Temporal changes of CT findings in 90 patients with COVID‐19 pneumonia: a longitudinal study. Radiology. 2020;296(2):E55‐E64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Bernheim A, Mei X, Huang M, et al. Chest CT findings in coronavirus disease‐19 (COVID‐19): relationship to duration of infection. Radiology. 2020;295(3):200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Li X, Fang X, Bian Y, Lu J. Comparison of chest CT findings between COVID‐19 pneumonia and other types of viral pneumonia: a two‐center retrospective study. Eur Radiol. 2020;30(10):5470‐5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sun Z, Zhang N, Li Y, Xu X. A systematic review of chest imaging findings in COVID‐19. Quant Imaging Med Surg. 2020;10(5):1058‐1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yoon SH, Lee KH, Kim JY, et al. Chest radiographic and CT findings of the 2019 novel coronavirus disease (COVID‐19): analysis of nine patients treated in Korea. Korean J Radiol. 2020;21(4):494‐500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kong W, Agarwal PP. Chest imaging appearance of COVID‐19 infection. Radiol: Cardiothoracic Imaging. 2020;2(1):e200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Bai HX, Hsieh B, Xiong Z, et al. Performance of radiologists in differentiating COVID‐19 from non‐COVID‐19 viral pneumonia at chest CT. Radiology. 2020;296(2):E46‐E54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Li L, Qin L, Xu Z, et al. Using artificial intelligence to detect COVID‐19 and community‐acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology. 2020;296(2):E65‐E71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Mei X, Lee H‐C, Diao K‐Y, et al. Artificial intelligence–enabled rapid diagnosis of patients with COVID‐19. Nat Med. 26(8), 2020:1224‐1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Harmon SA, Sanford TH, Xu S, et al. Artificial intelligence for the detection of COVID‐19 pneumonia on chest CT using multinational datasets. Nat Commun. 2020;11(1):4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Ko H, Chung H, Kang WS, et al. COVID‐19 pneumonia diagnosis using a simple 2D deep learning framework with a single chest CT image: model development and validation. J Med Internet Res. 2020;22(6):e19569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hu S, Gao Y, Niu Z, et al. Weakly supervised deep learning for COVID‐19 infection detection and classification from CT images. IEEE Access. 2020;8:118869‐118883. [Google Scholar]
- 76. Ouyang X, Huo J, Xia L, et al. Dual‐sampling attention network for diagnosis of COVID‐19 from community acquired pneumonia. IEEE Trans Med Imaging. 2020;39(8):2595‐2605. [DOI] [PubMed] [Google Scholar]
- 77. Bai HX, Wang R, Xiong Z, et al. Artificial intelligence augmentation of radiologist performance in distinguishing COVID‐19 from pneumonia of other origin at chest CT. Radiology. 2020;296(3):E156‐E165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Wang X, Deng X, Fu Q, et al. A weakly‐supervised framework for COVID‐19 classification and lesion localization from chest CT. IEEE Trans Med Imaging. 2020;39(8):2615‐2625. [DOI] [PubMed] [Google Scholar]
- 79. Xu X, Jiang X, Ma C, et al. A deep learning system to screen novel coronavirus disease 2019 pneumonia. Engineering. 2020;6(10):1122‐1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wu X, Hui H, Niu M, et al. Deep learning‐based multi‐view fusion model for screening 2019 novel coronavirus pneumonia: a multicentre study. Eur J Radiol. 2020;128:109041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ardakani AA, Kanafi AR, Acharya UR, Khadem N, Mohammadi A. Application of deep learning technique to manage COVID‐19 in routine clinical practice using CT images: results of 10 convolutional neural networks. Comput Biol Med. 2020;121:103795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z. Rethinking the inception architecture for computer vision. In: Paper Presented at 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), June 27–30, 2016.
- 83. Chen L‐C, Papandreou G, Schroff F, Adam H. Rethinking atrous convolution for semantic image segmentation. arXiv:170605587. 2017. http://arxiv.org/abs/1706.05587
- 84. Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder‐decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(12):2481‐2495. [DOI] [PubMed] [Google Scholar]
- 85. Devalla SK, Renukanand PK, Sreedhar BK, et al. DRUNET: a dilated‐residual U‐Net deep learning network to segment optic nerve head tissues in optical coherence tomography images. Biomed Opt Express. 2018;9(7):3244‐3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(4):640‐651. [DOI] [PubMed] [Google Scholar]
- 87. Lin T, Dollár P, Girshick R, He K, Hariharan B, Belongie S. Feature pyramid networks for object detection. In: Paper Presented at 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), July 21–26, 2017.
- 88. Rudyanto RD, Kerkstra S, van Rikxoort EM, et al. Comparing algorithms for automated vessel segmentation in computed tomography scans of the lung: the VESSEL12 study. Med Image Anal. 2014;18(7):1217‐1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Wang S, Shi J, Ye Z, et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur Respir J. 2019;53(3):1800986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Shan F, Gao Y, Wang J, et al. Lung infection quantification of COVID‐19 in CT images with deep learning. 2020. https://arxiv.org/abs/2003.04655
- 91. Ng M‐Y, Lee EY, Yang J, et al. Imaging profile of the COVID‐19 infection: radiologic findings and literature review. Radiol: Cardiothoracic Imaging. 2020;2(1):e200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Khan AI, Shah JL, Bhat MM. CoroNet: a deep neural network for detection and diagnosis of COVID‐19 from chest x‐ray images. Comput Methods Programs Biomed. 2020;196:105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Nambu A, Ozawa K, Kobayashi N, Tago M. Imaging of community‐acquired pneumonia: roles of imaging examinations, imaging diagnosis of specific pathogens and discrimination from noninfectious diseases. World J Radiol. 2014;6(10):779‐793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Paul NS, Roberts H, Butany J, et al. Radiologic pattern of disease in patients with severe acute respiratory syndrome: the Toronto experience. Radiographics. 2004;24(2):553‐563. [DOI] [PubMed] [Google Scholar]
- 95. Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265‐269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Phan LT, Nguyen TV, Luong QC, et al. Importation and human‐to‐human transmission of a novel coronavirus in Vietnam. N Engl J Med. 2020;382(9):872‐874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Feng H, Liu Y, Lv M, Zhong J. A case report of COVID‐19 with false negative RT‐PCR test: necessity of chest CT. Jpn J Radiol. 2020;38(5):409‐410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Brunese L, Mercaldo F, Reginelli A, Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID‐19 detection from X‐rays. Comput Methods Programs Biomed. 2020;196:105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Yoo SH, Geng H, Chiu TL, et al. Deep learning‐based decision‐tree classifier for COVID‐19 diagnosis from chest X‐ray imaging. Front Med. 2020;7:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Oh Y, Park S, Ye JC. Deep learning COVID‐19 features on CXR using limited training data sets. IEEE Trans Med Imaging. 2020;39(8):2688‐2700. [DOI] [PubMed] [Google Scholar]
- 101. Toraman S, Alakus TB, Turkoglu I. Convolutional capsnet: a novel artificial neural network approach to detect COVID‐19 disease from X‐ray images using capsule networks. Chaos, Solitons Fractals. 2020;140:110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ouchicha C, Ammor O, Meknassi M. CVDNet: a novel deep learning architecture for detection of coronavirus (Covid‐19) from chest X‐ray images. Chaos, Solitons Fractals. 2020;140:110245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Panwar H, Gupta PK, Siddiqui MK, Morales‐Menendez R, Singh V. Application of deep learning for fast detection of COVID‐19 in X‐rays using nCOVnet. Chaos, Solitons Fractals. 2020;138:109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Abbas A, Abdelsamea MM, Gaber MM. Classification of COVID‐19 in chest X‐ray images using DeTraC deep convolutional neural network. Appl Intell. 2020;51(2):854‐864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Canayaz M. MH‐COVIDNet: diagnosis of COVID‐19 using deep neural networks and meta‐heuristic‐based feature selection on X‐ray images. Biomed Signal Process Control. 2021;64:102257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Elaziz MA, Hosny KM, Salah A, Darwish MM, Lu S, Sahlol AT. New machine learning method for image‐based diagnosis of COVID‐19. PLOS One. 2020;15(6):e0235187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Jégou S, Drozdzal M, Vazquez D, Romero A, Bengio Y. The one hundred layers tiramisu: fully convolutional densenets for semantic segmentation. In: Paper Presented at 2017 IEEE Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), July 21–26, 2017.
- 108. Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception‐v4, inception‐ResNet and the impact of residual connections on learning. 2016. https://arxiv.org/abs/1602.07261
- 109. Redmon J, Farhadi A. YOLO9000: better, faster, stronger. In: Paper Presented at IEEE Conference on Computer Vision and Pattern Recognition (CVPR), July 21–26, 2017.
- 110. Peng Q‐Y, Wang X‐T, Zhang L‐N, Chinese G. Critical Care Ultrasound Study. Findings of lung ultrasonography of novel corona virus pneumonia during the 2019‐2020 epidemic. Intensive Care Med. 2020;46(5):849‐850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Tung‐Chen Y, Martí de Gracia M, Díez‐Tascón A, et al. Correlation between chest computed tomography and lung ultrasonography in patients with coronavirus disease 2019 (COVID‐19). Ultrasound Med Biol. 2020;46(11):2918‐2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Horry MJ, Chakraborty S, Paul M, et al. COVID‐19 detection through transfer learning using multimodal imaging data. IEEE Access. 2020;8:149808‐149824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zhu J, Shen B, Abbasi A, Hoshmand‐Kochi M, Li H, Duong TQ. Deep transfer learning artificial intelligence accurately stages COVID‐19 lung disease severity on portable chest radiographs. PLOS One. 2020;15(7):e0236621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Li Z, Zhong Z, Li Y, et al. From community‐acquired pneumonia to COVID‐19: a deep learning‐based method for quantitative analysis of COVID‐19 on thick‐section CT scans. Eur Radiol. 2020;30(12):1‐10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Xie S, Girshick R, Dollar P, Tu Z, He K. Aggregated residual transformations for deep neural networks. In: Paper Presented at the IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2017; New York.
- 116. Xiao L‐S, Li P, Sun F, et al. Development and validation of a deep learning‐based model using computed tomography imaging for predicting disease severity of coronavirus disease 2019. Front Bioeng Biotechnol. 2020;8:898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Pu J, Leader JK, Bandos A, et al. Automated quantification of COVID‐19 severity and progression using chest CT images. Eur Radiol. 2020;31(1):436‐446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Li MD, Arun NT, Gidwani M, et al. Automated assessment and tracking of COVID‐19 pulmonary disease severity on chest radiographs using convolutional Siamese neural networks. Radiol: Artif Intell. 2020;2(4):e200079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Carrer L, Donini E, Marinelli D, et al. Automatic pleural line extraction and COVID‐19 scoring from lung ultrasound data. IEEE Trans Sonics Ultrason. 2020;67(11):2207‐2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Melnikov V, Hüllermeier E. Learning to aggregate using uninorms. In: Paper Presented at: Machine Learning and Knowledge Discovery in Databases; 2016. Cham.
- 121. Tabatabaei SMH, Rahimi H, Moghaddas F, Rajebi H. Predictive value of CT in the short‐term mortality of Coronavirus Disease 2019 (COVID‐19) pneumonia in nonelderly patients: a case‐control study. Eur J Radiol. 2020;132:109298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Litjens G, Kooi T, Bejnordi BE, et al. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60‐88. [DOI] [PubMed] [Google Scholar]
- 123. Sahiner B, Pezeshk A, Hadjiiski LM, et al. Deep learning in medical imaging and radiation therapy. Med Phys. 2019;46(1):e1‐e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL. Artificial intelligence in radiology. Nat Rev Cancer. 2018;18(8):500‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Hassan M, Ali S, Alquhayz H, Safdar K. Developing intelligent medical image modality classification system using deep transfer learning and LDA. Sci Rep. 2020;10(1):12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sabottke CF, Spieler BM. The effect of image resolution on deep learning in radiography. Radiol: Artif Intell. 2020;2(1):e190015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D. Grad‐CAM: visual explanations from deep networks via gradient‐based localization. In: Paper Presented at 2017 IEEE International Conference on Computer Vision (ICCV), October 22–29, 2017, 2017.


