Abstract
The present study was conducted from July 1, 2020 to September 25, 2020 in a dedicated coronavirus disease 2019 (COVID‐19) hospital in Delhi, India to provide evidence for the presence of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) virus in atmospheric air and surfaces of the hospital wards. Swabs from hospital surfaces (patient's bed, ward floor, and nursing stations area) and suspended particulate matter in ambient air were collected by a portable air sampler from the medicine ward, intensive care unit, and emergency ward admitting COVID‐19 patients. By performing reverse‐transcriptase polymerase chain reaction (RT‐PCR) for E‐gene and RdRp gene, SARS‐CoV‐2 virus was detected from hospital surfaces and particulate matters from the ambient air of various wards collected at 1 and 3‐m distance from active COVID‐19 patients. The presence of the virus in the air beyond a 1‐m distance from the patients and surfaces of the hospital indicates that the SARS‐CoV‐2 virus has the potential to be transmitted by airborne and surface routes from COVID‐19 patients to health‐care workers working in COVID‐19 dedicated hospital. This warrants that precautions against airborne and surface transmission of COVID‐19 in the community should be taken when markets, industries, educational institutions, and so on, reopen for normal activities.
Keywords: airborne, COVID‐19, epidemiology, horizontal transmission, SARS‐CoV‐2, virus
1. INTRODUCTION
Since the outbreak of coronavirus disease‐19 (COVID−19) in Wuhan, China, humanity has been continuously engaged in a fight for controlling this pandemic. It is evident that we have become better at controlling the pandemic compared with previous pandemics. This is attributed to technological advancements. It took only about 2 weeks to identify and publish the gene sequence of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), the causative coronavirus for COVID‐19, and also to develop a diagnostic method for its detection using the reverse‐transcriptase polymerase chain reaction (RT‐PCR).1, 2, 3, 4 In addition, unprecedented progress has been made in other aspects of the disease. The roles of Angiotensinogen Converting Enzyme‐2 (ACE‐2) receptors and other molecules, cytokine storm, and coagulation dysregulation in its pathogenesis are established.5, 6, 7 Sero‐surveillance by immunoassay of antibodies has been developed.8, 9, 10 Many drug trials have been carried out that established the role of dexamethasone, anti‐retroviral agents, hydroxychloroquine (HCQ), ivermectin, tocilizumab, curcumin, and so on, in the treatment of COVID‐19 with some degree of success.11, 12, 13, 14, 15, 16, 17
Scientists have developed various vaccines against COVID‐19 and these are under evaluation.18, 19, 20, 21 However, we do not have clear knowledge about the routes followed by the virus during transmission from one individual to others.22, 23 As of now, the general notion is that the virus gets transmitted either through droplets or through direct contact with an infected person or by touching the surfaces/fomites where an infected person has shed the virus.22, 23, 24, 25, 26, 27, 28, 29, 30 The virus enters the human body through the nose, mouth, or eyes. However, whether the virus spreads through air is far from clear, although some reports are present, which have suggested considering air‐borne precautions.22, 23 At a virtual workshop entitled "Airborne Transmission of SARS‐CoV‐2" conducted by the National Academies of Science, Engineering, and Medicine, various field experts reviewed the available data related to transmission routes of the virus along with its historical context. 31 The evidence for airborne transmission of SARS‐CoV‐2 is admittedly incomplete. They also stated that the evidence for it not being airborne is also incomplete. The evidence for transmission of this virus through air remains inconclusive. It is therefore imperative to address this issue and provide evidence of its being air‐borne if it is so.
In India, the index case was detected in Thrissur, Kerala on January 30, 2020 and by the end of October 2020, the total number of confirmed cases crossed 81 million. 32 Various measures are being taken here to manage this pandemic. Mild cases are isolated at home or in isolation centers created for this purpose. Few hospitals with intensive care unit (ICU) care facilities are dedicated to treating moderate and severe cases of COVID‐19 patients only. Lok Nayak Hospital at New Delhi, India, where the study has been conducted, is one among those hospitals. Despite the best precautions, many health‐care workers working in these hospitals got infected and a few of them died.33, 34, 35 Air‐borne transmission or transmission by touching surfaces is suspected in the transmission of COVID‐19 in the hospital set up.36, 37, 38 Hence, this study was conducted to collect the data from this COVID‐19 dedicated hospital to find out whether the SARS‐CoV‐2 virus spreads beyond 1 m from COVID‐19 patients through air and if the viral concentration in air decreases with increase in distance from the active patients. The study also explored the presence of SARS‐CoV‐2 viral RNA on different surfaces of wards admitting COVID‐19 patients.
2. METHODOLOGY
2.1. Study sites
The collaborative study was conducted by the Departments of Biochemistry, Microbiology, Maulana Azad Medical College in collaboration with the Departments of Medicine and Anaesthesia, Lok Nayak Hospital (LNH), and CSIR‐National Physical Laboratory, New Delhi, India from July 1, 2020 to September 25, 2020. Delhi, the capital city of India having more than 20 million population is listed as one of the most populated megacities of the world. Delhi according to India Meteorological Department (IMD) has four different seasons, that is, winter (January–February), summer or pre‐monsoon (March–May), monsoon (June–September), and post‐monsoon (October–December). Winters are chilly (temperature can drop to ~ 2°C) and observe intense fog and haze events. Summers are generally very hot and dry (temperature can go up to ~ 47°C) and with frequent dust storms. Swabs and particulate matter from the air were collected from (a) emergency ward, (b) medicine ward, and ICU of LNH which was declared as a dedicated COVID‐19 hospital.
2.2. Study design
It was an in vitro study.
2.3. Ethical approval
Written informed consent was obtained from the participants ≥18 years of age before they were recruited for the study. The study protocol was approved [no: F.1/IEC/MAMC/(77/05/2020/No.143) dated June 19, 2020] by the Institutional Ethics Committee (IEC) of Maulana Azad Medical College (MAMC) and associated Lok Nayak Hospital (LNH), University of Delhi, New Delhi, India.
2.4. Selection of cases
Six patients per day who were suffering from COVID‐19 (RT‐PCR confirmed from their nasopharyngeal and nasal swab) with moderate illness were selected randomly among the COVID‐19 patients admitted to the medicine ward within the last 48 h. This protocol was repeated on 6 different days not necessarily consecutive. Moderate illness was evidenced by the presence of lower respiratory disease on clinical examination and/or imaging (X‐Ray or computed tomography (CT) scan of chest) during this period. These patients had high‐grade fever, cough and sneezing, and SpO2 levels above 94% in room air at the time of sample collection. They were asked to breathe out, talk, cough, or sneeze on a 47 mm diameter polyvinylidene difluoride (PVDF) membrane filter having 100 nm pore size (M/s. Millipore Corp) fixed on a petri dish by always keeping the petri dish within one foot from his mouth for 15 min. Immediately, the filter was placed with the help of forceps into a 15 ml centrifuge tube containing 5 ml of viral transport media (VTM) and then transported in an icebox to the laboratory for detection of SARS‐CoV‐2 by RT‐PCR. The positive report confirmed that these patients were releasing the SARS‐CoV‐2 virus from their respiratory tract and was defined as active cases. A swab from the bed and floor and particulate matter from the air nearby the area of the patient were collected only if the patient turned out to be an active case.
Similarly, on 6 different days, six different patients suffering from RT‐PCR confirmed COVID‐19 with severe illness requiring admission to ICU within the last 48 h for high flow oxygen therapy or invasive ventilator support were selected randomly among the COVID‐19 patients admitted to ICU. As these patients were very sick and on noninvasive or invasive ventilator support to perform the above‐mentioned activity, it was not checked if they were active cases or not. In the emergency ward, the COVID‐19 patients were getting clinically evaluated, receiving immediate management, and were sent to either the medicine ward or ICU depending on the treatment need. So the patients were in transit and were staying for few minutes to few hours in the emergency ward. Whether these cases were active or not was not checked when particulate matter from the air was collected from the emergency ward.
2.5. Collection of particulate matters from environmental airì
Total suspended particulate (TSP) air sampler, (M/s. Vayuvodhan, Okhla Industrial Area, New Delhi) which was calibrated as per national standards by CSIR‐NPL, India was used for collecting suspended particulate matter from the air. The air sampler used in this study was portable and handy with three sample collection passages for which flow rate was fixed at 1.5, 16.7, and 27 litre per minute (LPM). The air sampler consisted of an anodized aluminum body fitted with a vacuum motor that pulled the ambient air onto the 47 mm diameter PVDF membrane filters having 100 nm pore size (M/s. Millipore Corp) placed on sample collection passages.
Particulate matters from the ambient air of the medicine ward were collected by using the above‐mentioned air sampler keeping it at a distance of 3 m from the head end of the selected active COVID cases. Then the process was repeated by keeping the air sampler at 1‐m distance from the selected patients. The air sampler was placed on a table to cover up the height of the patients' bed.
From ICU, suspended particulate matters were collected similarly by keeping the air sampler at 1‐ and 3‐m distance from the selected cases who were on noninvasive ventilator support.
In the emergency ward, particulate matter from the air was collected by keeping the air sampler at the center of the ward. The TSP samples were collected for a 1‐h period at every instance.
After a collection period of 1 h, PVDF membranes were removed from the air sampler, placed with forceps into 15 ml centrifuge tubes containing 5 ml VTM and sent similarly for RT‐PCR.
For collection of negative control samples (n = 3), particulate matter from the air was collected from the green zone (area without any known COVID‐19 patients) of Maulana Azad Medical College, New Delhi.
2.6. Collection of swabs from surfaces
Swabs were collected from various surfaces to check the presence of SARS‐CoV‐2. On the surfaces of ward and ICU. The swab samples were collected randomly from 2.0 square feet area of the selected patient's bed, ward floors (within 1 m from selected patients’ bed), and tables placed in the nursing working station of the medicine ward and ICU. The collected swabs were placed in 5 ml of VTM and transported to the laboratory in an icebox for RT‐PCR testing.
For negative controls, we collected swab samples (n = 3 each) from floors and tables placed in the green zone of Maulana Azad Medical College, New Delhi.
2.7. Detection of SARS‐CoV‐2 virus by RT‐PCR
The PVDF membranes and swabs added to VTM were sent to the microbiology laboratory for the detection of the SARS‐CoV‐2 virus by using RT‐PCR. A tube containing swab/membrane was mixed thoroughly for 3 min by using vortex mixture, centrifuged at 3000 rpm in a clinical centrifuge for 3 min and 200 µl of supernatant VTM was used for extraction of RNA by using a fully automated nucleic acid extraction system (Magna Pure, Roche) utilizing Magna Pure 96 viral RNA Large Volume Kit (Roche) according to the manufacturer's instructions. The RT‐PCR technique, which specifically targets E‐gene for common coronavirus and RNA‐dependent RNA polymerase (RdRp) gene for SARS‐CoV‐2 was used for the detection of SARS‐CoV‐2 (SD Biosciences). The details of primers and probes used were as described by Afzal 2 are depicted in Table 1. This STANDARD M nCoV Real‐Time Detection kit was clinically evaluated by the manufacturer and independently for SARS‐CoV‐2 detection. The performance of the kit in terms of positive predictive value (PPV) and negative predictive value (NPV) are 100% (95% confidence interval [CI]: 88.65%–100%) and 100% (95% CI: 88.65%–100%) by the manufacturer and 100% (95% CI: 93.0%–100%) 99% (95% CI: 95.0%–100%) independently.2, 38
Table 1.
The details of the target genes and their primers, probes, and sequences used for detection of the SARS‐CoV‐2 virus
| Target gene | Size and position of amplicon | Primers and probes (Dye) | Sequence (5’−3’) |
|---|---|---|---|
| ORF 1ab (RdRp) gene | 101 bp | ORF gene‐forward primers | 5′GTGARATGGTCATGTGTGGCGG3’ |
| (15,431–15,330) | |||
| ORF gene‐reverse primers | 5′CARATGTTAAASACACTATTAGCATA3’ | ||
| ORF gene probe (FAM) | 5′CAGGTGGAACCTCATCAGGAGATGC3’ | ||
| E‐gene | 112 bp | E ‐ gene‐forward primers | 5′ACAGGTACGTTAATAGTTAATAGCGT3’ |
| (26,269–26,381) | |||
| E gene‐reverse primers | 5′ATATTGCAGCAGTACGCACACA3’ | ||
| E ‐ gene probe {JOE (VIC or HEX)} | 5′ACACTAGCCATCCTTACTGCGCCTTCG3’ |
Abbreviations: ORF, open reading frame; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Briefly, 25 µl reaction was set up which contained 4 µl of the template RNA, 14 µl of a primer and probe mixture (2019‐nCoV Reaction Solution) containing “a mixture of Taq polymerase, deoxyribonucleotide triphosphates (dNTPs), and magnesium chloride”, 6 µl of RTase mix, 0.5 µl of carboxy‐X‐rhodamine used as reference dye and 0.5 µl of internal control A. The thermal cycling profile consisted of one cycle of 50°C for reverse transcription for 15 min, one cycle of 95°C for initial denaturation for 3 min and five cycles of preamplification consisted of 95°C for 5 s and 60°C for 40 s. The final amplification profile consisted of forty (40) cycles of denaturation at 95°C for 5 s, 60°C for 40 s. The results of RT‐PCR were expressed as cycle threshold (C t) values. A cycle threshold less than 35 was interpreted as positive for SARS‐CoV‐2. If RT‐PCR gave a C t value of <35 for both the E‐gene and RdRp gene, the sampling was considered positive for SARS‐CoV‐2.
2.8. Statistical analysis
Collection of particulate matter from the air was carried out on 6 different days using different patients and similarly, swabs were collected from surfaces on three different occasions. Quantitative data (Ct value) was presented as mean and standard deviation. Qualitative data (+ve RT‐PCR) were presented as a percentage. Ct values were compared by unpaired t test or one‐way analysis of variance followed by Tukey's post hoc test. Ap < .05 was considered statistically significant.
3. RESULTS AND DISCUSSION
As shown in Table 2, the RNAs of SARS‐CoV‐2 were detected in atmospheric air at a distance of 1 and 3 m from the active COVID‐19 patients of the medicine ward and ICU and as well as in the emergency ward of the dedicated COVID‐19 hospital. RT‐PCR positivity rate was higher when particulate matter was collected at a 1‐m distance than that collected at a 3‐m distance from COVID‐19 patients. (Table 3).
Table 2.
Results of RT‐PCR for E‐gene and RdRp gene of SARS‐CoV‐2 virus performed with suspended particulate matter obtained from atmospheric air of medicine ward, ICU, and emergency ward of a dedicated COVID hospital
| Location | Distance from the COVID + ve patient | Different days | Flow rate in TSP air sampler in liter/minute | E‐gene | RdRp‐gene | ||
|---|---|---|---|---|---|---|---|
| +/− | C t value | +/− | C t value | ||||
|
Medicine ward admitting ~ 50 mild to moderate COVID‐19 patients. (area 1000 × 600 Feet2) |
One meter from the head end of the patients | Day 1 | 1.5 | Positive | 32.14 | Positive | 29.6 |
| 16.7 | Positive | 23.12 | Positive | 26.5 | |||
| 27 | Positive | 26.11 | Positive | 16.11 | |||
| Day 2 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Positive | 27.51 | Positive | 29.1 | |||
| 27 | Positive | 27.2 | Positive | 23.12 | |||
| Day 3 | 1.5 | Positive | 28.69 | Positive | 31 | ||
| 16.7 | Positive | 22.08 | Positive | 26.3 | |||
| 27 | Positive | 24.65 | Positive | 22.41 | |||
| Day 4 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Positive | 25.3 | Negative | Negative | |||
| 27 | Positive | 23.6 | Positive | 18.2 | |||
| Day 5 | 1.5 | Positive | 31 | Negative | Negative | ||
| 16.7 | Positive | 23.2 | Positive | 24.09 | |||
| 27 | Positive | 18.32 | Positive | 21.82 | |||
| Day 6 | 1.5 | Positive | 32.08 | Positive | 29.4 | ||
| 16.7 | Positive | 26 | Positive | 25.9 | |||
| 27 | Positive | 16.11 | Positive | 19.3 | |||
| Three meters from the head end of the patients | |||||||
| Day 1 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 2 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Positive | 28.1 | Positive | 31.8 | |||
| Day 3 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 4 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Positive | 21.11 | Positive | 29.85 | |||
| 27 | Positive | 29.7 | Positive | 34.1 | |||
| Day 5 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 6 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
|
ICU admitting ~20 patients with severe COVID‐19. (area: 200 × 50 Feet2) |
One meter from the head end of the patients | Day 1 | 1.5 | Positive | 30.1 | Positive | 29.81 |
| 16.7 | Positive | 29.32 | Positive | 27.51 | |||
| 27 | Positive | 23 | Positive | 18.41 | |||
| Day 2 | 1.5 | Positive | 26.3 | Negative | Negative | ||
| 16.7 | Positive | 29.2 | Positive | 24.15 | |||
| 27 | Positive | 24.3 | Positive | 17.26 | |||
| Day 3 | 1.5 | Positive | 27.5 | Positive | 30.2 | ||
| 16.7 | Positive | 30.2 | Positive | 26.87 | |||
| 27 | Positive | 21.68 | Positive | 19.47 | |||
| Day 4 | 1.5 | Positive | 26.7 | Positive | 30.26 | ||
| 16.7 | Positive | 19.8 | Positive | 25.51 | |||
| 27 | Positive | 19.11 | Positive | 22.4 | |||
| Day 5 | 1.5 | Positive | 30.7 | Positive | 29.11 | ||
| 16.7 | Positive | 25.3 | Positive | 24.98 | |||
| 27 | Positive | 20.3 | Positive | 16.84 | |||
| Day 6 | 1.5 | Positive | 30.4 | Negative | Negative | ||
| 16.7 | Positive | 27.02 | Positive | 24.52 | |||
| 27 | Positive | 21.66 | Positive | 20.1 | |||
| Three meters from the head end of the patients | Day 1 | 1.5 | Negative | Negative | Negative | Negative | |
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Positive | 31 | Negative | Negative | |||
| Day 2 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 3 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Positive | 30.2 | Positive | 32.89 | |||
| 27 | Positive | 32.5 | Positive | 30.5 | |||
| Day 4 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Positive | 29.9 | Positive | 33.67 | |||
| Day 5 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Positive | 31.9 | Positive | 33.25 | |||
| Day 6 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
|
Emergency Ward with variable number of patients ~3‐4/H. (area: 150 × 100 Feet2) |
Air sampler placed at the center of Emergency ward | Day 1 | 1.5 | Positive | 29.5 | Negative | Negative |
| 16.7 | Positive | 27.03 | Positive | 33.69 | |||
| 27 | Positive | 26.66 | Positive | 31.89 | |||
| Day 2 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Positive | 30 | Positive | 34.01 | |||
| 27 | Positive | 27.07 | Positive | 29.9 | |||
| Day 3 | 1.5 | Positive | 29.91 | Negative | Negative | ||
| 16.7 | Positive | 26.33 | Positive | 27 | |||
| 27 | Positive | 28.11 | Positive | 24.09 | |||
| Day 4 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Positive | 26.66 | Positive | 30 | |||
| 27 | Positive | 28.21 | Positive | 26.3 | |||
| Day 5 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Positive | 28 | Positive | 29.01 | |||
| 27 | Positive | 30.23 | Positive | 32.04 | |||
| Day 6 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
|
Nursing station area of medicine ward separated from the patients by glass wall. (area: 20 × 15 Feet2) |
Day 1 | 1.5 | Negative | Negative | Negative | Negative | |
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 2 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 3 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 4 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 5 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 6 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
|
Nursing station area of ICU separated from the patients by glass wall. (area: 20 × 20 Feet2). |
Day 1 | 1.5 | Negative | Negative | Negative | Negative | |
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 2 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 3 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 4 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 5 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
| Day 6 | 1.5 | Negative | Negative | Negative | Negative | ||
| 16.7 | Negative | Negative | Negative | Negative | |||
| 27 | Negative | Negative | Negative | Negative | |||
Abbreviations: COVID‐19, coronavirus disease 2019; ICU, intensive care unit; RT‐PCR, reverse‐transcriptase polymerase chain reaction; TSP, Total suspended particulate; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Table 3.
Frequency of SARS‐CoV‐2 detection in air samples collected at 01 and 03‐ m distance from COVID‐19 patients in medicine ward and ICU
| Site of sample collection | Distance from the patients | No. sample collected | SARS‐CoV‐2 positivity rate (%) | C t Value (RdRp gene) |
|---|---|---|---|---|
| Medicine ward | 1 m | 06 | 6/6 (100%) | 25.31 |
| 27.43 | ||||
| 24.56 | ||||
| 23.12 | ||||
| 28.92 | ||||
| 27.56 | ||||
| 3 m | 06 | 02/06 (33%) | 34.32 | |
| 33.09 | ||||
| ICU | 1 m | 06 | 06/06 (100%) | 22.32 |
| 23.12 | ||||
| 21.32 | ||||
| 25.79 | ||||
| 19.18 | ||||
| 19.23 | ||||
| 3 m | 06 | 03/06 (50%) | 24.54 | |
| 23.00 | ||||
| 21.76 |
Abbreviations: COVID‐19, coronavirus disease 2019; ICU, intensive care unit; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Viral load, in terms of cycle threshold (C t) values for both the RdRp gene and E‐gene of SARS‐CoV‐2 varied inversely with distance from the patients (Figure 1). It indicates that SARS‐CoV‐2 is present in atmospheric air of hospitals treating COVID‐19 patients and its concentration decreases with the increase in distance from the cases. Droplets produced during breathing, coughing, sneezing, or talking usually get settled within 1 m.22, 23 However, the presence of SARS‐CoV‐2 RNA at a 3‐m distance from the patients indicates that air‐borne transmission of virus, probably through micro‐droplets is possible, at least in the hospital environment, although viral load decreases with distance possibly due to (a) settlement of droplets within 1 m and (b) dilutional effect on micro‐droplets (that can move beyond 1 m) on their migration to distant places, which is now affirmed by World Health Organization. 22
Figure 1.

Bar diagram showing mean and standard deviation (SD) of cycle threshold (C t) values of RT‐PCR test for E‐gene and RdRp gene of SARS‐CoV‐2 virus conducted with particulate matter obtained from air at distance of 1 and 3‐m from COVID‐19 patients at medicine wards and ICU. Mean and SD were calculated from the samples, which were positive for both E‐gene and RdRp gene and collected through channels of air sampler that was set at air flow rate of 16.7 and 27 liters per minute. COVID‐19, coronavirus disease 2019; ICU, intensive care unit; RT‐PCR, reverse‐transcriptase polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
As shown in Table 4 and Figure 1, PCR positivity rate and the C t values of E‐gene and RdRp gene at 1 and 3‐m distances from COVID‐19 patients in ICU did not differ from those at the medicine ward. It indicates that viral concentration in air of ICU does not differ from that in the medicine ward despite various medical procedures (intubation, suction, etc.) that are conducted in ICU more frequently producing lots of aerosols.23, 39 However, one factor that might contribute to this observation is that number of patients and patient: ward area ratio in the ICU were always lower than that in the medicine ward.
Table 4.
Results of RT‐PCR for E‐gene and RdRp‐gene of SARS‐CoV‐2 virus performed with swabs collected from patients’ beds, floor, and nursing working stations at the medicine ward and ICU of a COVID‐dedicated hospital
| Different days | Location | Site of sample collection | CORONA (E‐gene) | COVID (RDRP gene) | ||
|---|---|---|---|---|---|---|
| +/− | C t value | +/− | C t value | |||
| Day 1 | ICU | Patients’ bed | Positive | 19.58 | Positive | 18.61 |
| ICU | Floor | Positive | 30.6 | Positive | 27.36 | |
| ICU | Nursing station | Negative | Negative | Negative | Negative | |
| Medicine ward | Patient bed | Positive | 28.9 | Positive | 22.52 | |
| Medicine ward | Floor | Positive | 33.8 | Negative | Negative | |
| Medicine ward | Nursing station | Negative | Negative | Negative | Negative | |
| Day 2 | ICU | Patients’ bed | Positive | 28.76 | Positive | 19.54 |
| ICU | Floor | Positive | 28.9 | Positive | 23.33 | |
| ICU | Nursing station | Positive | 34.02 | Negative | Negative | |
| Medicine ward | Patient bed | Positive | 29 | Positive | 19.55 | |
| Medicine ward | Floor | Positive | 32 | Positive | 23.43 | |
| Medicine ward | Nursing station | Negative | Negative | Negative | Negative | |
| Day 3 | ICU | Patients’ bed | Positive | 21.69 | Positive | 18.55 |
| ICU | Floor | Positive | 28.81 | Negative | Negative | |
| ICU | Nursing station | Positive | 30.12 | Negative | Negative | |
| Medicine ward | Patient bed | Positive | 26.72 | Positive | 22.13 | |
| Medicine ward | Floor | Positive | 32.54 | Negative | Negative | |
| Medicine ward | Nursing station | Negative | Negative | Negative | Negative | |
Abbreviations: COVID, coronavirus disease; ICU, intensive care unit; RT‐PCR, reverse‐transcriptase polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
The air in the emergency ward was also found to be contaminated with SARS‐CoV‐2 virus (Table 2). Athough the patients stay here for very short duration, yet the viral load in air was similar to that in atmospheric air of ICU and medicine ward (as found at 3‐m distance from the patients). Hence, we presume that chance of acquiring COVID‐19 by air‐borne route from the emergency ward is not less than that of ICU and medicine ward. Work stations in medicine ward and ICU had a glass separation from the area where patients were admitted and in the air of those work station areas, SARS‐CoV‐2 RNA was not detected (Table 4). Therefore, airborne transmission of COVID‐19 is not probable from these walled nursing work‐station areas. But in the emergency ward, the workstation did not have any wall or glass barrier to keep it separate from the environment where patients were admitted. Hence, the chance of air‐borne transmission for health‐care workers from work‐station at emergency ward might be present.
Patients’ bed and floor of the wards but not the tables in the work stations of nurses in medicine ward and ICU were found to contain RNA of SARS‐CoV‐2 (Table 4) indicating possible surface transmission of COVID‐19 through contaminated surfaces from patients’ beds and ward floors. It is a known mode of transmission of COVID‐19. 22 So, we surmise that hospital‐acquired COVID‐19 infection by health‐ care workers may occur through these contaminated surfaces.
As shown in Figure 2, it was found that chance of detecting SARS‐CoV‐2 viral RNA is less when flow rate of air sampler is fixed at 1.5 LMP for 1 h and that probability increased with increase in flow rate to 16.7 and 27 LMP. Therefore, we recommend that TSP collection should be done for 1 h at flow rate of 16.7 and/or 27 LPM for capturing SARS‐CoV‐2 from air for such studies.
Figure 2.
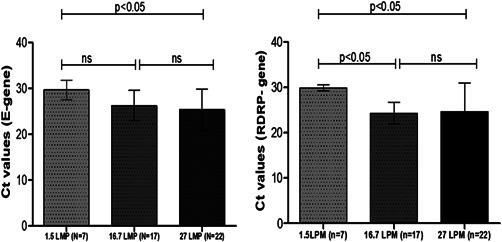
Bar diagram showing mean and standard deviation (SD) of cycle threshold (C t) values of RT‐PCR test for E‐gene and RdRp gene of SARS‐CoV‐2 virus conducted with particulate matter obtained from air of different wards admitting COVID‐19 patients when the flow rate of air sampler was adjusted to 1.5, 16.7, and 27 liter per minute (LPM). Mean and SD were calculated from the samples those were positive for both E‐gene and RdRp gene irrespective of distance from the patient at which the air sampler was placed. COVID‐19, coronavirus disease 2019; RT‐PCR, reverse‐transcriptase polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
RT‐PCR test results were negative with all negative air and swab control samples from green zone. This implies that SARS‐CoV‐2 virus detected by RT‐PCR from air suspended particulate matters and swabs from the wards of COVID‐19‐dedicated hospital samples are less likely to be due to false‐positive result. Three limitations of this study are: (a) detection of virus was done by RT‐PCR (which is a sensitive but less specific method than when accompanied with sequencing of PCR products) and not by viral culture as these facilities are not available in our set up. So, we cannot comment if the RNAs detected are from live virus or from dead ones, (b) we did not determine the corona species for the samples that showed positive test for corona virus (E‐gene) but negative test for SARS‐CoV‐2 (RdRp‐gene), and (c) the study was conducted in a hospital and not in the community, hence we need to be cautious while extrapolating these data to claim air‐borne transmission of COVID‐19 in the community.
4. CONCLUSIONS
Environmental air and surfaces of the hospitals treating COVID‐19 patients are contaminated with SARS‐CoV‐2 virus indicating potential air‐borne and surface transmission of SARS‐CoV‐2 from the patients. The viral load was similar in the atmospheric air of ICU and medicine ward indicating a similar chance of transmission at both the places. Similar kind of experimental data in localities having COVID‐19 patients would be useful for further policy making to prevent the spread of COVID‐19 infection in the community.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHORS CONTRIBUTIONS
Conceived and designed the experiments: Abhishek Dubey, Subash C. Sonkar, Vijay K. Singh, Tuhin K. Mandal, Bidhan C. Koner, and Binita Goswami. Carried out the experiments: Abhishek Dubey, Subash C. Sonkar, Vijay K. Singh, Garima Kotnala, Aastha Bansal, Monica Irungbam, Farah Husain, and Sameer A. Guru. Collection of clinical samples: Abhishek Dubey, Subash C. Sonkar, Vijay K. Singh, Monica Irungbam, Aastha Bansal, Garima Kotnala, and Sameer A. Guru. Performed the analysis: Binita Goswami, Sonal Saxena, Farah Husain, Sudhir K. Sharma, Ravindra K. Kotnala, Tuhin K. Mandal, Chhemendra Sharma, Kirti N. Saxena, Dinesh K. Aswal, Suresh Kumar, Vikas Manchanda, and Bidhan C. Koner. Analyzed the data: Bidhan C. Koner, Tuhin K. Mandal, Abhishek Dubey, Subash C. Sonkar, Vijay K. Singh, and Sameer A. Guru. Wrote and finalized the manuscript: Abhishek Dubey, Subash C. Sonkar, Sameer A. Guru, Binita Goswami, Bidhan C. Koner, and Tuhin K. Mandal. Contributed resources: Bidhan C. Koner, Sonal Saxena, Tuhin K. Mandal. All authors read and approved the final manuscript.
PEER REVIEW
The peer review history for this article is available at https://publons.com/publon/10.1002/jmv.27029
ACKNOWLEDGEMENT
The infrastructure of multidisciplinary research unit (MRU) at Maulana Azad Medical College, New Delhi which is funded by the Department of Health Research, Ministry of Health and Family Welfare, Govt. of India was used for carrying out the experiments for the study. Mr. Vijay Kumar Singh is a Senior Research Fellow (SRF) whose scholarship was provided by the Indian Council of Medical Research (ICMR), New Delhi, India.
Dubey A, Kotnala G, Mandal TK, et al. Evidence of the presence of SARS‐CoV‐2 virus in atmospheric air and surfaces of a dedicated COVID hospital. J Med Virol. 2021;93:5339–5349. 10.1002/jmv.27029
Contributor Information
Tuhin K. Mandal, Email: tuhin@nplindia.org.
Bidhan C. Koner, Email: bckoner@hotmail.com.
DATA AVAILABILITY STATEMENT
Research data are not shared.
REFERENCES
- 1. Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382(8):727‐733. 10.1056/NEJMoa2001017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Afzal A. Molecular diagnostic technologies for COVID‐19: limitations and challenges. J Adv Res. 2020;26:149‐159. 10.1016/j.jare.2020.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Saxena S, Manchanda V, Sagar T, et al. Clinical characteristic and epidemiological features of SARS CoV‐2 disease patients from a COVID‐19 designated hospital in New Delhi. J Med Virol. 2021;93(4):2487‐2492. 10.1002/jmv.26777 [DOI] [PubMed] [Google Scholar]
- 4. Islam KU, Iqbal J. An update on molecular diagnostics for COVID‐19. Front Cell Infect Microbiol. 2020;10:560616. 10.3389/fcimb.2020.560616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wiersinga WJ, Rhodes A, Cheng AC, Peacock SJ, Prescott HC. Pathophysiology, tansmission, diagnosis, and treatment of coronavirus disease 2019 (COVID‐19): a review. JAMA. 2020;324:782‐793. 10.1001/jama.2020.12839 [DOI] [PubMed] [Google Scholar]
- 6. Tsang HF, Chan LWC, Cho WCS, et al. An update on COVID‐19 pandemic: the epidemiology, pathogenesis, prevention and treatment strategies. Expert Rev Anti Infect Ther. 2020:1‐12. 10.1080/14787210.2021.1863146 [DOI] [PubMed] [Google Scholar]
- 7. Kordzadeh‐Kermani E, Khalili H, Karimzadeh I .Pathogenesis, clinical manifestations and complications of coronavirus disease 2019 (COVID‐19). Future Microbiol. 2020;15:1287‐1305. 10.2217/fmb-2020-0110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pisanic N, Randad PR, Kruczynski K, et al. COVID‐19 serology at population scale: SARS‐CoV‐2‐specific antibody responses in saliva. J Clin Microbiol. 2020. 59(1)(:e02204‐20). 10.1128/JCM.02204-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dhama K, Sharun K, Tiwari R, et al. COVID‐19, an emerging coronavirus infection: advances and prospects in designing and developing vaccines, immunotherapeutics, and therapeutics. Hum Vaccin Immunother. 2020;16(6):1232‐1238. 10.1080/21645515.2020.1735227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sil BK, Jahan N, Haq MA, et al. Development and performance evaluation of a rapid in‐house ELISA for retrospective serosurveillance of SARS‐CoV‐2. PLOS One. 2021;16:0246346. 10.1371/journal.pone.0246346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. World Health Organization . Coronavirus disease (COVID‐19): Dexamethasone. WHO; 2020. https://www.who.int/news-room/q-a-detail/coronavirus-disease-covid-19-dexamethasone [Google Scholar]
- 12. Villar J, Añón JM, Ferrando C, et al. Efficacy of dexamethasone treatment for patients with the acute respiratory distress syndrome caused by COVID‐19: study protocol for a randomized controlled superiority trial. Trials. 2020;21:717. 10.1186/s13063-020-04643-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Uzunova K, Filipova E, Pavlova V, Vekov T. Insights into antiviral mechanisms of remdesivir, lopinavir/ritonavir and chloroquine/hydroxychloroquine affecting the new SARS‐CoV‐2. Biomed Pharmacother. 2020;131:110668. 10.1016/j.biopha.2020.110668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Singh AK, Singh A, Singh R, Misra A. Hydroxychloroquine in patients with COVID‐19: A Systematic Review and meta‐analysis. Diabetes Metab Syndr Clin Res Rev. 2020;14:589‐596. 10.1016/j.dsx.2020.05.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heidary F, Gharebaghi R. Ivermectin: a systematic review from antiviral effects to COVID‐19 complementary regimen. J Antibiot (Tokyo). 2020;73:593‐602. 10.1038/s41429-020-0336-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang C, Wu Z, Li JW, Zhao H, Wang GQ. Cytokine release syndrome in severe COVID‐19: interleukin‐6 receptor antagonist tocilizumab may be the key to reduce mortality. Int J Antimicrob Agents. 2020;55:105954. 10.1016/j.ijantimicag.2020.105954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Soni VK, Mehta A, Ratre YK, et al. Curcumin, a traditional spice component, can hold the promise against COVID‐19? Eur J Pharmacol. 2020;886:173551. 10.1016/j.ejphar.2020.173551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Polack FP, Thomas SJ, Kitchin N, et al. Safety and efficacy of the BNT162b2 mRNA Covid‐19 vaccine. N Engl J Med. 2020;383:2603‐2615. 10.1056/nejmoa2034577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang NN, Li XF, Deng YQ, et al. A Thermostable mRNA vaccine against COVID‐19. Cell. 2020. 182(5):1271–1283. 10.1016/j.cell.2020.07.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dai L, Gao GF. Viral targets for vaccines against COVID‐19. Nat Rev Immunol. 2021;21:73‐82. 10.1038/s41577-020-00480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sharma O, Sultan AA, Ding H, Triggle CR. A review of the progress and challenges of developing a vaccine for COVID‐19. Front Immunol. 2020;11:585354. 10.3389/fimmu.2020.585354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. WHO . Coronavirus Disease (COVID‐19): How Is It Transmitted?. Geneva, Switzerland: WHO; 2020. https://www.who.int/news-room/commentaries/detail/modes-of-transmission-of-virus-causing-covid-19-implications-for-ipc-precaution-recommendations. [Google Scholar]
- 23. World Health Organization . Transmission of SARS‐CoV‐2: Implications for Infection Prevention Precautions. Scientific brief. Geneva, Switzerland: WHO; 2020. [Google Scholar]
- 24. Bourouiba L. Turbulent gas clouds and respiratory pathogen emissions: potential implications for reducing transmission of COVID‐19. JAMA. 2020. 323(18):1837–1838. 10.1001/jama.2020.4756 [DOI] [PubMed] [Google Scholar]
- 25. Morawska L. Droplet fate in indoor environments, or can we prevent the spread of infection? In: Indoor Air. 2006. 16(5):335–347. 10.1111/j.1600-0668.2006.00432.x [DOI] [PubMed] [Google Scholar]
- 26. Ong SWX, Tan YK, Chia PY, et al. Air, surface environmental, and personal protective equipment contamination by severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) from a symptomatic patient. JAMA. 2020;323:1610. 10.1001/jama.2020.3227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chia PY, Coleman KK, Tan YK, et al. Detection of air and surface contamination by SARS‐CoV‐2 in hospital rooms of infected patients. Nat Commun. 2020;11:2800. 10.1038/s41467-020-16670-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Liu Y, Ning Z, Chen Y, et al. Aerodynamic analysis of SARS‐CoV‐2 in two Wuhan hospitals. Nature. 2020;582(7813):557‐560. 10.1038/s41586-020-2271-3 [DOI] [PubMed] [Google Scholar]
- 29. Guo ZD, Wang ZY, Zhang SF, et al. Aerosol and surface distribution of severe acute respiratory syndrome coronavirus 2 in hospital wards, Wuhan, China, 2020. Emerg Infect Dis. 2020;26:1583‐1591. 10.3201/eid2607.200885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. National Academies of Sciences, Engineering and M . Virtual Workshop on Airborne Transmission of SARS‐CoV‐2. APIC,2020. https://apic.org/advocacy_update/virtual-workshop-on-airborne-transmission-of-sars-cov-2/ [PubMed]
- 31. Bindu Shajan Perappadan. India's first coronavirus infection confirmed in Kerala. The Hindu. 2020. https://www.thehindu.com/news/national/indias-first-coronavirus-infection-confirmed-in-kerala/article30691004.ece
- 32. Erdem H, Lucey DR. Healthcare worker infections and deaths due to COVID‐19: a survey from 37 nations and a call for WHO to post national data on their website. Int J Infect Dis. 2021;102:239‐241. 10.1016/j.ijid.2020.10.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dubey A, Bansal A, Sonkar SC, et al. In‐house assembled protective devices in laboratory safety against SARS‐nCoV‐2 in clinical biochemistry laboratory of a COVID dedicated hospital. medRxiv. 2020. 20155713. 10.1101/2020.08.24.20155713 [DOI] [Google Scholar]
- 34. Kapoor A, Kapoor KM. Covid‐19 related deaths among doctors in India. medRxiv. 2020;18:1559325820962600. 10.1101/2020.09.28.20202796 [DOI] [Google Scholar]
- 35. Morawska L, Tang JW, Bahnfleth W, et al. How can airborne transmission of COVID‐19 indoors be minimised? Environ Int. 2020;142:105832. 10.1016/j.envint.2020.105832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Morgenstern J. Aerosols, droplets, and airborne spread: everything you could possibly want to know; 2020. FIRST10EM.first10em.com/aerosols-droplets-and-airborne-spread
- 37. Wu S, Wang Y, Jin X, Tian J, Liu J, Mao Y. Environmental contamination by SARS‐CoV‐2 in a designated hospital for coronavirus disease 2019. Am J Infect Control. 2020;48:910‐914. 10.1016/j.ajic.2020.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Biosensor SD, Korea ROF. STANDARD M NCoV Real‐Time Detection Kit Instructions for Use (IFU). Vol 1; 2020. http://sdbiosensor.com/xe/product/7653. [Google Scholar]
- 39. Zhang XS, Duchaine C. SARS‐CoV‐2 and health care worker protection in low‐risk settings: a review of modes of transmission and a novel airborne model involving inhlable particles. Clin Microbiol Rev. 2020;34(1). e184–20. 10.1128/CMR.00184-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Research data are not shared.


