Abstract
As severe acute respiratory syndrome coronavirus‐2 (SARS‐CoV‐2) infections continue, there is a substantial need for cost‐effective and large‐scale testing that utilizes specimens that can be readily collected from both symptomatic and asymptomatic individuals in various community settings. Although multiple diagnostic methods utilize nasopharyngeal specimens, saliva specimens represent an attractive alternative as they can rapidly and safely be collected from different populations. While saliva has been described as an acceptable clinical matrix for the detection of SARS‐CoV‐2, evaluations of analytic performance across platforms for this specimen type are limited. Here, we used a novel sensitive RT‐PCR/MALDI‐TOF mass spectrometry‐based assay (Agena MassARRAY®) to detect SARS‐CoV‐2 in saliva specimens. The platform demonstrated high diagnostic sensitivity and specificity when compared to matched patient upper respiratory specimens. We also evaluated the analytical sensitivity of the platform and determined the limit of detection of the assay to be 1562.5 copies/ml. Furthermore, across the five individual target components of this assay, there was a range in analytic sensitivities for each target with the N2 target being the most sensitive. Overall, this system also demonstrated comparable performance when compared to the detection of SARS‐CoV‐2 RNA in saliva by the cobas® 6800/8800 SARS‐CoV‐2 real‐time RT‐PCR Test (Roche). Together, we demonstrate that saliva represents an appropriate matrix for SARS‐CoV‐2 detection on the novel Agena system as well as on a conventional real‐time RT‐PCR assay. We conclude that the MassARRAY® system is a sensitive and reliable platform for SARS‐CoV‐2 detection in saliva, offering scalable throughput in a large variety of clinical laboratory settings.
Keywords: MALDI‐TOF, real‐time RT‐PCR, saliva, SARS‐CoV‐2
1. INTRODUCTION
Accurate and rapid testing is vital to informing the response to the coronavirus disease 2019 (COVID‐19) pandemic. Since its inception, nucleic acid amplification testing (NAAT) for SARS‐CoV‐2 RNA in nasopharyngeal (NP) specimens has been the mainstay in diagnosis. Collection of such specimens requires trained healthcare professionals who need materials, such as swabs and viral transport medium (VTM), which may not be available in all settings.1, 2, 3
Saliva has garnered attention as an alternative specimen type given its minimal discomfort and its ability to be self‐collected. As of February 21, 2021, a total of 19 in vitro SARS‐CoV‐2 diagnostic tests utilizing saliva as a clinical matrix have received Emergency Use Authorization (EUA) by the U.S. Food and Drug Administration. 4 Indeed, recent reviews of studies demonstrated that saliva NAAT diagnostic performance is comparable to that of NP specimens.5, 6 While studies have compared detection of SARS‐CoV‐2 across matched NP and saliva specimens, there is large variability in specimen collection, processing methods, and testing platforms.5, 6, 7, 8, 9, 10, 11, 12, 13, 14 Moreover, studies that assess the analytical performance of detection in saliva across platforms are lacking.
Since the identification of SARS‐CoV‐2, large‐scale testing has been difficult to achieve given various hurdles, including instrument availability and supply chain limitations.15, 16, 17, 18 Recently, a novel multiplex reverse transcription (RT‐PCR)/MALDI‐TOF assay from Agena Bioscience has received EUA for SARS‐CoV‐2 detection in upper respiratory specimens. 19 The MassARRAY® SARS‐CoV‐2 Panel and MassARRAY® System have the potential to increase diagnostic capacity and complement standard NAAT technologies. This is particularly promising for utilizing saliva in large community‐based testing efforts. Here, we report the first evaluation, to date, of this platform (“Agena”) to detect SARS‐CoV‐2 RNA in saliva and compare it to the performance of the cobas® 6800/8800 SARS‐CoV‐2 real‐time RT‐PCR Test (“Roche”) for the same specimen type. Furthermore, we also compared the analytic sensitivity of each platform and its component targets.
2. MATERIALS AND METHODS
We undertook a direct comparison of saliva as a clinical specimen for detection of SARS‐CoV‐2 RNA across two platforms in the Clinical Microbiology Laboratory (CML) at the Mount Sinai Health System (MSHS), which is certified under Clinical Laboratory Improvement Amendments of 1988 (CLIA), 42 U.S.C. §263a and meets requirements to perform high‐complexity tests. Per the Human Research Protection Program, the study did not meet the definitions of human subject research and no IRB review/approval was required.
2.1. Saliva specimen collection and processing
Saliva specimens were collected from 60 deidentified patients who underwent molecular testing for SARS‐CoV‐2 in NP or anterior nares (AN) specimens collected in the previous 48 h. Saliva was collected in sterile containers (35 2070; Corning) and volumes ranged from 0.5 to 1.5 ml. Upon receipt in MSHS CML, 1 ml of VTM (R99; Hardy Diagnostics) was added to each. Mixtures were vortexed for 30 s, and 1 ml of each was incubated at 55°C for 15 min before SARS‐CoV‐2 testing.
2.2. SARS‐CoV‐2 testing
For the MassARRAY® SARS‐CoV‐2 Panel and MassARRAY® System (CPM384; Agena), RNA was extracted from 300 μl of processed specimens using the chemagicTM Viral DNA/RNA 300 Kit H96 (CMG‐1033‐S; PerkinElmer) on the automated chemagicTM 360 instrument (2024‐0020; PerkinElmer) per manufacturer's protocol. MS2 phage RNA internal control (IC) was included in all extraction steps. Extracted RNA underwent RT‐PCR with iPLEX® Pro chemistry to amplify different Agena targets per manufacturer's protocol. After inactivation of unincorporated dNTPs by treatment with shrimp alkaline phosphatase, a sequence‐specific primer extension step was performed, in which a mass‐modified terminator nucleotide was added to the probe, using supplied extension primers and iPLEX® Pro reagents.
Extension products (analytes) were desalted, transferred to a SpectroCHIP® Array (silicon chip with prespotted matrix crystal), and loaded into the MassARRAY® Analyzer (a MALDI‐TOF mass spectrometer). The analyte/matrix cocrystals were irradiated by a laser inducing desorption and ionization, and positively charged molecules accelerated into a flight tube toward a detector. Separation occurred by time‐of‐flight, which is proportional to molecular mass. After data processing, a spectral fingerprint was generated for each analyte that characterizes the mass/charge ratio and relative intensity of the molecules. Data acquired by the MassARRAY® Analyzer was processed with the MassARRAY® Typer software and SARS‐CoV‐2 Report software. The assay detects five viral targets: three in the nucleocapsid (N) gene (N1, N2, N3) and two in the ORF1ab gene (ORF1, Orf1ab). If the IC was detected, results were interpreted as positive if ≥2 targets were detected or negative if <2 targets were detected. If no IC and no targets were detected, the result was invalid and required rerunning of the specimen.
For the cobas® 6800/8800 SARS‐CoV‐2 real‐time RT‐PCR Test (09175431190; Roche), processed saliva specimens were run as previously described for NP specimens. 20 Briefly, the assay utilizes two targets to detect SARS‐CoV‐2 RNA: the SARS‐CoV‐2‐specific ORF1ab gene (T1) and the pan‐Sarbecovirus envelope E gene (T2). A result was positive for SARS‐CoV‐2 if both T1 and T2 were detected, or if T1 was detected alone. A result was presumptive positive if T2 was detected alone. A result was negative if neither T1 nor T2 was detected.
2.3. Limit of detection (LoD) of SARS‐CoV‐2 nucleic acid in saliva
The LoD was determined across both platforms using known concentrations of a SARS‐CoV‐2 standard spiked into saliva clinical matrix.
Briefly, an in‐house SARS‐CoV‐2 standard was generated by pooling 59 NP specimens that tested positive at MSHS CML (average cycle thresholds [Ct] T1 = 17.53, T2 = 17.59). To quantitate the standard, three dilutions of the pooled sample were made (e.g., 1:50 000 [D1], 1:100 000 [D2], 1:200 000 [D3]) and run alongside serial dilutions of a commercially‐available standard (ZeptoMetrix, NATSARS(COV2)‐ERC) on the Roche platform. Reactions were run in triplicate and the SARS‐CoV‐2‐negative NP matrix served as the diluent. Concentrations of each dilution were determined by extrapolation from standard curves generated across T1 and T2 targets (Figure S1) for each dilution. The stock concentration was calculated as the average of the extrapolated concentrations at each dilution. Aliquots of stock were stored at −80°C to prevent multiple freeze‐thaw cycles.
To simulate the collection of saliva for testing, saliva from healthy donors was combined one‐to‐one with VTM and spiked with the SARS‐CoV‐2 standard. Dilutions of the spiked specimens were generated over a range of 3125.0–97.7 copies/ml (cp/ml) and 12 500–195.3 cp/ml for testing on the Roche and Agena platforms, respectively. For each platform, ten replicates of each dilution were generated, and 10 replicates of saliva‐VTM spiked with the SARS‐CoV‐2‐negative NP diluent to serve as negative controls. Spiked saliva‐VTM specimens were run as described above. SARS‐CoV‐2 was not detected in any of the negative controls, and all results were valid across both platforms.
For each platform, the LoD of each assay and component targets were determined. The experimental LoD represents the lowest concentration with ≥95% detection. The probit LoD was determined by 95% detection based on a probit regression model.
2.4. Statistical analyses
For comparison of outcomes across both platforms, percent agreement and Cohen's kappa (κ) were calculated (Minitab Statistical Software 19.2020.2.0). Normality was assessed by D'Agostino and Pearson test for continuous variables (e.g., Ct values) (GraphPad Prism 9.0.2). The Student t test (two‐tailed) or Mann–Whitney test (two‐tailed) were performed for parametric and nonparametric data, respectively (GraphPad). Linear regression analyses were performed across Roche Ct values and serial dilutions. Probit regression modeling was performed if ≥2 probit points were available (e.g., not 100% or 0% detection) (Minitab). Where depicted, confidence intervals (CI) reflect the 95% level.
3. RESULTS
Sixty patients who underwent testing for SARS‐CoV‐2 by NAAT (NP or AN) at MSHS CML were provided with sterile containers for collection of saliva within 48 h of diagnosis. Saliva specimens were immediately processed and run on the Agena system. When compared to the paired NP or AN specimens, detection of SARS‐CoV‐2 had high sensitivity (97.14%, CI: 85.08%–99.93%) and specificity (100%, CI: 86.28%–100%). Of note, the same saliva specimens run side‐by‐side on the Roche platform demonstrated equivalent sensitivity and specificity.
The Agena platform detected SARS‐CoV‐2 RNA in 34/60 saliva specimens (Table 1). In comparison, the Roche platform detected SARS‐CoV‐2 RNA in 34/60 specimens. Two specimens resulted as presumptive positive on the latter and were considered not detected for this study. Of note, the Agena platform detected SARS‐CoV‐2 in one of the two presumptive positive specimens. In addition, the one specimen detected by Roche but not by Agena had the highest T1 Ct (31.62) and second‐highest T2 Ct (33.68) of all specimens tested. Overall, there was an almost perfect level of agreement across the two platforms (96.67% agreement; CI: 88.47–99.59; Cohen's κ = 0.9321; p = 2.6 × 10−13).
Table 1.
Detection of SARS‐CoV‐2 nucleic acids in saliva across Roche and Agena commercial systems
| Roche | ||||
|---|---|---|---|---|
| Positive | Negative | Total | ||
| Agena | Positive | 33 | 1a | 34 |
| Negative | 1 | 25 | 26 | |
| Total | 34 | 26 | 60 | |
Abbreviation: SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
Presumptive positive by Roche.
To preliminarily assess the sensitivity of the Agena platform, we evaluated the performance of component targets across the saliva clinical specimens. To determine how the number of Agena targets detected correlated with the quantity of SARS‐CoV‐2 RNA, we compared the number of targets to the Ct values from the Roche assay (Figure 1). At least one Agena target was detected for specimens whose Ct values ranged 18.80–31.62 for the Roche target T1 (ORF1ab gene) and 19.06–33.68 for Roche target T2 (E gene). Overall, the number of Agena targets detected in clinical saliva specimens progressively decreased with decreasing viral RNA quantity (e.g., increasing Ct values) across both Roche targets. Indeed, all five Agena targets were detected in specimens that had the lowest mean (±SD) Ct values on Roche T1 (24.64 ± 3.019) and T2 (25.26 ± 3.189) targets.
Figure 1.
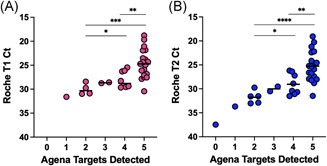
Quantitative comparison of SARS‐CoV‐2 targets detected in clinical saliva specimens. Scatter plots depict the number of SARS‐CoV‐2 targets on the Agena platform detected and the corresponding Roche Ct for each clinical saliva specimen. (A) Ct values for Roche target T1 (Orf1ab) and (B) Roche target T2 (E gene) are depicted for individual clinical saliva specimens. Medians are depicted in each column. Statistically significant differences are depicted (e.g., *p < 0.05; **p < 0.01; ***p < 0.001, ****p < 0.0001) based on the student t test or Mann–Whitney nonparametric test depending on whether data was normally distributed (see Sction 2). SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2
We next systematically measured the LoD of each platform and its component targets. We generated a SARS‐CoV‐2 standard from positive NP specimens collected from MSHS patients diagnosed at CML. The in‐house standard was quantitated by extrapolating concentrations of three dilutions run alongside serial dilutions of a commercial SARS‐CoV‐2 standard on the Roche platform (Figure S1).
The standard was spiked into saliva collected from healthy donors and ten replicates of serial dilutions were run side‐by‐side on each platform. On the Agena platform, the experimental LoD was 1562.5 cp/ml (Table 2), which is lower than the LoD reported by manufacturers for NP specimens (2500 cp/ml). 19 Across five Agena targets, the most sensitive was the N2 target (1562.5 cp/ml) followed by the N1 target (3125 cp/ml) (Table 2 and Figure 2a). The least sensitive was the Orf1ab target whose LoD could not be determined from the range of concentrations tested. This reflected a gradient in sensitivity across the individual Agena targets.
Table 2.
LoD of SARS‐CoV‐2 nucleic acids in spiked saliva on the Agena MassARRAY® platform
| No. detected/No. tested at viral concentrations (cp/ml): | Probit | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 12 500 | 6250 | 3125 | 1562.5 | 781.3 | 390.6 | 195.3 | 0.0 | Exp LoDa | LoDb | 95% CIc | |
| Overall | 10/10 | 10/10 | 10/10 | 10/10 | 1/10 | 0/10 | 0/10 | 0/10 | 1562.5 | NA | NA |
| N1 | 10/10 | 10/10 | 10/10 | 9/10 | 2/10 | 0/10 | 0/10 | 0/10 | 3125.0 | 1745.5 | (1336, 4069) |
| N2 | 10/10 | 10/10 | 10/10 | 10/10 | 1/10 | 0/10 | 0/10 | 0/10 | 1562.5 | NA | NA |
| N3 | 10/10 | 10/10 | 4/10 | 1/10 | 0/10 | 0/10 | 0/10 | 0/10 | 6250.0 | 5257.8 | (3989, 12801) |
| ORF1 | 10/10 | 10/10 | 8/10 | 8/10 | 0/10 | 0/10 | 0/10 | 0/10 | 6250.0 | 3544.7 | (2502, 8161) |
| Orf1ab | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | 0/10 | >12500 | NA | NA |
Abbreviations: LoD, limit of detection; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
Experimental (Exp) LoD determined by concentration at which detection is ≥95%.
LoD determined by probit analysis. “NA” reflects the inability to perform probit analyses due to lack of sufficient probit points.
95% fiduciary confidence interval.
Figure 2.
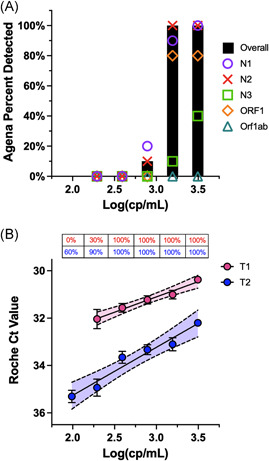
Evaluation of Roche and Agena SARS‐CoV‐2 target sensitivity. (A) Bar graph depicts the percent of spiked saliva specimens detected overall by the Agena MassARRAY® platform at five different concentrations (log). Overlaid are the individual sensitivities of the five Agena targets at each concentration. (B) Scatter plot of Ct values of Roche T1 (pink) and T2 (blue) targets across concentrations (log) of spiked saliva specimens at six different concentrations. Mean, SD of the mean, and line of best fit with 95% confidence intervals are depicted for each target. Above each concentration is the percent of replicates detected by T1 or T2 targets. SARS‐CoV‐2, severe acute respiratory syndrome coronavirus‐2.
On the Roche platform, the experimental LoD was lower than that of Agena (390.6 cp/ml). The Ct values for these saliva specimens demonstrated a linear correlation with the corresponding concentrations across both T1 (R 2 = 0.9760; p = 0.0016) and T2 (R 2 = 0.9534; p = 0.008) (Figure 2b). Overall, T2 Ct values were higher than T1 Ct values for specimens at the same concentration (p < 0.01) which is consistent with previous reports for NP specimens.21, 22 While the experimental LoD for T1 and T2 targets were equivalent, probit analyses suggest the LoD of T2 is lower (228.6 cp/ml). However, the CI for this value is broad (151.4–3.7 × 1010) given that the concentration at which no specimens were detected was not determined in our study.
4. DISCUSSION
Saliva represents an attractive specimen type for SARS‐CoV‐2 testing given its limited invasiveness, ability to be self‐collected, and its reduced need for limited supplies. These characteristics make this specimen type invaluable as it has the potential for scalable and cost‐effective SARS‐CoV‐2 screening and detection efforts across diverse groups of populations (e.g., young, old; symptomatic, asymptomatic). A number of groups have demonstrated that saliva is an acceptable specimen type when compared to other upper respiratory specimens.5, 8, 9, 12, 13, 14 However, the analytical performance of this specimen type has yet to be evaluated across the multitude of platforms utilized. Here, we demonstrate the utility of saliva as a diagnostic specimen on the Agena platform with a high agreement with a real‐time RT‐PCR diagnostic platform (Roche). Furthermore, saliva specimens collected within 2 days are equivocally sensitive and specific across both methods when compared to matched upper respiratory specimens.
It is important to note that these platforms tested differ by their technological basis. While the Roche system utilizes conventional real‐time RT‐PCR, the Agena platform utilizes mass spectrometry to detect targeted amplicons produced by RT‐PCR. Indeed, mass spectrometry represents a key technology in the clinical laboratory setting and has demonstrated its power in SARS‐CoV‐2 diagnostics. 23 Although distinct in platform technology, our findings demonstrate appropriate diagnostic capabilities of Agena as well as Roche systems for detection of SARS‐CoV‐2 RNA in saliva (Table 1).
The Agena platform also differs from the Roche platform in the SARS‐CoV‐2 viral targets probed. In contrast to a number of available real‐time RT‐PCR platforms, which are based on 2–3 target amplicons (e.g., ORF1ab (T1) and pan‐Sarbecovirus E genes (T2) on the Roche system), the Agena platform targets five regions across two viral genes (3 in the nucleocapsid gene [N1, N2, N3], 2 in the ORF1ab gene [ORF1, Orf1ab]). This redundancy in viral targets is vital to ensure robust sensitivity. When we assessed the analytic performance of each target in saliva specimens, we observed variation in target sensitivity with decreasing titers, particularly within the Agena platform (Figure 1). Specifically, the number of Agena targets detected progressively decreased with decreasing concentration. This reflects inherent analytic differences in the component targets that warrant further investigation.
To effectively utilize saliva as a clinical specimen for SARS‐CoV‐2 testing, it is essential to characterize the analytical sensitivity across diagnostic platforms. Most studies have not evaluated the LoD across platforms in a standardized method for saliva specimens. 6 In addition, the analytic sensitivity of component targets is not systematically evaluated, reported, or compared across platforms.6, 7, 24, 25, 26 Our study demonstrates a slightly lower sensitivity in the Agena platform when compared to that of the Roche for saliva specimens (Figure 2). Moreover, we find that there are differences in analytic sensitivity across Agena targets with the N2 as the most sensitive target (Figure 2 and Table 2), which is also seen on the Roche platform (e.g., T2) (Figure 2). These metrics are indispensable as they can inform how diagnostics address new circulating viral variants whose mutations may interfere with molecular methods.
Our study does have limitations in that our saliva collection methods did not occur at one time point but rather randomly within 2 days of initial NP/AN collection. While the utility of standardized collection methods (e.g., early morning) remains to be further assessed, this is not a variable we controlled. In addition, we utilized a pooled positive NP specimen to serve as our analyte to assess sensitivity. As a result, analytic sensitivities are based on a potentially heterogenous mixture of viral variants. To minimize the heterogeneity of our pooled positive NP specimen, we pooled specimens isolated from 2 consecutive days to ensure a sampling of viruses from the predominant clade at the given time.
Overall, we demonstrate that the novel Agena RT‐PCR/MALDI‐TOF mass spectrometry‐based system is capable of detecting SARS‐CoV‐2 RNA in saliva specimens. This platform is unique, has the potential for scalable throughput design, and is comparable in performance to the more ubiquitous real‐time RT‐PCR technology. Given the continued spread and rise of new SARS‐CoV‐2 variants, there is a critical need to understand the analytic capabilities of these technologies. This is especially necessary for large‐scale screening efforts where saliva has the potential to be further exploited for its utility. This understanding of assay and target sensitivity is essential to informing both effective detection efforts and broader public health measures to ultimately quell the COVID‐19 pandemic.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Matthew M. Hernandez, Radhika Banu, Paras Shrestha, Shelcie Fabre, Jessica Tan, Alberto E. Paniz‐Mondolfi, Heidi Lopez, Melissa R. Gitman, Michael D. Nowak, and Emilia Mia Sordillo: provided clinical samples for the study. Matthew M. Hernandez, Radhika Banu, Paras Shrestha, Armi Patel, Feng Chen, Liyong Cao, Heidi Lopez, Numthip Chiu, Giuliana Osorio, Biana Shifrin, Inessa Zapolskaya, Vanessa Flores, Pui Yiu Lee, and Alberto E. Paniz‐Mondolfi: accessioned clinical samples. Matthew M. Hernandez, Radhika Banu, Paras Shrestha, Armi Patel, Feng Chen, Liyong Cao, and Alberto E. Paniz‐Mondolfi: performed limit of detection studies. Matthew M. Hernandez, Juan David Ramírez, Jeffrey Jhang, David L. Reich, Carlos Cordon‐Cardo, Emilia Mia Sordillo, and Alberto E. Paniz‐Mondolfi: analyzed, interpreted, or discussed data. Matthew M. Hernandez. and Alberto E. Paniz‐Mondolfi: wrote the manuscript. Matthew M. Hernandez, Radhika Banu, and Alberto E. Paniz‐Mondolfi: conceived the study. Matthew M. Hernandez, Radhika Banu, and Alberto E. Paniz‐Mondolfi: supervised the study. David L. Reich: raised financial support.
Supporting information
Supporting information.
Supporting information.
ACKNOWLEDGMENTS
We thank the members of the MSHS CML for providing any assistance when needed throughout this study. We also would like to thank the patients and healthy donors for providing specimens to complete this study.
Hernandez MM, Banu R, Shrestha P, et al. RT‐PCR/MALDI‐TOF mass spectrometry‐based detection of SARS‐CoV‐2 in saliva specimens. J Med Virol. 2021;93:5481‐5486. 10.1002/jmv.27069
Contributor Information
Matthew M. Hernandez, Email: matthew.hernandez@mssm.edu.
Alberto E. Paniz‐Mondolfi, Email: alberto.paniz-mondolfi@mountsinai.org.
DATA AVAILABILITY STATEMENT
No new data were generated in this study; thus, sharing of data does not apply to this study.
REFERENCES
- 1. Lieberman JA, Pepper G, Naccache SN, Huang M‐L, Jerome KR, Greninger AL. Comparison of commercially available and laboratory‐developed assays for in vitro detection of SARS‐CoV‐2 in clinical laboratories. J Clin Microbiol. 2020;58(8):e00821‐20. 10.1128/JCM.00821-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kaul KL. Laboratories and pandemic preparedness: a framework for collaboration and oversight. J Mol Diagn. 2020;22(7):841‐843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zehnbauer B. Diagnostics in the time of coronavirus disease 2019 (COVID‐19): challenges and opportunities. J Mol Diagn. 2021;23(1):1‐2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. In Vitro Diagnostics EUAs. U.S. Food and Drug Administration. Published Febraury 17, 2021. Accessed February 21, 2021. https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas#individual-molecular
- 5. Butler‐Laporte G, Lawandi A, Schiller I, et al. Comparison of saliva and nasopharyngeal swab nucleic acid amplification testing for detection of SARS‐CoV‐2: a systematic review and meta‐analysis. JAMA Intern Med. Published online. 2021;181:353‐360. 10.1001/jamainternmed.2020.8876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. Performance of saliva, oropharyngeal swabs, and nasal swabs for SARS‐CoV‐2 molecular detection: a systematic review and meta‐analysis. J Clin Microbiol. 2021;59:e02881‐20. 10.1128/JCM.02881-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yee R, Truong TT, Pannaraj PS, et al. Saliva is a promising alternative specimen for the detection of SARS‐CoV‐2 in children and adults. J Clin Microbiol. 2021;59(2):e02686‐20. 10.1128/JCM.02686-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Procop GW, Shrestha NK, Vogel S, et al. A direct comparison of enhanced saliva to nasopharyngeal swab for the detection of SARS‐CoV‐2 in symptomatic patients. J Clin Microbiol. 2020;58(11):e01946‐20. 10.1128/JCM.01946-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wyllie AL, Fournier J, Casanovas‐Massana A, et al. Saliva or Nasopharyngeal swab specimens for detection of SARS‐CoV‐2. N Engl J Med. 2020;383(13):1283‐1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ceron JJ, Lamy E, Martinez‐Subiela S, et al. Use of saliva for diagnosis and monitoring the SARS‐CoV‐2: a general perspective. J Clin Med Res. 2020;9(5):1491. 10.3390/jcm9051491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Goldfarb DM, Tilley P, Al‐Rawahi GN, et al. Self‐collected Saline gargle samples as an alternative to healthcare worker collected nasopharyngeal swabs for COVID‐19 diagnosis in outpatients. J Clin Microbiol. 2021;59(4):e02427‐20. 10.1128/JCM.02427-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhu J, Guo J, Xu Y, Chen X. Viral dynamics of SARS‐CoV‐2 in saliva from infected patients. J Infect. 2020;81(3):e48‐e50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. To KK, Tsang OT, Leung W‐S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS‐CoV‐2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565‐574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Teo AKJ, Choudhury Y, Tan IB, et al. Saliva is more sensitive than nasopharyngeal or nasal swabs for diagnosis of asymptomatic and mild COVID‐19 infection. Sci Rep. 2021;11(1):3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Vandenberg O, Martiny D, Rochas O, van Belkum A, Kozlakidis Z. Considerations for diagnostic COVID‐19 tests. Nat Rev Microbiol. 2021;19(3):171‐183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lamprou DA. Emerging technologies for diagnostics and drug delivery in the fight against COVID‐19 and other pandemics. Expert Rev Med Devices. 2020;17(10):1007‐1012. [DOI] [PubMed] [Google Scholar]
- 17. Younes N, Al‐Sadeq DW, Al‐Jighefee H, et al. Challenges in laboratory diagnosis of the novel coronavirus SARS‐CoV‐2. Viruses. 2020;12(6):582. 10.3390/v12060582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sheridan C. Coronavirus and the race to distribute reliable diagnostics. Nat Biotechnol. 2020;38(4):382‐384. [DOI] [PubMed] [Google Scholar]
- 19. Agena Bioscience, Inc . MassARRAY® SARS‐CoV‐2 Panel Instructions for Use.; 2021. https://agenabio.com/wp-content/uploads/2020/04/GEN0027-02-CoV2-EUA-Product-Sheet-WEB.pdf
- 20. Hernandez MM, Gonzalez‐Reiche AS, Alshammary H, et al. Before the surge: molecular evidence of SARS‐CoV‐2 in New York city prior to the first report. bioRxiv. 2021. 10.1101/2021.02.08.21251303 [DOI] [Google Scholar]
- 21. Mostafa HH, Hardick J, Morehead E, Miller J‐A, Gaydos CA, Manabe YC. Comparison of the analytical sensitivity of seven commonly used commercial SARS‐CoV‐2 automated molecular assays. J Clin Virol. 2020;130:104578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nalla AK, Casto AM, Huang MW, et al. Comparative performance of SARS‐CoV‐2 detection assays using seven different primer‐probe sets and one assay kit. J Clin Microbiol. 2020;58(6):e00557‐20. 10.1128/JCM.00557-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nachtigall FM, Pereira A, Trofymchuk OS, Santos LS. Detection of SARS‐CoV‐2 in nasal swabs using MALDI‐MS. Nat Biotechnol. 2020;38(10):1168‐1173. [DOI] [PubMed] [Google Scholar]
- 24. SoRelle JA, Mahimainathan L, McCormick‐Baw C, et al. Evaluation of symptomatic patient saliva as a sample type for the Abbott ID NOW COVID‐19 assay. bioRxiv. 2020. 10.1101/2020.06.01.20119198 [DOI] [Google Scholar]
- 25. Becker D, Sandoval E, Amin A, et al. Saliva is less sensitive than nasopharyngeal swabs for COVID‐19 detection in the community setting. bioRxiv. 2020. 10.1101/2020.05.11.20092338 [DOI] [Google Scholar]
- 26. Pasomsub E, Watcharananan SP, Boonyawat K, et al. Saliva sample as a non‐invasive specimen for the diagnosis of coronavirus disease 2019: a cross‐sectional study. Clin Microbiol Infect. 2021;27(2):285.e1‐285.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.
Supporting information.
Data Availability Statement
No new data were generated in this study; thus, sharing of data does not apply to this study.


