Abstract
Objectives
To investigate the occurrence of Trichosporon asahii fungemia among critically ill COVID‐19 patients.
Methods
From 1 July to 30 September 2020, cases of T asahii fungemia (TAF) in a Brazilian COVID‐19 referral centre were investigated. The epidemiology and clinical courses were detailed, along with a mycological investigation that included molecular species identification, haplotype diversity analysis and antifungal susceptibility testing.
Results
Five critically ill COVID‐19 patients developed TAF in the period. All five patients had common risk conditions for TAF: central venous catheter at fungemia, previous exposure to broad‐spectrum antibiotics, prior echinocandin therapy and previous prolonged corticosteroid therapy. The average time of intensive care unit hospitalisation previous to the TAF episode was 23 days. All but one patient had voriconazole therapy, and TAF 30‐day mortality was 80%. The five T asahii strains from the COVID‐19 patients belonged to 4 different haplotypes, mitigating the possibility of skin origin and cross‐transmission linking the 5 reported episodes. The antifungal susceptibility testing revealed low minimal inhibitory concentrations for azole derivatives.
Conclusions
Judicious prescription of antibiotics, corticosteroids and antifungals needs to be discussed in critically ill COVID‐19 patients to prevent infections by hard‐to‐treat fungi like T asahii.
Keywords: Brazil, COVID‐19, fungemia, Trichosporon asahii
1. INTRODUCTION
Recent investigations have pointed out that hospitalised patients with COVID‐19 are at risk to develop fungemia. 1 Indeed, classical fungemia risk factors such as prolonged intensive care unit (ICU) stay, central venous catheters (CVC) and exposure to broad‐spectrum antibiotics have been reported in such patients. 1 Trichosporon asahii is a yeast‐like opportunistic pathogen that was initially associated with fungemia in neutropenic patients and more recently has been increasingly reported as a cause of sepsis in critically ill patients exposed to invasive devices, antibiotics, steroids and antifungals. 2 , 3 Unlike Candida spp., T asahii is resistant to the echinocandins, and fungemia episodes are frequently associated with pneumonia and have mortality rates usually above 50%. 3 In this scenario, the occurrence of T asahii fungemia (TAF) in COVID‐19 hospitalised patients is worrisome and may contribute to increasing the mortality in this population. We report the epidemiology, clinical course and microbiological findings of T asahii fungemia episodes in COVID‐19 patients hospitalised in an ICU from Salvador, Brazil.
2. MATERIAL AND METHODS
2.1. Epidemiological and clinical investigation
The number of TAF and candidemia episodes per/1.000 ICU admissions was collected, as well as information regarding demographics underlying conditions, body mass index (BMI), previous antimicrobial exposure, previous antifungal exposure, antifungal breakthrough infection as defined by Cornely et al, 4 use of mechanical ventilation, use of renal replacement therapy, laboratory tests on admission and at fungemia, presence of invasive devices, previous bacterial or fungal infections and Pitt bacteremia score at fungemia for each TAF episode. 5 , 6 Pitt bacteremia score was initially described for as a severity of acute illness in patients with bacteremia and also been applied as a prognostic evaluation in candidemia 6 and, recently, fungemia by Trichosporon spp. 7 Clinical management and 30‐day outcome were also collected. The concomitance of COVID‐19 pneumonia with TAF precluded any further analysis of lung involvement by the fungal pathogen. This study was approved by the Institutional Review Board, protocol number: 44674415.0.1001.5505.
2.2. COVID‐19 and Trichosporon fungemia diagnosis
Patients had SARS‐CoV‐2 detected in respiratory samples by RT‐PCR after 24–48 h of admission (AllplexTM 2019‐nCOV Assay).
Bactec (Becton and Dickinson) blood cultures and MALDI‐TOF mass spectrometry (Vitek MS, bioMérieux) were used for the aetiologic diagnosis of sepsis. Molecular identification of Trichosporon spp. was confirmed by sequencing the IGS1 region from the rDNA. 8
2.3. Mycological investigation
To investigate the possibility of T asahii horizontal transmission among COVID‐19 critically ill patients, we conducted a molecular investigation by multilocus DNA sequence analysis including the five T asahii epidemiologic‐related clinical isolates and nine controls (2 reference strains and 7 unrelated isolates from the same hospital). Housekeeping genes from T asahii genomes (CBS2479 and CBS8904) available in GenBank (https://www.ncbi.nlm.nih.gov/) were aligned, and in silico analyses were carried out to design and evaluate the new primers. After PCR reactions and DNA sequence analysis, four loci with higher haplotypic diversity were selected for the multilocus phylogenetic analysis along with the IGS1 region from the rDNA: topoisomerase 1 (A1Q1_08199), phosphate carrier protein (A1Q1_08315), beta‐1‐tubulin (A1Q1_04313) and copper‐exporting ATPase (A1Q1_05365). Primers and PCR conditions are provided in Table 1. DNA sequences from the five investigated loci were concatenated, and haplotype distribution was constructed by phylogenetic analysis using software MEGA X. 9 The five COVID‐19‐associated T asahii isolates had antifungal susceptibility testing (AST) of fluconazole (FLC), voriconazole (VRC), posaconazole (POS) and amphotericin B (AMB, Sigma) performed by using the CLSI broth microdilution method. 10 The minimal inhibitory concentration (MIC) values were determined after 48 h of incubation. 11
TABLE 1.
Primers developed for the multilocus analysis
| Locus | Primer name | Primer sequence | Melting temp. | Product size (bp) |
|---|---|---|---|---|
| Β‐1‐tubulin | BTUB‐F | 5'‐GCCCCGACAACTTTGTCTTT‐3' | 55℃ | 636 |
| BTUB‐R | 5'‐TCTTGGCGTCGAACATTTGC‐3' | 56℃ | ||
| Copper‐exporting ATPase | ATP‐F | 5'‐CTTCCATCGCAATGCTGGTT‐3' | 55℃ | 551 |
| ATP‐R | 5'‐TGATGTCATCGCCTTCGAGT‐3' | 55℃ | ||
| Phosphate carrier protein | PHCP‐F | 5'‐CAGCAATCATGTCCGACAGA‐3' | 54℃ | 664 |
| PHCP‐R | 5'‐CGAACTTGGCCATGGTGTA‐3' | 54℃ | ||
| Topoisomerase‐1 | TOP1‐F | 5'‐CGCACTTCTCAAGGCTGGTAAT‐3' | 57℃ | 380 |
| TOP1‐R | 5'‐GGACGTCAAGCCGAATGTCA‐3' | 57℃ |
3. RESULTS
From 1 July to 30 September 2020, a total of 183 patients were admitted to the COVID‐19 ICU from the São Rafael Hospital, Salvador, Bahia State, Brazil. In that lapse of time, five patients had TAF (27.3/1.000 ICU admissions) and 18 had candidemia (98.3/1.000 ICU admissions). The TAF patients had a mean age of 71 years (range 57–75), and three (60%) of them were severely obese (body mass index ≥35). Two (40%) had diabetes mellitus and were under insulin therapy. All five patients had common risk conditions for TAF: CVC at fungemia, previous exposure to broad‐spectrum antibiotics, prior echinocandin therapy (anidulafungin, average 8 days) and previous corticosteroid therapy (average 22 days). Three among the five TAF episodes were anidulafungin breakthrough infections. Corticosteroid therapy consisted of either methylprednisolone 1 mg/kg/day or prednisone 40 mg q12h. All five patients received piperacillin‐tazobactam before the TAF episode. The average time of ICU hospitalisation previous to the TAF episode was 23 days (range 11–31 days). All patients were under mechanical ventilation (11–30 days before TF), and four of them need renal replacement therapy (9–16 days before TF). Candidemia was diagnosed in three patients before the TAF, two by Candida parapsilosis and one by Candida tropicalis. One of the patients had concomitant TAF and C parapsilosis candidemia. All Candida isolates were susceptible to AMB, FLC, VRC and echinocandins (data not shown). Four patients developed diarrhoea during hospitalisation, and all four had Clostridioides difficile A/B toxin and glutamate dehydrogenase antigen tests negative in faeces samples (C DIFF QUICK CHECK COMPLETE, Abbott). Three of them had diarrhoea that persisted until fungemia was diagnosed. One patient had Enterococcus faecalis bacteremia before the TAF diagnosis, and another one, without diarrhoea, had concomitant bloodstream infection by T asahii and E faecalis. Concordant T asahii blood and catheter tip cultures were diagnosed in one patient. Trichosporon deep‐seated infections were investigated, including eye fundoscopy, heart echocardiogram and abdominal ultrasound, but all cases were considered isolated fungemia. None of the patients had neutropenia (<500 cells/mm3) before or at the TAF episode. One patient had a low blood lymphocyte count when TAF was diagnosed (300 cells/mm3). The average Pitt bacteremia score at fungemia diagnosis was 11 (range 7–14). All but one patient had VRC therapy, either in monotherapy (n = 1) or combined with liposomal AMB (n = 3). One patient died before receiving antifungal therapy for the TAF episode. The overall 30‐day mortality was 80%. The clinical course of the COVID‐19 patients that developed TAF is summarised in Figure 1 and Table 2.
FIGURE 1.
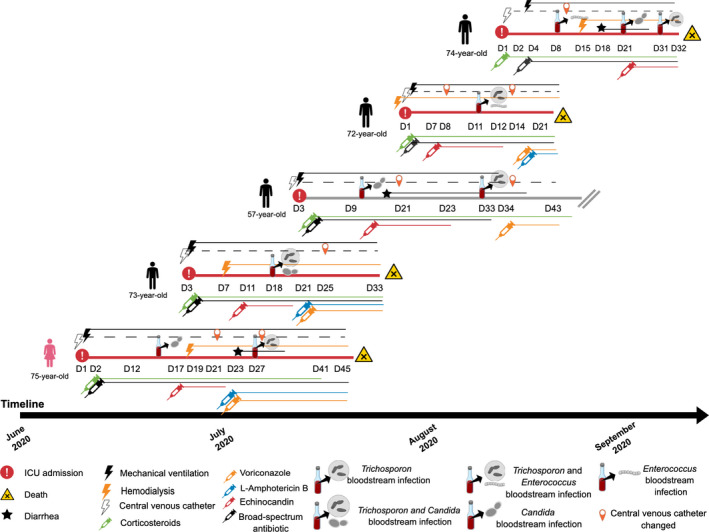
Clinical course of the five patients with severe COVID‐19 that develop Trichosporon fungemia, with red horizontal lines representing the patients that deceased and grey horizontal line representing the patient that survived
TABLE 2.
Clinical and Epidemiological data of the five patients with severe COVID‐19 that developed Trichosporon asahii fungemia
| Condition | Patient Data | ||||
|---|---|---|---|---|---|
| Age | 57 | 74 | 75 | 73 | 72 |
| Sex | Male | Male | Female | Male | Male |
| Body Mass Index ≥35 | No | No | Yes | Yes | Yes |
| Diabetes mellitus | Yes | No | No | Yes | No |
| ICU length of stay before fungemia (days) a | 30 | 31 | 27 | 15 | 11 |
| Mechanical ventilation duration before fungemia (days) | 30 | 27 | 27 | 15 | 11 |
| Renal replacement therapy duration before fungemia (days) | 0 | 16 | 9 | 11 | 11 |
| Broad‐spectrum antibiotic therapy duration with anti‐anaerobe activity before Trichosporon fungemia (days) | 30 | 30 | 26 | 15 | 10 |
| Diarrhoea duration before Trichosporon fungemia (days) | 16 | 12 | 4 | 0 | 0 |
| Corticosteroid therapy duration before fungemia (days) | 30 | 30 | 26 | 15 | 10 |
| Corticosteroid exposure during fungemia | Yes | Yes | Yes | Yes | Yes |
| Enteroccus spp. bacteremia before or at Trichosporon fungemia | No | Yes | No | No | Yes |
| Candidemia before Trichosporon fungemia | Yes | Yes | Yes | No | No |
| Echinocandin exposure before Trichosporon fungemia (days) | 13 | 9 | 6 | 9 | 5 |
| Echinocandin breakthrough infection | No | Yes | No | Yes | Yes |
| CVC at fungemia b | Yes | Yes | Yes | Yes | Yes |
| Pitt bacteremia score at fungemia | 7 | 14 | 10 | 10 | 14 |
| Blood neutrophils count at fungemia (cells/mm3) | 7200 | 2750 | 18,200 | 1260 | 970 |
| Blood Lymphocyte count at fungemia (cells/mm3) | 1440 | 300 | 1500 | 950 | 550 |
| Voriconazole therapy | Yes | No | Yes | Yes | Yes |
| CVC removal after Trichosporon fungemia | Yes | No | Yes | Yes | Yes |
| 7‐day outcome | Alive | Dead | Alive | Alive | Alive |
| 14‐day outcome | Alive | Dead | Alive | Dead | Dead |
| 30‐day outcome | Alive | Dead | Dead | Dead | Dead |
ICU, Intensive care unit.
CVC, central venous catheter.
The five T asahii strains from the COVID‐19 patients belonged to 4 different haplotypes, mitigating the possibility of skin origin and cross‐transmission linking the 5 reported episodes (Figure 2). GenBank accession numbers are available as appendix data (Table S1). The antifungal susceptibility testing revealed low MICs for azole derivatives, that is 1‐2 mcg/mL for FLC, 0.125 mcg/mL for POS and 0.03 mcg/mL for VRC, and high MICs for AMB (1‐16 mcg/mL).
FIGURE 2.
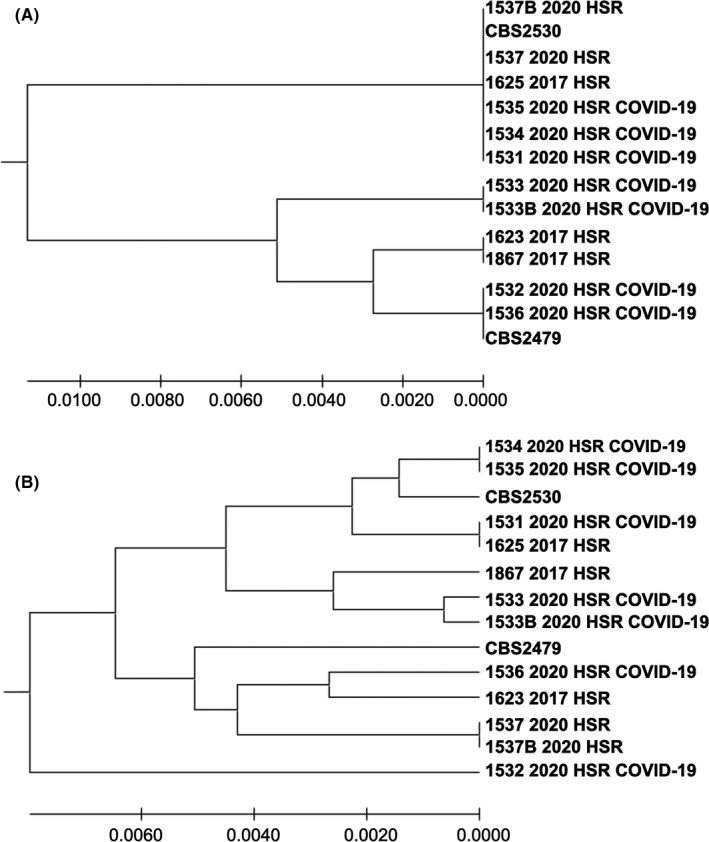
(A) UPGMA phylogenetic tree showing the evolutionary relationship among 14 IGS1 nucleotide sequences, from two reference strains (CBS2479 and CBS2530) and 12 clinical strains from 10 patients from the same hospital. The strains 1533 2020 HSR and 1533B 2020 HSR were recovered from the same patient, as well as the strains 1537 2020 HSR and 1537B 2020 HSR. There were a total of 562 nucleotides positions in the final data set. The final tree with the sum of branch length = 0.0304 is shown. Three strains from different COVID‐19 patients had identical sequences (1531 2020 HSR, 1534 2020 HSR, 1535 2020 HSR); (B) UPGMA phylogenetic tree showing the evolutionary relationship among 14 concatenated (five loci) nucleotide sequences, from two reference strains (CBS2479 and CBS2530) and 12 clinical strains from 10 patients from the same hospital. The strains 1533 2020 HSR and 1533B 2020 HSR were recovered from the same patient, as well as the strains 1537 2020 HSR and 1537B 2020 HSR. There were a total of 2467 nucleotides positions in the final data set. The final tree with the sum of branch length = 0.456 is shown. The strains recovered from the patients hospitalised at the COVID‐19 ICU showed that only two patients had phylogenetic closely related strains (1534 2020 HSR and 1535 2020 HSR strains belong to the same haplotype)
4. DISCUSSION
T asahii may be part of the human skin and digestive tract microbiome, and the role of gut and skin colonisation as the portal of entrance for TAF is controversial. 12 In the present cohort, all patients were under prolonged antimicrobial selective pressure, which certainly contributed to the overgrowth of this species in the skin and the digestive tract of the SARS‐CoV‐2‐infected patients. All patients had a CVC in place, which may have been the source of the candidemia and TAF episodes. However, despite catheter removal allied to voriconazole seen in four patients, three died in the first 14 days after the fungemia diagnosis. The prolonged corticosteroid‐induced immunosuppression, the intense dysbiosis caused by long periods of antimicrobial therapy associated with the impairment of the intestinal barrier and the multi‐organic damage characteristic of severe COVID‐19 may have contributed to the poor outcomes. Corticosteroid therapy has shown to be beneficial for the treatment of severe COVID‐19. However, prolonged corticosteroid treatment may be a double‐edged sword and detrimental for these patients, facilitating thrombotic events, 13 and also superinfections by opportunistic pathogens, including Candida and Trichosporon. 14 Translocation from the digestive tract has been pointed out as a relevant source for bacteremia and fungemia in patients with severe COVID‐19. 1 Of note, one interesting report from Greece described two patients with COVID‐19 that developed Saccharomyces cerevisiae fungemia after 4–6 days of probiotic treatment for diarrhoea. 15 Recently, an experimental model of SARS‐CoV‐2 infection with Syrian hamsters demonstrated enterocytes infection and apoptosis by the virus. 16 Thus, one has to keep in mind that direct enterocytes lesions by the SARS‐CoV‐2 may facilitate bacterial and fungal translocation from the digestive tract of some of these severely infected patients.
Despite the previous reports associating T asahii with horizontal transmission, 17 this event unlikely happened in this cohort. Some key elements against the hypothesis of horizontal transmission can be delineated as follows: patients had facilitating conditions for gut translocation and were exposed to echinocandins, which might have led to Trichosporon selection in the digestive tract; the molecular analysis showed a high haplotype diversity among the clinical isolates. The haplotype diversity translates genetic differences among the isolates, meaning that clonal dissemination which is suggestive of horizontal transmission did not occur in this cohort. These important observations raise the concern that critically ill COVID‐19 patients may have a higher risk to develop Trichosporon fungemia and not only candidemia. Indeed, the incidence of Trichosporon fungemia in this study was at least six times higher than we previously reported in Brazilian centres. 18 For every 3.6 episodes of candidemia, one episode of TAF occurred in this cohort.
5. CONCLUSIONS
We added evidence to the emerging issue regarding superinfections in severe COVID‐19 patients. Judicious prescription of broad‐spectrum antibiotics, corticosteroids and antifungals needs to be discussed in critically ill COVID‐19 patients to prevent infections not only by multidrug‐resistant bacteria but also by hard‐to‐treat fungi like T asahii.
CONFLICTS OF INTEREST
ALC has received honoraria from Pfizer, United Medical, Eurofarma, MSD, TEVA. All other authors have no conflicts to declare.
AUTHOR CONTRIBUTION
Joao Nobrega de Almeida Júnior: Conceptualization (equal); Methodology (lead); Visualization (lead); Writing‐original draft (equal); Writing‐review & editing (equal). Lis Moreno: Resources (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Elaine Cristina Francisco: Methodology (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Gabriela Noronha Marques: Resources (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Ana Verena Mendes: Resources (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Maria Goreth M. de Andrade Barberino: Resources (equal); Writing‐original draft (equal); Writing‐review & editing (equal). Arnaldo Lopes Colombo: Conceptualization (equal); Funding acquisition (equal); Methodology (supporting); Writing‐original draft (equal); Writing‐review & editing (equal).
Supporting information
Table S1
ACKNOWLEDGEMENTS
The study was partially supported by a grant received from Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 484020‐2013‐7 and CNPq 307510/2015‐8). JNAJ has received a research grant from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP 2018/19347‐4).
Nobrega de Almeida J Jr, Moreno L, Francisco EC, et al. Trichosporon asahii superinfections in critically ill COVID‐19 patients overexposed to antimicrobials and corticosteroids. Mycoses. 2021;64:817–822. 10.1111/myc.13333
REFERENCES
- 1. Arastehfar A, Carvalho A, Nguyen MH, et al. COVID‐19‐associated candidiasis (CAC): an underestimated complication in the absence of immunological predispositions? J Fungi (Basel). 2020;6(4):211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chagas‐Neto TC, Chaves GM, Melo AS, Colombo AL. Bloodstream infections due to Trichosporon spp.: species distribution, Trichosporon asahii genotypes determined on the basis of ribosomal DNA intergenic spacer 1 sequencing, and antifungal susceptibility testing. J Clin Microbiol. 2009;47(4):1074‐1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. de Almeida Júnior JN , Hennequin C. Invasive Trichosporon infection: a systematic review on a re‐emerging fungal pathogen. Front Microbiol. 2016;7:1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cornely OA, Hoenigl M, Lass‐Flörl C, et al. Defining breakthrough invasive fungal infection‐position paper of the mycoses study group education and research consortium and the European confederation of medical mycology. Mycoses. 2019;62(9):716‐729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Al‐Hasan MN, Lahr BD, Eckel‐Passow JE, Baddour LM. Predictive scoring model of mortality in Gram‐negative bloodstream infection. Clin Microbiol Infect. 2013;19(10):948‐954. [DOI] [PubMed] [Google Scholar]
- 6. Vaquero‐Herrero MP, Ragozzino S, Castaño‐Romero F, et al. The pitt bacteremia score, Charlson comorbidity index and chronic disease score are useful tools for the prediction of mortality in patients with candida bloodstream infection. Mycoses. 2017;60(10):676‐685. [DOI] [PubMed] [Google Scholar]
- 7. Nobrega de Almeida J, Francisco EC, Holguín Ruiz A, et al. Epidemiology, clinical aspects, outcomes and prognostic factors associated with Trichosporon fungaemia: results of an international multicentre study carried out at 23 medical centres. J Antimicrob Chemother. 2021;dkab085. [DOI] [PubMed] [Google Scholar]
- 8. Sugita T, Nakajima M, Ikeda R, Matsushima T, Shinoda T. Sequence analysis of the ribosomal DNA intergenic spacer 1 regions of trichosporon species. J Clin Microbiol. 2002;40(5):1826‐1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547‐1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. CLSI . Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. 4th ed. CLSI standard M27. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 11. Francisco EC, de Almeida Junior JN , de Queiroz TF , et al. Species distribution and antifungal susceptibility of 358 Trichosporon clinical isolates collected in 24 medical centres. Clin Microbiol Infect. 2019;25(7):909.e1‐909.e5. [DOI] [PubMed] [Google Scholar]
- 12. Colombo AL, Padovan ACB, Chaves GM. Current knowledge of Trichosporon spp. and Trichosporonosis. Clin Microbiol Rev. 2011;24(4):682‐700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mishra GP, Mulani J. Corticosteroids for COVID‐19: the search for an optimum duration of therapy. Lancet Respir Med. 2021;9(1):e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rodriguez JY, Le Pape P, Lopez O, Esquea K, Labiosa AL, Alvarez‐Moreno C. Candida auris: a latent threat to critically ill patients with COVID‐19. Clin Infect Dis. 2020;ciaa1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ventoulis I, Sarmourli T, Amoiridou P, et al. Bloodstream infection by saccharomyces cerevisiae in two COVID‐19 patients after receiving supplementation of saccharomyces in the ICU. J Fungi (Basel). 2020;6(3):98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chan JF‐W, Zhang AJ, Yuan S, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID‐19) in a golden Syrian hamster model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71(9):2428‐2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fanfair RN, Heslop O, Etienne K, et al. Trichosporon asahii among intensive care unit patients at a medical center in Jamaica. Infect Control Hosp Epidemiol. 2013;34(6):638‐641. [DOI] [PubMed] [Google Scholar]
- 18. Francisco EC, de Almeida Junior JN , Queiroz‐Telles F, et al. Correlation of Trichosporon asahii genotypes with anatomical sites and antifungal susceptibility profiles: data analyses from 284 isolates collected in the last 22 years across 24 medical centers. Antimicrob Agents Chemother. 2020;65(3):e01104‐e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1


