Abstract
The immune response plays a critical role in the pathophysiology of SARS-CoV-2 infection ranging from protection to tissue damage and all occur in the development of acute respiratory distress syndrome (ARDS). ARDS patients display elevated levels of inflammatory cytokines and innate immune cells, and T and B cell lymphocytes have been implicated in this dysregulated immune response. Mast cells are abundant resident cells of the respiratory tract and are able to release different inflammatory mediators rapidly following stimulation. Recently, mast cells have been associated with tissue damage during viral infections, but their role in SARS-CoV-2 infection remains unclear. In this study, we examined the profile of mast cell activation markers in the serum of COVID-19 patients. We noticed that SARS-CoV-2-infected patients showed increased carboxypeptidase A3 (CPA3) and decreased serotonin levels in their serum when compared with symptomatic SARS-CoV-2-negative patients. CPA3 levels correlated with C-reactive protein, the number of circulating neutrophils, and quick SOFA. CPA3 in serum was a good biomarker for identifying severe COVID-19 patients, whereas serotonin was a good predictor of SARS-CoV-2 infection. In summary, our results show that serum CPA3 and serotonin levels are relevant biomarkers during SARS-CoV-2 infection. This suggests that mast cells and basophils are relevant players in the inflammatory response in COVID-19 and may represent targets for therapeutic intervention.
Keywords: carboxypeptidase A3, COVID-19, Mast cell, SARS-CoV-2, serotonin
Graphical Abstract
Serum levels of CPA3 and serotonin are affected during SARS-CoV-2 infection and can be considered as biomarkers during COVID-19.
Graphical Abstract.
INTRODUCTION
Coronavirus disease 2019 (COVID-19) is caused by the SARS-CoV-2 virus, which was identified for the first time in Wuhan, China in late 2019. Since then, the virus has spread across the globe infecting more than 100 million people and causing the death of more than 2.1 million infected individuals.1
SARS-CoV-2 infects cells of the mucosa that express the angiotensin-converting enzyme II (ACE2), which is particularly abundant in type II alveolar cells of the lung and in other tissues.2 The COVID-19 infection has an incubation period of 3–7 days, generating moderate symptoms, such as head and muscle aches, sore throat, nasal congestion, dry cough, fatigue, and fever. However, 5–10% of infected patients develop acute respiratory distress syndrome (ARDS), a serious complication that causes respiratory failure leading to high mortality.3
A relevant factor for the development of ARDS in patients with COVID-19 is the exacerbated immune response triggered by the infection, reflected as a cytokine storm in which different cytokines such as: IL-1β, IL-2, IL-4, IL-6, IL-10, IFN-γ, and TNF-α are elevated.4 Besides, COVID-19 patients are known to show profound alterations in cell populations associated with the immune response against viruses, such as monocytes, macrophages, neutrophils, NK cells, B lymphocytes, CD8+ T lymphocytes, and memory and regulatory CD4+ lymphocytes.5 However, it is unclear how SARS-CoV-2 affects mast cells (MC).
MC are tissue resident leukocytes derived from hematopoietic precursors, distributed throughout the body and abundantly found along the respiratory tract.6 These cells are characterized by having many cytoplasmic granules that contain different chemical mediators released after activation. Among the molecules abundant in MC granules are tryptase, carboxypeptidase, chymase, serotonin, histamine and TNF-α. MC also produce other types of inflammatory mediators including prostaglandins, leukotrienes and reactive nitrogen species. In addition, activated MCs secrete different de novo synthesized cytokines and chemokines.7
MC are well known for their role in mediating allergic reactions, but recent evidence indicates an important role in the innate immune response to different pathogens including viruses, bacteria, protozoa, fungi and nematodes.7 The ability of MC to participate in viral infections is mediated by different Pattern Recognition Receptors, such as TLR-3, −7, RIG-I, MDA-5, etc., which are essential in the innate antiviral response.8 Recently, it has been shown that MCs are associated with tissue damage induced by an excessive inflammatory response during viral infections.9 For instance, in influenza virus infections, the observed lung tissue damage is due to an excessive inflammatory response characterized by the overproduction of cytokines and chemokines or “cytokine storm.”10 In mouse models of infection with the H5N1 influenza virus, treatment of mice with ketotifen, an inhibitor of MC activation, reduces damage to lung tissue. Furthermore, the combination of ketotifen with oseltamivir, an antiviral drug, significantly increased mice survival.11 On the other hand, it has been observed that mice deficient in MC show less lung damage when infected with influenza A virus, compared with wild-type mice. Interestingly, this effect was associated with a decreased production of TNF-α, CCL2, CCL3, CCL4, CXCL2, and CXCL10, suggesting a crucial role of MC in the “cytokine storm” triggered by influenza infection.12 Considering that MC activation plays a crucial role during the damage induced by viral infections and that a cytokine storm is a crucial feature during SARS-CoV-2 infection, this study was conducted to evaluate MC activation markers in the serum of patients with COVID-19 and to determine if they are associated with disease severity.
MATERIAL AND METHODS
Patients and control
Patients with COVID-19 and a control group were enrolled at Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, between March and April 2020. All participants in this study had COVID-19 suggestive symptoms, but only those showing a positive PCR test for SARS-CoV-2 were considered infected, whereas PCR test-negative patients with respiratory symptoms were selected as control. Based on clinical records, samples from asthmatic, HIV, cancer, autoimmune, or pregnant patients were not included in this study. Serum samples were taken on the first day of admission. Demographic and clinical parameters were collected, and laboratory indicators were tested with conventional methods in COVID-19 patients (Supplemental Table 1). Disease severity was classified as mild/moderate when patients showed fever, signs of airway disease, with or without a tomographic image indicating pneumonia. Severe COVID-19 patients showed either respiratory failure, an increased respiratory rate (>30 bpm), decreased oxygen saturation at rest (<92%), or decreased PaO2/FiO2 (<300 mmHg). This study was approved by the Research and Ethics Committee of the Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán (Ref. 3341). All protocols were in accordance with the Declaration of Helsinki. All participants of this study provided informed written consent to participate in this study.
Assessment of mast cell activation markers
Patient serum was analyzed by the ELISA, using commercial kits for histamine (Cat. RE59221; IBL International, Hamburg, Germany), human carboxypeptidase A3 (Cat. OKCD01671; Aviva Systems Biology, San Diego, CA, USA), serotonin (Cat. ab133053; Abcam, Cambridge, UK), and heparin (Cat. abx 258893; Abbexa, Houston, TX, USA), according to the manufacturer's instructions.
Nitric oxide was evaluated using a colorimetric method that measures the levels of the breakdown product NO2− (Griess Reagent System, Cat. G2930; Promega, Madison, WI, USA).
IL-6 quantification
IL-6 was evaluated with the Milliplex Map Human Cytokine/Chemokine Bead Panel-Premixed 29 Plex (Cat. HCYTMAG-60K-PX29; Millipore, USA) following manufacturer's instructions.
Images and statistical analysis
To identify potential organ or tissue sources of CPA3, protein and mRNA expression were analyzed in Human Protein Atlas (www.proteinatlas.org/ENSG00000163751-CPA3/tissue) and mRNA expression was obtained from HPA RNA-sEquation (www.ncbi.nlm.nih.gov/gene/1359#gene-expression).
All statistical analyses were performed with SigmaPlot software version 14.0 (Systat Software, San Jose, CA, USA). Data normality was assessed by Kolmogorov–Smirnov with Lilliefors correction. Data are shown as mean or median ± range, as appropriate. For comparisons between 2 groups, Student's t-test or Mann–Whitney rank sum test with Yates correction were used. For comparisons of 3 groups, one-way ANOVA with Student–Newman–Keuls post-hoc or Kruskal–Wallis test followed by a Dunn's post-hoc test were used. Results were considered significant at a P value < 0.05. For correlations of 2 variables, Spearman Rank Order Correlation was used. The r values for each of the correlations were plotted in a bubble chart generated with Microsoft Excel 2019 (https://office.microsoft.com/excel). Receiver operating characteristic (ROC) curves were generated to find the accuracy of biomarkers to distinguish infected individuals and the severity of COVID19.
RESULTS AND DISCUSSION
Several MC-derived biomarkers are used to diagnose and predict outcomes in allergic and infectious diseases. Among them, histamine, heparin, carboxypeptidase A3 (CPA3), serotonin, and nitric oxide have been studied.13,14 To address whether these markers are affected during COVID-19, serum samples were collected from patients admitted at a tertiary care center in Mexico City. Demographic and clinical characteristics of patients are shown in Supplemental Table 1. We noticed that COVID-19 patients had increased serum levels of CPA3 in serum when compared with the control group (Fig. 1A). Interestingly, serotonin showed decreased levels in sera from SARS-CoV-2-infected patients compared with those not infected (Fig. 1B). Serum levels of histamine, heparin, and nitric oxide were not affected in COVID-19 patients (Supplemental Fig. 1).
FIGURE 1.
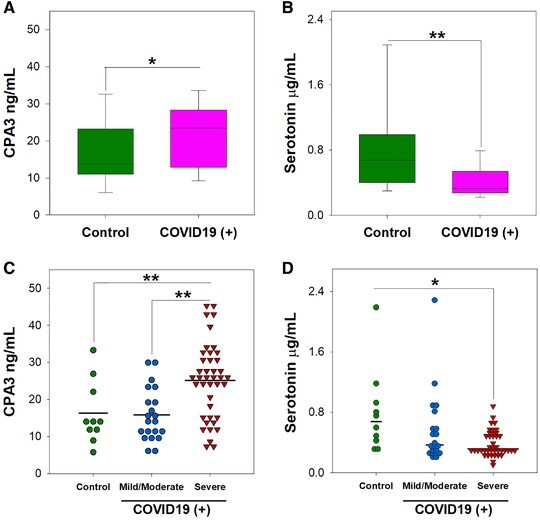
Serotonin and carboxypeptidase A3 are altered in COVID-19 patients.
The serum concentration of (A and C) carboxypeptidase A3 (CPA3), and (B and D) serotonin were measured upon patient admission by ELISA. Data from 21 patients with mild/moderate disease, 41 patients with severe COVID-19, and 10 control individuals is shown. Data are presented as mean or median ± range, as appropriate. *P < 0.05; **P < 0.01. Student's t test (A) Mann–Whitney test (B), one-way ANOVA test (C), and Kruskal–Wallis test (D)
Previous reports indicate that different immune markers are differentially expressed during severe COVID-19.15 Therefore, we investigated whether MC-derived biomarkers were altered depending on COVID-19 severity. We observed that patients with severe disease showed increased levels of CPA3 when compared with those with mild/moderate disease, or individuals in the control group (Fig. 1C). Serotonin levels were decreased in severe COVID-19 (Fig. 1D). No other MC-associated biomarker evaluated showed significant differences in patients with severe COVID-19, compared with patients with mild/moderate symptoms or controls (Supplemental Fig. 2).
Because CPA3 and serotonin levels were altered in severe COVID-19 patients, we next assessed their correlation with clinical and laboratory parameters. A matrix with Spearman's r coefficient values and representing correlation showed that CPA3 had a stronger correlation with an increased number of clinical and laboratory parameters in comparison with serotonin (Fig. 2A). Furthermore, CPA3 showed a significant positive correlation with inflammation-associated markers indicated by the level of circulating neutrophils (r = 0.291, P = 0.0447) (Fig. 2B) and C-reactive protein (r = 0.390, P = 0.00703) (Fig. 2D). Remarkably, CPA3 also associated with the disease severity score quick Sepsis-related Organ Failure Assesment (qSOFA), (r = 0.335, P = 0.00862) (Fig. 2C).
FIGURE 2.
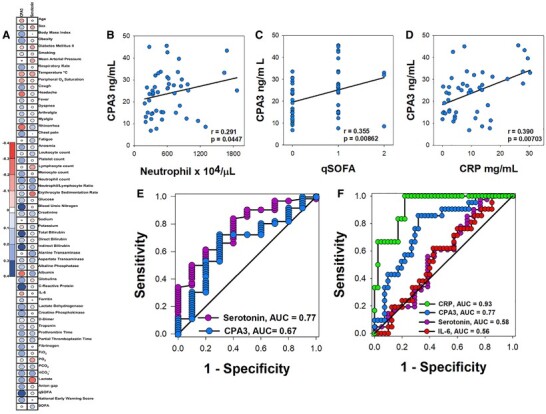
Serum levels of carboxypeptidase A3 correlates with clinical parameters of disease severity in COVID-19 patients.
(A) Correlation matrix representing correlation carboxypeptidase A3 or serotonin serum levels with clinical and laboratory parameters used to determine COVID-19 severity. Spearman's coefficient value r is used as correlation descriptor, and the size of each circle symbolizes correlation strength (color scale of red and blue indicates negative or positive correlation, respectively). Correlation between serum concentration of carboxypeptidase A3 and (B) blood neutrophils, (C) qSOFA, (D) C-reactive protein. Value of Spearman's correlation (r) and significant P values (P < 0.05) are shown. (E) Receiver-operator characteristics (ROC) curves of carboxypeptidase A3 (CPA3) and serotonin serum levels for the prediction of SARS-CoV-2 infection. (F) ROC curve of C-reactive protein (CRP), carboxypeptidase A3 (CPA3), serotonin, and IL-6 in the prediction of COVID-19 severity
Finally, to evaluate the potential clinical utility of serotonin and CPA3, ROC curves were performed to distinguish SARS-CoV-2 infection. Serotonin showed an acceptable AUC (area under the curve) values that allow to distinguish between SARS-CoV-2 infected from noninfected individuals (AUC 0.77) (Fig. 2E). When ROC curves were analyzed to differentiate between mild/moderate and severe COVID-19 patients, CPA3 (AUC 0.77) was more reliable than serotonin (AUC 0.58) or IL-6 (AUC 0.56) to detect patients with severe disease. This was close to the predictive values seen for C-reactive protein (AUC 0.93) (Fig. 2F). As a whole, these results suggest a relationship between MC activation, as reflected by CPA3 levels in serum, and severe COVID-19.
To the best of our knowledge, this is the first evidence of alteration in serum CPA3 levels during COVID-19. The source of this CPA3 in serum is unknown; however, CPA3 is abundantly expressed in different organs, including human lungs,16 with MC being the main cell source, and releases this enzyme after degranulation17 (Supplemental Fig. 3). Remarkably, MC granules contain other abundant proteases, such as chymase and tryptase, which are relevant in clinical pathologies such as hemorrhagic dengue18 and mastocytosis.19 Further work is needed to ascertain whether other MC proteases are modified during COVID-19. Interestingly, a second potential cellular source of CPA3 are basophils, which are found in blood and migrate to inflamed tissues.20 CPA3 is an enzyme that cleaves C-terminal amino acid residues from proteins and is an abundant protein in MC granules (16 μg CPA3 per 106 MC). This enzyme is released after cell degranulation and is associated with allergic pathologies of the respiratory tract.21 Identified substrates for CPA3 include neurotensin, kinetensin, neuromedin N, angiotensin I, and endothelin-1, which is associated with pulmonary fibrosis,22 a sequela observed in COVID-19 patients.23 Interestingly, we noticed that CPA3 correlated with clinical parameters associated with systemic inflammation during COVID-19. Remarkably, CPA3 correlated with circulating neutrophils and CPR, which are associated with an exacerbated inflammatory response during COVID-1915; however, CPA3 potential as a disease severity biomarker needs to be further analyzed in larger prospective cohorts. Previous studies have noticed the importance of MC activation for the recruitment of neutrophils to sites of infection.24 Furthermore, this increase in tissue neutrophil is proposed to be one mechanism of tissue damage and organ failure during COVID-19.25 Our results are in agreement with a recent histologic study, where an increased number of MC in the lungs of COVID-19 patients was observed.26 These results suggest an important role of MC and basophils in SARS-CoV-2 infection, but further work is needed to understand the mechanisms involved.
The second marker that was modified during SARS-CoV-2 infection was serotonin. Traditionally serotonin is considered as an important neurotransmitter regulating several neuronal activities in the central nervous system. However, recent evidence indicates that systemic serotonin distributed by the blood plays a more complex function in the organism. Blood serotonin is produced by different cell lineages, including MC, but is mainly produced by enterochromaffin cells of the intestine. Once in the blood, serotonin levels are primarily regulated by platelets that capture and store in dense granules that are secreted after cell activation.27 Previous studies have shown that infections can decrease, increase, or maintain serotonin levels in blood.28–30 For instance, in dengue infection, decreased levels of serum serotonin were associated with disease severity. Moreover, decreased serotonin levels correlate with thrombocytopenia, a clinical feature of severe dengue, associating decreased serum serotonin with decreased numbers of platelets.28,30 However, we did not observe a difference in platelet count in COVID-19 patients (Supplemental Table 1), implying other mechanisms could be involved in this decrease. A recent report noticed that platelet numbers are not affected in COVID-19, but their gene expression profile is different when compared with those from healthy individuals.31 Because platelets usually introduce serotonin from blood through a transporter (SERT or SLC6A4),32 an overexpression of this protein could explain the decrease of serotonin in blood. However, Manne et al.31 showed that SERT is not modified in platelets from COVID-19 patients. Another explanation could be related to an alteration in enterochromaffin cells functions by SARS-CoV-2, a phenomenon that is observed in other viral infections,33 or to increased serotonin degradation by monoamine oxidase,27 which is overexpressed by platelets in COVID-19 patients.31 How this serotonin decrease can affect the pathophysiology of SARS-CoV-2 infection is unknown; however, among the diverse functions of blood serotonin, one involves modulation of the immune response. Several cells of the immune system express serotonin receptors, including those that participate in innate and adaptive immune response.27 Activation of serotonin receptor 5-H2TA diminishes inflammation induced by TNF-α.34 Furthermore, serotonin decreases IL-6 and TNF-α production by macrophages and lymphocytes,35 suggesting the importance of serotonin in regulating exacerbated inflammation. Interestingly, recent reports have shown increased levels of serotonin in the serum of COVID-19 patients in relation to healthy subjects.36,37 This discrepancy with our results can be explained by the experimental design, where we compared serotonin levels from SARS-CoV-2-infected patients with noninfected patients, but with symptoms of other uncharacterized respiratory disease. Furthermore, in our study, serum serotonin levels in samples from COVID-19 patients are increased (median mild/moderate 366.36 ng/ml, severe 313.12 ng/ml) when compared with serotonin reference level in serum (283 ng/ml).38
In conclusion, our results demonstrate that serum levels of CPA3 and serotonin are affected during SARS-CoV-2 infection and can be considered as potential biomarkers during COVID-19. We suggest that MC and basophils play an important role in this disease and that these cells are potential therapeutic targets.
AUTHORSHIP
R.S.-C., Y.G.M.-P., and G.M.R.-L. contributed equally to this work, designed, performed experiments, analyzed data, and drafted the manuscript. S.R.-R., V.A.S.-H., R.C.-D., A.P.-F., J.J.T.-R., D.G.-M. assisted in processing and preservation of patient samples, collected patient data, generated, and organized clinical database. M.C.-N., V.D.A.-J., S.M.P.-T., A.D.C.-B., S.E.-P., J.L.M.-M., and R.C.-S. analyzed, interpreted data, and drafted the manuscript. J.L.M.-M. and R.C.-S. designed and supervised the study, and obtained funding. All authors critically revised and approved the final version of this manuscript.
Supplementary Material
Figure S1
Figure S2
Figure S3
Supplementary Table 1. Demographics and clinical characteristics of COVID-19 patients and control group.
Supplementary Figure 1. Serum levels of heparin, histamine, and nitrites are not significantly altered in COVID-19 patients.
Supplementary Figure 2. Serum levels of heparin, histamine, and nitrites are not significantly altered in severe COVID-19 patients.
Supplementary Figure 3. Source organ or tissues of CPA3 humans.
ACKNOWLEDGMENTS
We want to thank Dr Eduardo Ramírez San Juan for his invaluable advice for the statistical analysis of our study. Erick Kenneth Parkinson, David Baker, and Fabián Flores-Borja for critical reading of the manuscript, and Araceli Olvera G. for secretarial assistance. This study was supported by CONACyT (F0005-2020-01-313252) to J.L.M.-M. and (F0005-2020-01-312326) to R.C.-S., and Secretaría de Investigación y Posgrado del IPN (SIP-IPN).
Abbreviations
- ACE2
angiotensin-converting enzyme II
- ARDS
acute respiratory distress syndrome
- AUC
area under the curve
- COVID-19
coronavirus disease 2019
- CPA3
carboxypeptidase A3
- CRP
C-reactive protein
- MC
mast cells
- qSOFA
quick Sepsis-related Organ Failure Assesment
- ROC
receiver operating characteristic
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
Contributor Information
Rodolfo Soria-Castro, Departamento de Inmunología, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, ENCB-IPN, Mexico City, Mexico.
Yatsiri G Meneses-Preza, Departamento de Inmunología, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, ENCB-IPN, Mexico City, Mexico.
Gloria M Rodríguez-López, Departamento de Inmunología, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, ENCB-IPN, Mexico City, Mexico.
Sandra Romero-Ramírez, Red de Apoyo a la Investigación, Universidad Nacional Autónoma de México e Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico; Facultad de Medicina, Universidad Nacional Autónoma de México, Mexico City, Mexico.
Víctor A Sosa-Hernández, Red de Apoyo a la Investigación, Universidad Nacional Autónoma de México e Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico; Departamento de Biomedicina Molecular, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, Mexico City, Mexico.
Rodrigo Cervantes-Díaz, Red de Apoyo a la Investigación, Universidad Nacional Autónoma de México e Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico; Facultad de Medicina, Universidad Nacional Autónoma de México, Mexico City, Mexico.
Alfredo Pérez-Fragoso, Departamento de Inmunología y Reumatología, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
José J Torres-Ruíz, Departamento de Atención Institucional Continua y Urgencias, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
Diana Gómez-Martín, Departamento de Inmunología y Reumatología, Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
Marcia Campillo-Navarro, Research Coordination, Centro Médico Nacional 20 de Noviembre, ISSSTE, Mexico City, Mexico.
Violeta D Álvarez-Jiménez, Lab. de Biología Molecular y Bioseguridad Nivel 3. Centro Médico Naval-SEMAR, Mexico City, Mexico.
Sonia M Pérez-Tapia, Departamento de Inmunología, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, ENCB-IPN, Mexico City, Mexico; Unidad de Desarrollo e Investigación en Bioprocesos (UDIBI), Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, ENCB-IPN, Mexico City, Mexico.
Alma D Chávez-Blanco, División de Ciencia Básica, Instituto Nacional de Cancerología (INCan), Mexico City, Mexico.
Sergio Estrada-Parra, Departamento de Inmunología, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, ENCB-IPN, Mexico City, Mexico.
José L Maravillas-Montero, Red de Apoyo a la Investigación, Universidad Nacional Autónoma de México e Instituto Nacional de Ciencias Médicas y Nutrición Salvador Zubirán, Mexico City, Mexico.
Rommel Chacón-Salinas, Departamento de Inmunología, Escuela Nacional de Ciencias Biológicas, Instituto Politécnico Nacional, ENCB-IPN, Mexico City, Mexico.
DISCLOSURES
The authors declare no conflicts of interest. The data supporting the conclusions of this article will be made available by the authors upon reasonable request.
REFERENCES
- 1. World Health Organization Coronavirus Disease (COVID-19) Dashboard. 2021. https://covid19.who.int/, January 28, 2021.
- 2. Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367:1444–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Han H, Ma Q, Li C, Liu R, Zhao L, Wang W et al. Profiling serum cytokines in COVID-19 patients reveals IL-6 and IL-10 are disease severity predictors. Emerg Microbes Infect. 2020;9:1123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao X. COVID-19: immunopathology and its implications for therapy. Nat Rev Immunol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Andersson CK, Mori M, Bjermer L, Lofdahl CG, Erjefalt JS. Novel site-specific mast cell subpopulations in the human lung. Thorax. 2009;64:297–305. [DOI] [PubMed] [Google Scholar]
- 7. Campillo-Navarro M, Chavez-Blanco AD, Wong-Baeza I, Serafin-Lopez J, Flores-Mejia R, Estrada-Parra S et al. Mast cells in lung homeostasis: beyond type I hypersensitivity. Curr Respir Med Rev. 2014;10:115–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marshall JS, Portales-Cervantes L, Leong E. Mast cell responses to viruses and pathogen products. Int J Mol Sci. 2019;20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rathore AP, St John AL. Protective and pathogenic roles for mast cells during viral infections. Curr Opin Immunol. 2020;66:74–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobasa D, Jones SM, Shinya K, Kash JC, Copps J, Ebihara H et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–23. [DOI] [PubMed] [Google Scholar]
- 11. Hu Y, Jin Y, Han D, Zhang G, Cao S, Xie J et al. Mast cell-induced lung injury in mice infected with H5N1 influenza virus. J Virol. 2012;86:3347–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graham AC, Hilmer KM, Zickovich JM, Obar JJ. Inflammatory response of mast cells during influenza A virus infection is mediated by active infection and RIG-I signaling. J Immunol. 2013;190:4676–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mendez-Enriquez E, Hallgren J. Mast cells and their progenitors in allergic asthma. Front Immunol. 2019;10:821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Swindle EJ, Metcalfe DD. The role of reactive oxygen species and nitric oxide in mast cell-dependent inflammatory processes. Immunol Rev. 2007;217:186–205. [DOI] [PubMed] [Google Scholar]
- 15. Velavan TP, Meyer CG. Mild versus severe COVID-19: laboratory markers. Int J Infect Dis. 2020;95:304–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 17. Siddhuraj P, Clausson CM, Sanden C, Alyamani M, Kadivar M, Marsal J et al. Lung mast cells have a high constitutive expression of carboxypeptidase A3 mRNA that is independent from granule-stored CPA3. Cells. 2021;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rathore APS, Senanayake M, Athapathu AS, Gunasena S, Karunaratna I, Leong WY et al. Serum chymase levels correlate with severe dengue warning signs and clinical fluid accumulation in hospitalized pediatric patients. Sci Rep. 2020;10:11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schwartz LB, Sakai K, Bradford TR, Ren S, Zweiman B, Worobec AS et al. The alpha form of human tryptase is the predominant type present in blood at baseline in normal subjects and is elevated in those with systemic mastocytosis. J Clin Invest. 1995;96:2702–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lilla JN, Chen CC, Mukai K, BenBarak MJ, Franco CB, Kalesnikoff J et al. Reduced mast cell and basophil numbers and function in Cpa3-Cre; Mcl-1fl/fl mice. Blood. 2011;118:6930–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Pejler G, Knight SD, Henningsson F, Wernersson S. Novel insights into the biological function of mast cell carboxypeptidase A. Trends Immunol. 2009;30:401–8. [DOI] [PubMed] [Google Scholar]
- 22. Pejler G. The emerging role of mast cell proteases in asthma. Eur Respir J. 2019;54. [DOI] [PubMed] [Google Scholar]
- 23. George PM, Wells AU, Jenkins RG. Pulmonary fibrosis and COVID-19: the potential role for antifibrotic therapy. Lancet Respir Med. 2020;8:807–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Malaviya R, Ikeda T, Ross E, Abraham SN. Mast cell modulation of neutrophil influx and bacterial clearance at sites of infection through TNF-alpha. Nature. 1996;381:77–80. [DOI] [PubMed] [Google Scholar]
- 25. Chen XY, Huang MY, Xiao ZW, Yang S, Chen XQ. Lactate dehydrogenase elevations is associated with severity of COVID-19: a meta-analysis. Crit Care. 2020;24:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Motta Junior JDS, Miggiolaro A, Nagashima S, de Paula CBV, Baena CP, Scharfstein J et al. Mast cells in alveolar septa of COVID-19 patients: a pathogenic pathway that may link interstitial edema to immunothrombosis. Front Immunol. 2020;11:574862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Herr N, Bode C, Duerschmied D. The effects of serotonin in immune cells. Front Cardiovasc Med. 2017;4:48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cui L, Lee YH, Thein TL, Fang J, Pang J, Ooi EE et al. Serum metabolomics reveals serotonin as a predictor of severe dengue in the early phase of dengue fever. PLoS Negl Trop Dis. 2016;10:e0004607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bennuru S, Lustigman S, Abraham D, Nutman TB. Metabolite profiling of infection-associated metabolic markers of onchocerciasis. Mol Biochem Parasitol. 2017;215:58–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cui L, Fang J, Ooi EE, Lee YH. Serial metabolome changes in a prospective cohort of subjects with influenza viral infection and comparison with dengue fever. J Proteome Res. 2017;16:2614–22. [DOI] [PubMed] [Google Scholar]
- 31. Manne BK, Denorme F, Middleton EA, Portier I, Rowley JW, Stubben C et al. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136:1317–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robson MJ, Quinlan MA, Blakely RD. Immune system activation and depression: roles of serotonin in the central nervous system and periphery. ACS Chem Neurosci. 2017;8:932–42. [DOI] [PubMed] [Google Scholar]
- 33. Hagbom M, Istrate C, Engblom D, Karlsson T, Rodriguez-Diaz J, Buesa J et al. Rotavirus stimulates release of serotonin (5-HT) from human enterochromaffin cells and activates brain structures involved in nausea and vomiting. PLoS Pathog. 2011;7:e1002115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nau F Jr, Yu B, Martin D, Nichols CD. Serotonin 5-HT2A receptor activation blocks TNF-alpha mediated inflammation in vivo. PLoS One. 2013;8:e75426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kubera M, Maes M, Kenis G, Kim YK, Lason W. Effects of serotonin and serotonergic agonists and antagonists on the production of tumor necrosis factor alpha and interleukin-6. Psychiatry Res. 2005;134:251–8. [DOI] [PubMed] [Google Scholar]
- 36. Ha S, Jin B, Clemmensen B, Park P, Mahboob S, Gladwill V et al. Serotonin is elevated in COVID-19-associated diarrhoea. Gut. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zaid Y, Guessous F, Puhm F, Elhamdani W, Chentoufi L, Morris AC et al. Platelet reactivity to thrombin differs between patients with COVID-19 and those with ARDS unrelated to COVID-19. Blood Adv. 2021;5:635–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McPherson RA, Pincus MR. Henry's clinical diagnosis and management by laboratory methods. 21st. Philadelphia: Elsevier; 2006. pages cm p. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Figure S2
Figure S3
Supplementary Table 1. Demographics and clinical characteristics of COVID-19 patients and control group.
Supplementary Figure 1. Serum levels of heparin, histamine, and nitrites are not significantly altered in COVID-19 patients.
Supplementary Figure 2. Serum levels of heparin, histamine, and nitrites are not significantly altered in severe COVID-19 patients.
Supplementary Figure 3. Source organ or tissues of CPA3 humans.


