Abstract
Timing of detection of immunoglobulin G (IgG), immunoglobulin A (IgA), and immunoglobulin M (IgM) antibodies against severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2), and their use to support the diagnosis are of increasing interest. We used the Gold Standard Diagnostics ELISA to evaluate the kinetics of SARS‐CoV‐2 IgG, IgA, and IgM antibodies in sera of 82 hospitalized patients with polymerase chain reaction (PCR)‐confirmed coronavirus disease 2019 (COVID‐19). Serum samples were collected 1–59 days post‐onset of symptoms (PoS) and we examined the association of age, sex, disease severity, and symptoms' duration with antibody levels. We also tested sera of 100 ambulatory hospital employees with PCR‐confirmed COVID‐19 and samples collected during convalescence, 35–57 days PoS. All but four of the admitted patients (95.1%) developed antibodies to SARS‐CoV‐2. Antibodies were detected within 7 days PoS; IgA in 60.0%, IgM in 53.3%, and IgG in 46.7% of samples. IgG positivity increased to 100% on Day 21. We did not observe significant differences in the rate of antibody development in regard to age and sex. IgA levels were highest in patients with a severe and critical illness. In multiple regression analyses, only IgA levels were statistically significantly correlated with critical disease (p = .05) regardless of age, sex, and duration of symptoms. Among 100 ambulatory hospital employees who had antibody testing after 4 weeks PoS only 10% had positive IgA antibodies. The most frequently isolated isotype in sera of employees after 30 days PoS was IgG (88%). IgA was the predominant immunoglobulin in early disease and correlated independently with a critical illness. IgG antibodies remained detectable in almost 90% of samples collected up to two months after infection.
Keywords: COVID‐19, IgA, SARS‐CoV‐2, serology
Highlights
IgA was the predominant immunoglobulin detected in early COVID‐19 disease.
IgA levels were highest in patients with a severe and critical illness.
IgG antibodies remained detectable in almost 90% of samples collected up to 2 months after infection.
1. INTRODUCTION
Detection of severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) virus by nucleic acid amplification of viral RNA is the gold standard for the diagnosis of this newly emerging pathogen that causes coronavirus disease‐19 (COVID‐19), that has led to the largest pandemic of this century. The detection of antibodies can be helpful in confirming the infection,1, 2, 3 understanding the timing of the infection, 4 and determining the presence of neutralizing antibodies that may aid in the elimination of the virus and perhaps protection against re‐infection.5, 6
Serology methods employed for COVID‐19 have utilized several immunogenic antigens of SARS‐CoV‐2 that include the S (Spike) protein, its subunits S1 and S2, and its receptor‐binding domain (RBD) as well as the Nucleocapsid protein (N). 7 Although assays using the S antigen have shown less cross‐reactivity with human coronaviruses, including the related Sarbecovirus group, SARS‐CoV, and MERS‐CoV, the N antigen is more immunoreactive. 8 Evaluation of these assays, mostly from studies abroad, have determined that antibodies to SARS‐CoV‐2 appear early in the disease course. 9 Studies investigating the kinetics of the antibody response have reported the early appearance of immunoglobulin A (IgA) antibodies regardless of the antigens used in the immunoassay. 10
In the present study, we used the Gold Standard Diagnostic ELISA to assess the presence of immunoglobulin G (IgG), IgA, and immunoglobulin M (IgM) antibodies to the N antigen of SARS‐CoV‐2 in sera of hospitalized patients and ambulatory hospital employees with polymerase chain reaction (PCR)‐confirmed COVID‐19 infection. Samples were collected from 1 to 59 days post‐onset of symptoms (PoS) from hospitalized patients and during convalescence (35–57 PoS) from hospital employees. We studied the timing of the appearance of IgG, IgA, and IgM antibodies and the association of the antibodies' levels with disease severity. We also examined the specificity and cross‐reactivity of these newly commercially available enzyme‐linked immunosorbent assay (ELISA) assays using stored sera collected before the COVID pandemic and serum samples of patients with other viral infections.
2. MATERIALS AND METHODS
2.1. Study design, setting, and participants
The study was conducted at New York University Langone Health hospitals in New York City and included serum samples collected between April and May 2020. Serum samples were obtained from two groups of patients with PCR‐confirmed COVID‐19 infection, 82 inpatients and 100 outpatients. The 82 hospitalized patients were chosen based on the availability of serum samples submitted to the laboratory. The outpatients were 100 ambulatory hospital employees who consented to sample collection. Samples from the employees were obtained without identifiers and the only available data were the date of onset of symptoms and the date of sample collection.
Ninety‐three (93) serum samples were collected between 1 and 59 days PoS from the 82 hospitalized patients, and a single convalescent sample between 32 and 57 days PoS from the 100 employees who were all managed in the ambulatory setting.
The aims of this study were to verify the performance of a newly available commercial antibody assay in patients with PCR‐confirmed COVID‐19 infection on serum samples collected at different time intervals after disease onset and to assess the detection of IgG, IgA, and IgM antibodies according to disease severity.
The disease severity was classified according to the World Health Organization guidelines on a four‐point ordinal scale, consisting of the following categories: (1) mild disease, no evidence of viral pneumonia or hypoxia; (2) moderate disease, clinical signs of pneumonia (fever, cough, dyspnea, tachypnea) but no signs of severe pneumonia, including SpO2 ≥ 90% on room air; (3) severe disease, clinical signs of pneumonia (fever, cough, dyspnea, tachypnea) plus one of the following: respiratory rate greater than 30 breaths/min; severe respiratory distress; or SpO2 < 90% on room air; and (4) critical illness, acute respiratory distress syndrome, severe sepsis or septic shock. 11 This study was approved with a waiver of informed consent for hospitalized patients and Institutional Review Board approval for employees' samples. Additional antibody studies on the employee sera are being presented in separate publications.
2.1.1. Enzyme‐linked immunosorbent assay
The SARS‐CoV‐2 IgG, IgA, and IgM ELISA kits manufactured by Virotech Diagnostics for Gold Standard Diagnostics are qualitative assays that detect separately IgG, IgA, and IgM antibodies to the Nucleocapsid protein (N) of the SARS‐CoV‐2 virus. The assays were conducted according to the manufacturer's instructions. The assays use a 1:100 serum dilution, controls, and calibrator, and the reactions are read at a wavelength of 450 nm with a reference wavelength of 620nm. Tests are reported in units and are considered positive if the sample optical density/cut off 10× is greater than 11.0 units; equivocal if 9.0–11.0 units; and negative less than 9.0 units. For analysis of our data, we included samples in the equivocal category as positive. Although the assays are qualitative, we use the units obtained to compare intensities of reactivity. Specificity provided in the manufacturer's package insert was 100% for IgG and IgA, and 100% and 98.7% for IgM when testing healthy U.S. and German blood donors, respectively. Cross‐reactivity was observed by the manufacturer in 1 and 8 of 110 samples from patients with viral and bacterial respiratory pathogens for IgG and IgM, respectively, whereas no cross‐reactivity was observed in IgA ELISA. These assays have been submitted by the manufacturer to the FDA for EUA.
2.1.2. Non‐COVID‐19 sera
To investigate the specificity of the ELISA assays we included 54 serum samples that were collected before the COVID‐19 pandemic or from patients testing negative for COVID‐19 but confirmed with other bacterial, viral infections or autoimmune diseases. These samples had been stored frozen at −70°C. The samples included sera testing positive for Epstein‐Barr virus (n = 1), Cytomegalovirus (n = 4), Influenza A (n = 5), Influenza B (n = 6), Parainfluenza virus (n = 9), Mycoplasma pneumoniae (n = 3), Respiratory syncytial virus (n = 3), Borrelia burgdorferi (n = 6), Human granulocytic anaplasmosis (HGA) (n = 3), Treponema pallidum (n = 1), Rheumatoid arthritis (n = 3), Systemic lupus erythematosus (n = 1). Additionally, nine sera samples were obtained from patients with PCR‐confirmed human coronavirus infections, not SARS‐CoV‐2.
2.1.3. Statistics
All calculations were performed using the Stata v15.0 software package (Stata Corporation). Categorical variables were presented as percentages and were compared by the χ 2 or Fischer exact test using an alpha of 0.05. Continuous variables were presented as means with SD and range and were compared by Student's t‐test. Univariate analysis was performed to study the association of the development and levels of different antibody isotypes with age and sex. Also, a multiple regression analysis was performed to examine the association of the age, sex, duration of symptoms, and disease severity with the IgG, IgM, and IgA antibody levels as determined by ELISA units. The Strengthening the Reporting of Observational Studies in Epidemiology guidelines were followed in the preparation of this manuscript.
3. RESULTS
The study population consisted of 82 hospitalized patients who were diagnosed with COVID‐19 infection by reverse transcription PCR (RT‐PCR) between March and May 2020. Samples of 17 out of 82 (20.7%) patients were collected at autopsy. Among the remaining 65 patients, 16 died during the hospitalization (24.6%). The median age of the patients was 61 years (IQR: 49–74, range: 22–97) and 50 (61.0%) were male (Table 1). Eleven (13.6%) patients had mild disease, 15 (18.5%) had moderate and 55 (67.9%) had the severe or critical disease. All 100 hospital employees who were diagnosed with COVID‐19 were managed on an outpatient basis.
Table 1.
Patient characteristics
| Demographics/patient characteristics | No. of patients (%), mean (SD; range) |
|---|---|
| Age | 61 (49–74) |
| Gender | |
| Female | 32 (39.0%) |
| Male | 50 (61.0%) |
| Race | |
| White | 26 (31.7%) |
| African American/Black | 15 (18.3%) |
| Asian | 9 (11.0%) |
| Other/unknown | 32 (39.0%) |
| Ethnicity | |
| Hispanic | 22 (26.8%) |
| Non‐Hispanic | 44 (53.7%) |
| Other/unknown | 16 (19.5%) |
| Days of symptoms before antibody testing (mean, SD) | 17.3 (10.9; 1–59) |
| Disease severity | |
| Mild | 11 (13.6%) |
| Moderate | 15 (18.5%) |
| Severe | 55 (67.9%) |
3.1. Antibodies in early disease in hospitalized patients
Fifteen samples were collected between 1 and 7 days PoS. Antibodies were detected in 13 samples: IgA antibodies were detected in 9 of 15 (60.0%) samples, IgM and IgG were detected in 8 (53.3%) (p = 1.0) and 7 (46.7%) (p = .72), respectively (Table 2). The most frequent combination of antibodies in these samples was the simultaneous detection of IgA and IgM in four samples. IgA had the highest levels in these samples with an average of 27.1 units (SD: 24.6; range: 0.42–81.6), followed by IgM with an average of 22.6 units (SD: 20.7; range: 0.31–51.4) (Figure 1). Thirty‐five of 37 (94.6%) samples collected between days 8 and 14 PoS had detectable antibodies; two patients with single samples collected at Days 9 and 12 PoS did not have detectable antibodies. All 15 (100%) samples collected between 22 and 28 days PoS had detectable IgG and IgA antibodies; 11 (73.3%) had IgM antibodies. Although IgG antibodies were still detectable in all 11 (100%) samples collected between 29 and 59 days PoS, IgA, and IgM were detected in 7 (63.6%) (p = .09) patients each. The most frequent antibody combination on samples collected after the first‐week post symptoms was the simultaneous detection of all three immunoglobulin isotypes. Overall, 78 of 82 (95.1%) patients developed antibodies during the period of observation. Four patients with only one sample collected between 2 and 19 days, did not have detectable antibodies.
Table 2.
IgG, IgA, and IgM antibodies to SARS‐CoV‐2 according to days post‐onset of symptoms (PoS) on 92 samples from 81 hospitalized patients
| Days PoS | No. samples | IgG no. positive (%) | IgG mean units (SD) | IgA no. positive (%) | IgA mean units (SD) | IgM no. positive (%) | IgM mean units (SD) |
|---|---|---|---|---|---|---|---|
| 1–7 | 15 | 7 (46.7%) | 16.4 (16.2) | 9 (60.0%) | 27.1 (24.6) | 8 (53.3%) | 22.6 (20.7) |
| 8–14 | 37 | 33 (89.2%) | 30.5 (14.4) | 30 (81.1%) | 39.0 (29.2) | 28 (75.7%) | 25.4 (18.9) |
| 15–21 | 13 | 12 (92.9%) | 31.9 (12.5) | 11 (78.6%) | 34.0 (23.3) | 11 (78.6%) | 23.0 (14.5) |
| 22–28 | 15 | 15 (100.0%) | 34.8 (11.0) | 15 (100.0%) | 30.8 (22.2) | 11 (73.3%) | 18.1 (13.6) |
| 29–59 | 11 | 11 (100.0%) | 35.9 (11.1) | 7 (63.6%) | 18.1 (16.2) | 7 (63.6%) | 15.5 (15.2) |
| Total | 92 | 79 (85.9%) | 29.8 (14.7) | 72 (78.3%) | 32.6 (25.7) | 65 (70.7%) | 22.2 (17.4) |
Abbreviations: IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Figure 1.
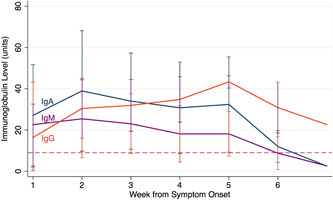
Kinetics of immunoglobulin G (IgG), immunoglobulin A (IgA), and immunoglobulin M (IgM) SARS‐CoV‐2 immunoglobulin levels among 82 hospitalized patients with PCR confirmed COVID‐19 infection. COVID‐19, coronavirus diease 2019; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
3.2. Correlation between antibody isotype frequency and levels according to age, gender and disease severity
The rate of antibody isotype development did not show significant differences between patients greater and less than 65 years of age and between male and female patients. The IgA levels increased with age (p = .03), but no statistically significant differences were noted between the IgG and IgM levels and age. When analyzing the antibodies according to gender, we observed higher levels of the IgG (p < .01) and IgA isotypes (p = .03) in male compared to female patients.
Patients with mild or moderate disease developed IgG, IgA, and IgM antibodies with similar frequency, 76.9% (20/26), 73.1% (19/26) (p = 1.0), and 57.7% (15/26), respectively (p = .24). The most frequent immunoglobulin isotypes seen in patients with the severe or critical disease were IgG (50/55, 90.9%) and IgA (46/55, 83.6%). When analyzing the immunoglobulin levels as determined by ELISA units, IgA had the highest levels in the group of patients with severe or critical disease compared to those with mild or moderate disease (Figure 2). The IgA level was on average 39.3 units (SD: 26.1) in serum of patients with severe or critical disease compared with 21.4 units (SD: 17.0) for IgM (p < .001). In univariate analyses, IgA (p = .02) and IgG (p < .01) levels correlated significantly with disease severity.
Figure 2.
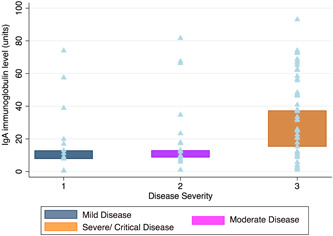
IgG, IgA, IgM immunoglobulin levels by disease severity. IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M
In multiple regression analyses, only IgA levels were found to be associated with critical disease (p = .05) after adjustment for age, sex, and duration of symptoms (Table 3). A significant association was also found between IgA levels and age (p = .05) as well as male gender (p = .03). On the other hand, increased IgG levels were only significantly associated with male gender (p = .01), with no statistically significant association noted with age (p = .49), critical disease (p = .23) and duration of symptoms (p = .08). The results of the multiple regression analysis examining the association of the age, sex, duration of symptoms, and disease severity with the IgG levels remained unchanged when patients who were within the first 2 weeks PoS were excluded.
Table 3.
Results of multiple regression analyses regarding factors associated with SARS‐CoV‐2 IgG, IgA, and IgM levels
| Patient characteristic | IgG levels (regression coefficient; p value) | IgA levels (regression coefficient; p value) | IgM levels (regression coefficient; p value) |
|---|---|---|---|
| Age | 0.07 (0.49) | 0.37 (0.05) | −0.01 (0.91) |
| Gender (male) | 8.3 (0.01) | 13.3 (0.03) | 3.8 (0.38) |
| Duration of symptoms | 0.30 (0.08) | −0.61 (0.06) | −0.33 (0.15) |
| Disease severity (critical disease) | 6.2 (0.23) | 19.1 (0.05) | 9.1 (0.19) |
Note: Bold values indicate statistical significance.
Abbreviations: IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.3. Serology on hospital employees samples collected during convalescence after 30 days post‐onset of symptoms
100 samples from 100 ambulatory hospital employees with PCR‐confirmed COVID‐19 were collected in April 2020. Samples were collected at a mean of 40.5 days (SD: 5.3; rage: 32–57 days) from the onset of symptoms. The most frequently detected antibodies were IgG in 88 patients (88.0%) followed by IgM in 42 patients (42.0%) (Table 4). Only 10 patients (10.0%) had detectable IgA antibodies; two of them were in the 5th week, six in the 6th week, and two in the 8th week PoS. Among the patients with detectable antibodies, IgG had the highest levels with an average of 26.9 units (SD: 11.4; range: 10.9–56.7) followed by IgM levels with an average of 19.9 units (SD: 11.6; range: 9.3–52.7).
Table 4.
IgG, IgA, and IgM antibodies to SARS‐CoV‐2 according to days PoS on 100 samples from 100 hospital employees
| Days PoS | No. samples | IgG no. positive (%) | IgA no. positive (%) | IgM no. positive (%) |
|---|---|---|---|---|
| 29‐42 | 71 | 62 (87.3%) | 8 (11.3%) | 31 (43.7%) |
| 43‐63 | 29 | 26 (89.7%) | 2 (6.9%) | 11 (37.9%) |
| Total | 100 | 88 (88.0%) | 10 (10.0%) | 42 (42.0%) |
Abbreviations: IgA, immunoglobulin A; IgG, immunoglobulin G; IgM, immunoglobulin M; PoS, post‐onset of symptom; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
3.4. ELISA specificity and cross‐reactivity
One of 24 samples from negative controls (specificity 95.8%) and 3 of 54 sera from patients with autoimmune or infectious diseases, not COVID, tested positive in the SARS‐CoV‐2 IgM ELISA. Most of these samples tested in the low positive and equivocal range. All tested negative for IgG and IgA antibodies to SARS‐CoV‐2. None of the samples from patients with human coronavirus infections tested positive in these assays. The overall specificity in our evaluation was 94.9% for IgM and 100% for IgG and IgA.
4. DISCUSSION
In this study, we examined the timing of appearance of IgG, IgA, and IgM antibodies to the N antigen of SARS‐CoV‐2 in sera of hospitalized patients with PCR‐confirmed COVID‐19 infection as detected by the ELISA kits by Gold Standard Diagnostics as well as the association of the antibodies' levels with disease severity. We found that IgA antibodies appeared early in the disease course and their levels correlated with critical disease after adjusting for age, gender, and duration of symptoms. Importantly, IgG antibodies were invariably present by the third week of illness, whereas IgM antibodies did not seem to offer any additional information to what IgA and IgG already provide. Interestingly, among the 100 ambulatory hospital employees who had antibody testing after 4 weeks PoS only 10% had positive IgA antibodies.
Our study findings on the early timing of IgA antibody appearance have important implications in determining the chronology of SARS‐CoV‐2 infection and might aid in diagnosis of COVID‐19 in patients who repeatedly test negative by RT‐PCR despite the high clinical suspicion. Interestingly, we observed IgA in only a small minority in the sera of employees that were tested after Week 4 PoS, a finding that can be attributed to the time in convalescence when the samples were collected. This has a potential implication in reappearance or increased levels of IgA in cases of SARS‐CoV‐2 reinfection that we might observe as the pandemic evolves. One of the limitations of our study was the unavailability of serum samples of ambulatory patients with mild symptoms during the early stages of the infection to compare with the antibody kinetics observed in hospitalized patients. As such, it is also possible that patients with a milder disease whose COVID‐19 disease does not warrant hospitalization develop lower levels of IgA antibodies that might become undetectable in early convalescence, our finding on the association of IgA levels and disease severity notwithstanding.
Prior studies conducted in Europe and China support our findings. Cervia et al. using commercial ELISA using S1 protein as antigen showed that IgA antibodies in serum appeared within 3–4 days post‐onset of symptoms among patients with severe disease. 12 Ma et al. 10 used chemiluminescence immunoassays (CLIA) to detect IgG, IgA, and IgM antibodies to the N and RBD antigens on sera of 87 Chinese patients diagnosed with COVID‐19. They also found that IgA antibodies improved the performance of immunoassays in early diseases showing higher sensitivity than IgG or IgM. Another study from China using CLIA to detect IgG, IgA, and IgM to the S protein also found that IgA antibodies were detected earlier, at 2 days PoS, with a higher rate of positivity compared to IgG or IgM. 13 Similarly, in the study by Guo et al., 14 92.7% and 85.4% of patients with confirmed or probable COVID‐19, had IgA and IgM antibodies within seven days after symptom onset, whereas IgG antibodies appeared at a median of 14 days from symptom onset with a positivity rate of 77.9%.
Antibody studies conducted on samples from patients diagnosed with SARS‐CoV infection that led to the emerging SARS in 2003, also showed an early IgA response. SARS‐CoV and SARS‐CoV‐2 have significant antigenic similarities making the antibody reactivity comparison against both pathogens very relevant. The Nucleocapsid N antigen of SARS‐CoV has been determined to be the most antigenic of the viral structural proteins in studies of the antibody profile of infected patients. 15 In addition, this antigen was found to elicit IgA antibodies to SARS‐CoV earlier than IgM and IgG. 16 Reactivities to the Spike (S) protein appeared later in the infection and were mostly IgG and IgA in this study. These findings can explain the different antibody reactivities and kinetics observed in SARS‐CoV‐2 by different investigators, and our own experience.
Multiple serologic assays for SARS‐CoV‐2 have become commercially available in the US and most of them have included the detection of IgG.17, 18 A recent evaluation of four commercial platforms detecting IgG to the N or S antigens of SARS‐CoV‐2 showed that these assays have a high sensitivity in convalescence. 19 However, only a few studies in the U.S. have investigated the use of IgA antibodies to SARS‐CoV‐2. Beavis et al. evaluated the commercial Euroimmun IgG and IgA assay that uses the S1 domain as antigen source in an ELISA format. They tested 82 samples of patients with PCR‐confirmed SARS‐CoV‐2 infection, as well as 86 samples collected during the same time period from patients who tested negative for SARS‐CoV‐2, and cross‐reactive populations. 20 They found that 82.9% of patients with PCR‐confirmed SARS‐CoV‐2 infection tested positive for IgA and 67.1% tested positive for IgG, with the sensitivity increasing to 90.5% and 100% for IgA and IgG, respectively, among samples collected 4 days after PCR positivity. However, as the actual time elapsed between illness onset and the sample collection is unknown in these patients, the determination of the timing of antibody development is unknown. 20 These authors, however, found that the Euroimmun IgA assay had lower specificity than the IgG assay. Ten samples from patients not diagnosed with COVID‐19 had IgA reactivity. Similarly, Okba et al. 21 found that commercial assays using S1 as antigen exhibited lower specificity of IgA antibodies as compared with IgG assays. The source of antigen used for antibody assay detection is of importance when analyzing performance and cross‐reactivities.
A notable finding in the present study is our observation that IgA antibody levels correlated with disease severity and this remained significant after adjusting for age, gender, and duration of symptoms. The fact that IgA levels correlated with disease severity might be attributed to higher immune responsiveness of the respiratory system facing a severe lower respiratory infection or that IgA has a pathogenetic role in the development of severe disease. Similar to our observation, Ma et al. 10 reported that IgA antibody levels were higher in patients with more severe disease. The authors postulated that IgA could contribute to the antibody‐dependent enhancement of infection seen in COVID‐19. They explained that the predominance of IgA over IgM antibodies in COVID‐19 infection differs compared to SARS‐CoV infection where the presence of IgM antibodies might be secondary to the viremia caused by this virus which is not observed in SARS‐CoV‐2. Cervia et al. 12 observed that among healthcare workers with possible SARS‐CoV‐2 exposure, the titers of IgA antibodies measured in mucosal sites were higher than those in serum with an inverse correlation noted between the IgA levels in mucosal sites and age. Further studies in this area are needed to understand this new clinical entity and the role antibodies play in the disease process and potentially guiding the development of therapeutic or preventive modalities.
Importantly, IgG antibodies also seemed to appear early in the disease course and were consistently detectable by three weeks PoS. All samples collected from patients between weeks four and eight PoS and all employees samples collected after 45 days PoS had detectable IgG antibodies. Long et al. 6 followed symptomatic and asymptomatic patients with confirmed SARS‐CoV‐2 infection and observed a decreasing trend in both IgG levels and neutralizing antibodies 2–3 months after infection. Further investigation is needed to determine the time antibodies will remain detectable and if detection is correlated with neutralizing antibodies capable of protecting from re‐infection with the SARS‐CoV‐2 virus. On the other hand, IgM antibodies in our study did not seem to contribute to support a clinical diagnosis, as opposed to IgA or IgG. Similar low IgM reactivity was found by Wu et al. 16 in their study of SARS‐CoV infected patients; this was more evident against the S protein. The lack of early IgM detection and decreased levels in early infection in our study could be also attributed to the inability of this particular commercial assay and antigen source to detect IgM antibodies, therefore further evaluation is needed. It should be noted that at the conclusion of this study, the manufacturer discontinued the production of the IgM assay as it had been formulated and it will be releasing a reformulated assay.
In summary, our study has shown the timing of detection of the IgG, IgA, and IgM antibodies in two populations of patients with PCR‐confirmed COVID‐19 infection and provides additional evidence to help guide the use of serologic testing in the diagnosis and management of COVID‐19 infection. Further studies on the potential reappearance and pattern of antibody development in SARS‐CoV‐2 re‐infection, is of significant interest and are warranted.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Fainareti N. Zervou and Maria E. Aguero‐Rosenfeld conceptualized and designed the study. Fainareti N. Zervou, Ping Louie, Anna Stachel, Ioannis M. Zacharioudakis, Yadira Ortiz‐Mendez, Kristen Thomas, and Maria E. Aguero‐Rosenfeld participated in data extraction. Fainareti N. Zervou, Anna Stachel, and Maria E. Aguero‐Rosenfeld analyzed the data. Fainareti N. Zervou, Ioannis M. Zacharioudakis, and Maria E. Aguero‐Rosenfeld drafted the original manuscript. All authors critically edited and accepted the final manuscript.
ACKNOWLEDGMENTS
The authors would like to sincerely acknowledge the help and support of the medical technologists from the Tisch Diagnostic Immunology laboratory (Rita Law, Andre Fidelia, Alice Lu, Mary Lau, Connie Belandres, and Christine Longobardi), Gerard Kick and his team in the Clinical Chemistry section for assistance in retrieving serum samples. They also would like to thank Sarah Hochman, Jennifer Lighter, and Stephanie Sterling for helping to obtain clinical data, our colleagues and providers caring for COVID‐19 patients, and those employees who provided their serum samples.
Zervou FN, Louie P, Stachel A, et al. SARS‐CoV‐2 antibodies: IgA correlates with severity of disease in early COVID‐19 infection. J Med Virol. 2021;93:5409‐5415. 10.1002/jmv.27058
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Peeling RW, Wedderburn CJ, Garcia PJ, et al. Serology testing in the COVID‐19 pandemic response. Lancet Infect Dis. 2020;20(9):e245‐e249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tang YW, Schmitz JE, Persing DH, Stratton CW. Laboratory diagnosis of COVID‐19: current issues and challenges. J Clin Microbiol. 2020;58(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Zhao R, Li M, Song H, et al. Early detection of SARS‐CoV‐2 antibodies in COVID‐19 patients as a serologic marker of infection. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Deeks JJ, Dinnes J, Takwoingi Y, et al. Antibody tests for identification of current and past infection with SARS‐CoV‐2. Cochrane Database Syst Rev. 2020;6:CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ibarrondo FJ, Fulcher JA, Goodman‐Meza D, et al. Rapid decay of anti‐SARS‐CoV‐2 antibodies in persons with mild COVID‐19. N Engl J Med. 2020;383(11):1085‐1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Long QX, Tang XJ, Shi QL, et al. Clinical and immunological assessment of asymptomatic SARS‐CoV‐2 infections. Nat Med. 2020;26(8):1200‐1204. [DOI] [PubMed] [Google Scholar]
- 7. Premkumar L, Segovia‐Chumbez B, Jadi R, et al. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS‐CoV‐2 patients. Sci Immunol. 2020;5(48). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dutta NK, Mazumdar K, Gordy JT. The nucleocapsid protein of SARS‐CoV‐2: a target for vaccine development. J Virol. 2020;94(13). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hanson KE, Caliendo AM, Arias CA, et al. Infectious Diseases Society of America Guidelines on the diagnosis of COVID‐19: serologic testing. Clin Infect Dis. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma H, Zeng W, He H, et al. Serum IgA, IgM, and IgG responses in COVID‐19. Cell Mol Immunol. 2020;17(7):773‐775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. WHO . Clinical management of COVID‐19. Interim guidance. 2020. https://www.who.int/publications/i/item/clinical-management-of-covid-19. Accessed September 29, 2020.
- 12. Cervia C, Nilsson J, Zurbuchen Y, et al. Systemic and mucosal antibody secretion specific to SARS‐CoV‐2 during mild versus severe COVID‐19. bioRxiv. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu HQ, Sun BQ, Fang ZF, et al. Distinct features of SARS‐CoV‐2‐specific IgA response in COVID‐19 patients. Eur Respir J. 2020;56(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guo L, Ren L, Yang S, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID‐19). Clin Infect Dis. 2020;71(15):778‐785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tan YJ, Goh PY, Fielding BC, et al. Profiles of antibody responses against severe acute respiratory syndrome coronavirus recombinant proteins and their potential use as diagnostic markers. Clin Diagn Lab Immunol. 2004;11(2):362‐371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wu HS, Hsieh YC, Su IJ, et al. Early detection of antibodies against various structural proteins of the SARS‐associated coronavirus in SARS patients. J Biomed Sci. 2004;11(1):117‐126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pickering S, Betancor G, Galao RP, et al. Comparative assessment of multiple COVID‐19 serological technologies supports continued evaluation of point‐of‐care lateral flow assays in hospital and community healthcare settings. PLOS Pathog. 2020;16(9):e1008817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herroelen PH, Martens GA, De Smet D, Swaerts K, Decavele AS. Humoral immune response to SARS‐CoV‐2. Am J Clin Pathol. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Theel ES, Harring J, Hilgart H, Granger D. Performance characteristics of four high‐throughput immunoassays for detection of IgG antibodies against SARS‐CoV‐2. J Clin Microbiol. 2020;58(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Beavis KG, Matushek SM, Abeleda APF, et al. Evaluation of the EUROIMMUN Anti‐SARS‐CoV‐2 ELISA assay for detection of IgA and IgG antibodies. J Clin Virol. 2020;129:104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okba NMA, Muller MA, Li W, et al. Severe acute respiratory syndrome coronavirus 2‐specific antibody responses in coronavirus disease patients. Emerg Infect Dis. 2020;26(7):1478‐1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


