Abstract
Rapid diagnostics for severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) are paramount for reducing the spread of the current pandemic. During additional seasonal epidemics with influenza A/B and respiratory syncytial virus (RSV), the clinical signs and symptoms cannot be distinguished easily from SARS‐CoV‐2. Therefore, a new assay combining four targets in the form of the new Xpert Xpress SARS‐CoV‐2/Flu/RSV assay was evaluated. The assay was compared to the Xpert Xpress SARS‐CoV‐2, Xpert Xpress Flu/RSV, Seegene Flu/RSV, influenza A/B r‐gene® and RSV/hMPV r‐gene®. A total of 295 nasopharyngeal and throat swabs were tested at four institutes throughout Europe including 72 samples positive for SARS‐CoV‐2, 65 for influenza A, 47 for influenza B, and 77 for RSV. The sensitivity of the new assay was above 95% for all targets, with the highest for SARS‐CoV‐2 (97.2%). The overall correlation of SARS‐CoV‐2 Ct values between Xpert Xpress SARS‐CoV‐2 assay and Xpert Xpress SARS‐CoV‐2/Flu/RSV assay was high. The agreement between Ct values above 30 showed the multiplex giving higher Ct values for SARS‐CoV‐2 on average than the singleplex assay. In conclusion, the new assay is a rapid and reliable alternative with less hands‐on time for the detection of not one, but four upper respiratory tract pathogens that may circulate at the same time.
Keywords: COVID‐19, influenza, multiplex PCR, pandemic, respiratory syncytial virus, SARS‐CoV‐2
1. INTRODUCTION
A year into the current severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) pandemic, the number of cases remain high and with new mutations resulting in increased infectiousness and transmission, this will most likely remain of concern at least until a significant percentage of the population is vaccinated or naturally immunized. 1 , 2 A fast and accurate SARS‐CoV‐2 diagnostic remains important to identify infected persons, reduce transmission and gather data for surveillance purposes. A shortage of consumables, protective materials, and polumerase chain reaction (PCR) platform capacity is thus seen. During winter, other respiratory viruses such as influenza A/B and respiratory syncytial virus (RSV), which are usually epidemic in Europe between November and April, may coincide with SARS‐CoV‐2. 3 , 4 However, recent data from Australia show that those seasonal epidemics have been reduced during the SARS‐CoV‐2 pandemic, possibly as a result of isolation, restricted travel, and social distancing. 5 These respiratory viruses often show initial similar symptoms and cannot be differentiated on clinical presentation alone. However, they do require different isolation regimens, depending on the patient category, such as children, elderly, and the immunocompromised and the course of the diseases evolve differently according to each virus. 6 , 7 , 8 , 9 Additionally, treatment options differ and surveillance of individual pathogens is relevant for public health decision making. 10 , 11 To distinguish between four of the most common and clinically relevant seasonal airway pathogens and to be able to identify coinfections rapidly, a multiplex PCR, the Xpert Xpress SARS‐CoV‐2/Flu/RSV (multiplex) has been made available. 12 , 13 The aim of this study was to evaluate this new multiplex PCR assay in a European multicenter setting.
2. MATERIALS AND METHODS
2.1. Study sites and samples selection
Samples from archived collections were selected at four institutes throughout Europe. The Radboud University Medical Center in Nijmegen the Netherlands, the Medical University of Graz in Austria, the Institute of Medical Virology, the University of Zurich/University Hospital Zurich in Switzerland, and the University Hospital of Rennes, in France. Each institute selected samples positive for SARS‐CoV‐2, influenza‐A, influenza B, and RSV. Those samples have previously been tested on a range of different platforms. A total of 295 clinical samples were included. The range of Ct values is shown in Table 1.
Table 1.
Number of samples tested at each institute, sample type, and reference platform
| Number of samples tested using the reference method | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SARS‐CoV‐2+ | Influenza A+ | Influenza B+ | RSV+ | Negative | ||||||||
| Study site | Sample type and medium | Reference platform | N | Range Ct (average) | N | Range Ct (average) | N | Range Ct (average) | N | Range Ct (average) | N | Range Ct (average) |
| Radboudumc, Nijmegen | NPS in UTM/GLY |
|
21 |
N2: 18–42 (30) E: 15–41 (22) |
20 |
A1: 18–37 (26) A2: 19–32 (24) |
10 | Unknown | 22 | 18–39 (26) | 10 | n.a. |
| Medical University of Graz | OPS in Copan UTM™ |
|
20 |
N2: 14–38 (26) E: 12–34 (24) |
19 | 19–33 (26) | 12 | 17–27 (24) | 24 | 17–38 (28) | 10 | n.a. |
| Institute of Medical Virology, University of Zurich | NPS/throat |
|
11 |
N2: 16–44 (27) E: 18–42 (29) |
10 |
A1: 23–39 (31) A2: 25–45 (35) |
10 | 20–37 (30) | 10 | 27–39 (34) | 9 | n.a. |
| Rennes University Hospital | NPS in TranSwab of eSwab |
|
20 |
N2: 14–38 (23) E: 12–35 (21) |
16 | 21–30 (23) | 15 | 14–36 (27) | 21 | 14–31 (19) | 20 | n.a. |
| Total 72 | Total 65 | Total 47 | Total 77 | Total 49 |
Note: For SARS‐CoV‐2 the reference method Xpert Xpress SARS‐CoV‐2 reports two target genes (E‐ and N2‐gene), similar for influenza A (reporting A1 and A2) in Xpert Xpress Flu/RSV.
Abbreviations: NPS, nasopharyneal swab; RSV, respiratory syncytial virus; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2; n.a., not available.
2.2. Reference method
2.2.1. Radboud University Medical Center
Samples consisted of nasopharyneal and/or throat swabs in UTM or GLY medium (Table 1) and stored at −80°C. SARS‐Cov‐2 was tested using Xpert Xpress SARS‐CoV‐2 (singleplex) and Xpert Xpress Flu/RSV was used for the influenza A, B, and RSV positive samples. All samples underwent one freeze‐thaw cycle before use in the validation of the SARS‐CoV‐2/Flu/RSV cartridge. Three samples included were co‐infections. Two of the samples selected as influenza A positive were also RSV positive. One influenza A sample was also SARS‐CoV‐2 positive.
2.2.2. Medical University of Graz
Left‐over routine samples that had been obtained by a deep oropharyngeal swab with the Copan UTM™ (Copan) collection system and had been stored at −80°C were used for this study. All SARS‐CoV‐2 diagnostics were performed using the Xpress Xpress SARS‐CoV‐2 cartridge. Influenza A and B was tested using the Influenza A/B R‐GENE® (bioMerieux SA) assay, RSV using the RSV/hMPV R‐GENE® (bioMerieux) assay. After extraction on the NicliSENS® EMAG® (bioMerieux) platform using the specific B protocol, amplification and detection were performed on the LC 480 II (Roche Diagnostics International Ltd.). Additionally, the CELL Control R‐GENE® (bioMerieux) kit was used for each sample. This assay includes an amplification premix detecting the human hypoxanthine phosphor‐ribosyl transferase 1 gene and thus checking for the presence of human cells in the sample. Four samples included were coinfections, three were influenza A with RSV and one sample was influenza B with RSV.
2.2.3. Institute of Medical Virology, University of Zurich/University Hospital Zurich
Samples of nasopharyngeal and throat swabs tested for SARS‐CoV‐2 were collected in an in‐house virus transport medium (HEPES, DMEM, FCS, antibiotics) and stored before use at −80°C, samples tested for influenza A/B and RSV were stored at −20°C. All samples underwent one freeze‐thaw cycle before use in the validation of the SARS‐CoV‐2/Flu/RSV cartridge. All samples, SARS‐CoV‐2, influenza A/B, and RSV were tested using Xpert Xpress SARS‐CoV‐2 or Xpert Xpress Flu/RSV. Three out of 10 RSV positive samples were tested previously using an in‐house PCR. 14 Samples with coinfections were not included.
2.2.4. Rennes University Hospital
All samples were nasopharyngeal swabs collected in either TranSwab or eSwab medium and stored at −80°C until use. SARS‐Cov‐2 was tested using Xpert Xpress SARS‐CoV‐2 (singleplex). influenza A/B and RSV were tested using Seegene Allplex respiratory panel 1 (Eurobio) as recommended by the manufacturer. Seven included samples had coinfections as determined using the Seegene technique. Five samples were both influenza A and RSV positive, one sample was influenza B and RSV positive. One sample was influenza A and B positive.
2.3. Xpert Xpress SARS‐CoV‐2/Flu/RSV
The Xpert SARS‐CoV‐2/Flu/RSV assay was performed at each participating institute according to the manufacturer's protocol and using research use only cartridges. Briefly, 300 µl of the sample was added to the cartridge, with a run time of 36 min on a GeneXpert platform. The cartridge performs sample preparation, RNA isolation, reverse transcription, and PCR in one single test, without additional hands‐on time. All discrepant results were repeated if sufficient sample material was available. Targets for SARS‐CoV‐2, influenza A/B, and RSV are similar to the individual cartridges (singleplex SARS‐CoV‐2 and the Flu/RSV). The multiplex assay reports four separate Ct values for SARS‐CoV‐2, Flu A, Flu B, and RSV. The main difference with Xpert SARS‐CoV‐2 is that the multiplex assay reports 1 Ct value for both N2 and E targets (mix) and the singleplex assay reports 2 Ct values (E and N2).
3. STATISTICAL ANALYSIS
Positive (PPA) and negative (NPA) percentage agreement of Xpert Xpress SARS‐CoV‐2/Flu/RSV was calculated using cross tables and analysed using IBM SPSS Statistics, version 25.0. To evaluate the agreement between tests, linear regression analysis with Passing‐Bablok fit, a Shapiro–Wilk test for normality of the difference, and a Bland–Altman plot were calculated using Analyse‐it Software, Ltd. and SPSS (version 25; IBM Corporation). For Xpert Xpress SARS‐CoV‐2 the average of E‐ and N2‐gene Ct values were calculated and compared to the Ct values of the Xpert Xpress SARS‐CoV‐2/Flu/RSV.
4. RESULTS
For all 121 SARS‐CoV‐2 positive samples tested with the new Xpert Xpress SARS‐CoV‐2/Flu/RSV, a PPA of 97.2% and NPA of 100% was observed compared to the reference assay performed at each institute (Table 2). Two discrepant results were found; both samples tested positive with the reference method (Xpert Xpress SARS‐CoV‐2) but negative with the multiplex assay. Upon retesting, one sample tested negative with both assays (Table 3). The other sample showed Ct values of 44.1 (E‐gene) and 42.9 (N2‐gene) with the singleplex assay.
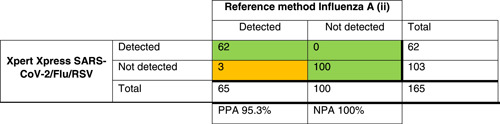
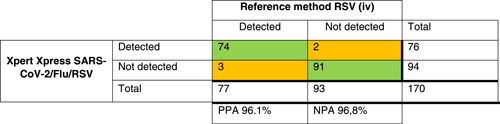
Table 2.
Crosstables comparing Xpert Xpress SARS‐CoV‐2/Flu/RSV to the local method for SARS‐COV‐2 (i), influenza A (ii), influenza B (iii), and RSV(iv) detection
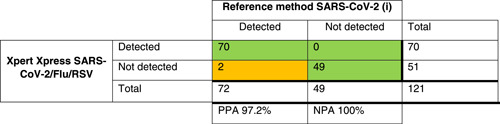
|
|

|
|
Note: The green/orange background indicate discrepancies in the agreement between tests.
Abbreviations: NPA, negative percentage agreement; PPA, positive percentage agreement; RSV, respiratory syncytial virus; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Table 3.
Samples (N = 11 out of 295) with discrepancies between Xpert Xpress SARS‐CoV‐2/Flu/RSV and comparator method
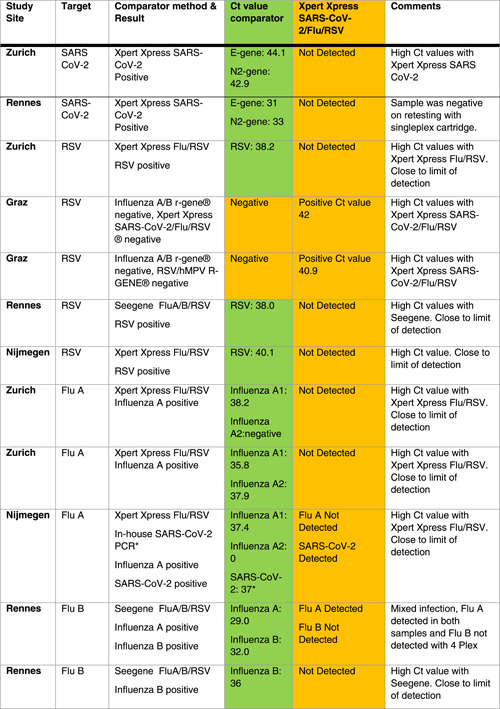
|
Note: Depicted for each target. Ct values of comparator method reported. The green/orange background indicate the details on individual samples showing discrepancies between tests. *One SARS‐CoV‐2 sample was detected using the in‐house PCR and Xpert Xpress SARS‐CoV‐2 as was done for all other samples, performed using COBAS 4800 with RT‐PCR described by Corman et al. 15
Abbreviations: RSV, respiratory syncytial virus; RT‐PCR, reverse‐transcription polymerase chain reaction; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2.
Agreement of SARS‐CoV‐2 Ct values for Xpert Xpress SARS‐CoV‐2 and Xpert Xpress SARS‐CoV‐2/Flu/RSV are shown in Figure 1. The correlation between both tests was high (R 2 = 0.89). Normal distribution of the differences was confirmed using the Shapiro–Wilks test to reliably construct the Bland–Altman plot. The plot depicted in zur2 showed a mean difference of 0.67 (95% CI: 0.0330–1.3133). For Ct values above 30, the multiplex assay revealed higher Ct values compared to the singleplex assay, whereas Ct values below 30 showed an equal distribution (Figure 2).
Figure 1.
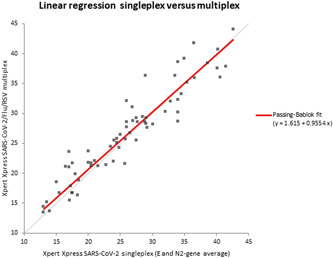
Linear regression of Ct values for SARS‐CoV‐2 singleplex and SARS‐CoV‐2/Flu/RSV multiplex. Slope of 0.9554 and correlation (r) of 0.943. RSV, respiratory syncytial virus; SARS‐CoV‐2, severe acute respiratory syndrome coronavirus 2
Figure 2.
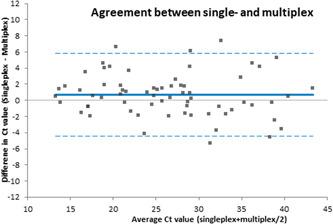
Bland–Altman plot with the difference in Ct value (multiplex–singleplex) on the Y‐axis and average of single‐ and multiplex on the X‐axis
For the other targets, the results are presented in Table 2. The PPA for influenza A was 95.3%, for influenza B 95.6%, and for RSV 96.1%. Discrepant results were found in samples with high Ct values (Table 3).
Fourteen samples showed co‐infections (eight influenza A plus RSV, one influenza A plus SARS‐CoV‐2, four influenza B plus RSV, and one influenza A plus B). Discrepant results were found in two samples, both with very high Ct values (Table 3).
The limit of detection for Influenza A/B R‐GENE®, RSV/hMPV R‐Gene, Seegene FluA/B/RSV is unavailable for these platforms.
5. DISCUSSION
Early and rapid detection of SARS‐CoV‐2 remains vital to reduce the current pandemic and optimize patient care. The Xpert Xpress SARS‐CoV‐2 test has been available since March 2020. In previous seasons, the Xpert Xpress Flu/RSV had been successfully used to rapidly identify influenza and RSV in patients with respiratory infections. 13 , 16 , 17 , 18
For seasons with high influenza and RSV incidence coinciding with the SARS‐CoV‐2 pandemic, the combined SARS‐CoV‐2/Flu/RSV cartridge was developed. Overall, we found the PPA to be above 95% for all tested targets, with the highest percentage for SARS‐CoV‐2 (97.2%). The correlation between testing was found to be high, which is to be expected when using two near‐identical platforms with similar targets. However, the Bland–Altman plot suggested that samples with Ct values above 30 show higher levels in the multiplex compared to the singleplex assay, and this is not equally distributed compared to samples below Ct 30. As the number of samples at this higher Ct value is relatively low, a greater sample size would be needed to identify if this is a common occurrence.
Two samples were discordant for detection of SARS‐CoV‐2 between the singleplex and multiplex assays. One of the samples was negative on retesting in the singleplex assay. This could either mean that freeze‐thawing steps degraded the sample although this would be relatively surprising as an initial Ct value of 33 is relatively low. Alternatively, misclassification or sample mix‐up on initial testing before selection of the sample could be the cause. The other discordant case occurred in a sample with a low viral load, which could be near the limit of detection of the Xpert assay. This finding was similar for samples positive for influenza A, B, and RSV by Xpert Flu/RSV but negative by Xpert SARS‐CoV‐2/Flu/RSV. These discordant cases could be caused by several freeze‐thaw steps as these samples were not retested using the reference method at the same time as testing with the multiplex assay.
In this study, throat and oropharyngeal swabs were used which are not part of the claim for the Xpert assay. However, these sample types are widely used in the field and are, in some countries, part of national guidelines, which is why it is important to assess the performance of the assay in this setting. Additionally, several types of transport medium were used for collection of samples. These were UTM/GLY medium, Copan UTM, and TranSwab or eSwab (Table 1). Validation of transport medium was not performed in the current assay however all transport media has previously been validated for its use on the GeneXpert platform at the individual sites, where it has been used in different cartridges or on other reference platforms (Table 1) without reduction of sensitivity.
Overall, the results obtained from the new SARS‐CoV‐2/Flu/RSV cartridge show good agreement with results obtained from previous testing. The faster turn‐around time of only half an hour is of major significance for fast and accurate treatment as well as for public health decisionmaking during the current SARS‐CoV‐2 pandemic.
CONFLICT OF INTERESTS
The authors have declared that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
Femke Wolters conducted the research, performed analysis, and wrote the manuscript. Maria Grünberg, Michael Huber, Harald H. Kessler, Florian Prüller, Lanja Saleh, Christine Fébreau, Janette Rahamat‐Langendoen, and Vincent Thibault conducted research, analyzed the data locally, and were responsible for proofreading the manuscript. Willem J. G. Melchers supervised the study and proofread the manuscript.
Wolters F, Grünberg M, Huber M, et al. European multicenter evaluation of Xpert® Xpress SARS‐CoV‐2/Flu/RSV test. J Med Virol. 2021;93:5798‐5804. 10.1002/jmv.27111
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- 1. Volz E, Hill V, McCrone JT, et al. Evaluating the effects of SARS‐CoV‐2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2020;184:64‐75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Korber B, Fischer WM, Gnanakaran S, et al. Tracking changes in SARS‐CoV‐2 spike: evidence that D614G increases infectivity of the COVID‐19 virus. Cell. 2020;182(4):812‐827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kong W‐H, Li Y, Peng MW, et al. SARS‐CoV‐2 detection in patients with influenza‐like illness. Nat Microbiol. 2020;5(5):675‐678. [DOI] [PubMed] [Google Scholar]
- 4. Bloom‐Feshbach K, Alonso WJ, Charu V, et al. Latitudinal variations in seasonal activity of influenza and respiratory syncytial virus (RSV): a global comparative review. PLOS One. 2013;8(2):e54445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sullivan SG, Carlson S, Cheng AC, et al. Where has all the influenza gone? The impact of COVID‐19 on the circulation of influenza and other respiratory viruses, Australia, March to September 2020. Euro Surveill. 2020;25(47):2001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fleming DM, Pannell RS, Cross KW. Mortality in children from influenza and respiratory syncytial virus. J Epidemiol Community Health. 2005;59(7):586‐590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ellis SE, Coffey CS, Mitchel EF Jr, Dittus RS, Griffin MR. Influenza‐ and respiratory syncytial virus‐associated morbidity and mortality in the nursing home population. J Am Geriatr Soc. 2003;51(6):761‐767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fleming D, Elliot A, Cross K. Morbidity profiles of patients consulting during influenza and respiratory syncytial virus active periods. Epidemiol Infect. 2007;135(7):1099‐1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kwon YS, Park SH, Kim MA, et al. Risk of mortality associated with respiratory syncytial virus and influenza infection in adults. BMC Infect Dis. 2017;17(1):785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abanses JC, Dowd MD, Simon SD, Sharma V. Impact of rapid influenza testing at triage on management of febrile infants and young children. Pediatr Emerg Care. 2006;22(3):145‐149. [DOI] [PubMed] [Google Scholar]
- 11. Alm E, Broberg EK, Connor T, et al. Geographical and temporal distribution of SARS‐CoV‐2 clades in the WHO European Region, January to June 2020. Euro Surveill. 2020;25(32):2001410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cuadrado‐Payán E, Montagud‐Marrahi E, Torres‐Elorza M, et al. SARS‐CoV‐2 and influenza virus co‐infection. Lancet (London, England). 2020;395(10236):e84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Loeffelholz MJ, Alland D, Butler‐Wu SM, et al. Multicenter evaluation of the cepheid Xpert Xpress SARS‐CoV‐2 test. J Clin Microbiol. 2020;58(8):e00926‐20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hu A, Colella M, Tam JS, Rappaport R, Cheng SM. Simultaneous detection, subgrouping, and quantitation of respiratory syncytial virus A and B by real‐time PCR. J Clin Microbiol. 2003;41(1):149‐154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Corman VM, Landt O, Kaiser M, et al. Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Euro Surveill. 2020;25(3):2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Salez N, Nougairede A, Ninove L, Zandotti C, de Lamballerie X, Charrel RN. Prospective and retrospective evaluation of the Cepheid Xpert® Flu/RSV XC assay for rapid detection of influenza A, influenza B, and respiratory syncytial virus. Diagn Microbiol Infect Dis. 2015;81(4):256‐258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cohen DM, Kline J, May LS, et al. Accurate PCR detection of Influenza A/B and respiratory syncytial viruses by use of Cepheid Xpert Flu+ RSV Xpress Assay in point‐of‐care settings: comparison to Prodesse ProFlu. J Clin Microbiol. 2018;56(2):e01237‐17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wolters F, van de Bovenkamp J, van den Bosch B, et al. Multi‐center evaluation of cepheid xpert® xpress SARS‐CoV‐2 point‐of‐care test during the SARS‐CoV‐2 pandemic. J Clin Virol. 2020;128:104426. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


