Abstract
Background
In Sweden, social restrictions to contain SARS‐CoV‐2 have primarily relied upon voluntary adherence to a set of recommendations. Strict lockdowns have not been enforced, potentially affecting viral dissemination. To understand the levels of past SARS‐CoV‐2 infection in the Stockholm population before the start of mass vaccinations, healthy blood donors and pregnant women (n = 5,100) were sampled at random between 14 March 2020 and 28 February 2021.
Methods
In this cross‐sectional prospective study, otherwise‐healthy blood donors (n = 2,600) and pregnant women (n = 2,500) were sampled for consecutive weeks (at four intervals) throughout the study period. Sera from all participants and a cohort of historical (negative) controls (n = 595) were screened for IgG responses against stabilized trimers of the SARS‐CoV‐2 spike (S) glycoprotein and the smaller receptor‐binding domain (RBD). As a complement to standard analytical approaches, a probabilistic (cut‐off independent) Bayesian framework that assigns likelihood of past infection was used to analyse data over time.
Setting
Healthy participant samples were randomly selected from their respective pools through Karolinska University Hospital. The study was carried out in accordance with Swedish Ethical Review Authority: registration number 2020–01807.
Participants
No participants were symptomatic at sampling, and blood donors were all over the age of 18. No additional metadata were available from the participants.
Results
Blood donors and pregnant women showed a similar seroprevalence. After a steep rise at the start of the pandemic, the seroprevalence trajectory increased steadily in approach to the winter second wave of infections, approaching 15% of all individuals surveyed by 13 December 2020. By the end of February 2021, 19% of the population tested seropositive. Notably, 96% of seropositive healthy donors screened (n = 56) developed neutralizing antibody responses at titres comparable to or higher than those observed in clinical trials of SARS‐CoV‐2 spike mRNA vaccination, supporting that mild infection engenders a competent B‐cell response.
Conclusions
These data indicate that in the first year since the start of community transmission, seropositivity levels in metropolitan in Stockholm had reached approximately one in five persons, providing important baseline seroprevalence information prior to the start of vaccination.
Keywords: antibody testing, COVID‐19, population immunity, SARS‐CoV‐2, serology, seroprevalence, Sweden
Introduction
Densely populated areas – such as the Stockholm region of 2.37 million people (975,000 within city limits) – have facilitated the spread of SARS‐CoV‐2. Evidence suggests that transmission can be curtailed by imposing restrictions on leisure and business activities, as well as by mask usage and contact tracing [1, 2, 3, 4]. In contrast to most comparable countries, Sweden has favoured a strategy in which individuals are encouraged to adhere to a set of basic public health recommendations, while society has remained largely open. Only recently – faced with a winter second wave – did the government opt for earlier closing hours for some businesses. The country has reported a significantly higher burden on the public healthcare system to date than its Scandinavian neighbours (Fig. 1a), and there are many reasons for why this might be the case.
Fig. 1.
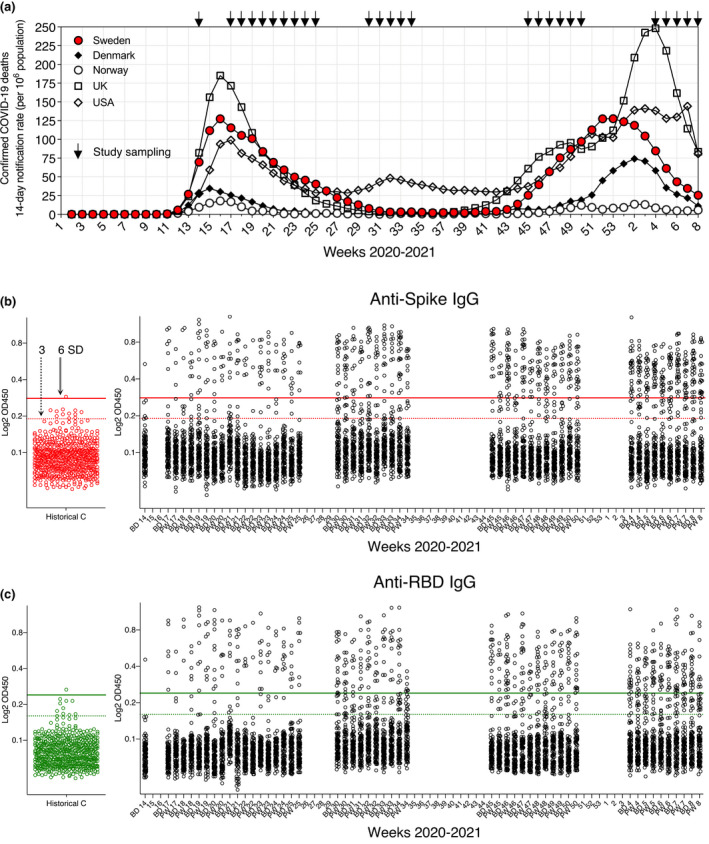
SARS‐CoV‐2 seropositivity estimates in Stockholm: March 2020–February 2021. (a) Population‐adjusted COVID‐19 deaths for selected countries during the pandemic. (b) Anti‐S IgG responses in blood donors (BD), pregnant women (PW) and n = 595 historical control (C) sera. 100 BD and 100 PW samples were analysed per sampling week alongside negative controls (cut‐out plot red points). Conventional 3 and 6 SD (from the mean of negative control values) assay cut‐offs are shown as dashed and solid lines, respectively. (c) Anti‐RBD IgG responses in BD, PW and n = 595 historical control sera (cut‐out plot green points). Conventional 3 and 6 SD assay cut‐offs are shown as dashed and solid lines, respectively
Serological enumeration of past infections is critical for estimating viral spread and understanding characteristics of the adaptive immune response to an emerging pathogen [5]. Antibody testing is also important for optimal planning of vaccination campaigns (especially when doses are limited) and could help demonstrate ‘immunity’ on official documentation. However, as illustrated by the current pandemic, not all antibody tests are of equal sensitivity and specificity (S&S), and closer scrutiny of assays is needed to improve public health measures and our understanding of COVID‐19 [6].
To monitor seropositivity in the Stockholm region, we developed highly sensitive and specific SARS‐CoV‐2 antibody tests based on the native‐like spike glycoprotein [7] (alongside a diagnostic clinical laboratory responsible for monitoring seropositivity during the pandemic [8]), as well as novel cut‐off‐independent statistical methods, and applied them to random, healthy adults in the region throughout 2020 and early 2021. Such approaches, which improve upon strictly thresholding a continuous variable, are critical for accurate seropositivity estimates at individual and population levels. This is especially important in the case of SARS‐CoV‐2, as asymptomatic/mild infections generate antibody responses of varying titres that can be difficult to classify [8, 9, 10, 11], and the knowledge that individual titres generally wane from peak levels over time [12, 13, 14, 15].
We chose to survey anti‐S IgG responses as these are the best indicators of past SARS‐CoV‐2 infection at the population level (i.e. present in >91% of PCR‐positive cases) [16, 17]. Spike harbours the ACE2 receptor‐binding domain (RBD) and is the major target of the neutralizing antibody response, central to vaccine efforts. For example, anti‐nucleocapsid (N) antibodies were not detectable in a subset of S‐seropositive individuals [8, 18, 19, 20]. For both S, N and RBD, the levels of antibodies in circulation were shown to increase with worsening disease severity [9, 11, 21], as also reported for SARS‐CoV and MERS [22, 23].
Blood donors and pregnant women, studied here, are important clinical groups, and pregnant women require further study in relation to infectious disease [24, 25]. Moreover, both groups represent good and accessible proxies for adult population health, being generally working age, mobile members of society – without being enriched for individuals at especially high‐risk of SARS‐CoV‐2 infection, such as healthcare workers or public transportation employees, where seroprevalence may be higher [26]. Similarly, seroprevalence may be higher in children and lower in the elderly – with complementary studies much needed.
All healthy individuals in this study (n = 5,100) were over the age of 18 and symptom‐free at sample collection. For ethical reasons, their ages and genders are not revealed, nor is it known whether any participant previously tested positive for the virus. Thus, while we here provide an unbiased assessment of past SARS‐CoV‐2 infection in two important groups in Stockholm in response to natural infection and before the start of mass adult vaccinations, future studies in other cohorts are needed to generate more integrated data to guide public health strategies.
Materials and methods
Human samples and ethical declaration
Anonymized samples (n = 5,100 total) from blood donors (n = 100/sampling week) and pregnant women (n = 100/sampling week) were randomly selected from their respective pools through the Department of Clinical Microbiology, Karolinska University Hospital (KUH). During 2020, 18,963 pregnant women were under the care of KUH, averaging 365 per week, of which 100 (~30%) were selected at random from each sampling week. Blood donations totalled 66,596 during 2020, averaging 1,281 per week and here weekly sampling approximated 8% of the pool. No study participant was analysed more than once over the course of the study. Blood donor and pregnant women samples were not collected during weeks 26–29, 35–45 and 51–53 of 2020 and 1–3 of 2021 for logistical reasons. Samples were collected from 26 weeks out of the 47 weeks the study ran. Blood donors (n = 595) collected through the same channels one year previously (Spring 2019) were randomly selected for use as assay negative serum controls.
No metadata, such as age or sex information, were available for the samples in the study. Blood donors must be over 50 kg and over the age of 18, while no upper age limit is established in Stockholm. Pregnant women were sampled as part of routine screening for infectious diseases during the first trimester of pregnancy. Whether a study participant had previously tested positive for SARS‐CoV‐2 PCR is unknown. Blood donors are required to be healthy for a minimum of two weeks before a donation and all participants reported symptom‐free at sample collection. We cannot exclude that a very small number of blood donor samples from 2021 were from individuals who had received a COVID‐19 vaccine. Healthcare workers in the region were receiving first doses at the time of writing, along with elderly persons; however, mass vaccinations of adults (including pregnant women) had not yet begun [27, 28].
The use of study samples was approved by the Swedish Ethical Review Authority (registration no. 2020–01807). Covid‐19 mortality data for Sweden was sourced from the European Centre for Disease Control and Prevention (European Union) (https://www.ecdc.europa.eu/en/covid‐19/data) and were current at the time of publication.
Serum sample processing
Blood samples were collected by the attending clinical team and serum isolated by the Department of Clinical Microbiology. Samples were anonymized, barcoded and stored at −20°C until use. Serum samples were not heat‐inactivated for ELISA protocols.
SARS‐CoV‐2 antigen generation
The plasmid for expression of the SARS‐CoV‐2 prefusion‐stabilized spike ectodomain with a C‐terminal T4 fibritin trimerization motif was obtained from Wrapp et al and produced as in Hanke et al [29]. Briefly, the plasmid was used to transiently transfect FreeStyle 293F cells using FreeStyle MAX reagent (Thermo Fisher Scientific). The ectodomain was purified from filtered supernatant on Streptactin XT resin (IBA Lifesciences), followed by size‐exclusion chromatography on a Superdex 200 in 5 mM Tris pH 8, 200 mM NaCl. The RBD domain (RVQ–QFG) of SARS‐CoV‐2 was cloned upstream of a sortase A recognition site (LPETG) and a 6xHIS tag and expressed in 293F cells as described above. RBD‐HIS was purified from filtered supernatant on His‐Pur Ni‐NTA resin (Thermo Fisher Scientific), followed by size‐exclusion chromatography on a Superdex 200.
Anti‐SARS‐CoV‐2 ELISA
96‐well ELISA plates (Nunc MaxiSorp) were coated with freshly prepared SARS‐CoV‐2 S trimers or the RBD (100 µl of 1 ng/µl) in PBS for 15 h at 4°C. Plates were washed six times with PBS–Tween‐20 (0.05%) and blocked using PBS‐5% no‐fat milk (Sigma). Human serum samples were thawed at room temperature, diluted, vortexed and incubated in blocking buffer for 1 h (4°C) before plating to block non‐specific binding. Serum samples were incubated for 15 h at 4°C to allow low‐affinity binding interactions, before washing as before. Secondary HRP‐conjugated anti‐human antibodies were diluted in blocking buffer and incubated with samples for 1 hour at 4°C. Plates were washed a final time before development with TMB Stabilized Chromogen kept at 4°C (Invitrogen). The reaction was stopped using 1 M sulphuric acid and optical density (OD) values were measured at 450 nm using an Asys Expert 96 ELISA reader (Biochrom Ltd.). Secondary antibodies (from Southern Biotech) and dilutions used were as follows: goat anti‐human IgG (2014‐05) at 1:10,000. All assays were developed for their fixed time, and negative control samples were run alongside test samples in all assays. Anti‐SARS‐CoV‐2 S and RBD IgG were detectable at up to 1:20,000 serum dilution using this assay [8], and all study samples were here run at 1:100 dilution.
In vitro virus neutralization assay
Pseudotyped viruses were generated by the co‐transfection of HEK293T cells with plasmids encoding the SARS‐CoV‐2 spike protein harbouring an 18 amino acid truncation of the cytoplasmic tail; a plasmid encoding firefly luciferase; a lentiviral packaging plasmid (Addgene 8455) using Lipofectamine 3000 (Invitrogen). Media was changed 12–16 hours post‐transfection and pseudotyped viruses harvested at 48 and 72 hours, filtered through a 0.45‐µm filter and stored at −80°C until use. Pseudotyped neutralization assays were adapted from protocols validated to characterize the neutralization of HIV, but with the use of ACE2‐expressing HEK293T cells. Briefly, pseudotyped viruses sufficient to generate ~100,000 RLUs were incubated with serial dilutions of heat‐inactivated serum for 60 min at 37°C. Approximately 15,000 HEK293T‐ACE2 cells were then added to each well and the plates incubated at 37°C for 48 hours. Luminescence was measured using Bright‐Glo (Promega) according to the manufacturer’s instructions on a GM‐2000 luminometer (Promega) with an integration time of 0.3s. The neutralization assay limit of detection was at 1:45 serum dilution.
Probabilistic seroprevalence estimations
Prior to analysis, each sample OD was log‐transformed and standardized by dividing by the mean OD of ‘no sample control’ wells on that plate or other plates run on the same day – to reduce batch variation. This resulted in more similar distributions for 2019 blood donor samples with 2020 blood donors and pregnant volunteers. Our Bayesian approach is presented in detail in Christian and Murrell [30]. Briefly, we used a logistic regression over anti‐RBD and anti‐S training data (n = 595 historical blood donor controls and n = 138 SARS‐CoV‐2 PCR+ individuals across the clinical spectrum – all of whom developed anti‐S IgG and 97% did so against the RBD) to model the relationship between the ELISA measurements and the probability that a sample is antibody‐positive. We adjusted for the training data class proportions and used these adjusted probabilities to inform the seroprevalence estimates for each time point. Given that the population seroprevalence cannot increase dramatically from one week to the next, we constructed a prior over seroprevalence trajectories using a transformed Gaussian Process and combined this with the individual class‐balance adjusted infection probabilities for each donor to infer the posterior distribution over seroprevalence trajectories. We compared our Bayesian approach to the output of more established probabilistic algorithms (specifically, an ensemble learner from support vector machines (SVM) and linear discriminant analysis (LDA) – SVM‐LDA) that we have previously developed for ELISA measurements.
The sensitivity and specificity (S&S) of our IgG ELISA assays were as follows:
Spike 3SD:100% (95% CI [97.5–100.0]) & 99.0% (95% CI [98.6–99.0])
Spike 6SD:100% (95% CI [97.5–100.0]) & 99.9% (95% CI [99.6–100.0])
RBD 3SD:100% (95% CI [97.5–100.0]) & 99.0% (95% CI [98.4–99.4])
RBD 6SD:98.0% (95% CI [94.2–99.3]) & 99.9% (95% CI [99.6–100.0])
SVM‐LDA:99.3% (95% CI [96.3–100.0) & 100.0% (95% CI [99.8–100.0])
To compute confidence intervals for S&S, we dichotomized predictions of seropositivity at prob > 0.5 or <= 0.5 and computed average sensitivity, specificity and 95% confidence intervals for each fold in the cross‐validation via Wilson's method before averaging over all folds. The Bayesian model does not make assignments of seropositivity to individual samples, but rather integrates over the uncertainty in relationship between infection and OD450, as well as the uncertainty due to sampling, to provide population‐level seroprevalence estimates. Thus, S&S cannot be calculated for this approach.
Results
Blood donor (BD) and pregnant women (PW) serum samples (n = 100 of each per sampling week) were selected at random from their respective pools and the IgG response against SARS‐ CoV‐2 S glycoprotein trimers and the smaller RBD subunit was measured in all sera using established ELISA assays extensively validated using sera from confirmed infections [8] (Fig. 1b,c); anti‐S and RBD responses are highly correlated, with lower titres generally observed for the smaller RBD.
Test samples were run alongside historical (SARS‐CoV‐2‐negative) control sera (n = 595 blood donors from spring 2019) over the course of the study. Seropositivity for S and RBD according to conventional 3 or 6 standard deviations (SD) (from the mean of negative control sera) assay cut‐offs is presented in Table 1 and Fig. 2a. Notably, seropositivity was not significantly different between blood donors and pregnant women over the study period (Fig. 2b,c).
Table 1.
IgG seropositivity to S and RBD in blood donors and pregnant women following virus emergence
| Weeks 2020–2021 | Bayesian estimate | S IgG (3SD) | RBD IgG (3SD) | S IgG (6SD) | RBD IgG (6SD) |
|---|---|---|---|---|---|
| 2020 | |||||
| 14: 30 March—5 April | 2.4 | 2.5 | 0.5 | 0.5 | 0.5 |
| Sampling gap | |||||
| 17: 20–27 April | 4.8 | 6.0 | 5.0 | 4.5 | 4.0 |
| 18: 27 April–3 May | 5.4 | 8.0 | 5.0 | 5.0 | 4.0 |
| 19: 4–10 May | 5.9 | 11.5 | 8.0 | 9.0 | 8.0 |
| 20: 11–17 May | 6.3 | 8.0 | 9.0 | 7.0 | 8.0 |
| 21: 18–24 May | 6.7 | 12.0 | 10.0 | 8.0 | 7.5 |
| 22: May 25–31 May | 7.0 | 4.5 | 3.5 | 4.0 | 3.5 |
| 23: 1–7 June | 7.3 | 10.5 | 9.0 | 9.5 | 7.0 |
| 24: 8–14 June | 7.8 | 8.0 | 8.0 | 6.5 | 7.0 |
| 25: 15–21 June | 8.3 | 8.5 | 7.0 | 7.5 | 7.0 |
| Sampling gap | |||||
| 30: 20–26 July | 11.4 | 22.0 | 20.5 | 17.0 | 14.5 |
| 31: 27 July–2 August | 11.9 | 14.5 | 13.5 | 10.5 | 9.0 |
| 32: 3–9 August | 12.3 | 18.0 | 16.0 | 13.5 | 10.5 |
| 33: 10–16 August | 12.5 | 15.5 | 12.5 | 12.5 | 10.0 |
| 34: 17–23 August | 12.7 | 20.0 | 20.0 | 16.5 | 9.5 |
| Sampling gap | |||||
| 45: 2–8 November | 14.1 | 15.5 | 14.0 | 12.5 | 11.0 |
| 46: 9–15 November | 14.2 | 21.0 | 17.0 | 16.5 | 13.5 |
| 47: 16–22 November | 14.3 | 17.5 | 16.0 | 17.0 | 12.0 |
| 48: 23–29 November | 14.5 | 20.5 | 18.0 | 16.0 | 12.5 |
| 49: 30 November–6 December | 14.7 | 16.5 | 14.5 | 12.5 | 12.0 |
| 50: 7–13 December | 14.8 | 20.0 | 17.5 | 18.0 | 15.5 |
| 2021 | Sampling gap | ||||
| 4: 25–31 January | 18.2 | 28.5 | 22.0 | 23.5 | 15.0 |
| 5: 1–7 February | 18.5 | 23.5 | 23.0 | 21.5 | 13.0 |
| 6: 8–14 February | 18.8 | 25.5 | 19.5 | 19.0 | 15.5 |
| 7: 15–21 February | 19.0 | 28.5 | 25.0 | 25.5 | 16.5 |
| 8: 22–28 February | 19.2 | 29.5 | 26.0 | 25.5 | 16.5 |
Fig. 2.
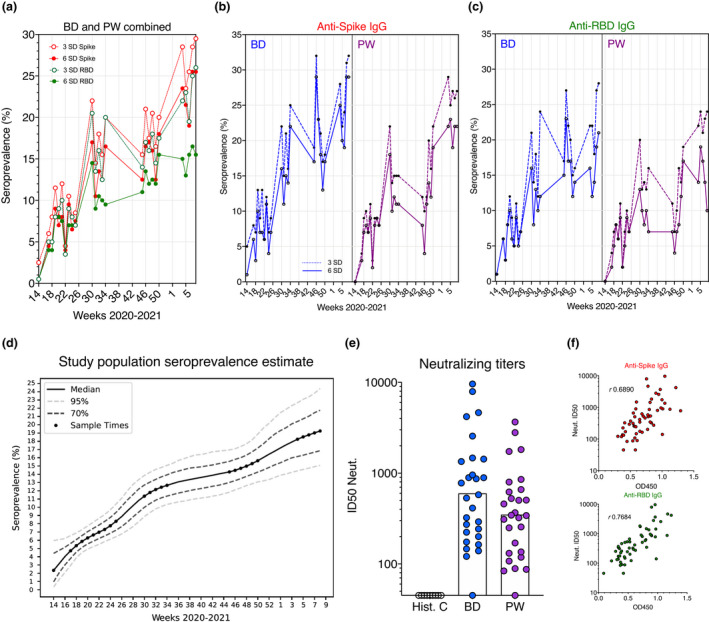
SARS‐CoV‐2 seropositivity estimates in Stockholm: March 2020–February 2021. (a) Seropositivity estimates in BD and PW combined, according to 3 and 6 SD assay cut‐offs. (b) Spike seropositivity in BD and PW according to 3 and 6 SD assay cut‐offs. (c) RBD seropositivity in BD and PW according to 3 and 6 SD assay cut‐offs. (d) Cut‐off‐independent Bayesian modelling of population seropositivity. (e) In vitro pseudotyped virus neutralizing titres in a subset (n =56) of antibody‐positive BD and PW. Bars represent the geometric mean. (f) Binding and neutralization – for samples in (e) – are highly correlated
However, the many measurements between the 3 and 6 SD cut‐offs for both or a single antigen (Fig. 1b,c) pose a problem when assigning case to low values, uncertainty that can significantly skew seroprevalence estimates and is undesirable at the individual level. Therefore, to provide an accurate seropositivity estimate for our population and to model seropositivity changes over time, we developed a cut‐off‐independent, probabilistic Bayesian framework that models the log odds that a sample is antibody‐positive based on anti‐S and anti‐RBD IgG responses in training data, in this case SARS‐CoV‐2 PCR+ COVID‐19 patients (n = 136) across the clinical spectrum (including asymptomatic/mild cases) [8].
Using this more quantitative approach that better considers the wide range of responses present in the population and shares information between sampling weeks, we found seropositivity to increase sharply at the start of the pandemic (Fig. 2d). By the time, the COVID‐19 death rate in the country was at low levels during August, following the first wave, the seroprevalence trajectory increased at a steady, but slower rate in approach to the winter second wave (Fig. 2d) – in agreement with continued viral spread in the Stockholm population during a summer recess in cases and mortality, and consistent with persistent individual antibody responses over a 9‐month period [31].
By week 50 (13 December 2020), our probabilistic approach identified 14.8% (95% Bayesian CI [12.2–18.0]) of the cohort to have been previously infected (Table 1 and Table S1). The seroprevalence level again increased more rapidly between mid‐December and the end of February 2021, consistent with the winter peak in mortality and infections, and reached 19.2% (95% Bayesian CI [15.1–24.4]) of the population by last sampling. Thus, approximately one in five healthy adults in Stockholm showed evidence of past natural SARS‐CoV‐2 infection before the start of mass adult vaccinations in the country. An equal‐weighted probabilistic SVM‐LDA learner that we had previously optimized for ELISA measurements showed highly consistent results (Table S2).
Importantly, 96% of seropositive blood donors and pregnant women randomly subsampled (n = 56 from March to May 2020) had virus‐neutralizing responses in their sera (ID50=600; 95% CI [357 – 1,010] and ID50=350; 95% CI [228–538], respectively, Fig. 2e,f), with titres comparable to those engendered by recently approved COVID‐19 mRNA vaccines that were shown to be protective in clinical phase 3 trials [32, 33]. These observations support that asymptomatic/mild infection generates an antibody response that provides a degree of protection upon re‐exposure, although inter‐individual heterogeneity, environmental factors and different SARS‐CoV‐2 variants will play a role in individual outcomes.
Discussion
To characterize immunological responses to SARS‐CoV‐2 and safeguard public health, it is critical to monitor the level of population immunity after natural infection, especially in settings with different public health measures – allowing for concurrent and retroactive evaluation of different strategies. As Sweden has taken a unique public health approach to mitigate the effects of the virus, data from the country provide an important contrast to comparable settings.
Serology is amenable to studies of large cohorts and remains the gold standard for determining previous exposure to pathogens. Several studies have highlighted the protective role antibodies play in controlling SARS‐CoV‐2 infection in humans [34, 35] and animal models [36, 37], and potent neutralizing and convergent antibodies were rapidly isolated from infected donor samples [38, 39, 40]. Indeed, mounting evidence suggests that most persons (>91%) previously infected with SARS‐CoV‐2 develop virus‐specific antibodies, including following mild infections [8, 16, 17, 31]. Critical ongoing and future research is required to determine the duration of serological responses and B‐cell memory in those infected, as well as following vaccination.
Early research has shown that the majority of mildly infected individuals maintain detectable virus‐specific B‐cell responses for at least 8–10 months [17, 31]. Together with the steadily increasing seropositivity observed in this study and elsewhere, such as New York City [41], these findings help allay early concerns that immunity waned in the short term, although individuals with mild infection can have antibody titres close to the assay boundary. Data suggest that ~10% of SARS‐CoV‐2 seropositive individuals can lose detectable titres 10 months post‐PCR test [12, 15, 17], although this proportion remains to be reported in larger and different cohorts. Therefore, the levels of past infection in this cohort may be slightly higher than we report. Anti‐SARS‐CoV antibodies have been reported to persist for 3 years following more severe infection [42, 43], while detectable responses to seasonal coronaviruses generally wane within 1–2 years and immunity is affected by rhythms in their circulation [44, 45, 46]. Importantly, as the activated B‐cell pool and antibody response contracts following viral clearance (and declines in function with age [47, 48]), the identification of low but persistent antibody responses (that may be highly effective) remains a challenge using conventional serological methods.
Notably, we observed virus‐neutralizing titres in our cohorts to be comparable to those engendered by the first mRNA vaccines of Pfizer/BioNTech [33] and Moderna [34], supporting that natural infection generates neutralizing immunity to the infectious strain, as do studies of healthcare workers [35] and elderly care home residents and staff [49], in whom re‐infection rates were substantially reduced. The potential endemicity and continued evolution of SARS‐CoV‐2 may alter the nature of protective immunity (e.g. possible re‐infection with a different strain), and knowledge of strain‐specific antibody responses should be used to optimally coordinate vaccine interventions and inform public health measures.
Epidemiologically, our data indicate that approximately one year since virus emergence in Sweden, approximately one in five adults in the categories studied here (blood donors and pregnant women) in Stockholm had been previously infected by the virus. The situation in Stockholm is like that observed in New York City, USA, where despite high rates of infection and mortality [50], herd immunity was not attained within six months of the outbreak, with seropositivity reaching ~20% of the general adult population [41]. Despite an estimated >60% IgG seroprevalence after the first wave [51], Manaus, Brazil, was also not spared a severe second wave of infections [52]. As SARS‐CoV‐2 variants continue to emerge [53], seroprevalence measurements reported from many countries highlight a continued need to curtail viral transmission through social restrictions and effective vaccination programmes.
Our study supports that Stockholm has suffered a higher number of infections (following the first wave) than most European locations studied [54, 55, 56, 57, 58, 59], in agreement with high per capita mortality. However, direct comparisons between sites are complicated by differences in the demographics of the study subjects as well as the S&S of the assay used for antibody detectiony [60]. Longitudinal studies over longer timescales and in different cohorts are urgently needed for improved understanding of population immune responses after SARS‐CoV‐2 infection and COVID‐19 vaccination.
Conclusions
These data highlight a relatively high SARS‐CoV‐2 seroprevalence in Stockholm, Sweden, following one year of community transmission. Population serology informs disease epidemiology and public health approaches and can help target vaccines where they are most efficacious.
Conflict of interest
The study authors declare no competing interests related to the work.
Supporting information
Table S1. Bayesian learner.
Table S2. SVM‐LDA learner.
Acknowledgements
We would like to thank the study participants and attending clinical teams for making the research possible. Funding for this work was provided by a Distinguished Professor grant from the Swedish Research Council (agreement 2017‐00968) and the National Institutes of Health, USA (agreement 400 SUM1A44462‐02), awarded to GKH. CW received funding from the Wellcome Trust (WT107881) and Medical Research Council, UK (MC_UP_1302/5). Additional funding was made possible by the European Union‐funded CoroNAb project (coordination number 101003653). For Open Access, the author has applied a CC BY public copyright licence to this manuscript.
Castro Dopico X, Muschiol S, Christian M, Hanke L, Sheward DJ, Grinberg NF, et al. Seropositivity in blood donors and pregnant women during the first year of SARS‐CoV‐2 transmission in Stockholm, Sweden. J Intern Med 2021; 290: 666–676.
Contributor Information
X. Castro Dopico, Email: xaquin.castro.dopico@ki.se.
G. B. Karlsson Hedestam, Email: Gunilla.Karlsson.Hedestam@ki.se.
Data availability statement
Data generated as part of the study, along with custom code for statistical analyses, are openly available via our GitHub repositories: https://github.com/MurrellGroup/DiscriminativeSeroprevalence/; https://github.com/chr1swallace/seroprevalence‐paper.
References
- 1. Ji T, Chen H‐L, Xu J, Wu L‐N, Li J‐J, Chen K, et al. Lockdown contained the spread of 2019 novel coronavirus disease in Huangshi City, China: Early epidemiological findings. Clin Infect Dis. 2020;71(6):1454–60. 10.1093/cid/ciaa390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chu DK, Akl EA, Duda S, Solo K, Yaacoub S, Schünemann HJ, et al. Physical distancing, face masks, and eye protection to prevent person‐to‐person transmission of SARS‐CoV‐2 and COVID‐19: a systematic review and meta‐analysis. Lancet. 2020;395(10242):1973–87. 10.1016/S0140-6736(20)31142-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gandhi M, Beyrer C, Goosby E. Masks do more than protect others during COVID‐19: Reducing the inoculum of SARS‐CoV‐2 to protect the wearer. J Gen Intern Med. 2020;35(10):3063–6. 10.1007/s11606-020-06067-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kucharski AJ, Klepac P, Conlan AJK, Kissler SM, Tang ML, Fry H, et al. Effectiveness of isolation, testing, contact tracing, and physical distancing on reducing transmission of SARS‐CoV‐2 in different settings: a mathematical modelling study. Lancet Infect. Dis. 2020;20:1151–60. 10.1016/S1473-3099(20)30457-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Winter AK, Hegde ST. The important role of serology for COVID‐19 control. Lancet Infect Dis. 2020;20(7):758–9. 10.1016/S1473-3099(20)30322-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Özçürümez MK, Ambrosch A, Frey O, Haselmann V, Holdenrieder S, Kiehntopf M, et al. SARS‐CoV‐2 antibody testing—questions to be asked. J Allergy Clin Immunol. 2020;146(1):35–43. 10.1016/j.jaci.2020.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh C‐L, Abiona O, et al. Cryo‐EM structure of the 2019‐nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–3. 10.1126/science.aax0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Castro Dopico X, Hanke L, Sheward DJ, Muschiol S, Aleman S, Grinberg NF, et al. Probability‐based approaches for identifying low‐titer antibody responses against SARS‐CoV‐2. medRxiv. 2021. 10.1101/2020.07.17.20155937. [DOI] [Google Scholar]
- 9. Cervia C, Nilsson J, Zurbuchen Y, Valaperti A, Schreiner J, Wolfensberger A, et al. Systemic and mucosal antibody responses specific to SARS‐CoV‐2 during mild versus severe COVID‐19. J Allergy Clin Immunol. 2021;147(2):545–57.e9. 10.1016/j.jaci.2020.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Marklund E, Leach S, Axelsson H, Nyström K, Norder H, Bemark M, et al. Serum‐IgG responses to SARS‐CoV‐2 after mild and severe COVID‐19 infection and analysis of IgG non‐responders. PLoS One. 2020;15(10):e0241104. 10.1371/journal.pone.0241104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shrock E, Fujimura E, Kula T, Timms RT, Lee I‐H, Leng Y, et al. Viral epitope profiling of COVID‐19 patients reveals cross‐reactivity and correlates of severity. Science. 2020;370(6520):eabd4250. 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS‐CoV‐2 infection in humans. Nat Microbiol. 2020;5(12):1598–607. 10.1101/2020.07.09.20148429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gaebler C, Wang Z, Lorenzi JCC, Muecksch F, Finkin S, Tokuyama M, et al. Evolution of antibody immunity to SARS‐CoV‐2. Nature. 2021;591(7851):639–44. 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Choe P, Kang CK, Suh HJ, Jung J, Song KH, Bang JH, et al. Waning antibody responses in asymptomatic and symptomatic SARS‐CoV‐2 infection. Emerg Infect Dis. 2021;27(1):327–9. 10.3201/eid2701.203515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ibarrondo FJ, Fulcher JA, Goodman‐Meza D, Elliott J, Hofmann C, Hausner MA, et al. Rapid decay of anti–SARS‐CoV‐2 antibodies in persons with mild covid‐19. N Engl J Med. 2020;383(11):1085–7. 10.1056/nejmc2025179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, et al. Humoral Immune Response to SARS‐CoV‐2 in Iceland. N Engl J Med. 2020;383(18):1724–34. 10.1056/nejmoa2026116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vanshylla K, Di Cristanziano V, Kleipass F, Dewald F, Schommers P, Gieselmann L, et al. Kinetics and correlates of the neutralizing antibody response to SARS‐CoV‐2. BioRxiv. 2021. 10.1101/2021.01.26.428207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Long QX, Tang X‐J, Shi Q‐L, Li Q, Deng H‐J, Yuan J, et al. Clinical and immunological assessment of asymptomatic SARS‐CoV‐2 infections. Nat Med. 2020;26(8):1200–4. 10.1038/s41591-020-0965-6. [DOI] [PubMed] [Google Scholar]
- 19. Weisberg SP, Connors TJ, Zhu Y, Baldwin MR, Lin W‐H, Wontakal S, et al. Distinct antibody responses to SARS‐CoV‐2 in children and adults across the COVID‐19 clinical spectrum. Nat Immunol. 2021;22(1):25–31. 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wu J, Liang B, Chen C, Wang H, Fang Y, Shen S, et al. SARS‐CoV‐2 infection induces sustained humoral immune responses in convalescent patients following symptomatic COVID‐19. Nat Comms. 2021;12(1):1813. 10.1038/s41467-021-22034-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Röltgen K, Powell AE, Wirz OF, Stevens BA, Hogan CA, Najeeb J, et al. Defining the features and duration of antibody responses to SARS‐CoV‐2 infection associated with disease severity and outcome. Sci Immunol. 2020;5(54):eabe0240. 10.1126/sciimmunol.abe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lee N, Chan PKS, Ip M, Wong E, Ho J, Ho C, et al. Anti‐SARS‐CoV IgG response in relation to disease severity of severe acute respiratory syndrome. J Clin Virol. 2006;35(2):179–84. 10.1016/j.jcv.2005.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Okba NMA, Raj VS, Widjaja IVY, GeurtsvanKessel CH, de Bruin E, Chandler FD, et al. Sensitive and specific detection of low‐level antibody responses in mild Middle East respiratory syndrome coronavirus infections. Emerg Infect Dis. 2019;25(10):1868–77. 10.3201/eid2510.190051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kourtis AP, Read JS, Jamieson DJ. Pregnancy and infection. N Engl J Med. 2014;370(23):2211–8. 10.1056/NEJMra1213566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jamieson DJ, Theiler RN, Rasmussen SA. Emerging infections and pregnancy. Emerg Infect Dis. 2006;12(11):1638–43. 10.3201/eid1211.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rudberg AS, Havervall S, Månberg A, Jernbom Falk A, Aguilera K, Ng H, et al. SARS‐CoV‐2 exposure, symptoms and seroprevalence in healthcare workers in Sweden. Nat Commun. 2020;11(1):5064. 10.1038/s41467-020-18848-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Folkhälsomyndigheten ( Public Health Agency of Sweden). Rekommendationer om prioritetsordning för vaccination mot covid‐19, 2021. https://www.folkhalsomyndigheten.se/smittskydd‐ber.
- 28. Folkhälsomyndigheten ( Public Health Agency of Sweden). Statistik för vaccination mot covid‐19, 2021. https://www.folkhalsomyndigheten.se/smittskydd‐ber.
- 29. Hanke L, Vidakovics Perez L, Sheward DJ, Das H, Schulte T, Moliner‐Morro A, et al. An alpaca nanobody neutralizes SARS‐CoV‐2 by blocking receptor interaction. Nat Commun. 2020;11(1):4420. 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Christian M, Murrell B. Discriminative bayesian serology: counting without cutoffs. BioRxiv. 2020. 10.1101/2020.07.14.202150. [DOI] [Google Scholar]
- 31. Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, et al. Immunological memory to SARS‐CoV‐2 assessed for greater than eight months after infection. Science. 2021;371(6529):eabf4063. 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mulligan MJ, Lyke KE, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Phase I/II study of COVID‐19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–93. 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 33. Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA Vaccine against SARS‐CoV‐2 — Preliminary Report. N Engl J Med. 2020;383(20):1920–31. 10.1056/nejmoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Addetia A, Crawford KHD, Dingens A, Zhu H, Roychoudhury P, Huang M‐L, et al. Neutralizing antibodies correlate with protection from SARS‐CoV‐2 in humans during a fishery vessel outbreak with a high attack rate. J Clin Microbiol. 2020;58(11):e02107–e02120. 10.1128/JCM.02107-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lumley SF, O'Donnell D, Stoesser NE, Matthews PC, Howarth A, Hatch SB, et al. Antibody Status and Incidence of SARS‐CoV‐2 Infection in Health Care Workers. N Eng J Med. 2021;384:533–40. 10.1101/2020.11.18.20234369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chandrashekar A, Liu J, Martinot AJ, McMahan K, Mercado NB, Peter L, et al. SARS‐CoV‐2 infection protects against rechallenge in rhesus macaques. Science. 2020;369(6505):812–7. 10.1126/science.abc4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Deng W, Bao L, Liu J, Xiao C, Liu J, Xue J, et al. Primary exposure to SARS‐CoV‐2 protects against reinfection in rhesus macaques. Science. 2020;369(6505):818–23. 10.1126/science.abc5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robbiani DF, Gaebler C, Muecksch F, Lorenzi JCC, Wang Z, Cho A, et al. Convergent antibody responses to SARS‐CoV‐2 infection in convalescent individuals. Nature. 2020;584(7821):437–42. 10.1101/2020.05.13.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wajnberg A, Amanat F, Firpo A, Altman DR, Bailey MJ, Mansour M, et al. Robust neutralizing antibodies to SARS‐CoV‐2 infection persist for months. Science. 2020;370(6521):1227–30. 10.1126/science.abd7728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ju B, Zhang Q, Ge J, Wang R, Sun J, Ge X, et al. Human neutralizing antibodies elicited by SARS‐CoV‐2 infection. Nature. 2020;584(7819):115–9. 10.1038/s41586-020-2380-z. [DOI] [PubMed] [Google Scholar]
- 41. Stadlbauer D, Tan J, Jiang J, Hernandez MW, Fabre S, Amanat F, et al. Repeated cross‐sectional sero‐monitoring of SARS‐CoV‐2 in New York City. Nature. 2021;590(7844):146–50. 10.1038/s41586-020-2912-6. [DOI] [PubMed] [Google Scholar]
- 42. Chan KH, Cheng VCC, Woo PCY, Lau SKP, Poon LLM, Guan Y, et al. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross‐reactivity with human coronaviruses 229E, OC43, and NL63. Clin Diagn Lab Immunol. 2005;12(11):1317–21. 10.1128/CDLI.12.11.1317-1321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu LP, Wang N‐C, Chang Y‐H, Tian X‐Y, Na D‐Y, Zhang L‐Y, et al. Duration of antibody responses after severe acute respiratory syndrome. Emerg Infect Dis. 2007;13(10):1562–4. 10.3201/eid1310.070576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Edridge AWD, Kaczorowska J, Hoste ACR, Bakker M, Klein M, Loens K, et al. Seasonal coronavirus protective immunity is short‐lasting. Nat Med. 2020;26(11):1691–3. 10.1038/s41591-020-1083-1. [DOI] [PubMed] [Google Scholar]
- 45. Dyrdak R, Hodcroft EB, Wahlund M, Neher RA, Albert J. Interactions between seasonal human coronaviruses and implications for the SARS‐CoV‐2 pandemic: A retrospective study in Stockholm, Sweden, 2009–2020. J Clin Virol. 2020. 10.1101/2020.10.01.20205096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kronfeld‐Schor N, Stevenson TJ, Nickbakhsh S, Schernhammer ES, Dopico XC, Dayan T, et al. Drivers of infectious disease seasonality: potential implications for COVID‐19. J Biol Rhythms. 2021;36(1):35–54. 10.1177/0748730420987322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bodogai M, O'Connell J, Kim K, Moritoh K, Chen C, Gusev F, et al. Commensal bacteria contribute to insulin resistance in aging by activating innate B1a cells. Sci Transl Med. 2018;10(467):eaat4271. 10.1126/scitranslmed.aat4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cancro MP, Hao Y, Scholz JL, Riley RL, Frasca D, Dunn‐Walters DK, et al. B cells and aging: molecules and mechanisms. Trends Immunol. 2009;30(7):313–8. 10.1016/j.it.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Jeffery‐Smith A, Iyanger N, Williams SV, Chow JY, Aiano F, Hoschler K, et al. Antibodies to SARS‐CoV‐2 protect against re‐infection during outbreaks in care homes, September and October 2020. Euro Surveill. 2021;26(5):2100092. 10.2807/1560-7917.es.2021.26.5.2100092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yang W, Kandula S, Huynh M, Greene SK, Van Wye G, Li W, et al. Estimating the infection‐fatality risk of SARS‐CoV‐2 in New York City during the spring 2020 pandemic wave: a model‐based analysis. Lancet Infect Dis. 2021;21(2):203–12. 10.1016/S1473-3099(20)30769-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Buss LF, Prete CA, Abrahim CMM, Mendrone A, Salomon T, de Almeida‐Neto C, et al. Three‐quarters attack rate of SARS‐CoV‐2 in the Brazilian Amazon during a largely unmitigated epidemic. Science. 2021;371(6526):288–92. 10.1126/science.abe9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sabino E, Buss LF, Carvalho MPS, Prete CA, Crispim MAE, Fraiji NA, et al. Resurgence of COVID‐19 in Manaus, Brazil, despite high seroprevalence. Lancet. 2021;397(10273):452–5. 10.1016/S0140-6736(21)00183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS‐CoV‐2 spike: Evidence that D614G increases infectivity of the COVID‐19 Virus. Cell. 2020;182(4):812–27.e19. 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rostami A, Sepidarkish M, Leeflang MMG, Riahi SM, Nourollahpour Shiadeh M, Esfandyari S, et al. SARS‐CoV‐2 seroprevalence worldwide: a systematic review and meta‐analysis. Clin Microbiol Infect. 2021;27(3):331–40. 10.1016/j.cmi.2020.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Pollán M, Pérez‐Gómez B, Pastor‐Barriuso R, Oteo J, Hernán MA, Pérez‐Olmeda M, et al. Prevalence of SARS‐CoV‐2 in Spain (ENE‐COVID): a nationwide, population‐based seroepidemiological study. Lancet. 2020;396(10250):535–44. 10.1016/S0140-6736(20)31483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fischer B, Knabbe C, Vollmer T. SARS‐CoV‐2 IgG seroprevalence in blood donors located in three different federal states, Germany, March to June 2020. Euro Surveill. 2020;25(28):2001285. 10.2807/1560-7917.ES.2020.25.28.2001285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Slot E, Hogema BM, Reusken CBEM, Reimerink JH, Molier M, Karregat JHM, et al. Low SARS‐CoV‐2 seroprevalence in blood donors in the early COVID‐19 epidemic in the Netherlands. Nat Commun. 2020;11(1):5744. 10.1038/s41467-020-19481-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Percivalle E, Cambiè G, Cassaniti I, Nepita EV, Maserati R, Ferrari A, et al. Prevalence of SARS‐CoV‐2 specific neutralising antibodies in blood donors from the Lodi Red Zone in Lombardy, Italy, as at 06 April 2020. Euro Surveill. 2020;25(24):2001031. 10.2807/1560-7917.ES.2020.25.24.2001031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stringhini S, Wisniak A, Piumatti G, Azman AS, Lauer SA, Baysson H, et al. Seroprevalence of anti‐SARS‐CoV‐2 IgG antibodies in Geneva, Switzerland (SEROCoV‐POP): a population‐based study. Lancet. 2020;396(10247):313–9. 10.1016/S0140-6736(20)31304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. GeurtsvanKessel CH, Okba NMA, Igloi Z, Bogers S, Embregts CWE, Laksono BM, et al. An evaluation of COVID‐19 serological assays informs future diagnostics and exposure assessment. Nat Commun. 2020;11(1):3436. 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Bayesian learner.
Table S2. SVM‐LDA learner.
Data Availability Statement
Data generated as part of the study, along with custom code for statistical analyses, are openly available via our GitHub repositories: https://github.com/MurrellGroup/DiscriminativeSeroprevalence/; https://github.com/chr1swallace/seroprevalence‐paper.


