Abstract
In the present study, the course of SARS‐CoV‐2 natural infection in two asymptomatic cats, which were negative for immunosuppressive retroviral infections, is investigated. The source of the virus for the cats was their COVID‐19‐affected owner, with whom they were in continuous proximity in a small household setting. The owner's signs included fatigue, sneezing, anosmia and loss of taste, and diagnosis was confirmed 4 days after symptom onset. Oropharyngeal and faecal swabs were collected from the cats, to investigate the course of SARS‐CoV‐2 RNA concentrations, as well as the directionality of the chain of virus transmission. Both infected cats were real‐time RT‐PCR‐positive on various time‐points. Pharyngeal shedding of at least 6 days was observed in them, with high SARS‐CoV‐2 titres (> 7 Log10 copies/swab) on the first sampling time‐point, that is, 7 days after the onset of owner's clinical signs. In one cat, after the initial decline, slightly increasing virus titres were measured 3 to 6 days after the first real‐time RT‐PCR‐positive swab. Serological testing of this cat revealed absence of seroconversion. The course of viral RNA concentrations in the faecal swabs of the other cat was similar to that in its pharynx. The detected SARS‐CoV‐2 strains, from both infected cats and their owner, underwent whole‐genome sequencing, revealing the absence of emergence of cross‐species adaptive mutations in cats. The results support the notion that human SARS‐CoV‐2 strains are relatively well‐adapted to cats. It is still unclear whether asymptomatic animals could play a role in COVID‐19 epidemiology, in case of interaction with naïve animals and/or people. Our findings highlight difficulties in SARS‐CoV‐2 transmission to cats, as neither the two infected cats nor their owner was able to transmit the virus to a third cat living in the same small flat, despite their very close contact during the days corresponding to high virus shedding.
Keywords: adaptation, cat, human, natural infection, SARS‐CoV‐2, transmission
1. INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS‐CoV‐2) is a novel member of the Coronaviridae family (Betacoronavirus genus), responsible for the coronavirus disease 2019 (COVID‐19) pandemic in humans, for which a spill‐over from wild animals is most likely (Boni et al., 2020; Dhama et al., 2020; Malik et al., 2020). This virus possesses a single‐stranded, positive‐sense RNA of ~30 kb that encodes approximately 9,860 amino‐acids. Two open reading frames (ORFs), ORF1a and ORF1b, located at the 5’ terminus of the genome encode 16 non‐structural proteins (NSPs). Four structural proteins [i.e. envelope (E), membrane (M), nucleocapsid (N) and spike (S) proteins] as well as several accessory proteins are produced through the expression of subgenomic mRNAs (Chan et al., 2020; Kim et al., 2020).
The S protein mediates receptor binding on target cells and determines host tropism. Similarly to SARS‐CoV‐1, the receptor utilized by SARS‐CoV‐2 to enter target cells is the angiotensin‐converting enzyme 2 (ACE2) (Lu et al., 2020). It has been identified that, besides humans, the receptor‐binding domain of the virus recognizes ACE2 from various animal species, including cats (Luan et al., 2020; Wan et al., 2020). Consequently, cats exposed to SARS‐CoV‐2 through contact with infected people, either asymptomatic virus carriers, or patients, can be affected, thus resulting in human‐to‐animal virus transmission, that is, a reverse zoonosis (Segalés et al., 2020). It is noteworthy, meanwhile, that coronaviruses have adapted species by species over relatively short to long periods of time, in human and animals. Domestic and wild cats are the hosts of feline coronavirus (FCoV), a highly prevalent virus with worldwide distribution, that is classified within the Alphacoronavirus 1 species (Alphacoronavirus genus), along with common cold‐causing coronaviruses of humans (Drechsler et al., 2011).
The capability of SARS‐CoV‐2 to infect cats has been indicated experimentally, as well as in clinically confirmed cases. Experimental infection of cats with a SARS‐CoV‐2 isolate originating from Wuhan indicated that the cats are permissive to the infection, since the virus was able to replicate in the nose and throat and caused inflammatory pathology in the lower respiratory tract (Shi et al., 2020). Cases of naturally infected cats with clinical signs of mild‐to‐moderate severity have been reported in Belgium, the USA, Spain, France and Brazil (Carlos et al., 2021; de Morais et al., 2020; Sailleau et al., 2020; Segalés et al., 2020). The clinical signs that have been described in cases involve mainly respiratory symptoms, such as breathing difficulty and cough. Gastrointestinal symptoms have also been reported (de Morais et al., 2020; Sailleau et al., 2020). In a single case, the developed signs were severe, leading to euthanasia, although post‐mortem examination implicated SARS‐CoV‐2‐unrelated hypertrophic cardiomyopathy with secondary thrombosis as the major cause of death (Segalés et al., 2020).
Reports for the detection of SARS‐CoV‐2 or antibodies specifically directed against this virus in asymptomatic cats also exist. A serological investigation which took place in Wuhan during early 2020 revealed detection of virus‐specific antibodies in naturally infected cats (Zhang et al., 2020). A more recent study which took place in Italian households indicates that cats can seroconvert under the normal conditions of pet ownership (Patterson et al., 2020). SARS‐CoV‐2 RNA was detected in the oral cavity, nasal and rectal swab samples obtained from a clinically healthy cat in Hong Kong (Abdel‐Moneim & Abdelwhab, 2020). However, the number of reports available for natural infection of cats within house environments is limited and, albeit the airborne virus transmission between infected and susceptible cats, has been experimentally demonstrated, the role of this host species in the natural transmission of SARS‐CoV‐2 remains unclear.
Since data currently available for natural infection of cats are scarce, the present study aims to investigate the course of SARS‐CoV‐2 infection in naturally exposed cats in a small household setting, in Greece. The owner of the animals was affected by COVID‐19 and was in continuous close proximity with three cats, during the days corresponding to high virus shedding and the quarantine period. Serial samplings were conducted from the cats, to determine the onset and duration of infection and viral shedding, as well as their antiviral humoural immune responses. In order to be able to detect introductions of known variants and/or any emerged and adapted variants, and to obtain a preliminary understanding of the directionality of the chain of virus transmission, whole SARS‐CoV‐2 genomes from the cats and their owner were sequenced and subjected to molecular characterization and quasispecies analysis.
2. MATERIALS AND METHODS
2.1. History, clinical examination, haematology and serum biochemistry
This study is focussed on the clinical and laboratory investigation of 3 European Shorthaired cats, which were residing exclusively indoors with their owner, a veterinary medicine undergraduate student affected by COVID‐19. The owner lives by herself, in the city of Thessaloniki (Central Macedonia, Northern Greece) in a 40 m2 flat of an apartment block. Owner's signs of worsening fatigue and intense sneezing first appeared on 12 November 2020 (day 0; D0), followed by anosmia and loss of taste two days later (D2). On D4, the owner was diagnosed with COVID‐19 after being subjected to a rapid SARS‐CoV‐2 antigen test, followed by real‐time RT‐PCR. Since the onset of clinical signs, the owner was quarantined at home, according to the guidelines issued by the National Public Health Organization of Greece (EODY). The duration of the owner's clinical signs was one week, followed by complete recovery. During the entire period (i.e. before the onset of signs and far beyond the time of apparent clinical recovery), the owner remained in close contact with all cats, sharing with them all the apartment's rooms, including the bedroom. The interaction pattern between the owner and each cat was similar (i.e. petting, giving kisses or licks) and did not change, even after the molecular confirmation of the SARS‐CoV‐2 infection of the owner.
One day after the confirmation of the diagnosis (D5), one of the cats, an 11‐year‐old neutered female (Cat #1; C1), showed mild signs of self‐limiting diarrhoea for 3 days, without exhibiting any other symptoms. Given the COVID‐19 infection status of the owner, this cat and the other two (Cat #2; C2: a 10‐year‐old neutered male and Cat #3; C3: a 10‐year‐old neutered female) that were living in the same environment and did not experience clinical signs were subjected to clinical examination and laboratory investigations for SARS‐CoV‐2 infection.
On D16, all cats were clinically evaluated in detail in their household environment, and blood samples were obtained for haematology and serum biochemistry. Complete blood counts (ADVIA® 120 hematology system; Siemens Healthcare Diagnostics, Eschborn, Germany), including 100‐cell manual differential leucocyte counts and evaluation for platelet aggregates, along with comprehensive blood serum biochemistry analysis (Vitalab Flexor E; Vital Scientific N.V., Spankeren/Dieren, The Netherlands) were performed for each cat.
2.2. Swab collection, RNA extraction and real‐time RT‐PCR
Oropharyngeal and faecal swabs were collected from the cats at various time‐points, to identify the period of viral shedding (Figure 1). All samples were obtained by means of 14.5‐cm snappable sterile polystyrene/Dacron swabs (Deltalab, Barcelona, Spain). Immediately after sample collection, the swab tips were being placed in 2‐mL microcentrifuge tubes containing 800 μl of guanidinium isothiocyanate‐based ‘Lysis buffer I’. Lysates were subjected to a phenol–chloroform‐based RNA extraction process coupled with silica column binding (Chaintoutis et al., 2019).
FIGURE 1.
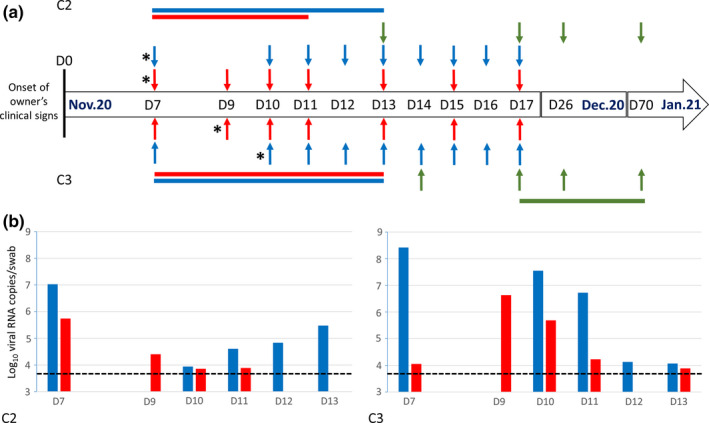
(a) Timeline of the samplings conducted in cats, for the laboratory investigation of SARS‐CoV‐2 infection status. Detailed sampling time‐points are presented for C2 (top row) and C3 (bottom row). Sampling time‐points for C1 were as same to those for C2, except for the fact that faecal swabs were not obtained on D9 and D11. Vertical arrows indicate oropharyngeal swab (blue), faecal swab (red), and serum sampling (green) time‐points. Horizontal bars of the same colours indicate time‐periods with real‐time RT‐PCR‐positive or ELISA‐positive results. The real‐time RT‐PCR‐positive RNA extracts that were subjected to next‐generation sequencing are marked with an asterisk on the left of the respective arrow. The numbering of the sampling time‐points refers to D0, that is, Nov. 12th, the date of onset of the owner's clinical signs. (b) SARS‐CoV‐2 titres in the real‐time RT‐PCR‐positive oropharyngeal (blue) and faecal (red) swabs of C2 and C3, expressed as Log10 viral RNA copies/swab. The dashed lines indicate the limit of detection
All swabs obtained from the 3 cats at all time‐points were tested for the presence of SARS‐CoV‐2 genome, via 2 separate real‐time RT‐PCR assays proposed as suitable for the laboratory diagnosis of COVID‐19 in humans by international agencies. Specifically, the protocol proposed by the European Centre for Disease Prevention and Control (ECDC) for the amplification of the genomic region that encodes the E protein was used (Corman et al., 2020). Similarly, the N2 method targeting the genomic region that encodes the N protein, which is proposed by the Centers for Disease Control and Prevention (CDC) within the ‘CDC 2019‐Novel Coronavirus (2019‐nCoV) Real‐Time RT‐PCR Diagnostic Panel’, was applied (Centers for Disease Control and Prevention (CDC), 2020). All oligonucleotides (primers and TaqMan probes) were synthesized by Integrated DNA Technologies (IDT, Coralville, IA, USA). Both assays were performed on a CFX96 Touch™ Real‐Time PCR Detection System (Bio‐Rad Laboratories, Hercules, CA, USA). Analysis of fluorescence data was performed using CFX Maestro™ software (version 4.1; Bio‐Rad Laboratories). RNA extracts with a Ct value >40 were considered as negative. Calibration curves for both targets were also generated for virus quantification, using the synthetic single‐stranded RNA standard ‘EURM‐019’ (Joint Research Centre, European Commission). Virus titres were Log10‐expressed as mean SARS‐CoV‐2 RNA copies per real‐time RT‐PCR‐positive swab.
2.3. Next‐generation sequencing (NGS)
RNA extracts from the cats (representing one real‐time RT‐PCR‐positive oropharyngeal and one faecal swab per animal; Figure 1) along with a pharyngeal swab from the owner (obtained on D7) were further processed for NGS‐based SARS‐CoV‐2 whole‐genome sequencing. Amplification of the complete viral genomes was performed via a commercially available oligonucleotide panel (Ion AmpliSeq™ SARS‐CoV‐2 Research Panel, ID: 05280253; Thermo Fisher Scientific, Vantaa, Finland). The Ion AmpliSeq™ Library Kit Plus was used for library preparation, according to the manufacturer's instructions. Briefly, the library preparation involved reverse transcription using the SuperScript™ VILO™ cDNA Synthesis Kit (Thermo Fisher Scientific), 16 cycles of PCR amplification, adapter ligation, library purification using Agencourt™ AMPure™ XP (Beckman Coulter, Brea, CA, USA), and library quantification using the Qubit™ Fluorometer dsDNA HS Assay Kit. An ion 540™ Chip was prepared using the Ion Chef™ instrument, and NGS reactions were run on an Ion GeneStudio™ S5 semiconductor benchtop sequencer (Thermo Fisher Scientific).
2.4. Bioinformatics
Quality control of the reads was performed within the Torrent Server of the sequencer, using the default settings. Reads were aligned using the bwa‐mem v. 0.7.17‐r1188 aligner (Li & Durbin, 2009). The complete genome sequence of the ‘Wuhan‐Hu‐1’ SARS‐CoV‐2 isolate, GenBank Acc. No. NC_045512 (Wu et al., 2020) was used as a reference. The aligned reads were subjected to both reference‐guided assembly and variant calling. Through the SAMtools software (Li et al., 2009), the % coverage on the reference sequence and the mean sequencing depth were calculated. In order to further identify differences that may point a direction of the chain of the transmission, quasispecies analysis was performed. The LoFreq v.2.1.4 software (Wilm et al., 2012) was used to identify low‐frequency variants (quasispecies) present in the sequenced samples, since it is able to detect variants that exist in a few aligned reads, while evaluating those based on quality metrics (cut‐off p = .01). Due to the fact that the SARS‐CoV‐2 genome contains long stretches of homopolymers, quasispecies analysis was focussed on single nucleotide polymorphisms (SNPs). SnpEff v.4.5covid19 (Cingolani et al., 2012) was used for annotation of variants, so as to be assigned to a specific viral protein. SnpEff utilizes a genomic feature file (gff) that contains all the information on the viral protein structure and intervals on the reference sequence. The positions of non‐synonymous (missense) variants were plotted onto the viral genome. The multiple sequence alignment was computed through Clustal Omega (Sievers & Higgins, 2014), using the default parameters.
The phylogenetic tree encompassing European and European‐related international isolates was implemented through the NextStrain platform (Hadfield et al., 2018). Tree calculations were based on the maximum‐likelihood method, with branch size taking into consideration temporal data (Price et al., 2010). Branch colours represent GISAID clade annotation, while branch names on the tree are the NextStrain clades (Bogner et al., 2006; Hadfield et al., 2018). Lineage assignment was achieved via the Phylogenetic Assignment of Named Global Outbreak LINeages (PANGOLIN) SARS‐CoV‐2 lineage assigner interface (Rambaut et al., 2020).
2.5. Serology
Serum samples were being collected at specific time‐points from all cats for detection of SARS‐CoV‐2‐specific antibodies (Figure 1). For this reason, the cats were mildly sedated with the alpha‐2 adrenoceptor agonist dexmedetomidine (10 μg/kg, im). Approximately 40 min after administration and once the procedures were completed, sedation was rapidly reversed by the administration of the specific alpha‐2 adrenoceptor antagonist, atipamezol (100 μg/kg, im).
The commercially available ID Screen® SARS‐CoV‐2 Double Antigen Multi‐species ELISA (ID.vet, Montpellier, France) kit was used. The antigen in this immunological assay is the N protein of SARS‐CoV‐2, and its format enables the detection of SARS‐CoV‐2‐specific antibodies in the tested sera, irrespective of their isotype. Testing was performed following the manufacturer's instructions. Optical density (OD) values were recorded using a Stat Fax 3,200 microplate photometer (Awareness Technology Inc., Palm City, FL, USA). For each sample, S/P% percentages were calculated as follows: (ODSample‐ODNegative Control)/(ODPositive Control‐ODNegative Control) × 100, with serum samples presenting S/P% ≥ 60% being considered as positive.
Additionally, anticoagulated blood samples from all cats obtained on D16 were tested for Feline immunodeficiency virus (FIV)‐specific antibodies and Feline leukaemia virus (FeLV) antigens, using a rapid enzyme immunoassay test (SNAP® FIV/FeLV Combo Test; IDEXX Laboratories Inc., Westbrook, ME, USA).
3. RESULTS
3.1. Clinical examination and laboratory testing
On D16, all cats and were found bright, alert and responsive, with no cardiorespiratory signs (including upper respiratory signs) and normal temperature. According to the owner, their clinical condition remained unaltered throughout the observation period. Haematology and serum biochemistry findings were unremarkable (Table 1), and testing results for immunosuppressive retroviral (FIV and FeLV) infections were negative for all 3 cats. Testing results of all oropharyngeal and faecal swabs obtained from C1 between D7 and D17 were negative via both SARS‐CoV‐2‐specific real‐time RT‐PCR methods. However, positive results against both targets were obtained by testing of the other two cats (C2 and C3), and specifically, from the first sampling time‐point (D7), and up to D13.
TABLE 1.
Haematology and serum biochemistry findings in the three healthy cats living in the same household with a symptomatic COVID‐19 human patient
| Parameter (reference interval), Unit | C1 | C2 a | C3 a |
|---|---|---|---|
| Haematology | |||
| Haematocrit (25–45) % | 39.5 | 39.7 | 37.5 |
| Haemoglobin (8–15) g/dl | 11.3 | 11.7 | 10.9 |
| Mean Corpuscular Volume (39–55) fl | 49.9 | 51.8 | 49 |
| Mean Corpuscular Haemoglobin Concentration (30–36) g/dl | 28.7 | 29.5 | 29 |
| Reticulocytes (<90) × 103/μl | 25.5 | 14.6 | 24.5 |
| White blood cells (5.5–19.6) × 103/μl | 7 | 8.3 | 6.3 |
| Neutrophils (3–13.4) × 103/μl | 4.3 | 5.3 | 3.2 |
| Bands (<0.3) × 103/μl | 0 | 0 | 0.1 |
| Lymphocytes (2–7.2) × 103/μl | 2.1 | 1.7 | 2.8 |
| Monocytes (0.1–1) × 103/μl | 0.3 | 0.3 | 0.1 |
| Eosinophils (0.3–1.7) × 103/μl | 0.3 | 1 | 0.1 |
| Platelets (300–800) × 103/μl | 90 b | 97 b | 131 b |
| Serum biochemistry | |||
| Total protein (6–8.8) g/dl | 8.3 | 7.2 | 6.8 |
| Albumin (2.9–4.8) g/dl | 3.8 | 4 | 3.5 |
| Alkaline phosphatase (15–125) U/L | 33 | 44 | 38 |
| Alanine aminotransferase (20–100) U/L | 56 | 47 | 29 |
| Creatinine (0.7–1.6) mg/dl | 0.8 | 0.8 | 0.6 |
| Blood urea nitrogen (9–32) mg/dl | 25 | 23 | 23 |
| Glucose (70–150) mg/dl | 121 | 187 | 65 |
| Phosphorus (3.5–6.7) mg/dl | 3.6 | 3.7 | 4.9 |
| Total calcium (8.5–11.4) mg/dl | 8.9 | 8.3 | 8.2 |
| Sodium (144–159) mEq/L | 152 | 148 | 150 |
| Potassium (3.4–5.4) mEq/L | 3.9 | 4 | 4.6 |
Infected cats.
Numerous large platelet aggregates were noticed in blood smear evaluation.
Daily monitoring of viral load in oropharyngeal and faecal swabs of the two SARS‐CoV‐2‐positive cats are summarized in Figure 1. High virus titres (Ct values <30) were observed in the pharynx of both C2 and C3 on the first sampling time‐point (D7). For C2, 7.03 Log10 RNA copies/swab were determined on D7. A considerably lower virus titre (3.94 Log10 RNA copies/swab) was measured on D10. During the subsequent 3 days, a slightly increasing viral load was measured (4.61, 4.83 and 5.48 Log10 RNA copies/swab, respectively) in this animal's pharynx, and it finally tested negative on D14. The oropharyngeal swab obtained from C3 on D7 was also characterized by a high viral load (8.42 Log10 RNA copies/swab). However, differences were observed in the kinetics of SARS‐CoV‐2 presence in the upper respiratory tract of this animal, since steadily declining virus titres were measured between D10 and D13 (7.55, 6.72, 4.13 and 4.07 Log10 RNA copies/swab, respectively). All subsequently collected swabs tested negative.
Faecal swabs obtained from C2 tested positive on the first day of sampling (D7, 5.31 Log10 RNA copies/swab) and up to D11 with progressively declining virus titres (3.89 Log10 RNA copies/swab). Faecal swabs collected on D12 (i.e. 5 days after the first real‐time RT‐PCR‐positive faecal swab) tested negative. Regarding C3, the kinetics of viral RNA concentrations in the faecal swabs was similar to that in its pharynx, although characterized by a time lag (Figure 1). A virus titre of 4.05 Log10 RNA copies/swab was measured on D7. Subsequently, a peak was observed on D9 (6.63 Log10 RNA copies/swab), followed by a progressive decline on D10, D11 and D13 (5.69, 4.23 and 3.89 Log10 RNA copies/swab, respectively). Faecal swabs collected on D15 (i.e. 8 days after the first real‐time RT‐PCR‐positive faecal swab) tested negative.
All sera obtained from C1 and C2 tested negative for SARS‐CoV‐2‐specific antibodies. On the contrary, serological testing of C3 on D17 (i.e. 10 days after the first real‐time RT‐PCR‐positive oropharyngeal swab) yielded a positive result (S/P% = 197.3%). Re‐testing on D26 indicated an even stronger positive result (S/P% = 256.7%), with the respective OD value having reached a plateau (2.934 AU). On D70, an S/P% of 217.6% was observed, indicating a decline in the respective SARS‐CoV‐2‐specific serum antibody titre.
3.2. Whole‐genome sequencing and bioinformatics
NGS processing yielded SARS‐CoV‐2 sequences for both C2 and C3 in oropharyngeal and faecal swabs, as well as in the oropharyngeal swab from the owner (Table 2). Multiple sequence alignment revealed an absolute overlap of amino‐acid sequences. Only one difference among 5 samples was observed, a synonymous T‐to‐C SNP in position 24544 in the oropharyngeal sample of C3, at a ratio of 0.57 (Table 3). A statistically significant imbalance of quasispecies abundance at the aforementioned nucleotide position was observed among samples. Specifically, the oropharyngeal swab of C3 showed an increased C variant ratio compared to the faecal swab of the same cat (Odds ratio: 11.96, p < .0001), since the presence of this variant in the faecal sample of this animal was also revealed, but the respective ratio was low (0.10). This variant was also identified in the owner's pharyngeal swab at an even lower ratio (0.07). Comparison with the oropharyngeal swab of C3 revealed an odds ratio of 17.72 (p < .0001). On the contrary, this variant was not identified neither in the oropharyngeal swab, nor the faecal swab from C2 and comparison with the oropharyngeal swab of C3 revealed an odds ratio of 1,061.57 (p < .0001). Two other sites (nucleotide positions 22685 and 23335) showed within‐sample T/C variability in at least one additional sample (Table 3).
TABLE 2.
NGS quality parameters. Per cent coverage of the ‘Wuhan‐Hu‐1’ SARS‐CoV‐2 reference genome alters alignment of the NGS reads from samples of C2 and C3, as well as from the cat owner. Mean depth is expressed in nucleotides, after alignment of the respective sample reads
| Sample | Coverage (%) | Mean depth (nucleotides) |
|---|---|---|
| C2—oropharyngeal | 99.16 | 367.2 |
| C2—faecal | 91.52 | 8.1 |
| C3—oropharyngeal | 99.72 | 1579.0 |
| C3—faecal | 88.79 | 20.9 |
| Owner | 97.69 | 27.5 |
TABLE 3.
Common quasispecies (low‐frequency variants) in the sequenced samples. Nucleotide positions refer to the position on the ‘Wuhan‐Hu‐1’ reference SARS‐CoV‐2 isolate. Next to the position the nucleotide present at any specific quasispecies variant is indicated. Numbers refer to the absolute number of reads that represented the respective variant. The ratio of the C variant (C ratio) per position is also presented (#)
| Sample | Owner | C2 | C3 | |||
|---|---|---|---|---|---|---|
| Position | Oropharyngeal | Faecal | Oropharyngeal | Faecal | ||
| 22685 | C | 2 | 62 | 1 | 151 | 2 |
| T | 7 | 166 | 0 | 559 | 5 | |
| C ratio# | 0.22 | 0.27 | 1.00 | 0.21 | 0.29 | |
| 23335 | C | 6 | 22 | 0 | 444 | 1 |
| T | 21 | 241 | 4 | 1658 | 30 | |
| C ratio# | 0.22 | 0.08 | 0.00 | 0.21 | 0.03 | |
| 24544 | C | 3 | 0 | 0 | 1,362 | 4 |
| T | 40 | 399 | 13 | 1,025 | 36 | |
| C ratio# | 0.07 | 0.00 | 0.00 | 0.57 | 0.10 | |
Comparison of the sequence obtained from the oropharyngeal swab of C3 against the ‘Wuhan‐Hu‐1’ reference isolate indicated 17 SNPs (Table 4). Eleven of the polymorphisms resulted in an amino‐acid substitution at the respective protein. Out of the 17 SNPs, 4 occurred in sequential nucleotides (28881–28884), with 3 of them being located at codon 203 of N and resulting in the N‐R203K amino‐acid substitution. The fourth SNP is associated with the N‐G204L amino‐acid substitution. In total, 6 out of 9 amino‐acid substitutions were located at the structural/accessory protein‐encoding moiety of the genome (Figure 2). Phylogenetic analysis using the GISAID and NextStrain platforms showed that the strains sequenced herein are placed in the GISAID GR clade, in the NextStrain 20B clade, and in the B.1.1 lineage, according to the PANGOLIN SARS‐CoV‐2 lineage assigner interface (Figure 3).
TABLE 4.
Nucleotide polymorphisms identified in the sequenced SARS‐CoV‐2 strains. Numbering of nucleotide positions refers to the ‘Wuhan‐Hu‐1’ reference isolate. Amino‐acid substitutions refer to amino‐acid positions on the respective protein of the reference isolate
| Nucleotide position | ‘Wuhan‐Hu−1’ reference | Nucleotide change | Amino‐acid substitution | ORF/Protein |
|---|---|---|---|---|
| 241 | C | T | – | – |
| 2091 | C | T | T429I | ORF1a/NSP2 |
| 3037 | C | T | – | ORF1a/NSP3 |
| 3117 | C | T | T133I | |
| 10507 | C | T | – | ORF1a |
| 14408 a | C | T | P323L | ORF1b/NSP12 |
| 19839 | T | C | – | ORF1b |
| 23403 | A | G | D614G | S |
| 24544 b | T | C | – | |
| 24872 | C | T | V1104L | |
| 25448 | A | G | E19G | ORF3a |
| 26951 | C | T | – | M |
| 27798 | C | T | A15S | ORF7b |
| 28881 | G | A | R203K | N |
| 28882 | G | A | ||
| 28883 | G | C | ||
| 28884 | G | T | G204L |
Nucleotide change not verified in the faecal swab of C2 due to reduced depth.
Nucleotide change only in the oropharyngeal swab of C3
FIGURE 2.
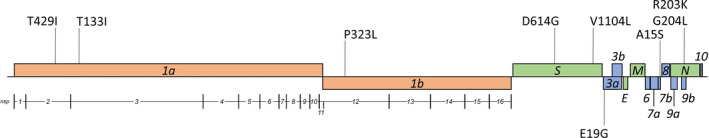
Graphical representation of amino‐acid substitutions on the ‘Wuhan‐Hu‐1’ SARS‐CoV‐2 isolates reference genome. Vertical lines project the position of amino‐acid substitutions on the viral genome
FIGURE 3.
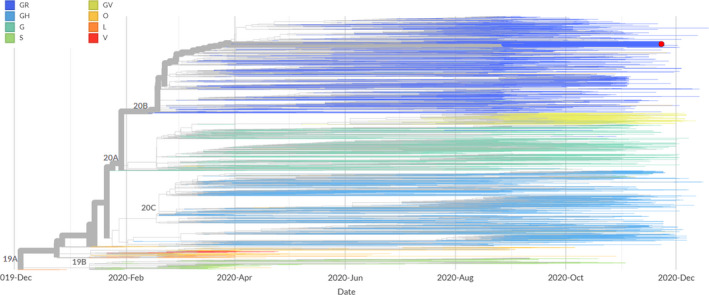
Phylogenetic tree based on temporal data of European isolates and their global links. The tree was adapted by NextStrain (3,891 complete genomes as of 29/12/2020). Branch colours represent GISAID clades (G: includes the S‐D614G amino‐acid substitution; GH: includes the S‐D614G and the ORF3a‐Q57H amino‐acid substitutions; GR: includes the S‐D614G and the N‐G204R amino‐acid substitutions; GV: includes the S‐D614G and the S‐A222V amino‐acid substitutions; S: includes the ORF8‐L84S amino‐acid substitution; V: includes the NSP6‐L37F and the ORF3a‐G251V amino‐acid substitutions; L: contains sequences close to the reference genome; O: contains sequences not clustered into any of the major clades). NextStrain clades are depicted on the branch junctions. The red circle represents the strain of the current study
4. DISCUSSION
Cats are popular companion animals in close contact with humans, and therefore, it is important to gain insight into their susceptibility to SARS‐CoV‐2 and their possible role in COVID‐19 epidemiology. The longitudinal monitoring of the cats in the present study indicates that, after the owner introduced the virus in the house environment, transmission occurred first to C2 and then to C3, while C1 was not infected. No clinical signs were observed in any of the infected cats, and haematology and serum biochemistry values were within normal limits. These findings support previous experimental infection/transmission studies (Gaudreault et al., 2020; Shi et al., 2020), as well as the limited reports of clinically affected cats around the globe, indicating that following exposure to SARS‐CoV‐2, cats are not invariably infected, and when infection is established, the course is mostly asymptomatic. Cats may thus be silent hosts of SARS‐CoV‐2, as they may not show any appreciable symptoms that might be recognized by their owners (Halfmann et al., 2020). It is possible that the outcome in companion animals may depend on several factors, such as the proximity of contact with humans shedding high viral loads, possible co‐morbidities, increased susceptibility to the virus (e.g. age‐dependent), or a combination of these factors (de Morais et al., 2020; Shi et al., 2020).
It has been reported that infected cats shed the virus for no more than 5 days following exposure (Bosco‐Lauth et al., 2020; Shi et al., 2020). The fact that viral RNA was detected in the pharynx of both infected cats of the present study, from D7 until D13, indicates that some cats may have a more prolonged shedding period. Interestingly, serological testing of C2 against SARS‐CoV‐2 was negative at all sampling time‐points. As previously suggested for infected humans, the absence of seroconversion may be indicative of mild infections involving only the respiratory mucosa, where secretory immune responses dominate, in conjunction with limited systemic IgG production (Staines et al., 2020). For this reason, the amount of virus‐specific secretory IgA in the respiratory mucosa could serve as an indicator of immune response in humans and in susceptible animal species (Chao et al., 2020). Another possible explanation for this condition is that exposure to SARS‐CoV‐2 can induce T‐cell‐mediated virus‐specific responses, without seroconversion, similar to what has been suggested for humans (Gallais et al., 2020; Staines et al., 2020). The fact that C2 did not seroconvert to SARS‐CoV‐2 may account for the continued presence of the virus in its pharynx, in low titres with a slightly increasing pattern (D10‐D13), after the initial decline which was observed between D7 and D10.
Faecal swabs obtained from both infected cats tested positive via real‐time RT‐PCR. A scenario of swallowing expectorated virus followed by a progressive tapering of SARS‐CoV‐2 viral load could interpret the presence of viral RNA in faecal swabs. Swallowed virus‐laden sputum could either passively pass through the intestine of these animals, or the virus could replicate in it. Rapid elimination of SARS‐CoV‐2 from the intestinal tract of cats has been reported in a case study (Sailleau et al., 2020). This rapid clearance has been also described in an experimental infection/transmission of cats, wherein SARS‐CoV‐2 titres as high as ~105 viral RNA copies/g were measured in the faeces of experimentally infected and exposed cats (Shi et al., 2020). In contrast to the aforementioned observations, the detection period of SARS‐CoV‐2 in the faecal swabs obtained from C3 of the present study was over 6 days long. The course of viral RNA concentrations in faecal swabs of this animal seemed to reflect the course in the oropharyngeal swabs, with a peak virus titre of ~6.5 Log10 viral RNA copies/swab being measured on D9 (i.e. 2 days after the first real‐time RT‐PCR‐positive faecal swab). A similar SARS‐CoV‐2 titre kinetics pattern in stool compared to sputum has been observed symptomatic COVID‐19 human patients (Wölfel et al., 2020). However, despite the fact that the observed virus titres in the stool of human patients exceeded 7 Log10 viral RNA copies/swab, isolation of SARS‐CoV‐2 from faeces was unsuccessful. Virus isolation results from viral RNA‐positive small intestines of experimentally infected cats were also negative (Shi et al., 2020). Further research is required to thoroughly investigate the factors sustaining the presence of SARS‐CoV‐2 in the gastrointestinal tract of cats with or without clinical signs, as well as the infectivity of the virus shed to the environment via faeces.
Coronaviruses are capable of adapting quickly to new hosts through the processes of genetic recombination and mutation in vivo (Boni et al., 2020; Sironi et al., 2020). Through the combined analysis of SARS‐CoV‐2 whole‐genome sequences obtained from the cats and their owner, changes in the viral genome indicating cross‐species adaptation of the virus were investigated. Our results support the notion that human SARS‐CoV‐2 strains are relatively well‐adapted to feline hosts and cross‐species adaptation is not initially required for the efficient virus replication. Whole‐genome sequencing of the detected SARS‐CoV‐2 strains revealed a T‐to‐C SNP in the nucleotide position 24544 in the oropharyngeal swab from C3. The fact that the difference between the C variant ratio in the oropharyngeal and in the faecal swab of the same cat was statistically significant is indicative of SARS‐CoV‐2 proliferation in this animal's intestine. Additionally, the presence of a significant fraction of 24544T reads in the oropharyngeal sample of C3 supports the notion that the transition from T‐to‐C was in the course to become dominant in the pharynx of this animal. Due to the high similarity among the viral genomes obtained from the owner and the cats, it cannot be ascertained whether the virus was transmitted directly from the owner to both cats, or that transmission from C2 to C3 took place. However, taking into consideration the chronological order of the real‐time RT‐PCR positivity in the two cats, as determined by the kinetics in virus titres, and the fact that the C variant in position 24544 was not observed in C2, it is suggested that the owner was the source of infection for C3 as well. This hypothesis is also supported by the very close contact of each cat with their owner and is in agreement with reports available so far that denote the human‐to‐feline transmission of the virus. Evidence‐based support in favour of a cat‐to‐human SARS‐CoV‐2 transmission is not currently available, and this specific report does not match the essential criteria to demonstrate cat‐to‐human transmission (de Morais et al., 2020; Totton et al., 2020).
The absolute overlap of amino‐acid sequences among the sequenced viral strains signifies the absence of emergence of cross‐species adaptive mutations. In a previous experimental infection study, SARS‐CoV‐2 variants possessing the S‐H655Y amino‐acid substitution have been reported to rapidly arise and become fixed following transmission between cats, suggesting that this site may be under positive selection in feline hosts (Braun et al., 2020). This substitution was not found in any of the cat samples which were sequenced in the present study. The S‐D614G amino‐acid substitution was present, which is characteristic of the GISAID G and G‐related clades and has been reported extensively in the past, as linked to increased SARS‐CoV‐2 infectivity and transmissibility (Hou et al., 2020). This substitution, along with the amino‐acid substitution NSP12‐P323L, which was also identified herein, has been previously retrieved from human isolates originating from all continents of the earth and seem to co‐evolve (Coppée et al., 2020). The rare variant S‐V1104L was also identified in the present report. This amino‐acid substitution has occurred independently before, in isolates belonging to three other B1 lineages (GISAID EPI_ISL_539372—Switzerland, B.1.160; GISAID EPI_ISL_722879—Italy, B.1.177; GISAID EPI_ISL_582648—United Arab Emirates, B.1.1.1). The E19G amino‐acid substitution in ORF3a is possibly indicative of the phylogenetic origin of the detected strains, as it was present only in one cluster of isolates within B.1.1 lineage.
Regarding C1, the mild diarrhoea signs were transient and completely subsided within a few days. This condition was reliably noticed by the owner, a veterinary medicine student, who was worried about the possibility that the diarrhoea could be due to SARS‐CoV‐2 infection, and thus notified us. The findings from the virological and serological testing showcase that the diarrhoea observed in C1 cannot be attributed to SARS‐CoV‐2. The cause of the diarrhoea signs could be linked to factors associated with the animal's diet, or less likely, to other infectious or parasitic agents. Results further support previous observations indicating that companion animals are not easily infected by SARS‐CoV‐2, even when they are in close contact with infected owners (Temmam et al., 2020). Additionally, our findings highlight difficulties in spontaneous cat‐to‐cat SARS‐CoV‐2 transmission, given that C1 was in continuous proximity to C2 and C3 during the peak of the viral shedding from them. The difficulty in cat‐to‐cat transmission is also supported by the findings of the previously discussed experimental infection/transmission study (Shi et al., 2020). The S‐D614G amino‐acid, which was identified in the viral strains of the present study, has been linked with increased SARS‐CoV‐2 replication in the upper airway of hamsters, suggesting its possible role in viral transmissibility (Plante et al., 2020). However, despite the prolonged contact of C1 with the other two infected cats and their owner, this cat remained uninfected.
In conclusion, the present study comprises the first report of SARS‐CoV‐2 natural infection of cats in Greece. The owner acted as a source of the virus for the animals, as COVID‐19 symptoms started one week before the first detection of the virus in the cats, confirming once again the reverse zoonotic character of SARS‐CoV‐2 transmission. Our results support the notion that human SARS‐CoV‐2 strains are relatively well‐adapted to cats, despite the absence of emergence of cross‐species adaptive mutations. It is still unclear whether the asymptomatic animals could play a role in COVID‐19 epidemiology, in case of interaction with naïve animals and/or people. Both infected cats and their owner were not able to transmit the virus to a third cat living in the same place, despite their very close contact and the high SARS‐CoV‐2 titres measured in the pharynx of the infected cats on the first sampling time‐point. It remains to be elucidated whether infected cats shed infective virus to the environment, and until then, keeping infected cats inside is a reasonable suggestion. Besides the risk of possible transmission of SARS‐CoV‐2 from infected cats to humans, it is advisable for COVID‐19 patients or asymptomatic virus carriers to avoid contact with cats, in order to minimize the possibilities of infecting them.
CONFLICT OF INTEREST
No potential conflicts of interest that could influence the results of the study exist.
ETHICAL APPROVAL
The procedures described were performed according to The Code of Ethics of the World Medical Association (Declaration of Helsinki). Informed consent was obtained for performing NGS analysis on the human specimen. The relevant study was approved by the Ethics Committee of the Department of Medicine, Democritus University of Thrace.
Chaintoutis SC, Siarkou VI, Mylonakis ME, et al. Limited cross‐species transmission and absence of mutations associated with SARS‐CoV‐2 adaptation in cats: A case study of infection in a small household setting. Transbound Emerg Dis. 2022;69:1606–1616. 10.1111/tbed.14132
Funding information
This study was partially funded by the European Union and Greek national funds, through the Operational Program Competitiveness, Entrepreneurship and Innovation, under the call RESEARCH‐CREATE‐INNOVATE (project code: T1EDK‐5000).
DATA AVAILABILITY STATEMENT
The nucleotide sequence of the strain "hCoV‐19/cat/Greece/2K/2020" is available at GISAID (EPI_ISL_717979).
REFERENCES
- Abdel‐Moneim, A. S. , & Abdelwhab, E. M. (2020). Evidence for SARS‐CoV‐2 infection of animal hosts. Pathogens, 9(7), 529. 10.3390/pathogens9070529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogner, P. , Capua, I. , Cox, N. J. , & Lipman, D. J. (2006). A global initiative on sharing avian flu data. Nature, 442(7106), 981. 10.1038/442981a [DOI] [Google Scholar]
- Boni, M. F. , Lemey, P. , Jiang, X. , Lam, T.‐Y. , Perry, B. W. , Castoe, T. A. , Rambaut, A. , & Robertson, D. L. (2020). Evolutionary origins of the SARS‐CoV‐2 sarbecovirus lineage responsible for the COVID‐19 pandemic. Nature Microbiology, 5(11), 1408–1417. 10.1038/s41564-020-0771-4 [DOI] [PubMed] [Google Scholar]
- Bosco‐Lauth, A. M. , Hartwig, A. E. , Porter, S. M. , Gordy, P. W. , Nehring, M. , Byas, A. D. , VandeWoude, S. , Ragan, I. K. , Maison, R. M. , & Bowen, R. A. (2020). Experimental infection of domestic dogs and cats with SARS‐CoV‐2: Pathogenesis, transmission, and response to reexposure in cats. Proceedings of the National Academy of Sciences of the United States of America, 117(42), 26382–26388. 10.1073/pnas.2013102117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun, K. M. , Moreno, G. K. , Halfmann, P. J. , Baker, D. A. , Weiler, A. M. , Haj, A. K. , Friedrich, T. C. (2020). Transmission of SARS‐CoV‐2 in 1 domestic cats imposes a narrow 2 bottleneck 3. BioRxiv, 2020.11.16.384917. 10.1101/2020.11.16.384917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlos, R. S. A. , Mariano, A. P. M. , Maciel, B. M. , Gadelha, S. R. , Melo Silva, M. , Belitardo, E. M. M. A. , Rocha, D. J. P. G. , Almeida, J. P. P. , Pacheco, L. G. C. , Aguiar, E. R. G. R. , Fehlberg, H. F. , & Albuquerque, G. R. (2021). First genome sequencing of SARS‐CoV‐2 recovered from an infected cat and its owner in Latin America. Transboundary and Emerging Diseases. 10.1111/tbed.13984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) (2020). CDC 2019‐Novel Coronavirus (2019‐nCoV) Real‐Time RT‐PCR Diagnostic Panel. Retrieved from https://www.cdc.gov/coronavirus/2019‐ncov/lab/rt‐pcr‐panel‐primer‐probes.html. Accessed December 31, 2020 [Google Scholar]
- Chaintoutis, S. C. , Papadopoulou, E. , Melidou, A. , Papa, A. , & Dovas, C. I. (2019). A PCR‐based NGS protocol for whole genome sequencing of West Nile virus lineage 2 directly from biological specimens. Molecular and Cellular Probes, 46, 101412. 10.1016/j.mcp.2019.06.002 [DOI] [PubMed] [Google Scholar]
- Chan, J. F. W. , Kok, K. H. , Zhu, Z. , Chu, H. , To, K. K. W. , Yuan, S. , & Yuen, K. Y. (2020). Genomic characterization of the 2019 novel human‐pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerging Microbes and Infections, 9(1), 221–236. 10.1080/22221751.2020.1719902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao, Y. X. , Rötzschke, O. , & Tan, E. K. (2020). The role of IgA in COVID‐19. Brain, Behavior, and Immunity, 87, 182–183. 10.1016/j.bbi.2020.05.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani, P. , Platts, A. , Wang, L. L. , Coon, M. , Nguyen, T. , Wang, L. , Land, S. J. , Lu, X. , & Ruden, D. M. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso‐2; iso‐3. Fly, 6(2), 80–92. 10.4161/fly.19695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppée, F. , Lechien, J. R. , Declèves, A. E. , Tafforeau, L. , & Saussez, S. (2020). Severe acute respiratory syndrome coronavirus 2: Virus mutations in specific European populations. New Microbes and New Infections, 36, 100696. 10.1016/j.nmni.2020.100696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman, V. M. , Landt, O. , Kaiser, M. , Molenkamp, R. , Meijer, A. , Chu, D. K. W. , Drosten, C. (2020). Detection of 2019 novel coronavirus (2019‐nCoV) by real‐time RT‐PCR. Eurosurveillance, 25(3). 10.2807/1560-7917.ES.2020.25.3.2000045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Morais, H. A. , dos Santos, A. P. , do Nascimento, N. C. , Kmetiuk, L. B. , Barbosa, D. S. , Brandão, P. E. , Guimarães, A. M. S. , Pettan‐Brewer, C. , & Biondo, A. W. (2020). Natural Infection by SARS‐CoV‐2 in companion animals: A review of case reports and current evidence of their role in the epidemiology of COVID‐19. Frontiers in Veterinary Science, 7, 1–10. 10.3389/fvets.2020.591216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhama, K. , Patel, S. K. , Sharun, K. , Pathak, M. , Tiwari, R. , Yatoo, M. I. , Malik, Y. S. , Sah, R. , Rabaan, A. A. , Panwar, P. K. , Singh, K. P. , Michalak, I. , Chaicumpa, W. , Martinez‐Pulgarin, D. F. , Bonilla‐Aldana, D. K. , & Rodriguez‐Morales, A. J. (2020). SARS‐CoV‐2 jumping the species barrier: Zoonotic lessons from SARS, MERS and recent advances to combat this pandemic virus. Travel Medicine and Infectious Disease, 37, 101830. 10.1016/j.tmaid.2020.101830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler, Y. , Alcaraz, A. , Bossong, F. J. , Collisson, E. W. , & Diniz, P. P. V. P. (2011). Feline coronavirus in multicat environments. Veterinary Clinics of North America: Small Animal Practice, 41(6), 1133–1169. 10.1016/j.cvsm.2011.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallais, F. , Velay, A. , Wendling, M.‐J. , Nazon, C. , Partisani, M. , Sibilia, J. , Fafi‐Kremer, S. (2020). Intrafamilial Exposure to SARS‐CoV‐2 Induces Cellular Immune Response without Seroconversion. MedRxiv, 2020.06.21.20132449. 10.1101/2020.06.21.20132449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaudreault, N. N. , Trujillo, J. D. , Carossino, M. , Meekins, D. A. , Morozov, I. , Madden, D. W. , Indran, S. V. , Bold, D. , Balaraman, V. , Kwon, T. , Artiaga, B. L. , Cool, K. , García‐Sastre, A. , Ma, W. , Wilson, W. C. , Henningson, J. , Balasuriya, U. B. R. , & Richt, J. A. (2020). SARS‐CoV‐2 infection, disease and transmission in domestic cats. Emerging Microbes and Infections, 9(1), 2322–2332. 10.1080/22221751.2020.1833687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadfield, J. , Megill, C. , Bell, S. M. , Huddleston, J. , Potter, B. , Callender, C. , Sagulenko, P. , Bedford, T. , & Neher, R. A. (2018). NextStrain: Real‐time tracking of pathogen evolution. Bioinformatics, 34(23), 4121–4123. 10.1093/bioinformatics/bty407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfmann, P. J. , Hatta, M. , Chiba, S. , Maemura, T. , Fan, S. , Takeda, M. , Kinoshita, N. , Hattori, S.‐I. , Sakai‐Tagawa, Y. , Iwatsuki‐Horimoto, K. , Imai, M. , & Kawaoka, Y. (2020). Transmission of SARS‐CoV‐2 in domestic cats. New England Journal of Medicine, 383(6), 592–594. 10.1056/nejmc2013400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou, Y. J. , Chiba, S. , Halfmann, P. , Ehre, C. , Kuroda, M. , Dinnon, K. H. , Leist, S. R. , Schäfer, A. , Nakajima, N. , Takahashi, K. , Lee, R. E. , Mascenik, T. M. , Graham, R. , Edwards, C. E. , Tse, L. V. , Okuda, K. , Markmann, A. J. , Bartelt, L. , de Silva, A. , … Baric, R. S. (2020). SARS‐CoV‐2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science, 370(6523), eabe8499. 10.1126/science.abe8499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Lee, J. Y. , Yang, J. S. , Kim, J. W. , Kim, V. N. , & Chang, H. (2020). The architecture of SARS‐CoV‐2 transcriptome. Cell, 181(4), 914–921.e10. 10.1016/j.cell.2020.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , & Durbin, R. (2009). Fast and accurate short read alignment with Burrows‐Wheeler transform. Bioinformatics, 25(14), 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Handsaker, B. , Wysoker, A. , Fennell, T. , Ruan, J. , Homer, N. , Marth, G. , Abecasis, G. , & Durbin, R. (2009). The Sequence alignment/map format and SAMtools. Bioinformatics, 25(16), 2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, R. , Zhao, X. , Li, J. , Niu, P. , Yang, B. O. , Wu, H. , Wang, W. , Song, H. , Huang, B. , Zhu, N. A. , Bi, Y. , Ma, X. , Zhan, F. , Wang, L. , Hu, T. , Zhou, H. , Hu, Z. , Zhou, W. , Zhao, L. I. , … Tan, W. (2020). Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. The Lancet, 395(10224), 565–574. 10.1016/S0140-6736(20)30251-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan, J. , Lu, Y. , Jin, X. , & Zhang, L. (2020). Spike protein recognition of mammalian ACE2 predicts the host range and an optimized ACE2 for SARS‐CoV‐2 infection. Biochemical and Biophysical Research Communications, 526(1), 165–169. 10.1016/j.bbrc.2020.03.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, Y. S. , Sircar, S. , Bhat, S. , Sharun, K. , Dhama, K. , Dadar, M. , Tiwari, R. , & Chaicumpa, W. (2020). Emerging novel coronavirus (2019‐nCoV)—current scenario, evolutionary perspective based on genome analysis and recent developments. Veterinary Quarterly, 40(1), 68–76. 10.1080/01652176.2020.1727993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, E. I. , Elia, G. , Grassi, A. , Giordano, A. , Desario, C. , Medardo, M. , Smith, S. L. , Anderson, E. R. , Prince, T. , Patterson, G. T. , Lorusso, E. , Lucente, M. S. , Lanave, G. , Lauzi, S. , Bonfanti, U. , Stranieri, A. , Martella, V. , Solari Basano, F. , Barrs, V. R. , … Decaro, N. (2020). Evidence of exposure to SARS‐CoV‐2 in cats and dogs from households in Italy. Nature Communications, 11(1), 6231. 10.1038/s41467-020-20097-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante, J. A. , Liu, Y. , Liu, J. , Xia, H. , Johnson, B. A. , Lokugamage, K. G. , Shi, P. Y. (2021). Spike mutation D614G alters SARS‐CoV‐2 fitness. Nature, 592(7852), 116–121. 10.1038/s41586-020-2895-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, M. N. , Dehal, P. S. , & Arkin, A. P. (2010). FastTree 2 ‐ Approximately maximum‐likelihood trees for large alignments. PLoS One, 5(3), e9490. 10.1371/journal.pone.0009490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut, A. , Holmes, E. C. , O’Toole, Á. , Hill, V. , McCrone, J. T. , Ruis, C. , du Plessis, L. , & Pybus, O. G. (2020). A dynamic nomenclature proposal for SARS‐CoV‐2 lineages to assist genomic epidemiology. Nature Microbiology, 5, 1403–1407. 10.1038/s41564-020-0770-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailleau, C. , Dumarest, M. , Vanhomwegen, J. , Delaplace, M. , Caro, V. , Kwasiborski, A. , Hourdel, V. , Chevaillier, P. , Barbarino, A. , Comtet, L. , Pourquier, P. , Klonjkowski, B. , Manuguerra, J.‐C. , Zientara, S. , & Le Poder, S. (2020). First detection and genome sequencing of SARS‐CoV‐2 in an infected cat in France. Transboundary and Emerging Diseases, 67(6), 2324–2328. 10.1111/tbed.13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segalés, J. , Puig, M. , Rodon, J. , Avila‐Nieto, C. , Carrillo, J. , Cantero, G. , Terrón, M. T. , Cruz, S. , Parera, M. , Noguera‐Julián, M. , Izquierdo‐Useros, N. , Guallar, V. , Vidal, E. , Valencia, A. , Blanco, I. , Blanco, J. , Clotet, B. , & Vergara‐Alert, J. (2020). Detection of SARS‐CoV‐2 in a cat owned by a COVID‐19‐affected patient in Spain. Proceedings of the National Academy of Sciences of the United States of America, 117(40), 24790–24793. 10.1073/pnas.2010817117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, J. , Wen, Z. , Zhong, G. , Yang, H. , Wang, C. , Huang, B. , Liu, R. , He, X. , Shuai, L. , Sun, Z. , Zhao, Y. , Liu, P. , Liang, L. , Cui, P. , Wang, J. , Zhang, X. , Guan, Y. , Tan, W. , Wu, G. , … Bu, Z. (2020). Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS‐coronavirus 2. Science, 368(6494), 1016–1020. 10.1126/science.abb7015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers, F. , & Higgins, D. G. (2014). Clustal omega. Current Protocols in Bioinformatics, 48(1), 1–3. 10.1002/0471250953.bi0313s48 [DOI] [PubMed] [Google Scholar]
- Sironi, M. , Hasnain, S. E. , Rosenthal, B. , Phan, T. , Luciani, F. , Shaw, M.‐A. , Sallum, M. A. , Mirhashemi, M. E. , Morand, S. , & González‐Candelas, F. (2020). SARS‐CoV‐2 and COVID‐19: A genetic, epidemiological, and evolutionary perspective. Infection, Genetics and Evolution, 84, 104384. 10.1016/j.meegid.2020.104384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staines, H. , Kirwan, D. , Clark, D. , Adams, E. , Augustin, Y. , Byrne, R. , Planche, T. (2020). Dynamics of IgG seroconversion and pathophysiology of COVID‐19 infections. MedRxiv, 2020.06.07.20124636. 10.1101/2020.06.07.20124636 [DOI] [Google Scholar]
- Temmam, S. , Barbarino, A. , Maso, D. , Behillil, S. , Enouf, V. , Huon, C. , Eloit, M. (2020). Absence of SARS‐CoV‐2 infection in cats and dogs in close contact with a cluster of COVID‐19 patients in a veterinary campus. BioRxiv, 2020.04.07.029090. 10.1101/2020.04.07.029090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totton, S. C. , Sargeant, J. M. , & O’Connor, A. M. (2020). How could we conclude cat‐to‐human transmission of SARS‐CoV‐2? Zoonoses and Public Health, 68(1), 67–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, Y. , Shang, J. , Graham, R. , Baric, R. S. , & Li, F. (2020). Receptor recognition by the novel coronavirus from Wuhan: An analysis based on decade‐long structural studies of SARS coronavirus. Journal of Virology, 94(7), 1–9. 10.1128/jvi.00127-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm, A. , Aw, P. P. K. , Bertrand, D. , Yeo, G. H. T. , Ong, S. H. , Wong, C. H. , Khor, C. C. , Petric, R. , Hibberd, M. L. , & Nagarajan, N. (2012). LoFreq: A sequence‐quality aware, ultra‐sensitive variant caller for uncovering cell‐population heterogeneity from high‐throughput sequencing datasets. Nucleic Acids Research, 40(22), 11189–11201. 10.1093/nar/gks918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel, R. , Corman, V. M. , Guggemos, W. , Seilmaier, M. , Zange, S. , Müller, M. A. , Niemeyer, D. , Jones, T. C. , Vollmar, P. , Rothe, C. , Hoelscher, M. , Bleicker, T. , Brünink, S. , Schneider, J. , Ehmann, R. , Zwirglmaier, K. , Drosten, C. , & Wendtner, C. (2020). Virological assessment of hospitalized patients with COVID‐2019. Nature, 581(7809), 465–469. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- Wu, F. , Zhao, S. U. , Yu, B. , Chen, Y.‐M. , Wang, W. , Song, Z.‐G. , Hu, Y. I. , Tao, Z.‐W. , Tian, J.‐H. , Pei, Y.‐Y. , Yuan, M.‐L. , Zhang, Y.‐L. , Dai, F.‐H. , Liu, Y. I. , Wang, Q.‐M. , Zheng, J.‐J. , Xu, L. , Holmes, E. C. , & Zhang, Y.‐Z. (2020). A new coronavirus associated with human respiratory disease in China. Nature, 579(7798), 265–269. 10.1038/s41586-020-2008-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Q. , Zhang, H. , Gao, J. , Huang, K. , Yang, Y. , Hui, X. , He, X. , Li, C. , Gong, W. , Zhang, Y. , Zhao, Y. A. , Peng, C. , Gao, X. , Chen, H. , Zou, Z. , Shi, Z.‐L. , & Jin, M. (2020). A serological survey of SARS‐CoV‐2 in cat in Wuhan. Emerging Microbes and Infections, 9(1), 2013–2019. 10.1080/22221751.2020.1817796 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The nucleotide sequence of the strain "hCoV‐19/cat/Greece/2K/2020" is available at GISAID (EPI_ISL_717979).


