Highlights
-
•
The role of autophagy in liver cancer is controversial.
-
•
Autophagy gene, Atg7 is a key regulator for autophagy process.
-
•
Transgenic mouse model for liver cancer can be generated via simple transgenic methodology called “Hydrodynamic Tail Vein Injection”.
-
•
Genetic suppression of Atg7 significantly suppressed development of liver cancer induced by activated RAS.
Keywords: Autophagy, Liver Cancer, Hydrodynamic Transfection, Murine Cancer Model, Transgenesis
Abstract
Hepatocellular Carcinoma (HCC) is the most common type of primary liver cancer in adults and a leading cause of cancer-related deaths worldwide. Studies have shown that autophagy is significantly involved in carcinogenesis, in particular, driven by activated RAS signaling. Autophagy related 7 (Atg7) is a critical component for the formation of autophagosome and required for autophagy processes. We investigated the role of autophagy in RAS-driven tumorigenesis in the liver, via the knockdown of Atg7 in the model. Transposon vectors encoding short hairpin RNAs targeting Atg7 (Atg7 shRNA) were constructed. Inhibition of autophagy via Atg7 knockdown was tested in Hep3B cells cultured in nutrient-starved medium. Formation of autophagosome was suppressed in nutrient-starved Hep3B cells expressing Atg7 shRNA, demonstrating that it efficiently inhibited autophagy in HCC cells. Transposons encoding Atg7 shRNA were mixed with those expressing HRASG12V and p53 shRNA, and subsequently used for hydrodynamic injection to 5-week-old C57BL/6 mice. Tumorigenesis in livers induced by HRASG12V and p53 shRNA was significantly suppressed by Atg7 knockdown. The inhibition of autophagy led to a decreased proliferation of cancer cells, as determined by Ki-67 staining. Our data indicate that knockdown of Atg7 led to a significant decrease in tumorigenesis in a murine HCC model induced by activated RAS. Inhibition of autophagosome formation is expected to be a therapeutic option for liver cancer.
Introduction
Hepatocellular Carcinoma (HCC) is the most common type of primary liver cancer in adults and causes about 800,000 deaths globally each year [1], [2], [3], [4], [5], [6]. Genetic studies have revealed that initiation and progression of HCC involves genetic and epigenetic changes in various genes which lead to activation of diverse oncogenic signaling pathways [7], [8], [9]. The RAS signaling pathway promotes cellular proliferation, growth, and survival, which is found frequently activated in most human cancers including HCC [10]. As well, the signaling pathway regulates cancer metabolism and autophagy [11], [12], [13].
Autophagy is a cellular recycling process by which macromolecules and organelles can be degraded in lysosomes to sustain metabolism during starvation [14]. During the process, an isolated membrane expands and wraps around portions of the cytoplasm to form an autophagosome, a double-membraned vesicle [15]. Autophagosome subsequently fuses with a lysosome to generate an autolysosome in which inner particles such as macromolecules and organelles are degraded by lysosomal proteases. In cancer cells with activated RAS signaling, autophagy is found frequently up-regulated, compared with that in normal cells, or cancer cells without activated RAS signaling [16,17]. Moreover, in murine xenograft and autochthonous models of RAS-driven pancreatic and lung cancers, inhibition of autophagy led to a substantial decrease in tumor growths [18,19]. In cancers showing activated RAS signaling, autophagy was found essential for cell survival by regulating cellular energy metabolism [20], [21], [22], [23].
Considering that the RAS signaling pathway is upregulated in more than 50% of HCC [10], we were curious about whether HCC with activated RAS signaling is also vulnerable to autophagic inhibition. For the purpose, we employed the hydrodynamics-based transfection method to develop a murine autochthonous HCC model driven by an activated RAS, and investigated effects of the inhibition of autophagy on the tumor development.
Materials and methods
Plasmids
Plasmids used in this study have been previously described [24,25]. The open reading frame (ORF) of murine Atg7 was PCR-amplified from murine cDNAs and cloned into pcDNA3 vector. The ORF encoding GFP-LC3 was PCR-amplified from pEGFP-LC3 (Addgene #24,920) and placed immediately before shRNA-coding region. The target sequence in the murine Atg7 mRNA recognized by each Atg7 shRNA was listed in Supplementary Table 1.
Animal experiment
All experiments using mice were approved by the institution's animal policy and welfare committee. Wild-type male C57BL/6 mice were purchased from Orient Bio (Korea). Animals were housed in an animal facility under a 12 h light/dark cycle and were provided food and water ad libitum. Hydrodynamic injection has also been previously described [25]. Briefly, DNA mixtures were suspended in lactated Ringer's solution and subsequently injected into the lateral tail veins of male 5–6-week-old mice (0.1 mL/g body weight). Mice were randomly assigned to hydrodynamic injection. Mice were treated with chloroquine (Sigma-Aldrich Inc., St Louis, MO) at a daily dose of 100 mg/kg body weight via intraperitoneal injection.
Transfection and Western blotting
Cells were plated 3 × 105 cells per well on 6 well plate one day prior to transfection so that the cells were approximately 80% confluent on the day of transfection. Cells were transiently transfected with 2 μg of DNA using 6μl FuGENE® HD Transfection Reagent (Promega, Madison, WI, USA) according to the manufacturer's instructions. Transfected cells were harvested 2 days post-transfection using the 10X RIPA buffer (Cell Signaling, Denvers, MA, USA) according to the manufacturer's instructions. Cells were briefly washed with PBS to remove residual media. 200 μl of 1X RIPA buffed and diluted in DW were added to each well and then were incubated on ice for 5 min. Afterwards, cells were scrapped with a scrapper and transferred to a new tube. For 30–60 min, cells were placed on ice, and extracts were centrifuged for 10 min at 14,000 RPM in a cold microcentrifuge. Finally, supernatants were removed for use. Western blot experiments were performed using standard methods. Primary antibodies used are anti-ATG7 (Wako 013–22,831, 1/1000 dilution), anti-GAPDH (CST #2118, 1/3000 dilution), anti-LC3B (CST #2775, 1/1000 dilution), and anti-p62 (ABCAM ab56416, 1/200 dilution) antibodies.
Fluorescence imaging
Fluorescence imaging of GFP was performed to confirm transfection of cells using an inverted microscope (IX71, Olympus, Japan) equipped with a 10X objective lens. For imaging of GFP-LC3 puncta within cells, transfected cells were fixed with 4% paraformaldehyde, stained with 4′,6-diamidino-2-phenylindole (DAPI) for two minutes at room temperature, and then imaged using an inverted laser-scanning microscope (Zeiss LSM 510), equipped with a 40X objective.
Liver harvesting and tissue processing
Mice were deeply anesthetized by intraperitoneal injection of Zoletil (30 mg/kg) and xylazine (10 mg/kg). A midline laparotomy incision was performed, and livers were removed and photographed. Pieces of extracted mouse liver samples were immersed in freshly prepared 10% neutral-buffered formalin overnight. Fixed tissue samples were embedded in paraffin and serially sectioned into 4 µm–thick slices. Slices were stained with hematoxylin & eosin (H&E).
Immunohistochemical analyses for GFP and KI-67
For immunohistochemistry, paraffin-embedded sections were deparaffinized in xylene and rehydrated through a gradual decrease in ethanol concentration. Antigen epitopes were then unmasked using a 10 mM sodium citrate buffer (pH 6.0) incubation procedure, after which sections were incubated overnight at 4 °C with the primary antibody against GFP (CST #2555). After incubation with primary antibodies, sections were incubated with the biotinylated secondary antibody followed by treatment with freshly prepared DAB substrates (Vector Laboratories, USA). Sections were lightly counter-stained with hematoxylin and mounted. For Ki-67 assay, the sections were incubated overnight at 4 °C using anti-Ki-67 primary antibodies (CST #9449). After primary antibody incubation, sections were incubated with the appropriate biotinylated secondary antibodies followed by the treatment with freshly prepared DAB substrates (SK-4100; Vector Laboratories). Sections were lightly counter-stained with hematoxylin and mounted. Positively stained nuclei in a field view under a 400 × magnification was counted. Area of view was randomly chosen. Slides were photographed using a microscope (Eclipse Ti; Nikon, Tokyo, Japan) equipped with a digital camera.
Statistical analysis
The liver weight/body weight ratio (LW/BW) data were expressed as the mean ± SEM with a sample size of n = 10. Statistical analyses were conducted via an unpaired parametric Student's t-test. Significant differences between two groups were noted by asterisks (*, p < 0.05; **, p < 0.01).
Results
Knockdown of Atg7 using short hairpin RNA
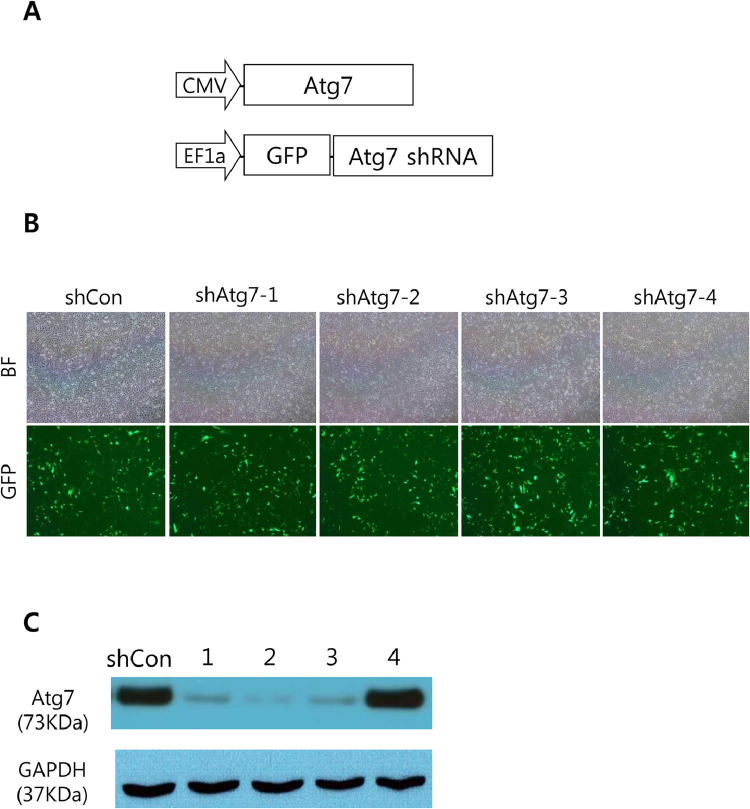
To investigate the effect of autophagy inhibition on the development of liver cancer, we chose to down-regulate Atg7 using short hairpin RNA (shRNA). Atg7 is an essential component for the autophagosome formation, and genetic abrasion of Atg7 successfully inhibited autophagy processes in murine models [26]. We selected 4 kinds of shRNAs targeting different locations in the murine Atg7 mRNA. To test knockdown efficiency of each Atg7 shRNA, NIH3T3 cells was co-transfected with plasmids encoding each Atg7 shRNA and those expressing murine Atg7 (Fig. 1A). Transfection of cells with plasmids expressing individual shRNA was verified using a co-expressed reporter protein, green fluorescent protein (GFP) (Fig. 1B). We found that the knockdown of Atg7 was efficiently achieved by the Atg7 shRNAs that we constructed, except for one case (Fig. 1C and Supplementary Figure 1). Among the three shRNAs that efficiently down-regulated Atg7, shAtg7–2 was the most effective, and thus was chosen for following study.
Fig. 1.
Efficiencies of Atg7 knockdown by shRNAs. (A) Plasmids used for the transfection experiment. (B) Bright field (upper panels) and fluorescence (lower panels) images of NIH3T3 cells transfected with indicated shRNAs. The shCon indicates control shRNA. (C) Western blots showing expression levels of Atg7 in cells transfected with plasmids encoding indicated shRNA (1, shAtg7–1; 2, shAtg7–2; 3, shAtg7–3; 4, shAtg7–4).
Knockdown of Atg7 inhibits autophagic processes in HCC cells upon starvation
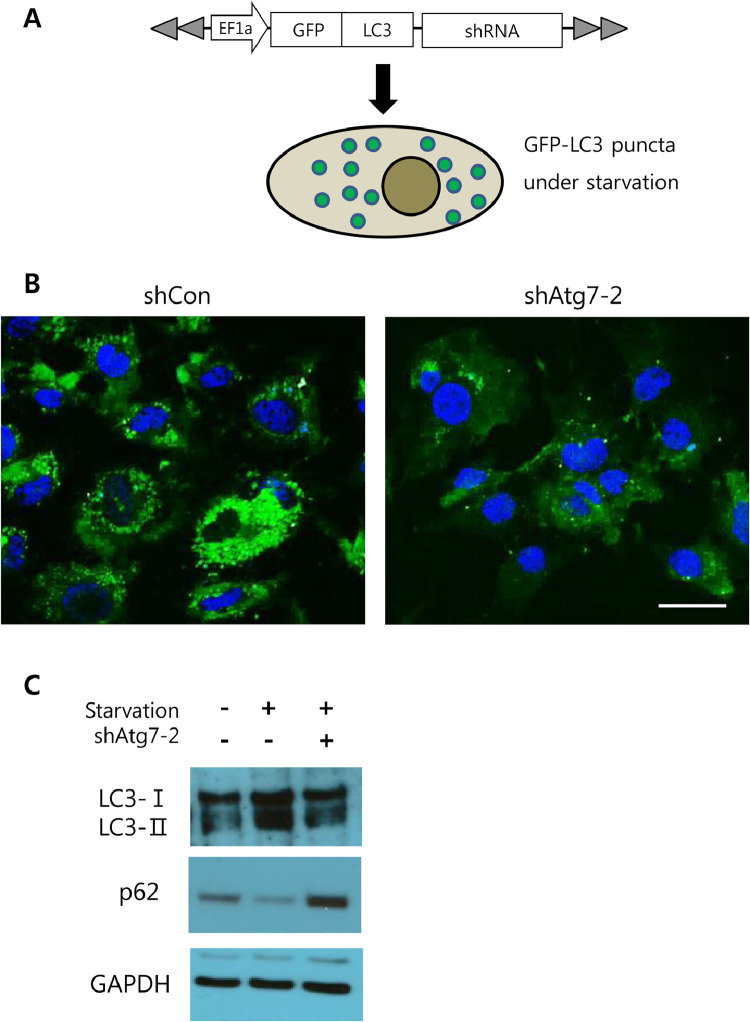
To verify that knockdown of Atg7 inhibits autophagy in HCC cells, Hep3B cells were transfected with plasmids simultaneously expressing Atg7 shRNA and GFP-fused LC3 (GFP-LC3). At 24 h after transfection, cells were subjected to starvation for 4 h to activate autophagy (Fig. 2A). GFP-LC3 is a marker protein for active autophagic processes which allows autophagosome structures to be visualized as green fluorescent punctate signals (also known as GFP-LC3 puncta) under fluorescence microscopy [27]. Following starvation, Hep3B cells expressing control shRNA (shCon) showed numerous GFP-LC3 puncta within cells, whereas those expressing Atg7 shRNA (shAtg7–2) rarely revealed green fluorescent puncta (Fig. 2B). As well, shAtg7–2 expression in nutrient-deprived Hep3B cells led to an elevated level of p62, an indicator for autophagy inhibition (Fig. 2C) [28,29]. Further, starvation led to an increase in the LC3-II level in Hep3B cells expressing control shRNA, which indicates up-regulation of autophagosome formation [29]. However, starvation failed to increase the LC3-II level in Hep3B cells expressing shAtg7–2, which strongly suggests that formation of autophagosome was efficiently suppressed by the Atg7 knockdown (Fig. 2C and Supplementary Figure 2). Overall, the data demonstrate that the Atg7 shRNA can efficiently suppress autophagy activity in HCC cells via down-regulation of Atg7.
Fig. 2.
Inhibition of autophagy by Atg7 shRNA (A) Schematic illustration of the experimental procedure. Hep3B cells were transfected with plasmids expressing GFP-LC3 and shRNA and then subjected to starvation (B) Representative fluorescence images of Hep3B cells transfected with plasmids encoding indicated shRNA upon starvation. Cells were stained with DAPI to reveal nuclei (blue). Scale bar, 10 μm. (C) Western blots showing expression levels of LC3-I, LC3-II, and p62 in Hep3B cells depending on starvation (+ indicates starvation) and expression of shAtg7–2 (+ indicates shAtg7–2 expression).
Knockdown of Atg7 suppresses hepatocarcinogenesis in mice
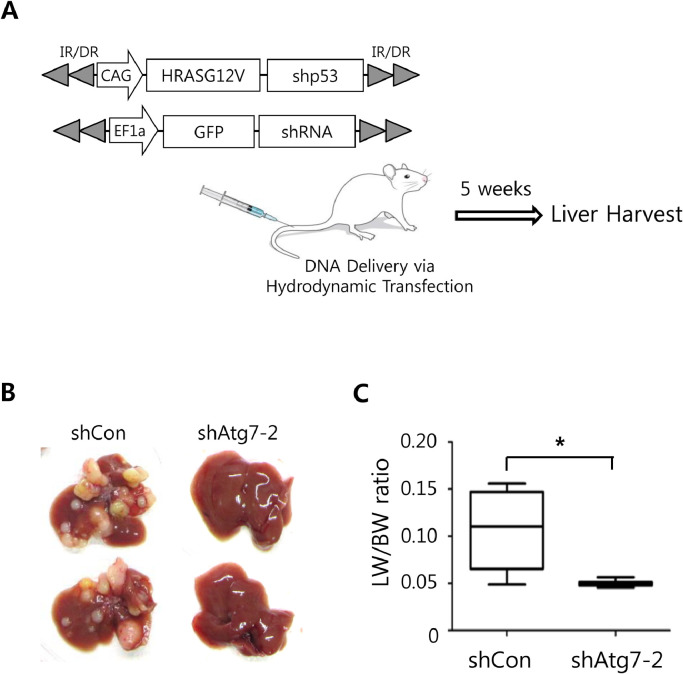
To investigate whether inhibition of autophagy affects tumor development driven by activated RAS signaling in the liver, we employed a simple liver-specific transgenic approach in which transposons encoding a constitutively active form of RAS (HRASG12V) and p53 shRNA were co-delivered to murine livers with those encoding Atg7 shRNA through hydrodynamic tail vein injection (HTVI) (Fig. 3A). Ten mice were assigned to each group. When livers were harvested at 5 weeks after the hydrodynamic injection, we found that expression of shAtg7–2 significantly inhibited hepatic tumorigenesis induced by HRASG12V and p53 shRNA, when compared with the control group (Fig. 3B). Numbers and sizes of tumors were notably reduced in the livers expressing shAtg7–2 compared with the control group (Table 1). Liver weight per body weight (LW/BW), often used to evaluate tumor burden in liver, was also significantly reduced in the Atg7 knockdown group, compared with the control (Fig. 3C).
Fig. 3.
Inhibition of autophagy led to tumor suppression in the liver. (A) Schematic illustration of the experimental procedure. Liver tumors were induced by HRASG12V and shp53. Atg7 shRNA (or control shRNA) was expressed in the tumor to investigate effects of Atg7 knockdown on liver cancer. (B) Gross morphology of representative livers expressing either control shRNA (shCon) and Atg7 shRNA (shAtg7–2). Livers were harvested at 5 weeks after hydrodynamic injection. (n = 10 for each group) (C) Liver weight/body weight (LW/BW) ratios of mice expressing shCon and shAtg7–2. The graph represents the mean ± SEM (n = 10 livers per group) (*, p < 0.05).
Table 1.
Tumor nodules in a mouse model of HCC following ATG7 knockdown.
| Group | Total # of tumors | # of tumors over 3 mm in diameter |
|---|---|---|
| shCon | TMTC | >10 |
| shAtg7–2 | 2 ± 0.5 | 0 |
TMTC, Too many to count.
To confirm that autophagy inhibition suppresses tumorigenesis induced by HRASG12V and p53 shRNA in the liver, mice were treated with an autophagy inhibitor, chloroquine following HTVI [30]. In line with the tumor suppression by the stable knockdown of Atg7 in the model, the treatment with chloroquine also suppressed tumor growth in the model (Supplementary Figure 3).
Knockdown of Atg7 suppresses cellular proliferation in HCC cells
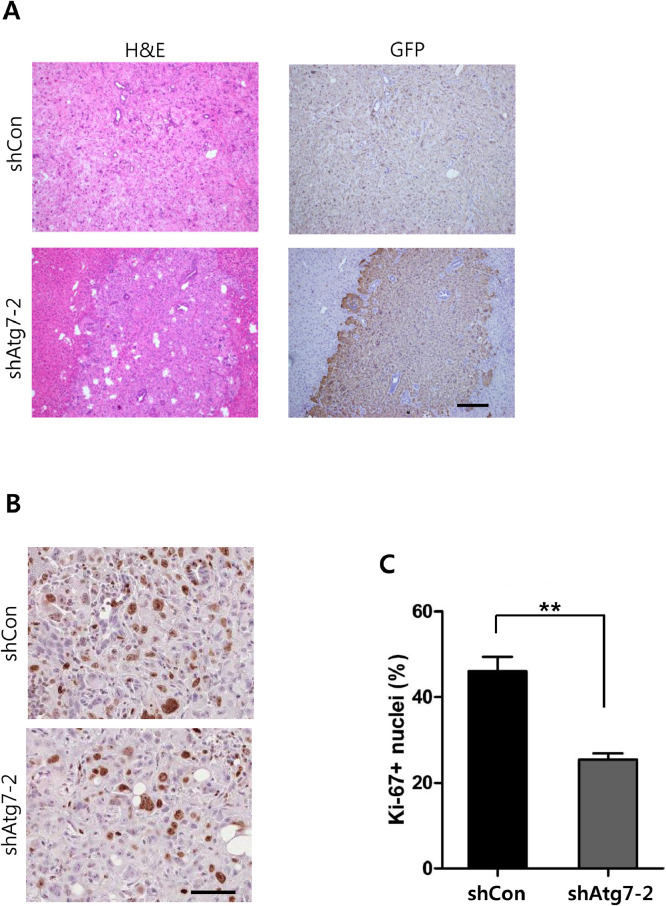
Morphological differences in cancer cells were not apparent between the shAtg7–2 group and control (Fig. 4A). All tumor nodules were GFP-positive, confirming shRNA expression in the tumors. To investigate the cellular mechanism underlying reduced tumor growths in the shAtg7–2 group, tumor tissues were analyzed for cellular proliferation and apoptosis. Of note, cellular proliferation was significantly lower in the shAtg7–2 group compared with that in the control, when determined by Ki-67 nuclear staining (Fig. 4B and C), while apoptosis levels were not significantly different between the two groups. Thus, the data suggest that in the liver cancer induced by activated RAS signaling, inhibition of autophagy suppressed tumor development via down regulating cellular proliferation. To further confirm that Atg7 knockdown can affect proliferation of HCC cells, MTT assay was performed using Hep3B cells stably expressing shAtg7–2. In line with the in vivo finding, inhibition of Atg7 in the HCC cells led to decreased cell proliferation compared with Hep3B cells expressing control shRNA (Supplementary Figure 4).
Fig. 4.
Microscopic examination of liver tumors. (A) H&E and IHC image of paraffin-embedded sections from shCon and shAtg7–2 tumors. GFP was used as a reporter for shRNA expression. Scale bar, 200 µm. Inhibition of autophagy led to tumor suppression in the liver. (B) Representative images of Ki-67 staining in the shCon and shAtg7–2 tumors. Scale bar, 50 µm. (C) Quantification of nuclei positive for Ki-67 staining in tumors of each group. The values represent percentages of Ki-67-positivity among the total nuclei. The graphs represent the mean ± SEM. (**, p < 0.01).
Discussion
The RAS signaling pathway is activated through receptor tyrosine kinases (RTKs) [31,32]. Ligand binding to the receptor induces receptor dimerization which leads to transphosphorylation at tyrosine residues of the cytoplasmic tails by the receptor's intracellular tyrosine kinase activity [33]. The phosphorylated tyrosine residues recruit the Grb2/Shc/SOS protein complex to plasma membrane, which converts GDP-bound inactive RAS to GTP-bound active RAS. This event leads to subsequent activation of the mitogen-activated protein (MAP) kinase signaling cascade through the RAF-MEK-ERK axis.
The importance of the RAS signaling pathway in human HCC has been neglected for a long time, understandably due to low frequencies of mutation in RAS and RAF in HCC [34]. However, based on MEK/ERK expression and phosphorylation, the RAS signaling pathway is found activated in approximately 50% of early-stage HCC and in most advanced-stage HCC [35,36]. The activity of the RAS signaling pathway in HCC can be elevated by hepatitis viral proteins, dysregulated RTKs, inactivation of Raf kinase inhibitor proteins, and etc. [37].
RAS signaling is involved in cell growth and proliferation [10]. The signaling pathway is also highly involved in regulating cancer metabolism [38,39]. The Ras signaling can induce activation of the glycolytic pathway and glutamine metabolism pathways which cancers preferentially utilize to meet their high metabolic needs.
Cancer with activated RAS signaling is heavily dependent on autophagy, as verified in murine models of RAS-driven pancreatic and lung cancers which showed high susceptibilities to autophagic suppression [18,19,40]. Autophagy can contribute to tumor growth during carcinogenesis by supplying neoplastic cells with nutrients such as amino acids, free fatty acids (FFA) and glucose. Consistent with cancer models of other tissues, our data showed that tumor development in murine livers driven by activated RAS signaling was significantly suppressed by the inhibition of autophagy through Atg7 knockdown.
Although our study supports pro-tumorigenic effects of autophagy, tumor-suppressive roles of autophagy have also been reported in HCC. Heterozygous deletion of the gene encoding Beclin1, another key molecule in the autophagy pathway, induced development of spontaneous tumors in the liver, [41,42]. Likewise, mice with systemic mosaic deletion of Atg5 or liver-specific deletion of Atg7 developed benign hepatic adenomas [43]. Thus, it is presumed that functional autophagy is required to suppress tumorigenesis in the liver and its deregulation can promotes neoplastic transformation of hepatocytes. Autophagy removes cellular wastes and damaged proteins, accumulation of which can cause oxidative damages to DNA, leading to genomic instability [44,45]. In line with this, mice with liver-specific deletion of Atg5 or Atg7 showed increased oxidative DNA damages in the livers [46].
On the contrary, autophagy contributes to survival of cancer cells by providing them with nutrients required for increased energy metabolism and removing toxic reactive oxygen radicals and mis-folded proteins [47,48]. Thus, inhibition of autophagy can render cancer cells susceptible to cytotoxic substances. It is tempting to think that autophagy inhibition initiates neoplastic transformation in normal hepatocytes by inducing oncogenic mutations, however, once oncogenic drivers have been established, sustained inhibition of autophagy will exert cytotoxic effects on tumor cells, hindering tumor growth. Further mechanistic studies are required to precisely determine the role of autophagy in HCC.
Authors’ contributions
KJC, SYS, HM, BKK, and SWR designed experiments, analyzed data, and wrote the manuscript. KJC and SYS performed cell culture-based experiments. HM performed animal experiments. BKK and SWR supervised the research. All authors read and approved the final manuscript.
Declaration of Competing Interest
The authors have declared that no competing interest exists.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grants (2018R1C1B6009434 awarded to BKK and 2019R1A2C2009518 awarded to SWR) which were funded by the Korea government (MSIT). This work was also supported by a faculty research grant of Yonsei University College of Medicine (6–2019–0072 awarded to BKK) and by a grant from Kyung Hee University in 2020 (KHU-20201114 awarded to SWR).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101158.
Contributor Information
Beom Kyung Kim, Email: beomkkim@yuhs.ac.
Simon Weonsang Ro, Email: simonro@khu.ac.kr.
Appendix. Supplementary materials
References
- 1.Llovet J.M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M., Gores G. Hepatocell. Carcinoma. Nat. Rev .Dis. Prim. 2016;2:16018. doi: 10.1038/nrdp.2016.18. [DOI] [PubMed] [Google Scholar]
- 2.Lee J.S., Kim B.K., Kim S.U., Park J.Y., Ahn S.H., Seong J.S., Han K.H., DY Kim. A survey on transarterial chemoembolization refractoriness and a real-world treatment pattern for hepatocellular carcinoma in Korea. Clin. Mol. Hepatol. 2020;26:24–32. doi: 10.3350/cmh.2018.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee S., Kim S.S., Chang D.R., Kim H., MJ Kim. Comparison of LI-RADS 2018 and KLCA-NCC 2018 for noninvasive diagnosis of hepatocellular carcinoma using magnetic resonance imaging. Clin. Mol. Hepatol. 2020;26:340–351. doi: 10.3350/cmh.2020.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee C.H., Lee Y.B., Kim M.A., Jang H., Oh H., Kim S.W., Cho E.J., Lee K.H., Lee J.H., Yu S.J. Effectiveness of nivolumab versus regorafenib in hepatocellular carcinoma patients who failed sorafenib treatment. Clin. Mol. Hepatol. 2020;26:328–339. doi: 10.3350/cmh.2019.0049n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim J., Kang W., Sinn D.H., Gwak G.Y., Paik Y.H., Choi M.S., Lee J.H., Koh K.C., SW Paik. Substantial risk of recurrence even after 5 recurrence-free years in early-stage hepatocellular carcinoma patients. Clin. Mol. Hepatol. 2020;26:516–528. doi: 10.3350/cmh.2020.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nishida N. Metabolic disease as a risk of hepatocellular carcinoma. Clin. Mol. Hepatol. 2021;27:87–90. doi: 10.3350/cmh.2020.0302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu M., Jiang L., XY Guan. The genetic and epigenetic alterations in human hepatocellular carcinoma: a recent update. Protein Cell. 2014;5:673–691. doi: 10.1007/s13238-014-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee S., Choi E.J., Cho E.J., Lee Y.B., Lee J.H., Yu S.J., Yoon J.H., YJ Kim. Inhibition of PI3K/Akt signaling suppresses epithelial-to-mesenchymal transition in hepatocellular carcinoma through the Snail/GSK-3/beta-catenin pathway. Clin. Mol. Hepatol. 2020;26:529–539. doi: 10.3350/cmh.2019.0056n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y., Wang X., Yang Y. Hepatic Hippo signaling inhibits development of hepatocellular carcinoma. Clin. Mol. Hepatol. 2020;26:742–750. doi: 10.3350/cmh.2020.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang S., Liu G. Targeting the Ras/Raf/MEK/ERK pathway in hepatocellular carcinoma. Oncol. Lett. 2017;13:1041–1047. doi: 10.3892/ol.2017.5557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldsmith J., Levine B., Debnath J. Autophagy and cancer metabolism. Methods Enzymol. 2014;542:25–57. doi: 10.1016/B978-0-12-416618-9.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimmelman A.C., White E. Autophagy and Tumor Metabolism. Cell Metab. 2017;25:1037–1043. doi: 10.1016/j.cmet.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim Y.S., Kim S.G. Endoplasmic reticulum stress and autophagy dysregulation in alcoholic and non-alcoholic liver diseases. Clin. Mol. Hepatol. 2020;26:715–727. doi: 10.3350/cmh.2020.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mizushima N. Autophagy: process and function. Genes. Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 15.Glick D., Barth S., Macleod K.F. Autophagy: cellular and molecular mechanisms. J. Pathol. 2010;221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lock R., Roy S., Kenific C.M., Su J.S., Salas E., Ronen S.M., Debnath J. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol. Biol. Cell. 2011;22:165–178. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J.Y., Karsli-Uzunbas G., Mathew R., Aisner S.C., Kamphorst J.J., Strohecker A.M., Chen G., Price S., Lu W., Teng X. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes. Dev. 2013;27:1447–1461. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karsli-Uzunbas G., Guo J.Y., Price S., Teng X., Laddha S.V., Khor S., Kalaany N.Y., Jacks T., Chan C.S., Rabinowitz J.D. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4:914–927. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosenfeldt M.T., O'Prey J., Morton J.P., Nixon C., MacKay G., Mrowinska A., Au A., Rai T.S., Zheng L., Ridgway R. p53 status determines the role of autophagy in pancreatic tumour development. NatureNature. 2013;504:296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 20.Guo J.Y., Chen H.Y., Mathew R., Fan J., Strohecker A.M., Karsli-Uzunbas G., Kamphorst J.J., Chen G., Lemons J.M., Karantza V. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes. Dev. 2011;25:460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M.J., Woo S.J., Yoon C.H., Lee J.S., An S., Choi Y.H., Hwang S.G., Yoon G., SJ Lee. Involvement of autophagy in oncogenic K-Ras-induced malignant cell transformation. J. Biol. Chem. 2011;286:12924–12932. doi: 10.1074/jbc.M110.138958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang S., Wang X., Contino G., Liesa M., Sahin E., Ying H., Bause A., Li Y., Stommel J.M., Dell'antonio G. Pancreatic cancers require autophagy for tumor growth. Genes. Dev. 2011;25:717–729. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim J.H., Kim H.Y., Lee Y.K., Yoon Y.S., Xu W.G., Yoon J.K., Choi S.E., Ko Y.G., Kim M.J., Lee S.J. Involvement of mitophagy in oncogenic K-Ras-induced transformation: overcoming a cellular energy deficit from glucose deficiency. Autophagy. 2011;7:1187–1198. doi: 10.4161/auto.7.10.16643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moon H., Ju H.L., Chung S.I., Cho K.J., Eun J.W., Nam S.W., Han K.H., Calvisi D.F., SW Ro. Transforming Growth Factor-β Promotes Liver Tumorigenesis in Mice via Up-regulation of Snail. Gastroenterology. 2017;153:1378–1391. doi: 10.1053/j.gastro.2017.07.014. e1376. [DOI] [PubMed] [Google Scholar]
- 25.Ju H.L., Ahn S.H., Kim D.Y., Baek S., Chung S.I., Seong J., Han K.H., SW Ro. Investigation of oncogenic cooperation in simple liver-specific transgenic mouse models using noninvasive in vivo imaging. PLoS ONE. 2013;8:e59869. doi: 10.1371/journal.pone.0059869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell. Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizushima N. Methods for monitoring autophagy using GFP-LC3 transgenic mice. Methods Enzymol. 2009;452:13–23. doi: 10.1016/S0076-6879(08)03602-1. [DOI] [PubMed] [Google Scholar]
- 28.Sikorski A.F. p62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell. Mol. Biol. Lett. 2016;21:29. doi: 10.1186/s11658-016-0031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.SR Yoshii, Mizushima N. Monitoring and measuring autophagy. Int. J .Mol. Sci. 2017;18:1865. doi: 10.3390/ijms18091865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.J., Coppes R.P., Engedal N., Mari M., Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito Y., Sasaki Y., Horimoto M., Wada S., Tanaka Y., Kasahara A., Ueki T., Hirano T., Yamamoto H., Fujimoto J. Activation of mitogen-activated protein kinases/extracellular signal-regulated kinases in human hepatocellular carcinoma. Hepatology. 1998;27:951–958. doi: 10.1002/hep.510270409. [DOI] [PubMed] [Google Scholar]
- 32.Delire B., Stärkel P. The Ras/MAPK pathway and hepatocarcinoma: pathogenesis and therapeutic implications. Eur. J. Clin. Invest. 2015;45:609–623. doi: 10.1111/eci.12441. [DOI] [PubMed] [Google Scholar]
- 33.Lemmon M.A., Schlessinger J. Cell signaling by receptor tyrosine kinases. CellCell. 2010;141:1117–1134. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taketomi A., Shirabe K., Muto J., Yoshiya S., Motomura T., Mano Y., Ikegami T., Yoshizumi T., Sugio K., Maehara Y. A rare point mutation in the Ras oncogene in hepatocellular carcinoma. Surg. Today. 2013;43:289–292. doi: 10.1007/s00595-012-0462-8. [DOI] [PubMed] [Google Scholar]
- 35.Llovet J.M., Villanueva A., Lachenmayer A., Finn R.S. Advances in targeted therapies for hepatocellular carcinoma in the genomic era. Nat. Rev. Clin. Oncol. 2015;12:408–424. doi: 10.1038/nrclinonc.2015.103. [DOI] [PubMed] [Google Scholar]
- 36.Neuzillet C., Tijeras-Raballand A., de Mestier L., Cros J., Faivre S., Raymond E. MEK in cancer and cancer therapy. Pharmacol. Ther. 2014;141:160–171. doi: 10.1016/j.pharmthera.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 37.Chen C., Wang G. Mechanisms of hepatocellular carcinoma and challenges and opportunities for molecular targeted therapy. World J. Hepatol. 2015;7:1964–1970. doi: 10.4254/wjh.v7.i15.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nagarajan A., Malvi P., Wajapeyee N. Oncogene-directed alterations in cancer cell metabolism. Trends Cancer. 2016;2:365–377. doi: 10.1016/j.trecan.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Papa S., Choy P.M., Bubici C. The ERK and JNK pathways in the regulation of metabolic reprogramming. OncogeneOncogene. 2019;38:2223–2240. doi: 10.1038/s41388-018-0582-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie X., Koh J.Y., Price S., White E., Mehnert J.M. Atg7 Overcomes Senescence and Promotes Growth of BrafV600E-Driven Melanoma. Cancer Discov. 2015;5:410–423. doi: 10.1158/2159-8290.CD-14-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu X., Yu J., Bhagat G., Furuya N., Hibshoosh H., Troxel A., Rosen J., Eskelinen E.L., Mizushima N., Ohsumi Y. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Invest. 2003;112:1809–1820. doi: 10.1172/JCI20039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yue Z., Jin S., Yang C., Levine A.J., Heintz N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. U S A. 2003;100:15077–15082. doi: 10.1073/pnas.2436255100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takamura A., Komatsu M., Hara T., Sakamoto A., Kishi C., Waguri S., Eishi Y., Hino O., Tanaka K., Mizushima N. Autophagy-deficient mice develop multiple liver tumors. Genes. Dev. 2011;25:795–800. doi: 10.1101/gad.2016211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee Y.J., Jang B.K. The role of autophagy in hepatocellular carcinoma. Int. J. Mol. Sci. 2015;16:26629–26643. doi: 10.3390/ijms161125984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee J.S. The mutational landscape of hepatocellular carcinoma. Clin. Mol. Hepatol. 2015;21:220–229. doi: 10.3350/cmh.2015.21.3.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ni H.M., Woolbright B.L., Williams J., Copple B., Cui W., Luyendyk J.P., Jaeschke H., WX Ding. Nrf2 promotes the development of fibrosis and tumorigenesis in mice with defective hepatic autophagy. J. Hepatol. 2014;61:617–625. doi: 10.1016/j.jhep.2014.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang A., Herter-Sprie G., Zhang H., Lin E.Y., Biancur D., Wang X., Deng J., Hai J., Yang S., Wong K.K. Autophagy sustains pancreatic cancer growth through both cell-autonomous and non-autonomous mechanisms. Cancer Discov. 2018;8:276–287. doi: 10.1158/2159-8290.CD-17-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chao X., Qian H., Wang S., Fulte S., WX Ding. Autophagy and liver cancer. Clin. Mol. Hepatol. 2020;26:606–617. doi: 10.3350/cmh.2020.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.