Abstract
The pseudogene legumain (LGMN) has been reported to regulate cancer cell biology. However, the role of LGMN in thyroid carcinoma remains unknown. Herein, Cell Counting Kit 8 and Transwell assays were performed to evaluate cellular proliferation and invasion capacity, respectively. In addition, a tube formation assay was performed to assess HUVEC angiogenesis. The results showed that LGMN depletion attenuated cellular proliferation, invasion and tube formation ability, and that LGMN expression was dysregulated in thyroid carcinoma tumors. Furthermore, patients with high LGMN expression levels exhibited a lower overall survival rate than those with low expression levels. LGMN and microRNA (miR)-495 modulated the expression levels of autophagy-related gene 3 (ATG3) and p62. Finally, ATG3 overexpression rescued the LGMN-regulated thyroid carcinoma phenotype. In conclusion, LGMN was found to promote thyroid carcinoma progression via the miR-495/autophagy axis, thus providing novel insights for understanding the pathogenesis of thyroid carcinoma.
Keywords: thyroid carcinoma, legumain, autophagy, microRNA-495
Introduction
Thyroid carcinoma is a tumor of the endocrine system with increasing incidence and morbidity (1,2). The most common type of thyroid carcinoma is papillary thyroid carcinoma (PTC), which accounts for ~80% of all thyroid carcinoma cases (3). Hormonotherapy and surgical resection are considered to be the two primary approaches for treating thyroid carcinoma (4,5).
Increasing evidence has suggested that pseudogenes consist of functional rather than junk DNA (6,7), and an estimated 18,000 pseudogenes are present in the human genome (8). Pseudogenes can regulate gene expression at multiple levels, and are involved in various biological processes, including cancer development and progression (9). The BRAF pseudogene has been shown to act as a competing endogenous RNA (ceRNA) in lymphoma (10). Additionally, other studies have demonstrated that both pituitary tumor-transforming 3 pseudogene and protein disulfide isomerase family A member 3 pseudogene 1 are upregulated in hepatocellular carcinoma (11,12), while double homeobox A pseudogene (DUXAP) 8 promotes non-small cell lung cancer (NSCLC) cell proliferation and invasiveness (13). Liao et al (14) revealed that pseudogene legumain (LGMN) promoted glioblastoma by sponging microRNA (miR)-495-3p. However, the effects of pseudogene LGMN on thyroid carcinoma progression remain elusive.
MicroRNAs (miRNAs/miRs) are small non-coding RNAs, 20–22 nucleotides in length (15,16). Pathologically, miRNAs can regulate gene expression during cancer progression (17,18). Ye et al (19) indicated that miR-204 inhibited thyroid cancer progression, and miR-338 was found to suppress thyroid carcinoma tumorigenesis by targeting Ras-associated binding protein 23 (20). Additional studies have also revealed that miR-495 regulates glioblastoma and colorectal cancer progression (14,21).
Autophagy is a lysosome-mediated cellular degradation process (22), exerting opposing roles in cancer. ATG3 and p62 serve keys roles in cancer by regulating the autophagy pathway (22). Autophagy has been reported to promote apoptosis, but also to enhance chemotherapeutic resistance (23–25). Another study suggested that autophagy could initiate and contribute to thyroid carcinoma (26). Therefore, in the current study, we hypothesized that autophagy may be a potential effector for LGMN in thyroid carcinoma.
The present study aimed to investigate the role of LGMN in thyroid carcinoma. Determining the molecular mechanism underlying the effects of LGMN on thyroid carcinoma progression may provide novel insights into the development of targeted therapies for this pathology.
Materials and methods
Cell culture
The human thyroid carcinoma cell lines, CAL-62 and SW579, were obtained from The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. The immortalized normal human thyroid follicular epithelial cell line, Nthy-ori3-1, was purchased from CoBioer Biosciences Co., Ltd. (cat. no. CBP61205). HUVECs were purchased from Shanghai Zhong Qiao Xin Zhou Biotechnology Co., Ltd. HUVECs, CAL-62 and SW579 cells were cultured in DMEM supplemented with 1% penicillin/streptomycin and 10% FBS, at 37°C (5% CO2) in a humidified atmosphere. Nthy-ori3-1 cells were maintained in RPMI-1640 with 10% FBS and 1% penicillin/streptomycin.
Construction of stable cell lines
293T cells (The Cell Bank of Type Culture Collection of The Chinese Academy of Sciences) were transfected with 1 µg pLKO.1-LGMN, 0.5 µg pVSVG (Addgene, Inc.) and 0.5 µg pPAX2 (Addgene, Inc.) plasmids, and the resulting 2nd generation lentiviruses were collected after 36–48 h of transfection. The viruses (MOI=4) were used to infect CAL-62 and SW579 cells. Following incubation for 24 h, cells were cultured in the presence of 2 µg/ml puromycin for 1 week to obtain resistant cells. Puromycin (1 µg/ml) were used for maintenance. The short hairpin RNA (shRNA) sequences used were as follows: Sh-negative control (NC), 5′-AUUCGGUUCAAGGUCCAUUGGG-3′; sh-LGMN-1, 5′-CCUGCCGGAUAACAUCAAU-3′; and sh-LGMN-2, 5′-CGUGGAAGAUCUGACUAAA-3′.
Transfection
Autophagy-related gene 3 (ATG3) cDNA (Genscript Biotech) or LGMN (Genscript Biotech) was subcloned into the pcDNA3.1 vector (Addgene, Inc.), 1 µg pcDNA3.1-ATG3 or pcDNA3.1-LGMN vector was used to transfect CAL-62 and SW579 cells with Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.) at room temperature. Following transfection for 24 h, the cells were harvested for further analysis. NC mimic (50 µM), miR-495 mimic (50 µM), NC inhibitor (50 µM), miR-495 inhibitor (50 µM) were transfected into CAL-62 and SW579 cells with Lipofectamine® RNAiMAX (Thermo Fisher Scientific, Inc.) at room temperature. The interval transfection and subsequent experiments was 48 h. NC mimic, 5′-CAUUCAUCCAUCAAUCGGGCAGGCCUU−3′; miR-495 mimic, 5′-UUACCGAUCCAAUUUCCGGACGGUUAC−3′; NC inhibitor, 5′-UUCAGGCAAUCCAAAUGCAGG−3′; and miR-495 inhibitor, 5′-AAUGGGACUUCCAUCGGAAUCCU-3′.
Colony formation assay
A total of 2×104 cells were seeded into each well of a 6-well plate, and cultured for ~1 week. Following colony formation, the cells were stained with 0.5% crystal violet at room temperature for 1–2 h.
Transwell invasion assay
CAL-62 and SW579 cells were seeded into the upper chambers of Matrigel-precoated inserts (BD Biosciences), at a density of 1×105 cells in 250 µl serum-free medium. In addition, 300 µl medium supplemented with 20% FBS was added to the bottom chamber as a chemoattractant. Following incubation at 37°C for 24 h, invasive cells were stained with 0.05% crystal violet solution at room temperature for 1–2 h. A light microscope (CKX53; Olympus Corporation) was used to observe migratory cells in lower chamber (magnification, ×100).
Cell Counting Kit 8 (CCK-8) assay
CAL-62 and SW579 cells were seeded into 96-well plates at a density of 1×103 cells/well. The cells were then transfected with the indicated oligonucleotides and incubated for different lengths of time. Subsequently, the cells were incubated with 10 µl CCK-8 reagent (Vazyme Biotech Co., Ltd.) for 3 h at 37°C (5% CO2), after which the absorbance of each well was determined at a wavelength of 450 nm.
Western blot analysis
Total protein were extracted by RIPA buffer (Beijing Solarbio Science & Technology Co. Ltd.) from CAL-62 and SW579 cells. Protein concentration was determined by bicinchoninic acid (BCA) method and ~50 µg/lane of protein was separated by 10% SDS-PAGE, prior to transfer onto PVDF membranes. Following blocking with 5% non-fat milk at room temperature for 1 h, the membranes were incubated with primary antibodies overnight at 4°C. The following primary antibodies were used: Anti-ATG3 (cat. no. PA5-17018; 1:1,000; Thermo Fisher Scientific, Inc.), anti-p62 (cat. no. ab155686; 1:1,000; Abcam) and anti-GAPDH (cat. no. 8884; 1:2,000; Cell Signaling Technology, Inc.). The membranes were then incubated with the following secondary antibodies at room temperature for 1 h: Anti-mouse HRP-conjugated IgG (cat. no. 7076; 1:3,000) or anti-rabbit HRP-conjugated IgG (cat. no. 7074; dilution, 1:3,000) (both Cell Signaling Technology, Inc.). Enhanced chemiluminescent (ECL) reagent (ThermoFisher Scientific Inc.) was used to probe protein bands, which was visualized by ChemiDoc™ MP and Image Lab v.4.1 software (Bio-Rad Laboratories, Inc.).
The Cancer Genome Atlas (TCGA) analysis
The gene expression data from thyroid carcinoma samples, including 512 tumor and 337 normal tissues, were downloaded from TCGA (https://tcga.xenahubs.net). Comparisons between the expression levels of LGMN were conducted using Student's t-test (unpaired, two-tailed).
Tube formation assay
Growth factor-reduced Matrigel was used to coat 24-well plates at 37°C for 4–5 h. Subsequently, HUVECs at a density of 3×103 cells/well were seeded into the coated plates in conditioned medium (Ham's F-12K + 0.1 mg/ml Heparin + 0.03–0.05 mg/ml ECGs + 10% FBS + 1% P/S). Tube formation was observed under a light microscope (magnification, ×200).
Reverse transcription-quantitative (RT-q) PCR
Total RNA was extracted from CAL-62 and SW579 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 1 µg RNA was reverse transcribed into cDNA using the PrimeScript RT reagent kit (Takara Bio, Inc.) according to the manufacturer's instructions. qPCR was carried out with SYBR Green kit (Roche Applied Science) to determine relative gene expression, and quantified using the 2−ΔΔCq method (27). The following thermocycling conditions were used: Initial denaturation at 95°C for 5 min, followed by 40 cycles of 95°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec. The primer sequences used were as follows: LGMN forward, 5′-GTTGAGAGGCGATGCAGAAG-3′ and reverse, 5′-GCAGGTGGTTCATCATGGAC-3′; ATG3 forward, 5′-CAATGGGCTACAGGGGAAGA-3′ and reverse, 5′-ATCCGCCATCACCATCATCT-3′; p62 forward, 5′-CACAGAGGAGAAGAGCAGCT-3′ and reverse, 5′-TGGAGTTCACCTGTAGACGG-3′; miR-495 forward, 5′-ACCTGAAAAGAAGTTGCCCA-3′ and reverse, 5′-GCACCATGTTTGTTTCGTCAC−3′; and β-actin forward, 5′-ACTCTTCCAGCCTTCCTTCC-3′ and reverse, 5′-CGTACAGGTCTTTGCGGATG-3′.
Statistical analysis
GraphPad Prism 8.0 software (GraphPad Software, Inc.) was used to compare the differences between two groups, using unpaired Student's t-test. The differences among multiple groups were assessed using one-way ANOVA followed by Tukey's post hoc test. The data are expressed as the mean ± SD, and P<0.05 was considered to indicate a statistically significant difference.
Results
LGMN depletion attenuates thyroid carcinoma progression
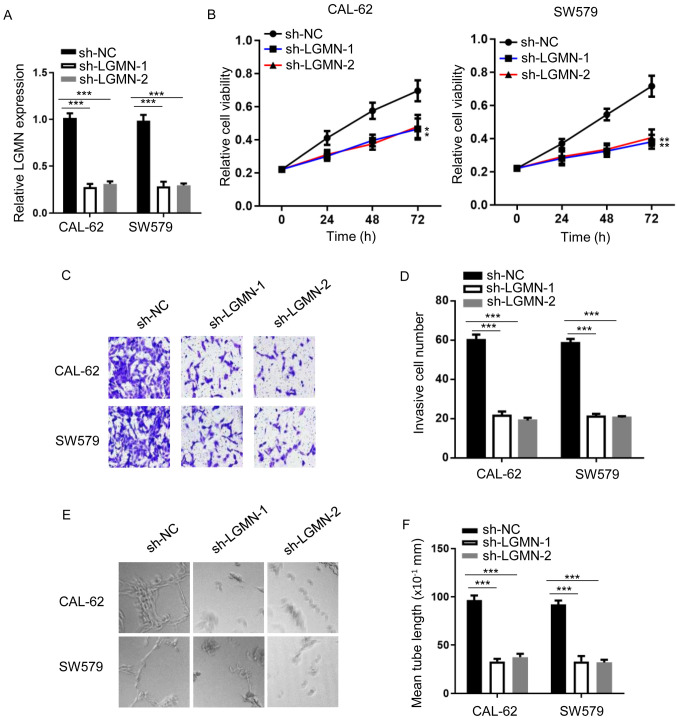
To investigate the tumorigenic role of LGMN in thyroid carcinoma, the shRNA gene knockdown approach was employed. shRNAs against LGMN were synthesized, and RT-qPCR analysis was used to confirm knockdown efficiency (Fig. 1A). LGMN-knockdown notably impaired CAL-62 and SW579 cell proliferation (Fig. 1B). Similarly, LGMN-knockdown decreased the invasive capacity of both cell lines (Fig. 1C and D). Furthermore, a tube formation assay was conducted to evaluate the tube formation ability of HUVECs treated with conditioned medium from thyroid carcinoma cells. The results showed that LGMN-knockdown attenuated the tube formation ability of HUVECs compared with the sh-NC group (Fig. 1E and F). These findings suggested that LGMN depletion inhibited the tumor phenotype of thyroid carcinoma cells.
Figure 1.
LGMN depletion attenuates the progression of thyroid carcinoma. (A) Reverse transcription-quantitative PCR analysis was conducted to determine the expression levels of LGMN in CAL-62 and SW579 cells transfected with sh-NC, sh-LGMN-1 or sh-LGMN-2. ***P<0.001. (B) Viability of CAL-62 or SW579 cells transfected with sh-NC, sh-LGMN-1 or sh-LGMN-2 was assessed by MTT assay. *P<0.05, **P<0.01. (C and D) Invasive ability of CAL-62 and SW579 cells transfected with sh-NC, sh-LGMN-1 or sh-LGMN-2 was evaluated by Transwell assay. ***P<0.001. (E and F) Tube length of HUVECs treated with conditioned medium from CAL-62 or SW579 cells transfected with sh-NC, sh-LGMN-1 or sh-LGMN-2. Magnification, ×200. ***P<0.001. Experiments were performed for three biological replicates. LGMN, pseudogene legumain; sh-NC, short hairpin RNA negative control.
LGMN is associated with thyroid carcinoma
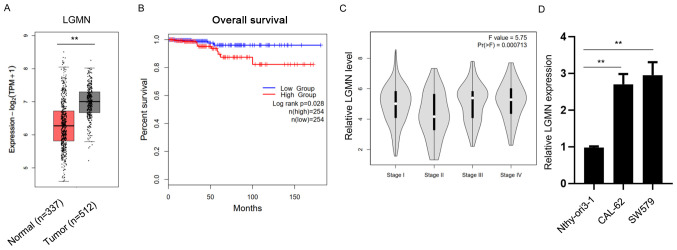
To investigate the potential effect of pseudogene LGMN on thyroid carcinoma, clinical TCGA datasets of thyroid carcinoma samples were obtained and analyzed using bioinformatics approaches. Bioinformatics analysis revealed that LGMN was significantly upregulated in thyroid carcinoma tissues compared with normal tissues (Fig. 2A). Furthermore, the overall survival (OS) rate was evaluated in the high- and low-LGMN expression groups. The results showed that the OS rate was higher in patients in the low-LGMN group compared with those in the high-LGMN group (Fig. 2B). Additionally, the analysis indicated that higher LGMN expression levels were associated with higher grades of thyroid carcinoma (Fig. 2C). Subsequently, RT-qPCR analysis was performed to detect the expression levels of LGMN in the normal thyroid epithelial cell line (Nthy-ori3-1) and thyroid carcinoma CAL-62 and SW579 cells. The results demonstrated that LGMN was significantly upregulated in thyroid carcinoma cell lines (Fig. 2D). The aforementioned findings suggested that LGMN promoted the development of thyroid carcinoma.
Figure 2.
LGMN is associated with the development of thyroid carcinoma. (A) Expression levels of LGMN in normal (n=337) and thyroid carcinoma (n=512) tissues. **P<0.01. (B) Overall survival rate of patients with thyroid carcinoma in high- (n=254) and low- (n=254) LGMN expression groups. (C) LGMN expression levels of patients at different stages of thyroid carcinoma. (D) mRNA expression levels of LGMN in the normal thyroid epithelial cell line (Nthy-ori3-1) and thyroid carcinoma cells (CAL-62 and SW579) were assessed by reverse transcription-quantitative PCR analysis. Experiments were performed for three biological replicates. **P<0.01. LGMN, pseudogene legumain; TPM, transcripts per million.
miR-495 modulates autophagy
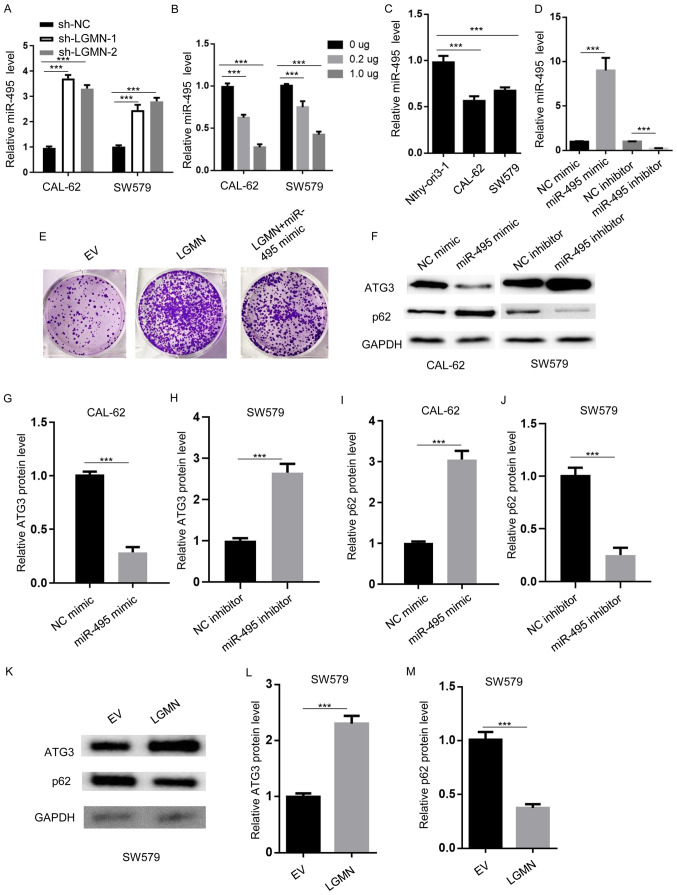
miRNAs have been reported to act as interacting factors for pseudogenes in various cancer types (28,29). A study reported that miR-495 could sponge LGMN in glioblastoma (14). Herein, RT-qPCR analysis was performed to determine the expression levels of miR-495 in CAL-62 and SW579 cells transfected with sh-NC, sh-LGMN-1 or sh-LGMN-2. The results revealed that miR-495 was significantly upregulated following LGMN-knockdown (Fig. 3A). In addition, following LGMN-overexpression in CAL-62 and SW579 cells, RT-qPCR showed that miR-495 levels were downregulated in a dose-dependent manner (Fig. 3B). Moreover, miR-495 expression levels were lower in thyroid carcinoma CAL-62 and SW579 cells compared with normal thyroid epithelial cells (Nthy-ori3-1) (Fig. 3C). miR-495 levels were upregulated in miR-495 mimic-transfected, and downregulated in miR-495 inhibitor-transfected CAL-62 cells (Fig. 3D). In order to determine whether miR-495 acts as key mediator for LGMN-regulated tumorigenesis, a colony formation assay was conducted to demonstrated that LGMN overexpression increased the number of CAL-62-cell colonies compared with the control, which was attenuated by miR-495 mimic transfection (Fig. 3E).
Figure 3.
miR-495 modulates autophagy. (A) RT-qPCR analysis was performed to determine the miR-495 expression levels in CAL-62 and SW579 cells transfected with sh-NC, sh-LGMN-1 or sh-LGMN-2. ***P<0.001. (B) RT-qPCR analysis of miR-495 expression levels in CAL-62 and SW579 cells transfected with the indicated amounts of pcDNA3.1-LGMN. ***P<0.001. (C) RT-qPCR was performed to establish the miR-495 expression levels in normal thyroid epithelial cells (Nthy-ori3-1) and thyroid carcinoma cells (CAL-62 and SW579). ***P<0.001. (D) RT-qPCR analysis of miR-495 expression levels in CAL-62 cells transfected with NC mimics, miR-495 mimics, NC inhibitor and miR-495 inhibitor. ***P<0.001. (E) Colony formation assay to determine the colony number of CAL-62 cells transfected with EV (pcDNA3.1), pcDNA3.1-LGMN, or pcDNA3.1-LGMN plus miR-495 mimics. (F-J) Western blot analysis was performed to measure the protein expression levels of ATG3 and p62 in CAL-62 cells transfected with NC or miR-495 mimics, and SW579 cells transfected with NC or miR-495 inhibitors. ***P<0.001. (K-M) Western blot analysis was carried out to determine the protein expression levels of ATG3 and p62 in SW579 cells transfected with EV (pcDNA3.1) or pcDNA3.1-LGMN. ***P<0.001. Experiments were performed for three biological replicates; western blotting was performed once (n=1). miR-495, microRNA-495; LGMN, pseudogene legumain; RT-q, reverse transcription-quantitative; sh-, short hairpin RNA; NC, negative control; ATG3, autophagy-related gene 3; EV, empty vector.
Subsequently, the present study aimed to identify the potential downstream effectors of miR-495. A previous report revealed that miR-495 attenuated ATG3 expression and promoted that of p62 (30). Therefore, western blot analysis was conducted to determine whether miR-495 could regulate the protein expression levels of ATG3 and p62 in thyroid carcinoma. To investigate whether miR-495 had a role in regulating autophagy in both CAL-62 and SW579 cells, western blotting was conducted in both cell lines using mimic or inhibitor. The results demonstrated that transfection with miR-495 mimics markedly reduced ATG3 and promoted p62 expression in CAL-62 cells, while the miR-495 inhibitor exerted opposing effects in SW579 cells (Fig. 3F-J). Furthermore, LGMN overexpression increased ATG3, and suppressed p62 expression in SW579 cells (Fig. 3K-M). Collectively, these results indicated that miR-495 was modulated by LGMN to regulate autophagy signaling.
Autophagy serves a crucial role in miR-495-regulated thyroid carcinoma progression
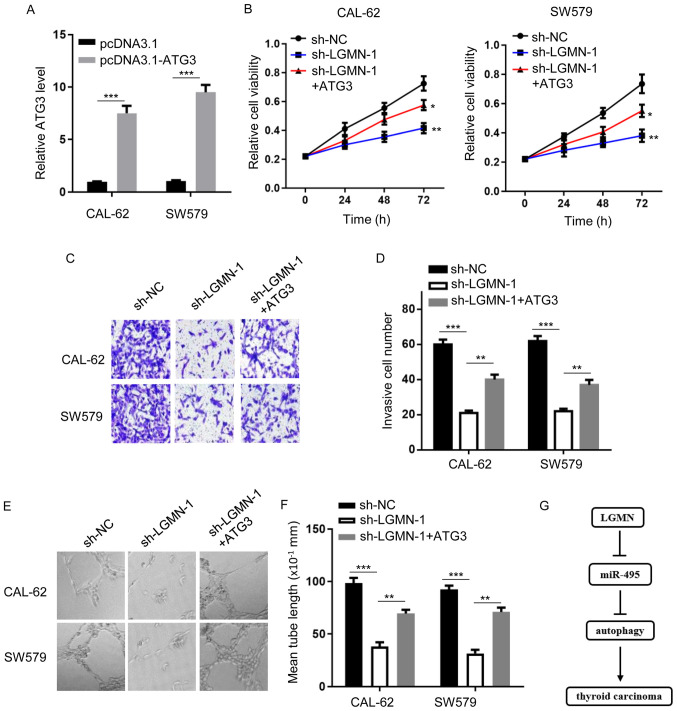
Since the miR-495/autophagy axis may act as an effector of LGMN-mediated thyroid carcinoma progression, the current study aimed to further investigate the role of autophagy in the LGMN-modulated thyroid carcinoma phenotype. Firstly, the expression level of ATG3 was evaluated in CAL-62 and SW579 cells transfected with or without an ATG3 overexpression plasmid (Fig. 4A). CCK-8 and Transwell assays showed that LGMN-knockdown attenuated cellular proliferation and invasiveness, respectively, whereas ATG3 overexpression rescued the effects of LGMN silencing, to a certain degree (Fig. 4B-D). In addition, impaired tube formation ability was observed in HUVECs transfected with sh-LGMN, which was reversed by ATG3 overexpression (Fig. 4E and F). Overall, the results indicate that autophagy serves a critical role in LGMN/miR-495-regulated thyroid carcinoma progression.
Figure 4.
Autophagy serves a crucial role in the progression of LGMN-regulated thyroid carcinoma. (A) Expression levels of miR-495 in CAL-62 and SW579 cells transfected with pcDNA3.1 or pcDNA3.1-ATG3 were determined by reverse transcription-quantitative PCR analysis. ***P<0.001. (B) Viability of CAL-62 and SW579 cells co-transfected with sh-NC, sh-LGMN-1 or sh-LGMN-1 and pcDNA3.1-ATG3 was evaluated by MTT assay. *P<0.05, **P<0.01. (C and D) Invasive ability of CAL-62 and SW579 cells co-transfected with sh-NC, sh-LGMN-1 or sh-LGMN-1 and pcDNA3.1-ATG3 was assessed by Transwell assay. **P<0.01 and ***P<0.001. (E and F) Tube length of HUVECs treated with conditioned medium from CAL-62 or SW579 cells co-transfected with sh-NC, sh-LGMN-1 or sh-LGMN-1 and pcDNA3.1-ATG3 was assessed by tube formation assay. Magnification, ×200. **P<0.01 and ***P<0.001. Experiments were performed for three biological replicates. LGMN, pseudogene legumain; miR-495, microRNA-495; sh-NC, short hairpin RNA negative control; ATG3, autophagy-related gene 3.
Discussion
The results of the present study suggested that LGMN plays a promotive role in the development of the thyroid carcinoma phenotype by modulating the proliferation, invasion and tube formation abilities of HUVECs. Notably, miR-495 and autophagy were identified as downstream effectors of LGMN in thyroid carcinoma cells (Fig. 4G). Therefore, the current study proposes a potential novel mechanism underlying thyroid carcinoma progression.
PTC accounts for ~80% of all thyroid carcinoma cases (10,31). Currently, the available diagnostic and therapeutic approaches for thyroid carcinoma remain ineffective, and limited targeted therapy approaches have been developed. The results of the present study suggested that LGMN may be used as a biomarker to facilitate the diagnosis of thyroid carcinoma, and that the RNA levels of LGMN may be used for patient classification, the patients with high and low levels of LGMN may be sensitive for different treatment options.
Studies have demonstrated that the pseudogenes BRAF and DUXAP10 are dysregulated in thyroid carcinoma (10,32). Consistent with these findings, the current study indicated that LGMN was upregulated in thyroid carcinoma, and that this increase in expression was associated with a poorer outcome. LGMN has also been shown to exert its function by interacting with miR-495 (14), and the present study confirmed that LGMN could negatively regulate miR-495.
miR-495 plays a crucial role in several types of cancer, including gastric cancer, NSCLC and lung cancer (33–35). Herein, miR-495 was found to induce the cellular proliferation, invasion and tube formation abilities of HUVECs. To the best of our knowledge, the present study was the first to report the role of miR-495 in thyroid carcinoma, and the results showed that miR-495 could modulate the expression of autophagy-related genes. However, it was hypothesized that miR-495 may also affect additional signaling pathways and downstream effectors to exert its function.
Emerging evidence has suggested that ATG3 and p62 serve a key role in autophagy. Herein, ATG3 was indicated to restore the LGMN-regulated tumorigenesis of thyroid carcinoma. In conclusion, the results of the current study suggest that LGMN may be considered an important regulator of thyroid carcinoma progression, while the miR-495/autophagy axis might serve as a downstream effector of LGMN. However, the study included a number of limitations that should be addressed in the future; for instance, the use of animal models to investigate the role of LGMN, and in addition to autophagy, other potential downstream targets could also be identified.
In conclusion, the findings of the present study also provide novel insights to increase the current understanding of thyroid carcinoma progression. Furthermore, these findings may aid the clinical diagnosis of thyroid carcinoma, and suggest a potential treatment by means of targeting the LGMN/miR-495/autophagy. High levels of LGMN and low expression of miR-495 may be beneficial for an improved treatment outcome and prognosis.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Authors' contributions
JS, YP and EY designed the present study. JS, YP, JL and QH performed the experiments. HZ, LS and EY analyzed the data and prepared the figures. EY drafted the manuscript. JS and EY confirmed the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Borran S, Ahmadi G, Rezaei S, Anari MM, Modabberi M, Azarash Z, Razaviyan J, Derakhshan M, Akhbari M, Mirzaei H. Circular RNAs: New players in thyroid cancer. Pathol Res Pract. 2020;216:153217. doi: 10.1016/j.prp.2020.153217. [DOI] [PubMed] [Google Scholar]
- 2.Rossi ED, Faquin WC, Pantanowitz L. Cytologic features of aggressive variants of follicular-derived thyroid carcinoma. Cancer Cytopathol. 2019;127:432–446. doi: 10.1002/cncy.22136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang KT, Lee CH. BRAF mutation in papillary thyroid carcinoma: Pathogenic role and clinical implications. J Chin Med Assoc. 2010;73:113–128. doi: 10.1016/S1726-4901(10)70025-3. [DOI] [PubMed] [Google Scholar]
- 4.Ito Y, Jikuzono T, Higashiyama T, Asahi S, Tomoda C, Takamura Y, Miya A, Kobayashi K, Matsuzuka F, Kuma K, Miyauchi A. Clinical significance of lymph node metastasis of thyroid papillary carcinoma located in one lobe. World J Surg. 2006;30:1821–1828. doi: 10.1007/s00268-006-0211-5. [DOI] [PubMed] [Google Scholar]
- 5.Zhao J, Xu C, Yao J, Yu C, Liao L, Dong J. Statins and thyroid carcinoma: A meta-analysis. Cell Physiol Biochem. 2018;47:1422–1431. doi: 10.1159/000490832. [DOI] [PubMed] [Google Scholar]
- 6.Stewart GL, Enfield KSS, Sage AP, Martinez VD, Minatel BC, Pewarchuk ME, Marshall EA, Lam WL. Aberrant expression of pseudogene-derived lncRNAs as an alternative mechanism of cancer gene regulation in lung adenocarcinoma. Front Genet. 2019;10:138. doi: 10.3389/fgene.2019.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shih JH, Chen HY, Lin SC, Yeh YC, Shen R, Lang YD, Wu DC, Chen CY, Chen RH, Chou TY, Jou YS. Integrative analyses of noncoding RNAs reveal the potential mechanisms augmenting tumor malignancy in lung adenocarcinoma. Nucleic Acids Res. 2020;48:1175–1191. doi: 10.1093/nar/gkz1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poliseno L. Pseudogenes: Newly discovered players in human cancer. Sci Signal. 2012;5:re5. doi: 10.1126/scisignal.2002858. [DOI] [PubMed] [Google Scholar]
- 9.Xiao-Jie L, Ai-Mei G, Li-Juan J, Jiang X. Pseudogene in cancer: Real functions and promising signature. J Med Genet. 2015;52:17–24. doi: 10.1136/jmedgenet-2014-102785. [DOI] [PubMed] [Google Scholar]
- 10.Lin JD, Fu SS, Chen JY, Lee CH, Chau WK, Cheng CW, Wang YH, Lin YF, Fang WF, Tang KT. Clinical manifestations and gene expression in patients with conventional papillary thyroid carcinoma carrying the BRAF(V600E) mutation and BRAF pseudogene. Thyroid. 2016;26:691–704. doi: 10.1089/thy.2015.0044. [DOI] [PubMed] [Google Scholar]
- 11.Huang JL, Cao SW, Ou QS, Yang B, Zheng SH, Tang J, Chen J, Hu YW, Zheng L, Wang Q. The long non-coding RNA PTTG3P promotes cell growth and metastasis via up-regulating PTTG1 and activating PI3K/AKT signaling in hepatocellular carcinoma. Mol Cancer. 2018;17:93. doi: 10.1186/s12943-018-0841-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong Y, Zhang L, Huang Y, He T, Zhang L, Zhao X, Zhou X, Zhou D, Yan Y, Zhou J, et al. Pseudogene PDIA3P1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Lett. 2017;407:76–83. doi: 10.1016/j.canlet.2017.07.031. [DOI] [PubMed] [Google Scholar]
- 13.Sun M, Nie FQ, Zang C, Wang Y, Hou J, Wei C, Li W, He X, Lu KH. The pseudogene DUXAP8 promotes non-small-cell lung cancer cell proliferation and invasion by epigenetically silencing EGR1 and RHOB. Mol Ther. 2017;25:739–751. doi: 10.1016/j.ymthe.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Liao K, Qian Z, Zhang S, Chen B, Li Z, Huang R, Cheng L, Wang T, Yang R, Lan J, et al. The LGMN pseudogene promotes tumor progression by acting as a miR-495-3p sponge in glioblastoma. Cancer Lett. 2020;490:111–123. doi: 10.1016/j.canlet.2020.07.012. [DOI] [PubMed] [Google Scholar]
- 15.Kim YW, Kim EY, Jeon D, Liu JL, Kim HS, Choi JW, Ahn WS. Differential microRNA expression signatures and cell type-specific association with Taxol resistance in ovarian cancer cells. Drug Des Devel Ther. 2014;8:293–314. doi: 10.2147/DDDT.S51969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan W, Kong X, Li J, Li P, Zhao Y, Liu T, Binang HB, Wang Y, Du L, Wang C. LncRNA AC010789.1 promotes colorectal cancer progression by targeting microRNA-432-3p/ZEB1 axis and the Wnt/β-catenin signaling pathway. Front Cell Dev Biol. 2020;8:565355. doi: 10.3389/fcell.2020.565355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kafshdooz L, Pourfathi H, Akbarzadeh A, Kafshdooz T, Razban Z, Sheervalilou R, Ebrahimi Sadr N, Khalilov R, Saghfi S, Kavetskyy T, et al. The role of microRNAs and nanoparticles in ovarian cancer: A review. Artif Cells Nanomed Biotechnol. 2018;46:241–247. doi: 10.1080/21691401.2018.1454931. [DOI] [PubMed] [Google Scholar]
- 18.Han D, Li J, Wang H, Su X, Hou J, Gu Y, Qian C, Lin Y, Liu X, Huang M, et al. Circular RNA circMTO1 acts as the sponge of microRNA-9 to suppress hepatocellular carcinoma progression. Hepatology. 2017;66:1151–1164. doi: 10.1002/hep.29270. [DOI] [PubMed] [Google Scholar]
- 19.Ye M, Dong S, Hou H, Zhang T, Shen M. Oncogenic role of long noncoding RNAMALAT1 in thyroid cancer progression through regulation of the miR-204/IGF2BP2/m6A-MYC signaling. Mol Ther Nucleic Acids. 2021;23:1–12. doi: 10.1016/j.omtn.2020.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shu T, Yang L, Sun L, Lu J, Zhan X. CircHIPK3 promotes thyroid cancer tumorigenesis and invasion through the Mirna-338-3p/RAB23 axis. Med Princ Pract. 2020 Oct 26; doi: 10.1159/000512548. (Epub ahead of Print) [DOI] [PubMed] [Google Scholar]
- 21.Qian J, Garg A, Li F, Shen Q, Xiao K. LncRNA LUNAR1 accelerates colorectal cancer progression by targeting the miR4953p/MYCBP axis. Int J Oncol. 2020;57:1157–1168. doi: 10.3892/ijo.2020.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer. 2017;17:528–542. doi: 10.1038/nrc.2017.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pagotto A, Pilotto G, Mazzoldi EL, Nicoletto MO, Frezzini S, Pastò A, Amadori A. Autophagy inhibition reduces chemoresistance and tumorigenic potential of human ovarian cancer stem cells. Cell Death Dis. 2017;8:e2943. doi: 10.1038/cddis.2017.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chude CI, Amaravadi RK. Targeting autophagy in cancer: Update on clinical trials and novel inhibitors. Int J Mol Sci. 2017;18:1279. doi: 10.3390/ijms18061279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wei W, Hardin H, Luo QY. Targeting autophagy in thyroid cancers. Endocr Relat Cancer. 2019;26:R181–R194. doi: 10.1530/ERC-18-0502. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Yan L, Yue C, Xu Y, Jiang X, Zhang L, Wu J. Identification of potential diagnostic and prognostic pseudogenes in hepatocellular carcinoma based on pseudogene-miRNA-mRNA competitive network. Med Sci Monit. 2020;26:e921895. doi: 10.12659/MSM.921895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hou Z, Wang Y, Xia N, Lv T, Yuan X, Song Y. Pseudogene KRT17P3 drives cisplatin resistance of human NSCLC cells by modulating miR-497-5p/mTOR. Cancer Sci. 2021;112:275–286. doi: 10.1111/cas.14733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li W, Yang Y, Hou X, Zhuang H, Wu Z, Li Z, Guo R, Chen H, Lin C, Zhong W, et al. MicroRNA-495 regulates starvation-induced autophagy by targeting ATG3. FEBS Lett. 2016;590:726–738. doi: 10.1002/1873-3468.12108. [DOI] [PubMed] [Google Scholar]
- 31.Somuncu E, Karatas A, Ferahman S, Saygili N, Yilmaz E, Ozturk O, Kapan M. The investigation of foxe1 variations in papillary thyroid carcinoma. Int J Clin Exp Pathol. 2015;8:13458–13464. [PMC free article] [PubMed] [Google Scholar]
- 32.Li J, Jiang L, Liu Z, Li Y, Xu Y, Liu H. Oncogenic pseudogene DUXAP10 knockdown suppresses proliferation and invasion and induces apoptosis of papillary thyroid carcinoma cells by inhibition of Akt/mTOR pathway. Clin Exp Pharmacol Physiol. 2020;47:1473–1483. doi: 10.1111/1440-1681.13310. [DOI] [PubMed] [Google Scholar]
- 33.Eun JW, Kim HS, Shen Q, Yang HD, Kim SY, Yoon JH, Park WS, Lee JY, Nam SW. MicroRNA-495-3p functions as a tumor suppressor by regulating multiple epigenetic modifiers in gastric carcinogenesis. J Pathol. 2018;244:107–119. doi: 10.1002/path.4994. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Xu Y, Liao X, Liao R, Zhang L, Niu K, Li T, Li D, Chen Z, Duan Y, Sun J. Plasma miRNAs in predicting radiosensitivity in non-small cell lung cancer. Tumour Biol. 2016;37:11927–11936. doi: 10.1007/s13277-016-5052-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halvorsen AR, Sandhu V, Sprauten M, Flote VG, Kure EH, Brustugun OT, Helland Å. Circulating microRNAs associated with prolonged overall survival in lung cancer patients treated with nivolumab. Acta Oncol. 2018;57:1225–1231. doi: 10.1080/0284186X.2018.1465585. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.