Abstract
The chaperone heat shock protein 70 (Hsp70) and its network of co-chaperones serve as a central hub of cellular protein quality control mechanisms. Domain organization in Hsp70 dictates ATPase activity, ATP dependent allosteric regulation, client/substrate binding and release, and interactions with co-chaperones. The protein quality control activities of Hsp70 are classified as foldase, holdase, and disaggregase activities. Co-chaperones directly assisting protein refolding included J domain proteins and nucleotide exchange factors. However, co-chaperones can also be grouped and explored based on which domain of Hsp70 they interact. Here we discuss how the network of cytosolic co-chaperones for Hsp70 contributes to the functions of Hsp70 while closely looking at their structural features. Comparison of domain organization and the structures of co-chaperones enables greater understanding of the interactions, mechanisms of action, and roles played in protein quality control.
Keywords: Hsp70, molecular chaperones, co-chaperones, protein quality control, J domain protein, nucleotide exchange factor, Hsp40, GrpE, BAG, Hsp110, CHIP, SMADs, Hop, Hip, Hsp90
Impact statement
Protein homeostasis is the cellular protein quality control machinery that maintains structural integrity of the cellular proteome. Genetic level mutations, errors occur at the transcriptional and translational levels, and conditions that contribute to cellular stress cause protein misfolding which ultimately leads to toxic protein aggregates resulting in wide range of diseases including Parkinson's disease, Alzheimer's disease, Huntington's disease, Creutzfeldt–Jakob disease, and type 2 diabetes. Hsp70 and its extensive network of co-chaperones serve as the principal components of the machinery that recognize misfolded proteins and direct them to the refolding or degradative pathways. An increased understanding of Hsp70 and co-chaperones, which serve as drug targets, is valuable for developing drugs to treat neurodegenerative diseases, diabetes, and cancer. This mini review seeks to serve as a curated resource of the structural and functional features of Hsp70 and its co-chaperone network.
Introduction
Cellular protein structural integrity is crucial for maintaining the normal functions and viability of cells. The process used by cells to maintain protein structural integrity is termed proteostasis or protein homeostasis. 1 The machinery that regulates protein homeostasis consists of a diverse network of chaperones and co-chaperones. The proteins that catalyze protein folding are simply known as chaperones, whereas non-client accessory proteins that aid fine-tuning of the folding process conducted by chaperones are known as co-chaperones. 2 Co-chaperones often act non-independently and require the assistance of a chaperone to be actively involved in the quality control process. However, there are exceptions, such as Hsp110 (HSPH) which is a chaperone by itself and also acts as a co-chaperone for Hsp70. 3
Chaperones are members of a large evolutionarily conserved protein family and are ubiquitous in both prokaryotic and eukaryotic cells.4–6 However, in eukaryotic cells, different chaperones, or different versions of the a particular chaperone, are localized in different cellular organelles and compartments.4,5 Chaperones play a vital part in every step of cellular metabolism including protein synthesis, folding, translocation, and rearrangements such as assembly and disassembly into complex organizations, and ultimately in protein degradation and apoptosis.4,5,7 A regulated balance among the above-mentioned processes are critical to maintain protein homeostasis in a cell. 8 Moreover, chaperones are equipped to deal with environmental, pathological and physiological stress conditions such as oxidative stress. 6 Chaperones and co-chaperones mediate when and where the unfolding and folding occurs and serve to direct misfolded and aggregated proteins to proteasomal degradation. 9 Although the interactions between non-native proteins, chaperones, and co-chaperones are transient, these interactions can prevent oligomerization and aggregation, promote folding, unfolding for translocation, or ultimately target a protein to proteasomal degradation.9–12 Deleterious outcomes are unavoidable if protein misfolding is not corrected, often leading to toxic aggregates in the cells and loss of function in the misfolded protein. 9
The central chaperone in eukaryotic cells is heat shock protein 70 (Hsp70), a 70 kDa protein which serves as a central hub for cellular protein quality control systems. The low specificity substrate recognition of Hsp70 enables binding to a broad range of proteins, 13 thereby providing the advantage of assisting in several different cellular processes.4,14 A list of processes that Hsp70 and Hsp70 homologs, broadly termed “Hsp70s”, are involved in are given in Table 1. The earliest implications of chaperone cooperation with co-chaperones were found in the early 1990s, with respect to DnaK (Hsp70 homolog of Escherichia coli). 4 In these studies, the replication cycle of bacteriophage λ was studied, and DnaK was shown to work hand in hand with DnaJ and GrpE.4,28 The network of co-chaperones that interact with Hsp70 can be divided into two main categories, termed J-domain protein co-chaperones (JDP) and nucleotide exchange factors (NEF), based on how they interact with Hsp70. 7 In this review, we will explore Hsp70 (HSPA) co-chaperones based on the co-chaperone function and the Hsp70 domains that mediate the Hsp70-co-chaperone interactions.
Table 1.
Summary of cellular functions conducted by Hsp70s.
| Cellular location | Function | Example(s) |
|---|---|---|
| Cytoplasm | Stabilization and folding of nascent polypeptides in translocation-competent conformations before assembly in the cytosol | Hsp70 in eukaryotes involve in nascent polypeptide-associated complex (NAC)15,16 |
| Ssb proteins of Saccharomyces cerevisiae bind to ribosome-bound nascent polypeptides 11 , 17 | ||
| ER/Mitochondria | Translocation and folding nascent polypeptides into organelles such as ER and mitochondria | Yeast Saccharomyces cerevisiae mitochondrial SSC1 (Mhsp70) aids translocation of nascent proteins to the matrix compartment of the mitochondria 18 , 19 |
| Bip (HSPA5) in eukaryotes helps in translocation process in the ER 16 , 17 | ||
| Lhs1p of yeast ER bind and protect the translocated nascent protein 20 | ||
| Plasma membrane | ATPase activity of Hsp70 facilitates assembly, rearrangement, and dissembling of protein oligomers | Hsc70 (Hsp70 homologue-heat shock cognate protein, HSPA8) involves in uncoating clathrin cages 21–23 |
| Plasma membrane integration and channel formation activity | Hsc/Hsp70 shown to have ATP dependent ion channel activity 24 , 25 | |
| Nucleus | Trafficking of nuclear hormone receptors | Direct binding of Hsp70 to nuclear import signal sequence (NLSs) in yeast 23 |
| Extracellular space | Antigen presentation in major histocompatibility complex (MHC) and function as a cytokine | Hsp70 in human monocytes have shown to activate signal transductions pathways 26 and contribute to antigen presentation in dendritic cells in mammals 27 |
Hsp70
The first identification of Hsp70 was in bacteria due to its induction of expression during the cellular response to elevated temperature, hence, the name heat shock protein.5,29 Hsp70 is highly conserved across domains of life as evidenced by 68% homology (53% identity) between Hsp70s from E. coli and humans (HSPA8). 29 Therefore, studies on Hsp70s from one organism are applicable to Hsp70s from other organisms, even if the organisms are from different domains of life. Below, we will be using the well-studied E. coli orthologue of Hsp70, DnaK, to discuss the structure and organization of Hsp70. Within this discussion, the term Hsp70 is therefore substituted for DnaK due to the structural, functional, and sequence homology.
Domain structure and organization
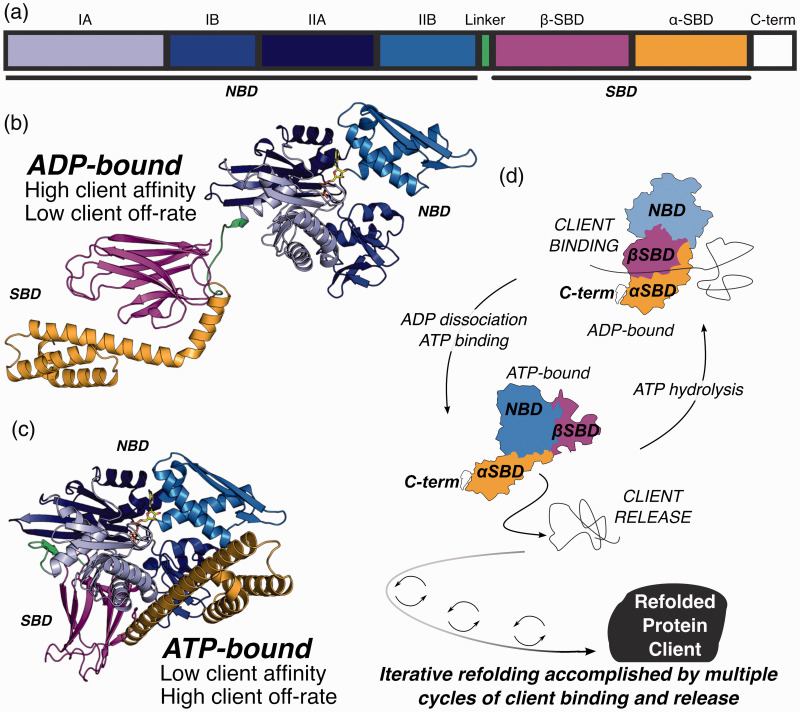
Hsp70 (HSPA) is composed of two large domains, the N-terminal nucleotide-binding domain (NBD) and the C-terminal substrate-binding domain (SBD), and two variably structured regions as shown in Figure 1.30,31 The region that connects the SBD and NBD domains, known as the interdomain linker, is composed of 10–12 highly conserved hydrophobic amino acids which adopt an unstructured conformation in the adenosine diphosphate (ADP) bound state 31 and a β-stranded conformation in the adenosine triphosphate (ATP) bound state.29,30,32 Overall, both the NBD (∼45 kDa) and SBD (∼25 kDa) exhibit differences in structure across the ATP hydrolysis cycle, including some flexibility in particular subdomains.33,34
Figure 1.
Domain organization of Hsp70 homolog of E. coli (DnaK), structure of Hsp70 and the ATP hydrolysis cycle. The domain organization of Hsp70 is shown according to length of sequence (a) divided into the nucleotide binding domain (NBD), interdomain linker region (linker), substrate binding domain (SBD), and the C-terminal tail region (C-term). Although the boundaries of subdomains IA and IIA are discontinuous in sequence space, blocks shown in (a) are accurately scaled by total number of amino acids in each subdomain: IA (1–37, 112–184, 363–383), IB (38–111), IIA (185–227, 310–362), and IIB (228–309). The SBD is further divided into β-SBD (396–507) and α-SBD (508–602). (b) Structure of ADP bound conformation of DnaK (PDB:2KHO). (c) Structure of ATP bound conformation of DnaK (PDB: 4B9Q). (d) Schematic of the ATP hydrolysis cycle of HSP70 and client folding.
The nucleotide-binding domain is divided into two lobes, termed I and II, and each lobe is further divided into two subdomain regions IA, IB, IIA, and IIB.18,35,36 The IA, IB, IIA, and IIB subdomains are arranged to form a V shape cleft within which the nucleotide binds. 37 The structural organization of NBD is identified as an actin-like fold where core α/β domains are connected by loops giving arise to a polymorphic structure.32,37,38 The NBD is responsible for the adenosine triphosphatase (ATPase) activity of Hsp70. 39 During nucleotide binding, the interface of subdomains IIA and IIB is docked with the adenosine and deoxyribose moieties. 40 Moreover, the γ-phosphate and a Mg2+ cofactor are coordinated by interactions with residues in subdomain IA of NDB, whereas α and β phosphates form contacts with subdomain IIA.41,42 Mutagenesis experiments have suggested that multiple residues, including Thr13 43 in subdomain IA and Lys70 (Lys 71 in Hsc70) 44 in subdomain IB are involved in the catalysis of the hydrolysis of ATP and that no single residue is responsible for the catalysis process. 29
The SBD domain is subdivided into two domains, termed α and β. 45 The α-SBD composed of a helical lid is connected to β-SBD via a short linker region that adopts different conformations in the ATP-bound and ADP-bound forms. 46 The β-SBD, containing the substrate-binding site used for binding misfolded proteins, is arranged in β sandwich organization made out of two sets of antiparallel β sheets arranged in sets of four strands.29,32 The substrate-binding site is comprised of two pairs of loop regions composed of hydrophobic residues and is lodged upward from two of the β sheets.29,47 The bound substrate is cradled in the substrate-binding cleft in which SBD binds to the client peptides/proteins using the β-SBD loops and the α-SBD helical lid (∼15 kDa) closed over the β-SBD. It is evident that SBD binds to exposed segments of five to seven amino acids, mostly composed of hydrophobic residues and flanked by positively charged residues.7,32 Promiscuity of the SBD domain substrate recruitment, enabled by the limited specificity of the binding site, allows for the interaction of a wide variety of polypeptides, which are not significantly homologous on the sequence level. 4 Thus, it is evident that the interactions are not fully dependent on the substrate’s unique linear amino acid sequence. The C-terminus of Hsp70 is composed of highly dynamic and disordered extended region, called the tail region. 48 There is evidence that suggests this region possesses a weak transient binding site that facilitates chaperone activity.49,50 Moreover, the four residues at the C-terminus of the tail region, EEVD, are vital for the interaction of several co-chaperones such as C-terminus of Hsc70 interacting protein (CHIP). 49
Allostery in Hsp70
Structural evidence, initially based on the Hsp70 homolog Hsp110, suggests allosteric regulation of Hsp70 where binding of ATP to the NBD and subsequent hydrolysis regulates substrate binding and release by the β-SBD domain.9,29,32 The binding and release of a client substrate are synergistically and strictly modulated by the ATPase activity of Hsp70 along with the JDP and NEF co-chaperones.9,34 Therefore, the thermodynamics and kinetics of substrate binding to the SBD are dependent on the ATPase cycle of NBD. 51 Structural evidence, obtained by X-ray crystallography and NMR spectroscopy, has vastly contributed to the understanding of the allosteric mechanism.
The main conformational states of Hsp70 are known as the open 52 and closed 31 conformations. The Hsp70 NBD has high affinity toward ATP 39 and the resultant ATP-bound state has a low affinity towards the client protein substrates. 27 Conversely, the ADP-bound Hsp70 state has higher affinity towards the substrate. 53 ATPase activity of Hsp70 alone is weak and hydrolysis of ATP to ADP is considered as a slow process. 54 Additionally, substrate binding to the SBD leads to enhancement in the rate of hydrolysis of ATP, resulting a stable substrate bound ADP-Hsp70. 54 When ADP is released and ATP binds again to the NBD, the binding of ATP initiates hydrophobic contacts between the NDB, the α-SBD lid domain, and the linker domain, opening the buried substrate binding site in the β-SBD domain. 55 This prompts formation of the open state of Hsp70 which has low affinity for client proteins. 32 The rate of association of substrate (kon) is higher in the SBD in the closed state whereas dissociation rate (koff) is low. 47 The kon values recorded for model client peptide substrates is about 3 s−1. 56 It has been suggested that the NBD and SBD are more tightly coupled in the presence of ATP than in the apo-state. 33 The secondary structure level rearrangements that occur in the β-SBD during the NBD-SBD contact is explained in detail in a 2015 study by Zhuravleva and Gierasch. 51 Additionally, the bidirectionality of allostery has been demonstrated through effects of peptide substrate binding upon ATPase activity.57–59
Once the substrate is bound to the SBD, ATPase activity is promoted resulting in hydrolysis of ATP to ADP. 33 Nucleotide hydrolysis then leads to a conformational change in the NBD which disrupts interactions with the α-SBD lid and the linker domains, resulting in closing of the α-SBD onto the β-SBD and relaxing the linker domain to a flexible conformation between the NBD and SBD domains. 32 The kinetics of ATP hydrolysis are altered through the allosteric regulation by binding of Hsp70 co-chaperones. Hsp70, which binds tightly to ATP with a dissociation constant (Kd) of 1 nM, and exhibits a slow rate of ATP hydrolysis, 3.3 × 10−4 s−1 at 25°C, without the enhancement of catalysis by a co-chaperone such as Hsp40. 56 Bacterial Hsp40 has been shown to increase the rate of ATP hydrolysis by 15,000 fold at 5°C.7,56 The Hsp70 ATP-bound open state features a lower kon and a higher koff for substrate binding in the SBD. 47 In contrast, koff values recorded for peptide binding to the ADP-bound closed state of bacterial Hsp70 ranges from 10−3 to 10−4 s−1 at 25°C. 56 Conformational studies based on hydrogen-deuterium exchange and mutagenesis suggest that changes in hydrogen bond patterns control the conformational dynamics between the open and closed states. 33 The affinities of Hsp70 for client substrates are also dependent on amino acid sequence of the client substrates where decreased hydrophobic content results in a lower affinity. 47
Even though a greater emphasis has been placed on studies of the allosteric regulation by the NBD domain, substrate binding and release are finely regulated by bidirectional heterotrophic allostery between the two domains of Hsp70. 59 Initially it was suggested that the energetic balance between the formation of two orthogonal interfaces of the NBD and SBD, and the domain conformations were important for the allosteric regulation. 60 However, until recently, the exact mechanistic details of how substrate release by the SBD domain affects the ATPase activity of NBD domain of Hsp70 were unclear. The existence of two distinct signal transmission pathways, via conserved hydrogen bond networks, that facilitate the bidirectional crosstalk between the NBD and β-SBD subdomains was illustrated by Kityk et al., 2015. 57 Furthermore, this study determined that these structurally and functionally distinctive signal transmissions inhibit the ATPase activity. 57 Interestingly, the study also implicated that substrate release induced by ATP binding is more significant than the stimulation of ATPase activity elicited by the binding of substrates to Hsp70s. 57
In addition to dependence on client substrate sequence, the substrate binding and release cycle depend upon the Hsp70 linker domain and presence of co-chaperones. The linker domain acts as the communicator between the NBD and SBD domains, enabling coupling of nucleotide binding, substrate binding, and substrate release during the functional cycle. 61 For example, exchange of ADP back to ATP in the NBD is required for and leads to adoption of the open state of Hsp70 and release of the client protein from the substrate-binding cleft. 32 While exchange of ADP for ATP is slow for Hsp70 alone, the presence of a nucleotide exchange factor (NEF) co-chaperone, such as GrpE, allosterically regulates Hsp70 by spurring the exchange of ADP to ATP, accelerating the exchange process 5000 fold.54,62 Therefore, the fine regulation of the Hsp70 cycle by co-chaperones is primarily achieved by inducing higher affinity toward ATP and altering the rates of ATP hydrolysis. As mentioned above, these effects are manifested as an increasing exchange to ATP by 5000-fold in the presence of NEFs, and as a 15,000-fold acceleration of ATP hydrolysis in the presence of JDPs. Once the ATP-bound Hsp70 state is achieved, the resultant conformational change promotes the release of the substrate from Hsp70 SBD leading Hsp70 to start a new cycle of ATP-hydrolysis and substrate binding (Figure 1).
Functions of Hsp70 and co-chaperones
Primary roles of Hsp70
Once a protein is synthesized as a linear polypeptide, it needs to be folded into a native, low-energy conformational state by passing through a complex energy landscape to achieve the normal function.63,64 Since the cytoplasm of cells is a crowded space filled with macromolecules, the presence of unfolded proteins in the cytoplasm could increase chances of aggregation. 65 Hsp70 is the central component of cellular chaperone machinery that prevents aggregation and corrects misfolding.56,64 In general, Hsp70 is involved in functions related to protein quality control as well as housekeeping and signal transduction machinery in a cell. A detailed list of functions is provided in Table 1. In this section, we focus on the protein quality control functions of Hsp70, including the molecular mechanisms behind Hsp70 functionalities.
Holdase, foldase, and disaggregase/unfoldase activity
Hsp70 coordinates with JDPs to prevent the aggregation or proteasomal degradation of non-native unfolded, aggregated, or misfolded substrates via binding of the Hsp70 SBD to exposed hydrophobic regions in a function is known as holdase activity. 10 JDPs such as Hsp40 (DNAJA) facilitate rapid ATP hydrolysis, thereby accelerating transient association of Hsp70 with client substrates and preventing aggregation. 10 Holdase activity sequesters misfolded or unfolded proteins and is also involved in the refolding process by sequestering non-native intermediates along the refolding pathway to the native folded state. This process is aided by Hsp40 which binds to the non-native denatured or unfolded substrates via hydrophobic loops on the substrate surface.66,67 Human Hsp40 has been shown to bind to larger aggregates ranging from 700 to 4000 kDa in molecular weight. 68 The substrate/Hsp40 complex will be bound by Hsp70 in the ATP-bound state forming a tertiary Hsp70/substrate/Hsp40 complex.69,70 A recent crystallographic study using a JDP-Hsp70 fusion protein suggested the molecular level interactions and the mechanism for the interaction of bacterial Hsp70 and Hsp40 homologs. 71 The Kityk et al. study suggests that hydrolysis of ATP and bipartite interactions occur nearly simultaneously. 71 The bipartite interactions include binding of the Hsp40 (DnaJ) J-domain to the Hsp70 NBD domain and transfer of substrate-bound to Hsp40 to the substrate-binding site within the β-SBD domain of Hsp70. Structures from the Kityk et al. study further indicated that the Hsp40 J domain interacts with the Hsp70 interdomain linker region, potentially aiding both ATP hydrolysis and formation of the closed conformation of Hsp70 by abstracting the Hsp70 interdomain linker from binding with the NBD.56,71 Despite the structural information provided by the Kityk et al. study, conclusive details regarding the mechanism by which the chaperones conduct holdase activity remain elusive, particularly given conflicting structural information from Alderson, et al.40,72
Foldase activity is tied to ATP hydrolysis and drives the folding of misfolded, aggregated, or unfolded substrates to the native state. 73 The function of Hsp70 as a foldase is often linked to interactions with the 90-kilo Dalton heat shock protein (Hsp90) by forming a complex capable of remodeling the substrate until it reaches the native state. 74 In this process, once a client is released from Hsp70 during the allosteric cycle, the co-chaperone Hsp70/Hsp90 organizing protein (HOP) transfers the substrate between Hsp70 and Hsp90 as folding progresses towards the native state of the substrate. 75 A closer look at the substrate binding determinants for Hsp70 and the predominant involvement of client substrate residues that are buried inside the substrate when folded in the native state seems to indicate that Hsp70 would act as a holdase. This holdase activity, while effective at preventing protein aggregation, would seem to lead Hsp70 to act as a folding preventer. However, binding of substrates during foldase activity is more consistent with disaggregase or unfoldase activity, activities that are an integral part of foldase activity. The disaggregase and unfoldase activities of Hsp70 aid foldase activity by preventing the irreversible aggregation of unfolded client substrates and kinetically biasing populations toward those conformations that can be readily shuttled back to a folded conformation. 76 Hsp70, along with the Hsp110, also exhibits disaggregase activity independent of foldase activity, such as situations where aggregated proteins were broken apart and subjected to a later refolding process.3,73 Moreover, the earliest evidence suggests that bacterial Hsp70 is involved in dissociating RNA polymerase aggregates and returning the activity of RNA polymerase. 28 However, in contrast to Hsp70, Hsp110 alone cannot perform protein folding3,7 and Hsp70 often passes along the disaggregated substrates to above explained foldase cycle involving Hsp90. 77
Hsp70 and co-chaperones directing the protein quality control process
Regulation of synthesis and the degradation of proteins is important for maintaining the structural and functional fidelity of proteins within a cell. Key players that contribute to the balance between synthesis and degradation of proteins are the molecular chaperones Hsp70, Hsp90, and Hop, and the E3-ubiquitin ligase CHIP. 78 If Hsp70 interacts with Hop and Hsp90, misfolded proteins recruited by Hsp70 are shuttled through a refolding pathway, whereas interaction of Hsp70 with CHIP co-chaperone will lead to CHIP-mediated ubiquitination which ultimately shuttles misfolded and ubiquitinated clients to proteasomal degradation. The choice of whether a misfolded protein is shuttled to the degradation pathway or refolding pathways, in other words the fate of a the protein, is known as the triage decision.78,79 Moreover, due to binding of CHIP and Hop to the same epitope on Hsp70, a single misfolded client protein will typically not be engaged in both pathways simultaneously. 78 It is also known that CHIP inhibits ATP hydrolysis of Hsp70, an activity normally promoted by Hsp40, in order to drive the triage decision towards degradation. 80
Hsp90 is a heat shock chaperone that exists as a dimer and each monomer contains a N-terminal NBD, a middle domain, and a C-terminal dimerization domain. 81 The NBD hosts the ATP binding site, whereas the middle domain interacts with the substrate. 78 The C-terminal domain of Hsp90 contains a recognition sequence known as the MEEVD motif. 82 Strikingly, Hsp70 and Hsp90 share very little sequence and structural homology with the exception of this conserved EEVD motif 83 which regulates binding to tetratricopeptide repeat (TPR) domains.
Co-chaperone Hop (∼60 kDa) contains three TPR domains (TPR1, TPR2A, and TPR2B), two DP domains (DP1 and DP2), and a linker region in their structure. 75 The TPR domains are made out of seven helices arranged in helix-turn-helix repeats which stack to form a superhelix within each TPR. 50 The DP1 and DP2 domains allow for refolding and activation of the susbtrates. 75 Along the refolding pathway, complexes consisting of Hop, Hsp70, and Hsp90 are formed. TPR1 or TPR2B binds to Hsp70 via while TPR2A is preferred for binding to Hsp90 via the MEEVD motif.50,75 Affinity for Hop TPR1 to the C-terminal IEEVD motif of Hsp70 is reported as 15 µM, whereas TPR2A exhibits an affinity of 6 µM towards Hsp90 C-terminus. 50 Combined evidence from crystallography and electron microscopy (EM) maps suggests a four-step mechanism for refolding mediated by Hsp70, Hop, and Hsp90. A misfolded substrate is recognized and bound by the SBD of Hsp70 in the first step, 75 typically mediated by aid of Hsp40 and Hsp70 ATPase activity. 74 Simultaneously, TPR2A of Hop binds to dimerized Hsp90 leading Hsp90 to adopt a semi-closed substrate binding conformation. 75 In the second step, the Hop/Hsp90 complex binds to the Hsp70/substrate complex via TPR1 domain of Hop. 75 The DP2 domain of Hop activates the client and passes the client off to dimerized Hsp90 76 resulting in substrate loaded Hsp90. Lastly, the Hsp70 NBD interacts with the Hsp90 NBD, while the Hsp70 SBD interacts with Hop TPR2B, inducing lateral reengagements in Hop that facilitate further folding of the client protein mediated by Hsp90.84,85
In contrast to Hop, which can form complexes with Hsp70 and Hsp90 simultaneously, CHIP (∼35 kDa) exists as a homodimer with each monomer capable of binding either Hsp70 or Hsp90, although not simultaneously. Each monomer is composed of an N-terminal TPR domain, a central coiled coil domain, and C-terminal U box domain. 79 The TPR domain mediates the binding of the EEVD motif of Hsp70 or Hsp90, whereas the central coiled coil and U-box domains aid in the dimerization of CHIP and the U-box serves as a ubiquitin ligase domain. Acting as an E3 ubiquitin ligase, CHIP catalyzes ubiquitination of Hsp70 or Hsp70-bound substrates. During this process, ubiquitin is transferred from an E2 ubiquitin-conjugating enzyme bound to CHIP to an ε-amino group of a lysine residue on Hsp70 or an Hsp70-bound substrate. 80 Ultimately, CHIP tags substrates with ubiquitin to direct proteasomal degradation; 78 however, not all ubiquitinated substrates are directed to degradation and the exact underlying mechanism behind this activity is not fully known. Binding of CHIP to Hsp70 is predominantly mediated by interactions between the CHIP TPR domain and the C-terminal EEVD of Hsp70. 81 The reported affinity of full length CHIP towards full length Hsc70 is 60 nM by biolayer interferometry (BLI). 86 Interestingly, the interaction between full length CHIP and the C-terminal EEVD of Hsp70 is weaker, determined to be 370 nM by BLI. Structural studies by our lab identified additional bipartite interactions between CHIP and Hsc70 where the TPR domain interacts with both the Hsc70 α-SBD lid domain as well as C-terminal tail via IEEVD that may explain the variation in affinities of CHIP for full length Hsp70 versus the C-terminal tail via IEEVD motif alone. 87 Moreover, it was found that the post-translational modifications on α-SBD lid domain are involved in the regulation of CHIP-mediated ubiquitination. 87 A recent study of posttranslational modifications of CHIP identified that serine phosphorylation of CHIP also serves to stabilize the interactions between CHIP and Hsp70 and positively influences CHIP-mediated ubiquitination and subsequent proteasomal degradation. 88
Interactions of co-chaperones and substrates with Hsp70
Hsp70 chaperones and the co-chaperone network are composed of components that are localized throughout the compartments and organelles of cells as well as in all domains of life. Although our review focuses on the cytosolic components of this network, Table 2 serves as a resource listing the different homologs of chaperone and co-chaperones found throughout different compartments of cells.
Table 2.
Cellular localization of chaperones and co-chaperones.
| Type | Common name/ class | Bacterial analog | Compartment | Yeast | Higher eukaryotes |
|---|---|---|---|---|---|
| Chaperone | Hsp70 system | DnaK | Cytosol | SSA1,SSA2 89 | Hsp70Hsc70 |
| Mitochondria | SSC1 19 | Mortalin(mtHsp70) 90 | |||
| Endoplasmic reticulum (ER) | Kar2/Bip 91 | Bip 92 | |||
| Co-chaperone | Hsp40 | DnaJ | Cytosol | YDJ1, SIS1 93 | Hsp40,Hdj1 79 , 93 |
| Mitochondria | MDJ1 18 | DnajA1 94 Tid1 95 | |||
| ER | SCJ1(lumen), Sec63(Membrane) 63 | ERdj4 and ERdj5 95 | |||
| GrpE a | GrpE | Mitochondria | MGE1 90 | hMge1 90 | |
| HspBP1 | HspBP1 96 | Cytosol | Sil1 97 , Fes1 98 | HspBP1 99 | |
| ER | Sls1p/Sil1p 100 , 101 | BAP 101 | |||
| HOP b | N/A | Cytosol | Sti1 102 | hHOP 75 | |
| Bag | N/A | Cytosol | Snl1-M and Snl1-S 103 | BAG1M, BAG1L, BAG-1S, BAG 2-6 104 , 105 | |
| Nucleus | BAG1L 105 | ||||
| ER membrane | Snl1-L 103 | ||||
| Hsp110(also known as Hsp105 106 ) | N/A 107 | Cytosol | Sse1p,Sse2p 108 , 109 | Hsp110Apg1-2 107 , 110 | |
| Hip | N/A | Cytosol | Hip (p48, St13) 111 , 112 | ||
| CHIP(also known as U-Box Containing gene 1-(STUB1) 113 ) | N/A | Cytosol | CHIP 80 |
Interactions with the Hsp70 NBD
Two main classes of proteins that interact with the NBD domain are the JDPs, such as Hsp40, and NEFs, such as GrpE, Bcl2-associated athanogene (BAG) proteins, Hsp70 binding protein 1 (HspBP1), and Heat shock protein 110 (Hsp110). The domain organizations and example structures of these proteins can be found in Figure 2 and Table 3, respectively.
Figure 2.
Structures of Hsp70 co-chaperones. The domain organization of Hsp70 (a) is shown according to length of sequence with the size of subdomain blocks scaled by total number of amino acids in each subdomain, as in Figure 1(a). Colored bars beneath the Hsp70 domain indicate approximate regions of Hsp70 that interact with each co-chaperone. NEF co-chaperones include (b) Hsp110 (dark blue) in complex with Hsp70 NBD (white); (c) HspBP1 (red) in complex with Hsp70 NBD subdomain IIB (white); (d) GrpE (teal and cyan) in complex with Hsp70 NBD (white); (e) Bag2 (purple and violet) Hsp70 NBD (white); and (f) Bag1 (orange) Hsp70 NBD (white). The JDP co-chaperone is (g) Hsp40 in complex with Hsp70 NBD/β-SBD/α-SBD (white, grey, charcoal). Other co-chaperones include (h) Hip (brown) in complex with Hsp70 NBD (white); (i) CHIP (lime green and forest green) in complex with Hsp70 α-SBD/C-term (charcoal and black); and (j) HOP (light blue) in complex with Hsp70 C-term (black).
Table 3.
Protein data bank IDs for structures of co-chaperones and Hsp70/co-chaperone complex structures.
| Co-chaperone | Structure of the co-chaperone (A) | Structure of the complex with Hsp70 (B) |
|---|---|---|
| Bag1 | 1HX1a | 1HX1a |
| Bag2 | 3D0Tb | 3CQXc |
| CHIP | 2C2Ld | 4KBQe |
| GrpE | 3A6Mf (from Thermus thermophilus)1DKGg (from E.coli) | 1DKGg |
| HOP | 2NC9h | 1ELWi |
| Hsp40 | 1XBLj | 5NROk |
| HspBP1 | 1XQRl | 1XQSm |
| Hsp110 | 2QXLn | 3D2Fo |
| Hip | 4J8D, 4J8Ep | 4J8Fq |
aComplex of the BAG domain (residues 151-264) of BAG1M with the NBD domain (residue 5-381) of Hsc70. 116
bBNB domain (residues 107–189) of murine BAG2.
cComplex of BNB domain (residues 107–189) of murine Bag2 with Hsc70 NBD domain (residues 1-381).
dNear full length structure of CHIP (residues 25-304) from mouse. 117
eComplex structure contains the CHIP-TPR domain (residues 21-154) and Hsp70 lid-tail domain (residues 541-646 Δ626-638). 87
fCrystal structure of GrpE from Thermus thermophilus HB8. 118
gComplex of GrpE (residues 1-197) and NBD domain of DnaK (residues 1-388) from E.coli. 119
hTPR2A domain (residues 220-350) of Hop.
iComplex of TPR1 domain (residues 1-118) of Hop with C-terminal Hsc70 peptide(residues 625-732). 50
jJ domain(residues 2-76) of DNAj. 120
kComplex of J domain (residues 3-65) of Hsp40 with DnaK (residues 2–604). 71
lCrystal structure of core domain of human HspBP1. 96
mCrystal structure of the HspBP1 core domain (residues 84-359) complexed with the lobe II of Hsp70 NBD domain (183–371 residues). 96
nYeast homolog Hsp110: Sse1 (residues 2–659). 108
oComplex of Yeast homolog Hsp110: Sse1p (residues 2–659 and Δ503–524) with NBD (residues 1–377) of yeast Hsp70 homolog Ssa1p. 121
pTPR domain (residues 78–247) of Hip. 111
qComplex of TPR domain (residues 78–247) of Hip and the NBD domain (residues 1–382, point mutation D110E) of Hsp70. 111
Hsp40
Broad sequence and structural diversity is observed among the J domain proteins despite all of the members sharing a conserved J domain. Conventionally, there are three classes of JDPs based on domain organization known as class A, B, and C.7,67,122 However, diversity exists within each class as well. Class A is more similar to E. coli Hsp40 homolog DnaJ and contains an N-terminal J domain, a glycine-phenylalanine-rich region, four cysteine-rich repeats arranged to resemble a zinc finger type motif, and a C-terminal extension. 67 Class B lacks the zinc finger motif, yet retains other domains found in class A JDPs. 123 JDPs that do not share similar structural organization, with the exception of the J domain, to either class A or B are classified as class C.67,122 Comprehensive details about different classes of JDPs can be found in the review by Kampinga and Craig. 67
Human Hsp40 (DNAJA) is a conserved class B 7 JDP and contains an N-terminal J domain, a conserved glycine-phenylalanine (GF) rich sequence, an approximately 30 residues long linker region, two C-terminal β-sandwich substrate-binding domains (CTDI and II), and a dimerization domain. 40 The J-domain contains four α-helices, and the second and third helices are connected by the linker region. 124 The functions of Hsp40 are discussed in detail above, concerning the allosteric cycle and foldase activity of Hsp70. Some controversy exists within the field as differences are observed between a crystal structure of the E. coli DnaJ/DnaK complex, 71 and an NMR-derived model of the DnaJ/DnaK complex. 72 However, the general features of the complex structure are similar and the effects of Hsp40s on Hsp70s are clearly mediated by the J domain.
GrpE
Many of the proteins that bind to the Hsp70 NBD domain fall within the category of NEFs. GrpE (∼22 kDa) is a prokaryotic homodimeric NEF that contains two distinct regions, a head and a tail. 125 The tail region is formed by an N-terminal unstructured region and a long-coiled coil α-helical dimerization domain. 125 The head region is composed of a four helical bundle and a C-terminal compact β-sheet domain.119,125 The functional importance of GrpE for Hsp70 activity is discussed above under the allosteric cycle of Hsp70 of this review. Dimeric GrpE has shown to interact with a single Hsp70. 119 Binding of GrpE results in insertion of the head region into the nucleotide-binding cleft of the Hsp70 NBD domain acting as a wedge between the SBD and NBD. 119 Conformational changes due to the interaction include a 14° rotation in the NBD IIB subdomain which opens the nucleotide-binding cleft and modulates the nucleotide binding pocket and promoting release of the bound ADP molecule, 119 thereby promoting the swapping of ADP to ATP in NDB during the allosteric cycle of Hsp70. This conformational change that opens up the NBD has shown to reduce its affinity to ADP by 200 fold, whereas affinity of ATP is increased by 5000 fold by fulfilling the nucleotide exchange activity of GrpE. 62 Moreover, the GrpE N-terminal tail region is suggested to form contacts as a pseudosubstrate with the Hsp70 substrate binding cleft promoting a conformational change that leads to substrate release from SBD of Hsp70. 125 Additional details of the interactions between Hsp70 and GrpE can be found in the Melero et al. electron microscopy study. 125
BAG
BAG domain proteins are one of the three main classes of eukaryotic NEFs, joined by HspBP1 and Hsp110s. These groups are structurally different, and there is little to no homology. 7 However, evidence suggests that all three classes interact with the ATP-bound NBD domain of Hsp70 in the open conformation. The BAG family of proteins includes six subclasses (BAG 1–6) based on domain organization. 105 The members of the BAG family of proteins characteristically contain at least one conserved BAG domain. 103 The C-terminal BAG domain is typically composed of three antiparallel helical bundles, each consisting of 30–40 amino acids.105,126 There are three isoforms of BAG1 known as BAG1L (∼50 kDa), BAG1M (∼46 kDa), and BAG1S (∼36 kDa). 105 Due to high expression levels of BAG1S itself is often referred to as BAG1.105,127 In addition to a BAG domain, BAG1S also contains an N-terminal nuclear localization signal (NLS) not found in BAG1L and BAG1M, a ubiquitin-like domain (UBL). 105 In contrast, BAG2 harbors an N-terminal coiled coil domain and a novel BAG domain termed the brand new bag (BNB) which composed of two long antiparallel alpha helixes (α1 and α2) connected by a linker comprise of a short helix and a disordered loop region.124,128 Moreover, the BAG2 BNB exists as nearly identical symmetric homodimers. 124 BAG4 has similar domain organization to BAG2, lacking the UBL domain found in BAG1 and only composed of canonical BAG domain and a N-terminal unspecific region. BAG3, also known as BIS/CAIR, contains an additional N-terminal WW domain (W stands for the amino acid Trp) and a proline-rich region called PXXP, besides the BAG domain. 129 BAG5 uniquely contains four additional putative BAG domains at the N-terminus. 104 BAG6 (also known as Scyther/BAT3) contains an N-terminal UBL domain in addition to the BAG domain and is the longest of the family. 130
All members of BAG family interact directly with the NBD domain of the Hsp70 via the BAG domain, whereas the other domains such as the PXXP and WW domains serve a range of diverse cellular functions.129,131 For example, BAG1 isoforms were indicated to form a link between proteasomal degradation and Hsc/Hsp70.127,132
Among the family, BAG1 and 3 are well characterized. Affinities for BAG1 and BAG 3 towards NBD of Hsp70 are about 12 nM and 10 nM, respectively, whereas BAG2 exhibits a significantly weaker affinity of 380 nM. 94 Crystallographic evidence of BAG1 with constitutively expressed cytosolic human Hsc70 and a bacterial Hsp70 homologue illustrate that second and third helices of the BAG domain of BAG1 bind to the IB and IIB subdomains of NBD inducing an outward 14° rotation of subdomain IIB.116,126 The induced conformational change ultimately result in higher rates of ADP exchange for ATP. 127 However, presence of both Hsp40 and BAG1 is required for both fast ATP hydrolysis and nucleotide exchange respectively as neither Hsp40 nor BAG domain alone are sufficient for both increasing ATP hydrolysis and nucleotide exchange rates. 116 Binding between the NBD and BAG1 is mediated by electrostatic interactions that involve highly conserved residues of all members of BAG family. These residues include Glu212, Gln245, Asp222, and Arg237 in BAG1, while the interacting residues in Hsp70 include Arg261 and Glu283, each of which are conserved among bacterial and eukaryotic cytosolic forms of Hsp70. 116 Moreover, a study using BAG1 and BAG3 demonstrated a non-canonical interaction outside the BAG domain with the Hsp70 β-SBD that is crucial for facilitating the release of the substrate bound to SBD of Hsp70. 126 Therefore, the entire mechanism of action for nucleotide exchange is suspected be driven by bidentate interactions that include the canonical BAG and NBD of Hsp70 and non-canonical interactions outside the BAG domain. However, the exact region of interaction with the SBD has not been identified. Intriguingly, the mechanism of canonical BAG proteins is very similar to GrpE, 116 with primary differences including stoichiometry of the NEF and the striking structural differences between GrpE and BAG domains. When the former is considered, GrpE interacts with the Hsp70 as a dimer, whereas BAG1 interacts as a monomer. As the latter, the Hsp70-interacting region of BAG1 is mainly composed of α-helices, while the Hsp70-interacting region of GrpE is primarily β-strands. 116
Since BAG2 contains the BNB, a domain distinctive from the rest of the BAG family proteins, the mechanism of action and the interactions involved in the nucleotide exchange process are significantly different. Even though the BNB domain of BAG2 binds to IB and IIB subdomains in NBD of Hsc70, the same domains bound by canonical BAG domains, the binding occurs in end-on-end fashion compared to parallel binding of canonical BAG domains. 124 Additionally, the rotation in subdomain II of NBD induced by the binding is classified as a rigid since the entire domain moves as one unit. 124 Additionally, the BNB dimer binds to the NBD and contacts are formed via linker loop and helix α2 of the BNB. 124
HspBP1
HspBP1 is a single domain protein composed of four concave α-helical armadillo repeats and a flexible N-terminal extension domain (RD). 98 The mechanism of action for HspBP1 consists of two steps. First, the HspBP1 armadillo domain forms contacts with lobe IIB of ADP-bound Hsp70 NBD domain resulting in movement of lobe I away from the nucleotide binding cleft thereby reducing the affinity for ADP which enables ADP release.96,98 Second, the conformational change in the NBD opens the SBD to facilitate release of substrate from the SBD. 98 Meanwhile, HspBP1 utilizes the RD to mimic a substrate and binds to the β-SBD to preclude rebinding of true substrates to the SBD. 98 Key secondary structure elements of the NBD serving as HspBP1 contact sites are identified as helix α7 which forms minor contacts with C-terminus of third armadillo repeat, and strands β16 and β17 which interact with armadillo repeats 1–3. 96 These contacts are facilitated by hydrogen bonding and van der Waals interactions due to the polarity and shape complementarity of the surfaces.
Although the mechanistic details differ, HspBP1 inherently resembles GrpE functionality in releasing the substrate from the SBD domain and the ADP from NBD of Hsp70. 97 However, the mechanism of action HspBP1 is different from the BAG proteins. BAG binds the Hsp70 NBD at the top of the cleft facilitating the interaction with subdomains IB and IIB, whereas HspBP1 interactions with lobe II of NBD interactions are rotated in comparison. 96 Moreover, HspBP1 does not form contacts with subdomain IB of NBD as in the case of BAG.
Hsp110
Hsp110 (HSPH2) is the most abundant amongst NEFs and serves as an NEF to canonical Hsp70s promoting protein disaggregation.9,98 Available mechanistic details are based on the studies conducted on the Saccharomyces cerevisiae Hsp110 homolog, Sse1p. Hsp110 (∼110 kDa) was initially classified as a member of Hsp70 family due to sequence similarity and contains a N-terminal NBD, a larger SBD, a linker region, and a C-terminal extension harboring three helical bundle domains (3HBD).108,110,121,133 The SBD of Hsp110 is divided into an α helical lid and a β-sandwich domain that contains an insertion called an acidic subdomain (AS). 133 The interaction points enabling the nucleotide exchange function are the NBD and 3HBD of Hsp110 and the NBD of Hsp70. The two NBDs arrange in head to head fashion forming contact between subdomains IB and IIB, IA and IIB, and IIB and IB of Hsp70 and Hsp110, respectively.121,134 Further contacts are formed between subdomain IIB of Hsp70 NBD and 3HBD of Hsp110 and the clamping of the subdomains121,134 elicits a 27° rotation of Hsp70 subdomain IIB, similar to the effect exerted by GrpE and BAG proteins,7,121 although the NBD domain of Hsp110 causes a comparatively higher degree of rotation. Ultimately the conformational change opens up the nucleotide binding site within the Hsp70 NBD, enabling release of ADP, allowing the exchange to ATP, and promoting substrate release from the Hsp70 SBD. 121 The affinity of Hsp110 toward Hsp70 reported using surface plasmon resonance is 100–150 nM with a kon of 2.3 mM−1s−1. 109 Moreover, it was proposed that in order to prevent aggregation of the multidomain substrates, both Hsp70 and Hsp110 work in concerted manner where one domain refolds, while other domains remain stay chaperone-bound. 135 Ultimately, ATP binding to Hsp70 releases both Hsp110 and the substrate, allowing for escape of the refolded substrate or another round of binding to Hsp70 for further folding.
Hip
All of the proteins discussed above which interact with the Hsp70 NBD belong to either the JDP or NEF families. However, Hsc70 interacting protein (Hip) is a protein that binds to NBD and yet does not belong in either the JDP or NEF classes. Hip (∼40 kDa) exists as a dimer composed of an N-terminal dimerization module, a tetratricopeptide repeat (TPR) domain, a charged region, GGMP peptide repeats, and a C-terminal aspartic-proline (DP) domain.111,136 The C-terminal domain of Hip is structurally similar to the yeast Hop homolog, Sti1.137,138 Interestingly, binding of the Hip stabilizes the ADP bound state of Hsp70 delaying the release of substrate.127,137 This is supported by isothermal titration calorimetry studies which found a higher affinity of 8 µM for Hip towards the ADP-bound state of Hsp70 compared to an affinity of 51 µM against nucleotide-free chaperone. 111 Moreover, Hip is known to inhibit the nucleotide exchange activity of BAG1 by competing for the binding to the NBD domain of Hsp70. 139 Intriguingly, Hip and BAG1 do not appear to interact directly with each other during the process. 139 The mechanism behind the process was uncovered by the X-ray crystal structure of rat Hip protein. 111 The structure features a bracket that is formed over the NBD by the TPR domain of Hip which serves to obstruct the dynamic nature of the NBD, thereby locking Hsp70 in the ADP bound state and delaying the substrate release by Hsp70. 111 Hip appears to be important for avoiding premature substrate release and for facilitating the foldase function of Hsp70.137,139
Interactions with the Hsp70 SBD
Substrates
In addition to interactions of the HOP and CHIP TPR domains described above, additional notable interactions are known to occur between other binding partners and regions of the Hsp70 SBD. The first structural evidence of the substrate Hsp70 complex was based on bacterial Hsp70 bound to a small model peptide, NRLLLTG.53,140 Several studies have been done over the years, and a non-exhaustive list of different substrates of Hsp70 that have been studied is given in Table 4. Binding of a substrate to the substrate-binding cleft of the β-SBD has shown to be promiscuous. Hsp70 binds to segments of five to seven amino acids, predominantly composed of hydrophobic residues and flanked by positively charged residues, and these nebulous recognition sequence determinants result in the ability to bind a wide array of proteins as substrates.141,152,153 Moreover, it has shown that the binding to the substrate occurs only at the unstructured exposed hydrophobic loops and linker regions while the rest of the substrate remains folded. 142
Table 4.
List of Hsp70 substrates that bind to the SBD.
| Type of substrate | Substrate | References |
|---|---|---|
| Peptides | NRLLLTG (NR), PL, PP,LYZ, Onc72 141 | 46,53, 140 , 142 |
| Non-native substrates | The glucocorticoid receptor (GR) -ligand binding domain | 96 |
| Immunoglobulin 27 (I27) domain of the human striated muscle protein titin | 143 | |
| Native substrates | Transcription factor σ32 | 144 |
| λ repressor | 117 | |
| Superoxide dismutase 1 (SOD1) | 145 | |
| Unfolded | Ribonuclease H (RNase H) | 146 |
| NCA-SNase- Staphylococcal nuclease, RCMLA | 147 | |
| Rhodanase | 148 | |
| A fragment of apomyoglobin | 149 | |
| Conformational ensemble of substrates | drkN SH3 from Drosophila | 150 |
| hTRF1 (human telomere repeat binding factor 1) | 151 | |
| Other | RepE54-Replication initiation protein | 13 |
Upon binding of the substrate, packing of the α-helical lid domain of SBD is subtly altered, compared to the β-SBD. 154 When the solution structures of free and NRLLLTG peptide substrate bound DnaK (E. coli Hsp70 homolog) are compared, the NBD, SBD, and linker appear to move relatively independently of each other in the ADP/substrate/Hsp70 complex. 31 However, movement of the SBD is restricted to a cone with about 70° angle. 31 A comparison of NMR structures of ADP-bound Hsp70 and nucleotide free Hsp70 illustrated that the NBD and SBD move independently from each other in the absence of a substrate, suggesting that the binding of a substrate restricts the movement of the SBD. 61 Interactions that correspond to allosteric communication between the NDB and SBD are primarily clustered on the NBD subdomain IA on the side of IA-IIA interface where the α-helical lid domain and β-SBD are docked. 31 Even though there several structures with both NBD and SBD domains of Hsp70 are available, they lack common ground. For example, when the structure of E213A/D214A human Hsc70 mutant that contains both NBD and SBD is considered, it is noted that SBD interacts with both subdomain IA and IIA of NBD, instead of just subdomain IA. 155 The source for the differences may be the lack of an actual substrate as this structure has intramolecular leucine residues bound to the substrate binding cleft of Hsp70 in place of an exogenous substrate. However, it may be difficult to obtain a structure without a bound substrate or substrate analog since the promiscuous binding determinants enable the substrate binding cleft to bind to a wide array of available hydrophobic residues. 154 Therefore, understanding the structural consequences of binding of a substrate to the Hsp70 SBD should be treated in a case by case manner to understand the interactions involved and identity of residues occupying the substrate binding cleft.
SMADs
SMADs, named for homology to the Caenorhabditis elegans’s “Sma” and Drosophila “Mad” proteins, are intracellular mediators of transforming growth factor-β (TGFβ) secretory proteins. 156 Human’s contain eight SMADs classified in to receptor regulated (R-SMADs), co-mediator (Co-SMADs), and inhibitory SMADs (I-SMADs).156,157 SMADs 1, 2, 3, 5, and 8 belong to R-Smads, while SMAD 4 is a Co-SMAD.156,158 SMADs 6 and 7 serve as inhibitory SMADs.158,159 SMADs contain two conserved domains, an N-terminal Mad homology (MH) domain 1 and a C-terminal MH2 connected by a proline-rich non-conserved linker region. 156 MH1 domains assume a compact globular fold and contain a β hairpin that mediates contacts with DNA during transcription activation. 156 The MH2 domain, composed of five α-helices and β-sandwich domain enclosed by three loops, does not interact with DNA and instead associates with variety of proteins.156,160,161
As intracellular effectors, tight regulation of SMADs is a necessity to maintain basal levels of effector concentrations. 162 In aiding this process, some members of SMADs such as SMAD3 are known to be ubiquitinated by CHIP as their first step in proteasomal degradation. 163 Furthermore, there are investigations showing that Hsp70 and Hsp90 are involved in the regulation of SMAD3 interactions with CHIP, thereby affecting the ubiquitination process.159,162 A study by Shang et al. has shown opposing effects of Hsp70 and Hsp90 on SMAD3 where Hsp70 facilitates the formation of CHIP/SMAD3 complex leading to SMAD3 ubiquitination. 162 Conversely, Hsp90 inhibits formation of the CHIP/SMAD3 complex thereby inhibiting the ubiquitination of SMAD3 by CHIP. 162
Although the mechanisms behind the above processes are still unclear, crystallographic evidence from one study found the CHIP TPR to directly interact with C-terminal ISSVS sequence within the MH2 domain of SMADs 1, 5, and 8. 159 Since the C-terminal tail peptide of Hsp70/Hsc70 also interacts with the same C-terminal groove of CHIP TPR domain as SMADs, it has implicated that SMAD1/5 and Hsp70 are in competition to bind to CHIP. 159 The same study indicated that in the absence of chaperone, CHIP can promote polyubiquitination of SMAD 1/5/8 suggesting that Hsp70 plays a regulatory role rather than merely facilitating CHIP-mediated ubiquitination. 159 Further studies have shown SMAD1 and SMAD3, along with CHIP and Hsp70, form a ternary complex indicating that CHIP might also mediate the ubiquitination of SMADs presented by Hsp70.157,162 Moreover, CHIP prefers to ubiquitinate phosphorylated SMADs, compared to non-phosphorylated SMADs, and polyubiquitination of SMADs is promoted by phosphorylation.157,159 Altogether these findings have implicated Hsp70 as playing a regulatory role in the TGFβ signaling pathway and in related diseases. 164
Insights on drug discovery and therapeutic targeting
There are numerous implications for the dysregulation of Hsp70 leading to diseases165,166 These include cancer, 162 central nervous system (CNS) disorders, 167 and cardiovascular diseases. 168 Moreover, the co-chaperones BAG,105,169,170 Hsp40,171,172 Hip, 138 HOP, 173 HspBP1, 174 HSP110,175,176 CHIP,177–179 and SAMDs162,164 are also implicated in wide variety of diseases. The implications of Hsp70 and co-chaperones in wide range of diseases have led to calls to target these proteins for drug discovery. However, a range of issues, including the number of Hsp70 and co-chaperone family members and level of sequence similarity among each family need to be considered. According to Kampinga and Craig, there are 11 Hsp70s localized in different compartments of the cells along with 41 JDPs and 13 NEFs in humans. 67 Thus, targeting Hsp70s, JDPs, or NEFs requires efforts to ensure specificity when needed. In general, targeting Hsp70 would typically be broad and universal, whereas targeting a specific co-chaperone could provide finer control over Hsp70. Depending on the target, co-chaperone targeting offers the opportunity for regulating specific portions of the Hsp70 allosteric cycle. Therefore, choosing the optimal target among Hsp70s, JDPs, or NEFs is a challenging, but potentially highly rewarding pursuit.
Conclusions and future perspectives
Hsp70 and its co-chaperone network play vital part in cellular protein quality control system. Understanding the structural features of the co-chaperone network of Hsp70 aids in understanding the mechanisms and roles the various proteins that comprise protein quality control machinery. However, knowledge regarding the mechanistic details and regulation of protein quality control in the instance of stress conditions and disease condition is still lacking. Moreover, the physiological roles of co-chaperones and the exact mechanisms by which chaperones and co-chaperones contribute to diseases should be more fully investigated as part of the pursuit of drug development. Thus, this review has been structured to provide an outline of key areas of importance for the structures and interactions of Hsp70 and co-chaperones and to provide a collated resource of structural data and corresponding functional consequences.
Footnotes
AUTHORS’ CONTRIBUTIONS: All authors participated in the writing and review of the manuscript.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The authors acknowledge support from National Institutes of Health grant R35 GM128595 (RCP).
ORCID iDs: Chamithi Karunanayake https://orcid.org/0000-0002-8502-5615
Richard C Page https://orcid.org/0000-0002-3006-3171
References
- 1.Morán Luengo T, Mayer MP, Rüdiger SGD. The Hsp70–Hsp90 chaperone Cascade in protein folding. Trends Cell Biol 2019; 29:164–77 [DOI] [PubMed] [Google Scholar]
- 2.Caplan AJ. What is a co-chaperone? Cell Stress Chaper 2003; 8:105–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu X, Sarbeng EB, Vorvis C, Kumar DP, Zhou L, Liu Q. Unique peptide substrate binding properties of 110-kDa heat-shock protein (Hsp110) determine its distinct chaperone activity. J Biol Chem 2012; 287:5661–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gething M-J. Molecular chaperones: individualists or groupies? Curr Opin Cell Biol 1991; 3:610–4 [DOI] [PubMed] [Google Scholar]
- 5.Craig EA, Gross CA. Is hsp70 the cellular thermometer. Trends Biochem Sci 1991; 16:135–40 [DOI] [PubMed] [Google Scholar]
- 6.Qu B, Jia Y, Liu Y, Wang H, Ren G, Wang H. The detection and role of heat shock protein 70 in various nondisease conditions and disease conditions: a literature review. Cell Stress Chaperones 2015; 20:885–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Faust O, Rosenzweig R. Structural and biochemical properties of Hsp40/Hsp70 chaperone system. Adv Exp Med Biol 2020; 1243:3–20 [DOI] [PubMed] [Google Scholar]
- 8.Fernández-Fernández MR, Gragera M, Ochoa-Ibarrola L, Quintana-Gallardo L, Valpuesta JM. Hsp70 – a master regulator in protein degradation. FEBS Lett 2017; 591:2648–60 [DOI] [PubMed] [Google Scholar]
- 9.Saibil HR. Chaperone machines in action. Curr Opin Struct Biol 2008; 18:35–42 [DOI] [PubMed] [Google Scholar]
- 10.Mayer MP, Bukau B. Hsp70 chaperones: cellular functions and molecular mechanism. Cell Mol Life Sci 2005; 62:670–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Craig EA. Hsp70 at the membrane: driving protein translocation. BMC Biol 2018; 16:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frydman J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 2001; 70:603–48 [DOI] [PubMed] [Google Scholar]
- 13.Schlecht R, Erbse AH, Bukau B, Mayer MP. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat Struct Mol Biol 2011; 18:345–51 [DOI] [PubMed] [Google Scholar]
- 14.Mayer MP, Gierasch LM. Recent advances in the structural and mechanistic aspects of Hsp70 molecular chaperones. J Biol Chem 2019; 294:2085–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kramer G, Boehringer D, Ban N, Bukau B. The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 2009; 16:589–97 [DOI] [PubMed] [Google Scholar]
- 16.Preissler S, Deuerling E. Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci 2012; 37:274–83 [DOI] [PubMed] [Google Scholar]
- 17.Pfund C, Huang P, Lopez-Hoyo N, Craig EA. Divergent functional properties of the ribosome-associated molecular chaperone ssb compared with other Hsp70s. Mol Biol Cell 2001; 12:3773–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horst M, Oppliger W, Rospert S, Schö NH-J. Sequential action of two hsp70 complexes during protein import into mitochondria. EMBO J 1997; 16:1842–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Voisine C, Craig EA, Zufall N, Von Ahsen O, Pfanner N., Voos W. The protein import motor of mitochondria: unfolding and trapping of preproteins are distinct and separable functions of matrix Hsp70. Cell 1999; 97:565–74 [DOI] [PubMed] [Google Scholar]
- 20.Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the endoplasmic reticulum: brothers in arms. Mol Cell 2010; 40:238–52 [DOI] [PubMed] [Google Scholar]
- 21.Schmid SL, Braell WA, Schlossman DM, Rothman JE. A role for clathrin light chains in the recognition of clathrin cages by 'uncoating ATPase'. Nature 1984; 311:228–31 [DOI] [PubMed] [Google Scholar]
- 22.Sousa R, Lafer EM. The role of molecular chaperones in clathrin mediated vesicular trafficking. Front Mol Biosci 2015; 2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sousa R, Liao HS, Cuéllar J, JS, Valpuesta JM, Jin AJ, Lafer EM. Clathrin-coat disassembly illuminates the mechanisms of Hsp70 force generation. Nat Struct Mol Biol 2016; 23:821–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arispe N, De M. A. ATP and ADP modulate a cation channel formed by Hsc70 in acidic phospholipid membranes. J Biol Chem 2000; 275:30839–43 [DOI] [PubMed] [Google Scholar]
- 25.Vega VL, Rodríguez-Silva M, Frey T, Gehrmann M, Diaz JC, Steinem C, Multhoff G, Arispe N, De Maio A. Hsp70 translocates into the plasma membrane after stress and is released into the extracellular environment in a membrane-associated form that activates macrophages. J Immunol 2008; 180:4299–307 [DOI] [PubMed] [Google Scholar]
- 26.Asea A, Kraeft SK, Kurt-Jones EA, Stevenson MA, Chen LB, Finberg RW, Koo GC, Calderwood SK. HSP70 stimulates cytokine production through a CD 14-dependant pathway, demonstrating its dual role as a chaperone and cytokine. Nat Med 2000; 6:435–42 [DOI] [PubMed] [Google Scholar]
- 27.Milani V, Noessner E, Ghose S, Kuppner M, Ahrens B, Scharner A, Gastpar R, Issels RD. Heat shock protein 70: role in antigen presentation and immune stimulation. Int J Hyperthermia 2002; 18:563–75 [DOI] [PubMed] [Google Scholar]
- 28.Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A 1991; 88:2874–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zuiderweg ERP, Bertelsen EB, Rousaki A, Mayer MP, Gestwicki JE, Ahmad A. Allostery in the Hsp70 chaperone proteins. Top Curr Chem 2012; 328:99–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi R, Sarbeng EB, Liu Q, Le KQ, Xu X, Xu H, Yang J, Wong JL, Vorvis C, Hendrickson WA. Allosteric opening of the polypeptide-binding site when an Hsp70 binds ATP. Nat Struct Mol Biol 2013; 20:900–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bertelsena EB, Chang L, Gestwicki JE, Zuiderweg ERP. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci U S A 2009; 106:8471–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim YE, Hipp MS, Bracher A, Hayer-Hartl M, Ulrich Hartl F. Molecular chaperone functions in protein folding and proteostasis. Annu Rev Biochem 2013; 82:323–55 [DOI] [PubMed] [Google Scholar]
- 33.Rist W, Graf C, Bukau B, Mayer MP. Amide hydrogen exchange reveals conformational changes in Hsp70 chaperones important for allosteric regulation. J Biol Chem 2006; 281:16493–501 [DOI] [PubMed] [Google Scholar]
- 34.Bhattacharya A, Kurochkin AV, Yip GNB, Zhang Y, Bertelsen EB, Zuiderweg ERP. Allostery in Hsp70 chaperones is transduced by subdomain rotations. J Mol Biol 2009; 388:475–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stevens FJ, Argon Y. Protein folding in the ER. Semin Cell Dev Biol 1999; 10:443–54 [DOI] [PubMed] [Google Scholar]
- 36.Parodi AJ. Protein glucosylation and its role in protein folding. Annu Rev Biochem 2000; 69:69–93 [DOI] [PubMed] [Google Scholar]
- 37.Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 1990; 346:623–8 [DOI] [PubMed] [Google Scholar]
- 38.Kabsch W, Holmes KC. The actin fold. FASEB J 1995; 9:167–74 [DOI] [PubMed] [Google Scholar]
- 39.Jeung-Hoi H, McKay DB. ATPase kinetics of recombinant bovine 70 kDa heat shock cognate protein and its amino-terminal ATPase domain. Biochemistry 1994; 33:14625–35 [DOI] [PubMed] [Google Scholar]
- 40.Alderson TR, Kim JH, Markley JL. Dynamical structures of Hsp70 and Hsp70–Hsp40 complexes. Structure 2016; 24:1014–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bukau B, Horwich AL. The Hsp70 and Hsp60 chaperone machines. Cell 1998; 92:351–66 [DOI] [PubMed] [Google Scholar]
- 42.Bork P, Sander C, Valencia A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc Natl Acad Sci U S A 1992; 89:7290–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sousa MC, McKay DB. The hydroxyl of threonine 13 of the bovine 70-kDa heat shock cognate protein is essential for transducing the ATP-induced conformational change. Biochemistry 1998; 37:15392–9 [DOI] [PubMed] [Google Scholar]
- 44.O'Brien MC, Flaherty KM, McKay DB. Lysine 71 of the chaperone protein Hsc70 is essential for ATP hydrolysis. J Biol Chem 1996; 271:15874–78 [DOI] [PubMed] [Google Scholar]
- 45.Zhu X, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science 1996; 272:1606–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zahn M, Berthold N, Kieslich B, Knappe D, Hoffmann R, Sträter N. Structural studies on the forward and reverse binding modes of peptides to the chaperone DnaK. J Mol Biol 2013; 425:2463–79 [DOI] [PubMed] [Google Scholar]
- 47.Mayer MP, Schröder H, Rüdiger S, Paal K, Laufen T, Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struct Biol 2000; 7:586–93 [DOI] [PubMed] [Google Scholar]
- 48.Bertelsen EB, Zhou H, Lowry DF, Flynn GC, Dahlquist FW. Topology and dynamics of the 10 kDa C-terminal domain of DnaK in solution. Protein Sci 2008; 8:343–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smock RG, Blackburn ME., Gierasch LM. Conserved, disordered C terminus of DnaK enhances cellular survival upon stress and DnaK in vitro chaperone activity. J Biol Chem 2001; 286:31821–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheufler C, Brinker A, Bourenkov G, Pegoraro S, Ulrich Hartl F, MoarefiIsmail M. Structure of TPR domain–peptide complexes: critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine clemens. Cell 2000; 101:199–210 [DOI] [PubMed] [Google Scholar]
- 51.Zhuravleva A, Gierasch LM. Substrate-binding domain conformational dynamics mediate Hsp70 allostery. Proc Natl Acad Sci U S A 112:E2865–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kityk R, Kopp J, Sinning I, Mayer MP. Structure and dynamics of the ATP-Bound open conformation of Hsp70 chaperones. Mol Cell 2012; 48:863–74 [DOI] [PubMed] [Google Scholar]
- 53.Stevens SY, Cai S, Pellecchia M, Zuiderweg ERP. The solution structure of the bacterial HSP70 chaperone protein domain DnaK(393-507) in complex with the peptide NRLLLTG. Protein Sci 2009; 12:2588–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarty JS, Buchberger A, Reinstein J, Bukau B. The role of ATP in the functional cycle of the DnaK chaperone system. J Mol Biol 1995; 249:126–37 [DOI] [PubMed] [Google Scholar]
- 55.Karzai AW, McMacken RA. Bipartite signaling mechanism involved in DnaJ-mediated activation of the Escherichia Coli DnaK protein. J Biol Chem 1996; 271:11236–46 [DOI] [PubMed] [Google Scholar]
- 56.Slepenkov SV., Witt SN. The unfolding story of the Escherichia Coli Hsp70 DnaK: is DnaK a holdase or an unfoldase? Mol Microbiol 2002; 45:1197–206 [DOI] [PubMed] [Google Scholar]
- 57.Kityk R, Vogel M, Schlecht R, Bukau B, Mayer MP. Pathways of allosteric regulation in Hsp70 chaperones. Nat Commun 2015; 6:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dores-Silva PR, Barbosa LRS, Ramos CHI, Borges JC. Human mitochondrial Hsp70 (mortalin): shedding light on ATPase activity, interaction with adenosine nucleotides, solution structure and domain organization. PLoS One 2015; 10:e0117170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.da Silva KP, Borges JC. The molecular chaperone Hsp70 family members function by a bidirectional heterotrophic allosteric mechanism. Protein Pept Lett 2011; 18:132–42 [DOI] [PubMed] [Google Scholar]
- 60.Zhuravleva A, Clerico EM, Gierasch LM. An interdomain energetic tug-of-War creates the allosterically active state in Hsp70 molecular chaperones. Cell 2012; 151:1296–307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell 2007; 26:27–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Packschies L, Theyssen H, Buchberger A, Bukau B, Goody RS, Reinstein J. GrpE accelerates nucleotide exchange of the molecular chaperone DnaK with an associative displacement mechanism. Biochemistry 1997; 36:3417–22 [DOI] [PubMed] [Google Scholar]
- 63.Ellis RJ, Hemmingsen SM. Molecular chaperones: proteins essential for the biogenesis of some macromolecular structures. Trends Biochem Sci 1989; 14:339–42 [DOI] [PubMed] [Google Scholar]
- 64.Balchin D, Hayer-Hartl M, Hartl FU. In vivo aspects of protein folding and quality control. Science 2016; 353:aac4354. [DOI] [PubMed] [Google Scholar]
- 65.Dobson CM, Karplus M. The fundamentals of protein folding: bringing together theory and experiment. Curr Opin Struct Biol 1999; 9:92–101 [DOI] [PubMed] [Google Scholar]
- 66.Fan C-Y, Lee S, Cyr DM. Mechanisms for regulation of Hsp70 function by Hsp40. Cell Stress Chaper 2003; 8:309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 2010; 11:579–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nillegoda NB, Stank A, Malinverni D, Alberts N, Szlachcic A, Barducci A, De Los Rios P, Wade RC, Bukau B. Evolution of an intricate J-protein network driving protein disaggregation in eukaryotes. Elife 2017; 6:3417–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pierpaoli EV, Sandmeier E, Schönfeld HJ, Christen P. Control of the DnaK chaperone cycle by substoichiometric concentrations of the co-chaperones DnaJ and GrpE. J Biol Chem 1998; 273:6643–9 [DOI] [PubMed] [Google Scholar]
- 70.Han W, Christen P. Mechanism of the targeting action of DnaJ in the DnaK molecular chaperone system. J Biol Chem 2003; 278:19038–43 [DOI] [PubMed] [Google Scholar]
- 71.Kityk R, Kopp J, Mayer MP. Molecular mechanism of J-domain-triggered ATP hydrolysis by Hsp70 chaperones. Mol Cell 2018; 69:227–37 [DOI] [PubMed] [Google Scholar]
- 72.Alderson TR, Kim JH, Cai K, Frederick RO, Tonelli M, Markley JL. The specialized Hsp70 (HscA) interdomain linker binds to its nucleotide-binding domain and stimulates ATP hydrolysis in both cis and trans configurations. Biochemistry 2014; 53:7148–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hall D. On the nature of the optimal form of the Holdase-Type chaperone stress response. FEBS Lett 2020; 594:43–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Genest O, Wickner S, Doyle SM. Hsp90 and Hsp70 chaperones: collaborators in protein remodeling. J Biol Chem 2019; 294:2109–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Alvira S, Cuéllar J, Röhl A, Yamamoto S, Itoh H, Alfonso C, Rivas G, Buchner J, Valpuesta JM. Structural characterization of the substrate transfer mechanism in Hsp70/Hsp90 folding machinery mediated by hop. Nat Commun 2014; 5:5484. [DOI] [PubMed] [Google Scholar]
- 76.Mayer MP. Hsp70 chaperone dynamics and molecular mechanism. Trends Biochem Sci 2013; 38:507–14 [DOI] [PubMed] [Google Scholar]
- 77.Velasco L, Dublang L, Moro F, Muga A. The complex phosphorylation patterns that regulate the activity of Hsp70 and its cochaperones. Int J Mol Sci 2019; 20:4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kundrat L, Regan L. Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry 2010; 49:7428–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stankiewicz M, Nikolay R, Rybin V, Mayer MP. CHIP participates in protein triage decisions by preferentially ubiquitinating Hsp70-bound substrates. FEBS J 2010; 277:3353–67 [DOI] [PubMed] [Google Scholar]
- 80.Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin L-Y, Patterson C. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol 1999; 19:4535–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quintana-Gallardo L, Martín-Benito J, Marcilla M, Espadas G, Sabidó E, Valpuesta JM. The cochaperone CHIP marks Hsp70- and Hsp90-bound substrates for degradation through a very flexible mechanism. Sci. Rep 2019; 9:1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carrello A, Ingley E, Minchin RF, Tsai S, Ratajczak T. The common tetratricopeptide repeat acceptor site for steroid receptor-associated immunophilins and hop is located in the dimerization domain of Hsp90. J Biol Chem 1999; 274:2682–9 [DOI] [PubMed] [Google Scholar]
- 83.Assimon V, Southworth D, Gestwicki J. Specific binding of tetratricopeptide repeat (TPR) proteins to heat shock protein 70 (Hsp70) and heat shock protein 90 (Hsp90) is regulated by affinity and phosphorylation. Biochemistry 2015; 54:7120–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schmid AB, Lagleder S, Gräwert MA, Röhl A, Hagn F, Wandinger SK, Cox MB, Demmer O, Richter K, Groll M. The architecture of functional modules in the Hsp90 co-chaperone Sti1/hop. EMBO J 2012; 31:1506–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Brinker A, Scheufler C, Von Der Mülbe F, Fleckenstein B, Herrmann C, Jung G, Moarefi I, Ulrich Hartl F. Ligand discrimination by TPR domains. Relevance and selectivity of EEVD-recognition in Hsp70·hop·Hsp90 complexes. J Biol Chem 2002; 277:19265–75 [DOI] [PubMed] [Google Scholar]
- 86.Smith MC, Scaglione KM, Assimon VA, Patury S, Thompson AD, Dickey CA, Southworth DR, Paulson HL, Gestwicki JE, Zuiderweg ERP. The E3 ubiquitin ligase CHIP and the molecular chaperone Hsc70 form a dynamic, tethered complex. Biochemistry 2013; 52:5354–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang H, Amick J, Chakravarti R, Santarriaga S, Schlanger S, McGlone C, Dare M, Nix JC, Scaglione KM, Stuehr DJ. A bipartite interaction between Hsp70 and CHIP regulates ubiquitination of chaperoned client proteins. Structure 2015; 23:472–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ranek MJ, Oeing C, Sanchez-Hodge R, Kokkonen-Simon KM, Dillard D, Aslam MI, Rainer PP, Mishra S, Dunkerly-Eyring B, Holewinski RJ. CHIP phosphorylation by protein kinase G enhances protein quality control and attenuates cardiac ischemic injury. Nat Commun 2020; 11:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sharma D, Masison DC. Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (hsp)-70 chaperones Ssa1p and Ssa2p. Proc Natl Acad Sci U S A 2011; 108:13665–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Iosefson O, Sharon S, Goloubinoff P, Azem A. Reactivation of protein aggregates by mortalin and Tid1 – the human mitochondrial Hsp70 chaperone system. Cell Stress Chaperones 2012; 17:57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Simons JF, Ferro-Novick S, Rose MD, Helenius A. BiP/Kar2p serves as a molecular chaperone during carboxypeptidase Y folding in yeast. J Cell Biol 1995; 130:41–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang J, Lee J, Liem D, Ping P. HSPA5 gene encoding Hsp70 chaperone BiP in the endoplasmic reticulum. Gene 2017; 618:14–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Shorter J. The mammalian disaggregase machinery: Hsp110 synergizes with Hsp70 and Hsp40 to catalyze protein disaggregation and reactivation in a cell-free system. PLoS One 2011; 6:e26319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rauch JN, Gestwicki JE. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J Biol Chem 2014; 289:1402–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dong M, Bridges JP, Apsley K, Xu Y, Weaver TE. ERdj4 and ERdj5 are required for endoplasmic reticulum-associated protein degradation of misfolded surfactant protein C. MBoC 2008; 19:2620–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shomura Y, Dragovic Z, Chang HC, Tzvetkov N, Young JC, Brodsky JL, Guerriero V, Hartl FU, Bracher A. Regulation of Hsp70 function by HspBP1: structural analysis reveals an alternate mechanism for Hsp70 nucleotide exchange. Mol Cell 2005; 17:367–79 [DOI] [PubMed] [Google Scholar]
- 97.Rosam M, Krader D, Nickels C, Hochmair J, Back KC, Agam G, Barth A, Zeymer C, Hendrix J, Schneider M. Bap (Sil1) regulates the molecular chaperone BiP by coupling release of nucleotide and substrate. Nat Struct Mol Biol 2018; 25:90–100 [DOI] [PubMed] [Google Scholar]
- 98.Gowda NKC, Kaimal JM, Kityk R, Daniel C, Liebau J, Öhman M, Mayer MP, Andréasson C. Nucleotide exchange factors Fes1 and HspBP1 mimic substrate to release misfolded proteins from Hsp70. Nat Struct Mol Biol 2018; 25:3–89 [DOI] [PubMed] [Google Scholar]
- 99.Gowda NKC, Kaimal JM, Masser AE, Kang W, Friedländer MR, Andréasson C. Cytosolic splice isoform of Hsp70 nucleotide exchange factor Fes1 is required for the degradation of misfolded proteins in yeast. Mol Biol Cell 2016; 27:1210–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kabani M, Beckerich J-M, Gaillardin C. Sls1p stimulates Sec63p-mediated activation of Kar2p in a conformation-dependent manner in the yeast endoplasmic reticulum. Mol Cell Biol 2000; 20:6923–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chung KT, Shen Y, Hendershot LM. BAP, a mammalian BiP-associated protein, is a nucleotide exchange factor that regulates the ATPase activity of BiP. J Biol Chem 2002; 277:47557–63 [DOI] [PubMed] [Google Scholar]
- 102.Bhattacharya K, Weidenauer L, Luengo TM, Pieters EC, Echeverría PC, Bernasconi L, Wider D, Sadian Y, Koopman MB, Villemin M. The Hsp70-Hsp90 co-chaperone hop/Stip1 shifts the proteostatic balance from folding towards degradation. Nat Commun 2020; 11:5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kumar N, Gaur D, Masison DC, Sharma D. The BAG homology domain of Snl1 cures yeast prion [URE3] through regulation of Hsp70 chaperones. G3 2014; 4:461–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takayama S, Reed JC. Molecular chaperone targeting and regulation by BAG family proteins. Nat Cell Biol 2001; 3:E237–41 [DOI] [PubMed] [Google Scholar]
- 105.Kabbage M, Dickman MB. The BAG proteins: a ubiquitous family of chaperone regulators. Cell Mol Life Sci 2008; 65:1390–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zuo D, Subjeck J, Wang XY. Unfolding the role of large heat shock proteins: new insights and therapeutic implications. Front Immunol 2016; 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Easton DP, Kaneko Y, Subjeck JR. The Hsp110 and Grp170 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaper 2000; 5:276–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 2007; 131:106–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Raviol H, Sadlish H, Rodriguez F, Mayer MP, Bukau B. Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J 2006; 25:2510–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Raviol H, Bukau B, Mayer MP. Human and yeast Hsp110 chaperones exhibit functional differences. FEBS Lett 2006; 580:168–74 [DOI] [PubMed] [Google Scholar]
- 111.Li Z, Hartl FU, Bracher A. Structure and function of hip, an attenuator of the Hsp70 chaperone cycle. Nat Struct Mol Biol 2013; 20:929–35 [DOI] [PubMed] [Google Scholar]
- 112.Gebauer M, Zeiner M., Gehring U. Proteins interacting with the molecular chaperone Hsp70/Hsc70: physical associations and effects on refolding activity. FEBS Lett 1997; 417:109–13 [DOI] [PubMed] [Google Scholar]
- 113.Ferreira JV, Soares AR, Ramalho JS, Ribeiro-Rodrigues T, Máximo C, Zuzarte M, Girão H, Pereira P. Exosomes and STUB1/CHIP cooperate to maintain intracellular proteostasis. PLoS One 2019; 14:e0223790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Harrison C. GrpE, a nucleotide exchange factor for DnaK. Cell Stress Chaper 2003; 8:218–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zapun A, Jakob CA, Thomas DY, Bergeron JJM. Protein folding in a specialized compartment: the endoplasmic reticulum. Structure 1999; 7:R173–82 [DOI] [PubMed] [Google Scholar]
- 116.Sondermann H, Scheufler C, Schneider C, Höhfeld J, Hartl FU, Moarefi I. Structure of a bag/Hsc70 complex: convergent functional evolution of Hsp70 nucleotide exchange factors. Science 2001; 291:1553–7 [DOI] [PubMed] [Google Scholar]
- 117.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation – crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol Cell 2005; 20:525–38 [DOI] [PubMed] [Google Scholar]
- 118.Nakamura A, Takumi K, Miki K. Crystal structure of a thermophilic GrpE protein: insight into thermosensing function for the DnaK chaperone system. J Mol Biol 2010; 396:1000–11 [DOI] [PubMed] [Google Scholar]
- 119.Harrison CJ, Hayer-Hartl M, Di Liberto M, Hartl FU, Kuriyan J. Crystal structure of the nucleotide exchange factor GrpE bound to the ATPase domain of the molecular chaperone DnaK. Science 1997; 276:431–5 [DOI] [PubMed] [Google Scholar]
- 120.Pellecchia M, Szyperski T, Wall D, Georgopoulos C, Wüthrich K. NMR structure of the J-domain and the gly/Phe-rich region of the Escherichia coli DnaJ chaperone. J Mol Biol 1996; 260:236–50 [DOI] [PubMed] [Google Scholar]
- 121.Polier S, Dragovic Z, Hartl FU, Bracher A. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 2008; 133:1068–79 [DOI] [PubMed] [Google Scholar]
- 122.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaper 1998; 3:28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ohtsuka K, Hata M. Molecular chaperone function of mammalian Hsp70 and Hsp40 – a review. Int J Hyperthermia 2000; 16:231–45 [DOI] [PubMed] [Google Scholar]
- 124.Xu Z, Page RC, Gomes MM, Kohli E, Nix JC, Herr AB, Patterson C, Misra S. Structural basis of nucleotide exchange and client binding by the Hsp70 cochaperone Bag2. Nat Struct Mol Biol 2008; 15:1309–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Melero R, Moro F, Pérez-Calvo MÁ, Perales-Calvo J, Quintana-Gallardo L, Llorca O, Muga A, Valpuesta JM. Modulation of the chaperone DnaK allosterism by the nucleotide exchange factor GrpE. J Biol Chem 2015; 290:10083–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rauch JN, Zuiderweg ERP, Gestwicki JE. Non-Canonical interactions between heat shock cognate protein 70 (Hsc70) and Bcl2-associated anthanogene (BAG) Co-Chaperones are important for client release. J Biol Chem 2016; 29:19848–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lüders J, Demand J, Höhfeld J. The Ubiquitin-Related BAG-1 provides a link between the molecular chaperones Hsc70/Hsp70 and the proteasome. J Biol Chem 2000; 275:4613–7 [DOI] [PubMed] [Google Scholar]
- 128.Lin EHB, Von Korff M, Peterson D, Ludman EJ, Ciechanowski P, Katon W. Structural basis of nucleotide exchange and client binding by the novel Hsp70-Cochaperone Bag2. Am J Manag Care 2014; 20:887–9325495109 [Google Scholar]
- 129.Stürner E, Behl C. The role of the multifunctional Bag3 protein in cellular protein quality control and in disease. Front Mol Neurosci 2017; 10:177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kuwabara N, Minami R, Yokota N, Matsumoto H, Senda T, Kawahara H, Kato R. Structure of a BAG6 (bcl-2-associated athanogene 6)-Ubl4a (ubiquitin-like protein 4a) complex reveals a novel binding interface that functions in tail-anchored protein biogenesis. J Biol Chem 2015; 290:9387–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Takayama S, Bimston DN, Matsuzawa SI, Freeman BC, Aime-Sempe C, Xie Z, Morimoto RI, Reed JC. BAG-1 modulates the chaperone activity of Hsp70/Hsc70. EMBO J 1997; 16:4887–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hantouche C, Williamson B, Valinsky WC, Solomon J, Shrier A, Young JC. Bag1 co-chaperone promotes TRC8 E3 ligase-dependent degradation of misfolded human ether a go-go-related gene (HERG) potassium channels. J Biol Chem 2017; 292:2287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Cabrera Y, Dublang L, Angel Fernández-Higuero J, Albesa-Jové D, Lucas M, Viguera AR, Guerin ME, Vilar JMG, Muga A, Moro F. Regulation of human Hsc70 ATPase and chaperone activities by Apg2: role of the acidic subdomain. J Mol Biol 2018; 431:444–61 [DOI] [PubMed] [Google Scholar]
- 134.Andréasson C, Fiaux J, Rampelt H, Druffel-Augustin S, Bukau B. Insights into the structural dynamics of the Hsp110-Hsp70 interaction reveal the mechanism for nucleotide exchange activity. Proc Natl Acad Sci U S A 2008; 105:16519–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J 2006; 25:2519–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Irmer H, Höhfeld J. Characterization of functional domains of the eukaryotic co-chaperone hip. J Biol Chem 1997; 272:2230–5 [DOI] [PubMed] [Google Scholar]
- 137.Höfeld J, Minami Y, Hartl FU. Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell 1995; 83:589–98 [DOI] [PubMed] [Google Scholar]
- 138.Shi Z-Z, Zhang J-W, Zheng S. What we know about ST13, a Co-Factor of heat shock protein, or a tumor suppressor? J Zhejiang Univ Sci B 2007; 8:170–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Höhfeld J, Jentsch S. GrpE-like regulation of the Hsc70 chaperone by the anti-apoptotic protein BAG-1. EMBO J 1997; 16:6209–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Buczynski G, Slepenkov SV, Sehorn MG, Witt SN. Characterization of a lidless form of the molecular chaperone DnaK: deletion of the lid increases peptide on- and off-Rate constants. J Biol Chem 2001; 276:27231–6 [DOI] [PubMed] [Google Scholar]
- 141.Clerico EM, Tilitsky JM, Meng W, Gierasch LM. How Hsp70 molecular machines interact with their substrates to mediate diverse physiological functions. J Mol Biol 2015; 427:1575–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Rüdiger S, Buchberger A, Bukau B. Interaction of Hsp70 chaperones with substrates. Nat Struct Biol 1997; 4:342–9 [DOI] [PubMed] [Google Scholar]
- 143.Nunes JM, Mayer-Hartl M, Hartl FU, Müller DJ. Action of the Hsp70 chaperone system observed with single proteins. Nat Commun 2015; 6:6307. [DOI] [PubMed] [Google Scholar]
- 144.Rodriguez F, Arsène-Ploetze F, Rist W, Rüdiger S, Schneider-Mergener J, Mayer MP, Bukau B. Molecular basis for regulation of the heat shock transcription factor Σ32 by the DnaK and DnaJ chaperones. Mol Cell 2008; 32:347–58 [DOI] [PubMed] [Google Scholar]
- 145.Serlidaki D, van Waarde MAWH, Rohland L, Wentink AS, Dekker SL, Kamphuis MJ, Boertien JM, Brunsting JF, Nillegoda NB, Bukau B. Functional diversity between HSP70 paralogs caused by variable interactions with specific co-chaperones. J Biol Chem 2020; 295:7301–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sekhar A, Santiago M, Lam HN, Lee JH, Cavagnero S. Transient interactions of a slow-folding protein with the Hsp70 chaperone machinery. Protein Sci 2012; 21:1042–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Palleros DR, Shi L, Reid KL, Fink L. Hsp70-protein complexes. J Biol Chem 1994; 268:13107–14 [PubMed] [Google Scholar]
- 148.Kellner R, Hofmann H, Barducci A, Wunderlich B, Nettels D, Schuler B. Single-Molecule spectroscopy reveals chaperone-mediated expansion of substrate protein. Proc Natl Acad Sci U S A 2014; 111:13355–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Chen Z, Kurt N, Rajagopalan S, Cavagnero S. Secondary structure mapping of DnaK-Bound protein fragments: chain helicity and local helix unwinding at the binding Site. Biochemistry 2006; 45:12325–33 [DOI] [PubMed] [Google Scholar]
- 150.Sekhar A, Lam HN, Cavagnero S. Protein folding rates and thermodynamic stability are key determinants for interaction with the Hsp70 chaperone system. Protein Sci 2012; 21:1489–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Rosenzweig R, Sekhar A, Nagesh J, Kay LE. Promiscuous binding by Hsp70 results in conformational heterogeneity and fuzzy chaperone-substrate ensembles. Elife 2017; 6:1–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fourie AM, Sambrook JF, Gething MJH. Common and divergent peptide binding specificities of Hsp70 molecular chaperones. J Biol Chem 1994; 269:30470–8 [PubMed] [Google Scholar]
- 153.Rüdiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J 1997; 16:1501–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Swain JF, Schulz EG, Gierasch LM. Direct comparison of a stable isolated Hsp70 substrate-binding domain in the empty and substrate-bound states. J Biol Chem 2006; 281:1605–11 [DOI] [PubMed] [Google Scholar]
- 155.Jiang J, Prasad K, Lafer EM, Sousa R. Structural basis of interdomain communication in the Hsc70 chaperone. Mol Cell 2005; 20:513–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Attisano L, Tuen Lee-Hoeflich S. The smads. Genome Biol 2001; 2:reviews3010.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Li RF, Shang Y, Liu D, Ren ZS, Chang Z, Sui SF. Differential ubiquitination of Smad1 mediated by CHIP: implications in the regulation of the bone morphogenetic protein signaling pathway. J Mol Biol 2007; 374:777–90 [DOI] [PubMed] [Google Scholar]
- 158.Imamura T, Oshima Y, Hikita A. Regulation of TGF-β family signalling by ubiquitination and deubiquitination. J Biochem 2013; 154:481–9 [DOI] [PubMed] [Google Scholar]
- 159.Wang L, Liu YT, Hao R, Chen L, Chang Z, Wang HR, Wang ZX, Wu JW. Molecular mechanism of the negative regulation of Smad1/5 protein by carboxyl terminus of Hsc70-Interacting protein (CHIP). J Biol Chem 2011; 286:15883–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Liu J, Bao J, Hao J, Peng Y, Hong F. HSP70 inhibits high Glucose-Induced Smad3 activation and attenuates epithelial-to-Mesenchymal transition of peritoneal mesothelial cells. Mol Med Rep 2014; 10:1089–95 [DOI] [PubMed] [Google Scholar]
- 161.Seo J, Han SY, Seong D, Han HJ, Song J. Multifaceted C-Terminus of HSP70-Interacting protein regulates tumorigenesis via protein quality control. Arch Pharm Res 2019; 42:63–75 [DOI] [PubMed] [Google Scholar]
- 162.Shang Y, Xu X, Duan X, Guo J, Wang Y, Ren F, He D, Chang Z. Hsp70 and Hsp90 oppositely regulate TGF-β signaling through CHIP/Stub1. Biochem Biophys Res Commun 2014; 446:387–92 [DOI] [PubMed] [Google Scholar]
- 163.Xin H, Xu X, Li L, Ning H, Rong Y, Shang Y, Wang Y, Fu XY, Chang Z. CHIP controls the sensitivity of transforming growth factor-β signaling by modulating the basal level of Smad3 through Ubiquitin-Mediated degradation. J Biol Chem 2005; 280:20842–50 [DOI] [PubMed] [Google Scholar]
- 164.Antognelli C, Gambelunghe A, Muzi G, Talesa VN. Glyoxalase i drives epithelial-to-Mesenchymal transition via Argpyrimidine-Modified Hsp70, MiR-21 and SMAD signalling in human bronchial cells BEAS-2B chronically exposed to crystalline silica Min-U-Sil 5: transformation into a neoplastic-like phenotype. Free Radic Biol Med 2016; 92:110–25 [DOI] [PubMed] [Google Scholar]
- 165.Hassan AQ, Kirby CA, Zhou W, Schuhmann T, Kityk R, Kipp DR, Baird J, Chen J, Chen Y, Chung F. The novolactone natural product disrupts the allosteric regulation of Hsp70. Chem Biol 2015; 22:87–97 [DOI] [PubMed] [Google Scholar]
- 166.Edkins AL, Price JT, Graham Pockley A, Blatch GL. Heat shock proteins as modulators and therapeutic targets of chronic disease: an integrated perspective. Philos Trans R Soc B Biol Sci 2018; 373:DOI: 10.1098/rstb.2016.0521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Geraci F, Turturici G, Sconzo G. Hsp70 and its molecular role in nervous system diseases. Biochem Res Int 2011; 2011:DOI: 10.1155/2011/618127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Patury S, Miyata Y, Gestwicki J. Pharmacological targeting of the Hsp70 chaperone. Curr Top Med Chem 2009; 9:1337–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kalia SK, Lee S, Smith PD, Liu L, Crocker SJ, Thorarinsdottir TE, Glover JR, Fon EA, Park DS, Lozano AM. BAG5 inhibits parkin and enhances dopaminergic neuron degeneration. Neuron 2004; 44:931–45 [DOI] [PubMed] [Google Scholar]
- 170.Behl C. Breaking BAG: the co-Chaperone BAG3 in health and disease. Trends Pharmacol Sci 2016; 37:672–88 [DOI] [PubMed] [Google Scholar]
- 171.Hasegawa T, Yoshida S, Sugeno N, Kobayashi J, Aoki M. DnaJ/Hsp40 family and Parkinson’s disease. Front Neurosci 2018; 11:743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Howarth JL, Kelly S, Keasey MP, Glover CPJ, Lee YB, Mitrophanous K, Chapple JP, Gallo JM, Cheetham ME, Uney JB. Hsp40 molecules that target to the Ubiquitin-Proteasome system decrease inclusion formation in models of polyglutamine disease. Mol Ther 2007; 15:1100–5 [DOI] [PubMed] [Google Scholar]
- 173.Lackie RE, Maciejewski A, Ostapchenko VG, Marques-Lopes J, Choy WY, Duennwald ML, Prado VF, Prado MAM. The Hsp70/Hsp90 chaperone machinery in neurodegenerative diseases. Front Neurosci 2017; 11:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zhao T, Hong Y, Yin P, Li S, Li XJ. Differential HspBP1 expression accounts for the greater vulnerability of neurons than astrocytes to misfolded proteins. Proc Natl Acad Sci U S A 2017; 114:E7803–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Taguchi YV, Gorenberg EL, Nagy M, Thrasher D, Fenton WA, Volpicelli-Daley L, Horwich AL, Chandra SS. Hsp110 mitigates α-Synuclein pathology in vivo. Proc Natl Acad Sci U S A 2019; 116:24310–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Dorard C, De Thonel A, Collura A, Marisa L, Svrcek M, Lagrange A, Jego G, Wanherdrick K, Joly AL, Buhard O, Gobbo J, Penard-Lacronique V, Zouali H, Tubacher E, Kirzin S, Selves J, Milano G, Etienne-Grimaldi MC, Bengrine-Lefevre L, Louvet C, Tournigand C, Lefvre JH, Parc Y, Tiret E, Fléjou JF, Gaub MP, Garrido C, Duval A. Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat Med 2011; 17:1283–9 [DOI] [PubMed] [Google Scholar]
- 177.Zhang S, Hu ZW, Mao CY, Shi CH, Xu YM. CHIP as a therapeutic target for neurological diseases. Cell Death Dis 2020; 11:1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hayer SN, Deconinck T, Bender B, Smets K, Züchner S, Reich S, Schöls L, Schüle R, De Jonghe P, Baets J, Synofzik M. STUB1/CHIP mutations cause gordon holmes syndrome as part of a widespread multisystemic neurodegeneration: evidence from four novel mutations. Orphanet J Rare Dis 2017; 12:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Synofzik M, Schüle R, Schulze M, Gburek-Augustat J, Schweizer R, Schirmacher A, Krögeloh-Mann I, Gonzalez M, Young P, Züchner S, Schöls L, Bauer P. Phenotype and frequency of STUB1 mutations: next-generation screenings in caucasian ataxia and spastic paraplegia cohorts. Orphanet J Rare Dis 2014; 9:57. [DOI] [PMC free article] [PubMed] [Google Scholar]