Abstract
Microphysiological systems (MPS) are promising in vitro tools which could substantially improve the drug development process, particularly for underserved patient populations such as those with rare diseases, neural disorders, and diseases impacting pediatric populations. Currently, one of the major goals of the National Institutes of Health MPS program, led by the National Center for Advancing Translational Sciences (NCATS), is to demonstrate the utility of this emerging technology and help support the path to community adoption. However, community adoption of MPS technology has been hindered by a variety of factors including biological and technological challenges in device creation, issues with validation and standardization of MPS technology, and potential complications related to commercialization. In this brief Minireview, we offer an NCATS perspective on what current barriers exist to MPS adoption and provide an outlook on the future path to adoption of these in vitro tools.
Keywords: Microphysiological systems, microfluidics, bioengineering, induced pluripotent stem cells, National Institutes of Health
Impact statement
This work is important to the field as it outlines the progress and current hurdles to widespread community adoption of MPS technology. This is useful information to be aware of for members of the MPS field, as well as end-users such as those in academic, industry and regulatory institutions, stakeholders, and future partners.
Introduction
The process of bringing a new drug or therapeutic to market is extremely complex and challenging, with the average R&D cost for each new medicine estimated at nearly $3 billion dollars and a timeframe of over 10 years from initial lead compound discovery to successful approval by the Food and Drug Administration (FDA).1–3 The extensive length of time and low efficiency of the drug development process is largely due to toxicity of compounds in early preclinical stages, and lack of efficacy within later clinical stages.4,5 The attrition rate of promising therapeutics is even higher for rare diseases,6,7 neural-based conditions, and diseases affecting pediatric populations. 7
Preclinical testing of new candidate therapeutics is conventionally performed in 2D cell culture systems, and in in vivo animal models consisting of at least a rodent species (mice and rats) and non-rodent species (non-human primates, pigs, dogs). Simple 2D systems comprising cells grown in monolayers on flat surfaces are used in mainstream drug development due to their ease of manipulation, rapid scalability, and high-throughput capabilities. 8 However, 2D cell culture systems often lack the complexity, structure, proper cellular composition, inter- and intracellular interactions, and microenvironmental niche of native human tissues, thereby inhibiting cells from reaching maturity and proper response. 9 In fact, 2D cell culture models rely on growing cells on hard, flat plastic substrates, which may not accurately mimic human physiologic response. 10 Animal models such as mice have advantages over other model organisms, including their short gestation period, availability of a large number of offspring, ease of genetic manipulation, and amenability for use in medium to high-throughput screening. 11 However, despite these many advantages, animal models may not replicate critical off-target effects and toxicities that could occur in humans due to specific species differences in metabolism, pathophysiology, immune response, microbiota, and drug response. 12 These current preclinical models can be poorly predictive of human responses, providing evidence that alternative tools and approaches are needed to speed up, potentially reduce cost, and increase the efficiency of developing new drugs that are safe and effective. 12 Microphysiological systems (MPS) or “tissue-chips”/“organs on chips” are one such new alternative method which could improve and expedite the drug development process by resolving the discrepancies in drug safety and efficacy observed between conventional models and human/clinical responses. 8 Perhaps in the future MPS will be considered a “third-species” for testing of candidate therapeutics. 13
As defined by the FDA’s Alternative Methods Working Group, MPS are in vitro platforms composed of human or animal cells, tissue/organ-derived explants, and/or self-assembling organoids contained in microenvironmental niches that facilitate proper biochemical, electrical, and mechanical organ/tissue level responses (https://www.fda.gov/science-research/about-science-research-fda/advancing-alternative-methods-fda). These 3D biomimetic devices, which combine advancements made in materials science, bioengineering, microscale technologies and human stem cell biology, aim to model specific biological properties that define organ and tissue function. 14 MPS platforms can provide controlled tissue perfusion via pressurized fluid reservoirs or microfluidic output devices such as pumps and valves to recreate relevant biomechanical forces, or other mechanical actuation in a controlled and explicit manner.15,16
Precise design of MPS platforms allows researchers to exert unparalleled control over tissue composition and architecture by providing an array of cellular and extracellular cues including molecular, structural, and physical cues found within the human in vivo organ system.17,18 Researchers are also able to precisely control nutrient diffusion and tissue oxygenation profiles, which allow significant prolongation of MPS viability from several weeks to several months. 19 The ultimate goal of MPS systems is not to build an entire living human organ, but rather to create a minimally functional organ/tissue unit that is able to accurately recapitulate specific relevant aspects of human physiology and response. 20 For example, MPS are able to model multicellular architectures and tissue interfaces, physicochemical microenvironments, vascular perfusion, and innervation of human organs, as well as maintain proper establishment of growth hormone gradients, cytokines, and other signaling pathways highly relevant to modeling any human organ system. 12 Through inclusion of biosensor technologies (sensors to measure oxygen, glucose, and other small molecules/chemicals) 21 and automated high-resolution real-time imaging and monitoring approaches,22,23 genetic, metabolic, and biochemical activities of living cells are able to be studied in the context of a functional human organ. 7 In addition, MPS are also responsive to the 3R principles of animal use in research – refinement, reduction, and replacement – and offer a way for significant reduction on the use of animals or animal models for preclinical studies.8,24
It is now becoming increasingly clear that MPS can serve as in vitro tools within many areas of drug development,25,26 including serving as physiologically-relevant platforms for early lead compound target validation and therapeutic safety and toxicity assessment during the preclinical phase.12,27–29 In addition, MPS can be used for modeling of both common and rare diseases, 7 with successful modeling of a number of rare disorders, such as Progeria,30,31 Barth syndrome, 32 and hereditary hemorrhagic telangiectasia. 33 MPS have also been employed to better understand drug response pathophysiology, stratification of patient subpopulations, 7 and patient-specific chips for personalized medicine. 34 In addition, MPS have found utility in assessment of environmental exposures and environmental toxicology,35–38 reproductive and developmental toxins, infectious diseases, 39 microbiome, 40 and countermeasure agents. 12 A useful prioritization of MPS applications can be found in Table 1 of the review by Watson, Hunziker, and Wikswo, 2017. 10 Currently, there are MPS of nearly every major human organ 20 including liver,16,41 vasculature,42,43 blood–brain barrier (BBB),44–46 skeletal 47–49 and cardiac muscle,50–57 kidney,58–61 female reproductive tissues (uterus, cervix, fallopian tubes), 62 testes, 63 brain,37,64 skin,65–67 gut, 40 bone, 68 and eye.69,70 Specific MPS organ systems have also been linked to form multi-organ systems to more accurately depict systemic responses to a variety of drugs and therapeutics.62,71–73
The US National Institutes of Health (NIH) MPS program, alongside a sister program at the US Defense Advanced Research Projects Agency (DARPA), began in 2012 and have helped to catalyze the development of MPS technology. 12 The programs focused on developing single organ human chips with viabilities of at least 28 days that could be interconnected or linked to form a more representative example of a “human-body-on-a-chip” and improve modeling of human inter-organ interaction. 12 Ultimately, this 5-year program provided proof of principle that tissue chips could accurately recapitulate human physiology and drug responses. 12 While DARPA funding ended in 2017, the NIH MPS program has continued and since evolved to look at various tissue chip technology end uses, including disease modeling and efficacy testing;74–79 modeling opioid use disorders; using tissue chips to improve clinical trial design and execution; and creating independent validation/testing centers 59,80–82 plus a publicly accessible database 83,84 for MPS data. In addition to the NIH and DARPA MPS programs, the US Environmental Protection Agency established the “Science to Achieve Results” (STAR) grant program, one goal of which is to further the development of organotypic culture models, including organs-on-chips (OoCs) for predictive toxicology.71,85,86
One of the major goals of the NIH MPS program now is to demonstrate the utility and disseminate the use of this emerging technology into a variety of communities including but not limited to academic organizations, industry, regulatory agencies such as the US FDA, and for profit entities such as contract research organizations. 12 However, adoption of this emerging technology is hampered by many confounding factors including biological challenges such as (1) cell sourcing and linkage of multiple platforms, (2) technical challenges such as platform fabrication and design, (3) issues surrounding validation and standardization of MPS technology; and (4) commercial considerations and potential complications. In this review we briefly address some of these pressing challenges, providing an outlook on the future of MPS technology adoption. However, we note that these challenges we address represent in no way an exhaustive list. For more information on pressing hurdles to MPS adoption, we recommend the following reviews: Watson et al., Low and Tagle, and Ewart et al.10,87,88
The need to define MPS context of use
Before delving into the diverse array of biological and technical hurdles hindering MPS adoption, it is important to discuss context of use (CoU) – or the specific use case where MPS can provide informative preclinical data. Currently, specific contexts of use for MPS platforms include toxicology, pharmacokinetics (ADME), pharmacodynamics, efficacy, and drug safety.61,89–91 CoU is extremely important, as no single MPS system is able to fully recapitulate a functional or integrated tissue/organ in its entirety.20,26 Currently, systems are designed to model important features of a tissue/organ, while mimicking specific morphological and functional phenotypes. Therefore, it is critical that end users of this technology select the appropriate MPS model as a tool to answer a specific set of questions germane to the disease being modeled or the intended use of the candidate therapeutic being tested in the system. 92
The International Consortium for Innovation and Quality in Pharmaceutical Development (also known as the IQ Consortium) has made significant progress in helping to define MPS CoU within the industrial sector. The IQ Consortium has established an IQ MPS Affiliate (https://www.iqmps.org/) to provide a venue for cross-pharma collaboration, data sharing, implementation, and qualification of MPS models. Recently, IQ MPS Affiliate scientists prepared a series of organotypic manuscripts for various key drug safety and disposition target tissues including lung, liver, kidney, cardiovascular, blood brain barrier/central nervous system, skin, and gastrointestinal systems.61,66,90,91,93–96 The goal of these organotypic manuscripts is to provide relevant information on MPS CoU and the key characterization data required to incorporate MPS into pharmaceutical safety screening. 91 For example, the initial CoU for MPS in applications of safety are likely to include testing compounds during the lead and candidate optimization stages; drug absorption, distribution, metabolism and excretion (ADME) applications; and identification of toxicity mechanisms in all clinical stages but particularly within Phase 1 clinical trials when adverse events in humans are first observed.88,91 In addition to the need for further MPS characterization, the IQ MPS Affiliate have identified a variety of other hurdles impeding MPS adoption in the pharmaceutical industry including the need for standards, bridging and comparing pre-clinical animal model data with tissue chip data, providing enough data to increase confidence in the technology, and increasing throughput for specific applications.25,91 For example, the level of throughput needed varies widely based on CoU, e.g. the quantity of actively investigated compounds and the degree of certainty required to inform data-driven decisions. 91
In addition to industry, regulatory bodies such as the FDA have also made significant progress in helping to guide CoU for MPS. For example, the FDA has signed collaborative agreements with various tissue chip companies to assess the utility of these systems in house. In 2017, the FDA signed a collaborative agreement with the MPS company Emulate, Inc., to study how their organ chips can be used as a toxicology testing platform for products affecting human health and safety (https://www.emulatebio.com/press/fda-collab-agreement-emulate). Recently, FDA researchers evaluated the effects of the known hepatotoxin, diglycolic acid (DGA), within Emulate’s Human Liver Organ-Chip in comparison to a 2D multi-well plate format. 97 They assessed the Liver-Chip’s capabilities, limitations, performance, and concordance with previous in vitro and in vivo studies, and found that the Liver-Chip (as well as the 2D 24-well plates) displayed similar toxicity upon DGA exposure, in concordance with in vivo studies. 97 Overall, the Liver-Chip platform had high specificity and sensitivity, good power, and low variability. 97 Studies such as this help define CoU for specific MPS devices, furthering regulatory acceptance of this technology. In addition, the FDA recently announced a new pilot program, “Innovative Science and Technology Approaches for New Drugs” (ISTAND), designed to expand types of Drug Development Tools (DDT) that are currently out of scope for current DDT qualification programs but that may prove beneficial for the drug development process. The use of tissue chips (MPS) to assess safety or efficacy questions are cited as an example of submissions that may be considered for ISTAND qualification as a novel DDT. In addition to pilot programs such as these, recommendations and a roadmap towards regulatory acceptance of MPS models from nearly 50 leading academic, regulatory, pharmaceutical and biotechnology experts was recently published by the t4 (the transatlantic think tank for toxicology) workshop. 98
Technical and biological challenges to MPS adoption
There are a variety of technical and biological challenges hindering MPS adoption. We briefly discuss a few major hurdles below.
Cell types used in microphysiological systems
To independently validate emerging tissue chip technology, the US NIH MPS program, led by the National Center for Advancing Translational Science (NCATS), established Tissue Chip Testing Centers 12 to independently onboard and validate chips developed under the NIH program (see section 2 below: Commercialization, validation and automation of MPS technology). A recent case study by one of these centers investigating a human kidney proximal tubule tissue chip revealed that overall reproducibility in data is greatly dependent on the cell source.59,80 The source, cell types, and microenvironment of cellular materials used within MPS devices are therefore of great importance. Various cell types are currently being used including primary cells, immortalized cell lines, and human pluripotent stem cells. 99 Primary cells, normally taken from discarded surgical tissues or biopsy specimens, have the advantages of being phenotypically and functionally mature. However, primary cells are difficult to obtain from the majority of human organs, finite in quantity, and at higher passages will decline in health and function, which can lead to variable data.100–103 Alternatively, the use of human induced pluripotent stem cells (hIPSCs) has specific advantages over the use of primary cells, including a potentially unlimited renewable cell source derived from blood or skin fibroblasts that are patient specific, and can be expanded and selectively differentiated into multiple cellular lineages.99,104,105 Use of a single iPSC line to generate a variety of tissue and organ systems within an MPS platform allows separation of genotype and phenotype effects, with genetic homogeneity being particularly advantageous for modeling drug ADME profiles in both individuals and patient groups.106,107 Additionally, the creation of genetically identical (isogenic) cell lines from iPSCs, using technologies such as CRISPR-Cas9 to introduce or remove disease-modifying mutations, allows modeling of monogenic disorders. This use of isogenic lines allows mechanistic studies of the disease process and allows target-specific development of new drugs. 92 While the use of iPSCs allows scientists to further understand the critical differences in patients’ genetic diversity, sex, age, and ethnicity, 77 there are disadvantages to using pluripotent stem cells. Not all iPSC lineages are able to be effectively derived from the same iPSC line (have varying differentiation potentials 108 ) and lineages commonly lose epigenetic markers during the derivation process. 109 In addition, pluripotent stem cells often fail to mature phenotypically or functionally. 110 In addition, the generation of iPSCs is very time consuming and expensive. Currently, our incomplete understanding of iPSC biology slows the development of robust and reliable iPSC differentiation and maturation protocols and presents a major hurdle to all sectors utilizing iPSC technology, including the MPS field. 111 Ultimately, each organ and tissue system is unique and will benefit from different approach methodologies, including the choice of cell type – i.e. primary, iPSC, etc.
MPS platform fabrication
MPS platforms or OoCs rely on material support for proper cellular organization and tissue attachment. Many commercially available MPS devices rely on replica molding processes using soft lithography for microfabricated design. 112 Elastomeric replicas are created using polydimethylsiloxane (PDMS) 113 for precise designs which permits spatiotemporal control over cell growth and maturation, rates of fluid flow, and compound/drug exposures. Soft lithography is popular as it is a fast, cheap, and easy method of prototyping. Unfortunately, soft lithography is an extremely manual process and is often difficult to automate. In addition to lithography, 3D-printing is also being used to fabricate microfluidic systems, as 3D-printing allows automated, assembly-free 3D fabrication at higher throughput.114,115 PDMS is one of the most common elastomeric organic materials used for microfabrication of MPS platforms due to its excellent biocompatibility, low stiffness, elasticity, and optical transparency making it amenable to real-time optical imaging. 116 Additionally, it is gas permeable, its surface chemistry can be modified, and it is sterilizable by autoclave. However, PDMS absorbs small hydrophobic molecules, meaning large amounts of drug can bind to the platform material and affect the concentration reaching the tissues, which must be taken into account when designing drug studies. 26 In addition, use of PDMS can lead to cross-contamination of chambers or adjacent channels leading to reduction of drug predictivity and reliability of the system. 26 Methods have been developed to improve PDMS qualities, such as fabrication of coatings that reduce permeability 117 and design considerations that account for or reduce absorption and unintended mixing of drugs and compounds. 118 Alternative materials for chip fabrication include silicon, glass, thermoplastics such as cyclic olefin copolymer and poly(methyl methacrylate), polyurethane, and tetrafluoroethylene-propylene elastomer.20,26,119,120 Often the material choice is a trade-off between the platform requirements and the affordability, availability, and/or ease of fabrication of the material in question. 26 Regardless of the choice of fabrication material(s), all MPS platforms require extensive testing and characterization of drug absorption and biocompatibility. 26 In addition, the biocompatibility of all platform materials should be assessed for potential biotoxicity. 121
Multi-organ MPS platform integration
Individual MPS “organs” can be integrated or linked to one another to allow assessment of disease or drugs in more human-relevant systems. Integration of linked organ systems provides new ways to research the pharmacokinetics/pharmacodynamics (PK/PD) profile of a drug. It also allows modeling of hormone, cytokine and immune responses to compound exposure, and invites analysis of human systemic physiological responses to drugs and their associated metabolites. MPS platform integration is particularly advantageous for studying diseases that involve multi-organ pathologies, such as specific types of cancers. The integration of MPS organ systems also allows researchers to study off-target drug effects, as well as unexpected metabolism of drugs by a non-target organ. Recently, researchers at Columbia developed a bone cancer tumor (Ewing Sarcoma) and heart muscle integrated system, which they subjected to the clinically used dosage of the novel anti-cancer drug, Linsitinib. 79 They measured the anti-tumor efficacy as well as the cardiotoxicity of the drug and compared these results with recent clinical trial results. Linsitinib treatment in the integrated platform matched the clinical trial results, with poor tumor response and mild cardiotoxicity, indicating that the integrated tissue platform was able to accurately predict both the direct and off-target effects of the drug. 79 Integrated model systems such as these could be used preclinically to allow better and earlier prediction of clinical outcomes.
The easiest way to integrate systems is through functional coupling, whereby one takes effluent from one MPS and transfers it to another MPS to simulate passage of drug/compound from one tissue/organ type to another. 71 Linkage of systems to create in vitro models of human body subsystems enables organ crosstalk and communication through secreted factors such as exosomes, soluble molecules, and immune cells.122,123 The main advantages to functional coupling are technical ease and flexibility to integrate individual specialized single-organ chambers created in different laboratories into functional multi-organ platforms that can be configured and re-configured for a variety of study designs. 20 However, this approach fails to mimic the physiological flow occurring between organ systems, and because of this, physical linkage of systems may be a better approach, depending on the intended CoU of the MPS. Physical linkage of individual systems, while more physiologically accurate and biologically meaningful, brings about major biological challenges such as organ scaling and the need for a universal/common medium. Organ scaling, or how best to represent the size and/or cell number of linked organ platforms in a physiologically relevant way, continues to be a hurdle. Some investigators have proposed that MPS scaling should be based on human organ sizes, i.e. respective mass. 124 However, others have discussed the need for functional scaling based on blood flow or metabolic rates, which may be more appropriate in determining correct ratios between individual organs with the organ volume determined from levels which support functionality.20,124,125 For example, an appropriate level of drug metabolism per fluidic pass in the liver could be created by combining multiple functional liver organ chambers. Allometric scaling is another strategy to quantify the relationship between differently sized organs; however, this scaling strategy fails to consider specific size differentials and complexities between MPS and in vivo tissues.124,126
One of the first simple integrated proto-MPS devices consisted of a three-chamber – lung-liver-“other” – microscale cell culture platform with a dissolved oxygen sensor for real-time measurements/readouts. 127 The device, which fits on a one-inch square silicon chip, contained compartments connected by recirculating tissue culture media (blood surrogate), with a flow rate that was able to approximate physiological liquid-to-cell ratios and hydrodynamic shear stress. Over the last decade, a variety of interconnected MPS have been developed such as the three-organ chip “first pass” model of the liver, gut, and kidney or bone marrow, which demonstrates quantitative prediction of human drug PK with two different drug compounds via both oral and IV administration routes. 128 In addition, a female reproductive system on a chip containing organ modules of the ovaries, fallopian tubes, uterus, cervix, and liver within a single MPS platform has also been engineered. 62 This “EVATAR” device could simulate a 28-day human female menstrual cycle and hormonal profile. 62 Recently, two publications from DARPA’s MPS program were published, including a human physiome-on-a-chip 73 and an automated “Interrogator” device able to culture up to 10 human organ chips. 72 The physiome-on-a-chip, containing interconnected liver (immune), gut (immune), brain, pancreas, kidney, skin, heart, endometrium, and skeletal muscle 73 is able to generate complex molecular distribution profiles and has been found suitable for PD/PK and PBPK modeling using quantitative systems pharmacology (QSP) approaches. 12 The “Interrogator” device maintains fluidic coupling for 3 weeks between eight vascularized, two-channel cultured organ chips (intestine, liver, kidney, heart, lung, BBB, brain, and skin) (Figure 1). 72
Figure 1.
Example of a linked multi-organ system. (a) The ‘Interrogator’ device maintains fluidic coupling for 3 weeks between eight vascularized, two-channel cultured organ chips (intestine, liver, kidney, heart, lung, blood–brain barrier, brain, and skin). Scale bar, 5 mm. (b) These individual organ constructs can be linked to form a ‘human body-on-a-chip’. Individual organ chips were connected through vascular endothelial channels, allowing a variety of linkage possibilities and multiple sampling points. (A color version of this figure is available in the online journal.)
Source: Adapted with permission from Novak et al. 72
Although there has clearly been major progress in linkage of MPS devices, there are still many challenges on the road ahead. These include more basic technical challenges such as inhibiting air bubble formation which can impede fluid flow, maintaining sterility when physically linking separately cultured platforms, preventing leakage during the integration process, and controlling oxygenation levels between different organ systems. 8 Fluidic integration of multiple tissue chips also creates the need for a common universal medium capable of keeping the heterogeneous cell types within the platform healthy and functional. This medium, which would function either as a blood mimetic or as interstitial fluid, would contain all growth factors, chemokines, and nutrients required to support all tissue types in the system. 129 Currently, no medium has been developed that can function as a universal medium, although it is possible to circumvent this issue through inclusion of organ-specific endothelial barriers between tissues, with a common basic circulating medium that allows some tissue crosstalk as well as inclusion of immune cells and other circulating factors.20,26 Incorporation of immune cells into MPS platforms, particularly to better understand innate and adaptive immunity, will be a key step in wider adoption of MPS models.130,131
Commercialization, validation and automation of MPS technology
MPS are becoming increasingly commercially available, with many organ-on-a-chip companies advancing products to market. 132 In fact, a variety of MPS start-up companies have been established, each bringing unique device constructs and ways to perform tissue assembly. 132 NIH has supported a number of these MPS small business entities through Small Business Innovation Research (SBIR) and Small Business Technology Transfer (STTR) funding programs. These programs allow US-owned and operated small businesses to receive early-stage capital for innovative technology commercialization in the US. Phase II SBIR funding from NCATS was awarded to the MPS company Hesperos, Inc, to commercialize their “body-on-a-chip” technology. Hesperos offers pumpless and serum-free multi-organ systems with built-in mechanical and chemical sensors for bioanalysis and systemic toxicology. 132 Hesperos has constructed MPS representing many major human organ systems including cardiac, skeletal muscle, liver, vasculature, neuronal, BBB, neuromuscular junctions, and the gastrointestinal tract. A number of these organ constructs can be combined and integrated into single platforms to probe the tissues and systemic responses to drugs and their associated metabolites. Another MPS company that was awarded NIH SBIR/STTR funding for further commercialization of their platforms is Nortis, Inc. Nortis was awarded Phase-II SBIR funding for commercialization of a microfluidic platform for modeling drug transport and cell trafficking across the BBB. While initially developing vessel-on-a-chip 133 and kidney-on-a-chip 134 platforms for drug testing, currently, Nortis offers pre-seeded organ platforms such as vasculature, kidney, liver, BBB, heart, immune system, and pancreatic islets in plug-and-play settings for drug screening and precision medicine applications. SBIR/STTR funding of MPS companies such as Nortis and Hesperos helps push the technology along the path to mainstream adoption.
In addition to NIH SBIR and STTR funding, NCATS and NIH funding of MPS developers in the NIH Tissue Chip Program has also led to the formation of start-up companies, such as TARA Biosystems, Inc. Founded in 2014, TARA Biosystems’ Biowire™ platform is being used for cardiovascular drug testing and is based on initial work performed in the Vunjak-Novakovic laboratory to create functional cardiac tissue. 135 A recent validation study showed that treatment of TARA’s in vitro human cardiac tissues with eight drugs from several inotrope classes (therapeutic agents which increase the strength of cardiac muscle contraction) mimicked contractile responses observed in native human heart tissue. 136 These results demonstrate the potential of TARA’s Biowire™ platform in identifying inotropes with different mechanisms and ultimately as an in vitro model for evaluating potential cardiac therapeutics. 136 Despite promising results from companies such as TARA Biosystems, there are still commercial hurdles that remain. One major commercial hurdle is how to scale up MPS platform manufacturing to reach an industrial pace. 26 Because many early MPS platform designs are created in academic labs and university institutions, fabrication is often limited by cost and equipment and personnel availability. 26 Therefore, it would be helpful for academic laboratories to focus on quality control very early on within device development to ensure reproducibility and reliability before scale-up in manufacturing. 26 Developers must carefully compile standard operating procedures for design and fabrication of tissue chip, as well as providing extremely clear quality control practices and procedures that are able to be easily followed by outside manufacturers or laboratories. 26 Indeed, all manufacturers should maintain Good Manufacturing Practices, such as those issued by the FDA. These Good Manufacturing practices include issues such as equipment verification, sanitation, cleanliness of manufacturing facilities, appropriate personnel training, and validation of collective processes. 26
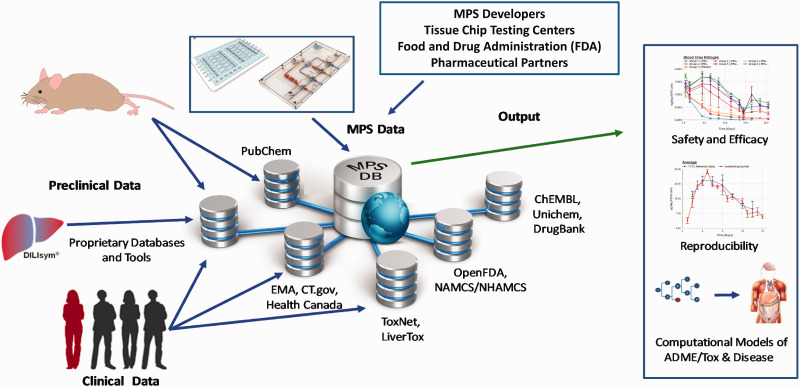
As commercial availability of MPS devices increases and this technology evolves, so comes the need for independent reproducibility and validation of these systems at multiple sites. Validation studies are absolutely critical to provide quantitative proof that organ-on-a-chip devices are faithfully able to reproduce adult human organ functional characteristics, for example action potential recordings, enzyme function, barrier permeability, gene expression, protein translation, metabolomic profile, and ultimately systemic response.132,137 Ultimately, any technology that will be used to understand disease and test drugs should be robust, reliable, and reproducible. However, there are no agreed upon “gold standard” validation methods that fit all tissue and cell types within the field, which poses a variety of problems. To attempt to address this, the NIH-funded Tissue Chip Testing Centers were tasked with the goal of onboarding a variety of tissue chips while monitoring and validating assay reproducibility and investigating parameters set forth by the community. 8 These Testing Centers collaborate with the FDA and pharmaceutical industry partners to validate assays and outcomes from MPS platform developers, and a variety of platforms have been tested to date.59,80,87,138 Data collected from both developers and the Testing Centers are deposited into a central, publicly accessible MPS Database, where users are able to access assay data as well as compare chip data with preclinical data from other modalities 83,84 (Figure 2). In addition to these validation efforts, the Tissue Chips in Space Program are working to develop robust automated MPS platforms with small laboratory benchtop footprints. 139 The miniaturization of the instrumentation supporting the chips along with a more turn-key MPS platform is expected to catalyze the adoption of technology by minimizing the need for highly specialized infrastructure and highly trained personnel currently required for MPS implementation. 139 In addition, more automated, miniaturized MPS platforms will be critical to increasing throughput and the number of replicates available per platform, which will be potentially advantageous within the early stages of the drug development process.
Figure 2.
The microphysiological systems (MPS) database is Key for the development and application of MPS. (a) The NCATS-funded MPS Database is a web-based system that aggregates experimental data taken from tissue chip developers, the NCATS-funded Tissue Chip Testing Centers, the FDA and the IQ Consortium (pharmaceutical partners). These MPS data are then aggregated with preclinical and clinical data taken from a variety of databases to enable analysis and comparison. The Database contains built-in tools to enable assessment of reproducibility and transferability of MPS experimental models and platforms, while additional computational models are being developed to enable utilization of MPS experimental models to better understand disease mechanism(s), drug and compound toxicity, and PK/PD drug predictions. (A color version of this figure is available in the online journal.)
Source: Adapted from Schurdak et al. 84 CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
FDA: Food and Drug Administration; MPS: microphysiological systems; NCATS: National Center for Advancing Translational Sciences.
Current status and future directions for MPS technology
MPS technology is uniquely positioned to play a role in precision medicine, particularly with the ability to seed patient-specific primary or iPS cells into MPS models (“patient-on-a-chip”). 26 These patient-specific chips could help stratify patients into different subpopulations based on response to various drugs or treatments. In addition, personalized MPS will also lead to the development of “clinical-trials-on-a-chip” (CToCs), particularly advantageous for patients with neural diseases, rare diseases or cancers, or diseases affecting pediatric populations. 26 Rare disease sufferers in particular are extremely limited in the number of clinical trials they can participate in, and are often disqualified from new emergent trials due to involvement in previous trials. 6 CToCs are advantageous as platforms can be made from individuals and banked cell lines from deceased patients, which helps increase the number of “subjects” in initial safety trials. 7 In addition, CToCs allow for the designing of patient-centric trials, where trials are designed with the feedback of affected patient groups, and patient-related data and outcome information are collected. 7 Ultimately, if tissue chip data can predict which patient populations may benefit the most from investigational new drugs and therapeutics, this data could potentially improve clinical trial success rates.
Although tissue chips hold much promise for applications including precision medicine; improving the clinical trial process; disease modeling; drug safety and toxicity testing; and understanding human systemic drug responses, many challenges remain to widespread adoption of MPS. 10 To help support the implementation and adoption of tissue chips in the drug development process, there must be continued collaborations between tissue chip developers, end users, and regulatory bodies such as the FDA. 26 The MPS field is rapidly evolving, and diverse stakeholders have expressed interest in the broad potential applications of this technology. These applications include therapeutic safety, toxicity, and efficacy testing within preclinical stages of drug development, plus common and rare disease modeling and mechanistic dissection, as well as for precision medicine e.g. improving clinical trial design and execution. Despite challenges, tissue chips could contribute positive changes in drug discovery and development. Ultimately, the type of platform utilized will likely depend on the stage of drug development, for example high-throughput plate-based platforms with simplistic but cheap tissue constructs may be useful in the target identification, lead selection, or lead optimization phases of pre-clinical drug discovery. 26 Alternatively, lower to medium throughput platforms capable of modeling tissue–tissue/organ–organ interactions with more complexity could also be used in preclinical single or double-organ efficacy and toxicity studies. 26 For example, convergence of self-organizing organoids with microfluidics/organ-on-chip based technologies through “synergistic engineering” may prove useful for preclinical drug toxicity studies. 140 Multi-organ systems could be useful in reducing the need for animal studies (perhaps as a “third species”), 13 and could be used in parallel with phase I and II clinical trials. 26 Patients with rare diseases, neurological diseases, rare cancers, and pediatric disorders could benefit from incorporation of MPS into clinical trial planning and execution.6,7 One day, the safety and efficacy of drug candidates may be able to be evaluated in individualized, human MPS platforms, with less need for often-times dangerous “first-in-human” testing. 10 To increase adoption and implementation of this technology, continued cooperation, coordination, and engagement between end-users such as MPS developers, industry, CROs, and regulatory bodies is critical. Ultimately, all end-users must work together to push this paradigm-shifting technology forward to bring more drugs to more patients faster.
Footnotes
AUTHORS’ CONTRIBUTIONS: All authors participated in the preparation of the manuscript.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the Cures Acceleration Network, through the National Center for Advancing Translational Sciences.
ORCID iDs: Passley Hargrove-Grimes https://orcid.org/0000-0002-7644-0462
Lucie A Low https://orcid.org/0000-0001-6082-8625
References
- 1.DiMasi JA, Grabowski HG, Hansen RW. Innovation in the pharmaceutical industry: new estimates of R&D costs. J Health Econ 2016; 47:20–33 [DOI] [PubMed] [Google Scholar]
- 2.Wagner J, Dahlem AM, Hudson LD, Terry SF, Altman RB, Gilliland CT, DeFeo C, Austin CP. A dynamic map for learning, communicating, navigating and improving therapeutic development. Nat Rev Drug Discov 2018; 17:150. [DOI] [PubMed] [Google Scholar]
- 3.Wagner JA, Dahlem AM, Hudson LD, Terry SF, Altman RB, Gilliland CT, DeFeo C, Austin CP. Application of a dynamic map for learning, communicating, navigating, and improving therapeutic development. Clin Transl Sci 2018; 11:166–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arrowsmith J. Trial watch: phase II failures: 2008-2010. Nat Rev Drug Discov 2011; 10:328–9 [DOI] [PubMed] [Google Scholar]
- 5.Arrowsmith J. Trial watch: phase III and submission failures: 2007-2010. Nat Rev Drug Discov 2011; 10:87. [DOI] [PubMed] [Google Scholar]
- 6.Low LA, Tagle DA. Tissue chips to aid drug development and modeling for rare diseases. Expert Opin Orphan Drugs 2016; 4:1113–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blumenrath SH, Lee BY, Low L, Prithviraj R, Tagle D. Tackling rare diseases: clinical trials on chips. Exp Biol Med (Maywood) 2020; 245:1155–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Low LA, Sutherland M, Lumelsky N, Selimovic S, Lundberg MS, Tagle DA. Organs-on-a-chip. Adv Exp Med Biol 2020; 1230:27–42 [DOI] [PubMed] [Google Scholar]
- 9.Breslin S, O'Driscoll L. Three-dimensional cell culture: the missing link in drug discovery. Drug Discov Today 2013; 18:240–9 [DOI] [PubMed] [Google Scholar]
- 10.Watson DE, Hunziker R, Wikswo JP. Fitting tissue chips and microphysiological systems into the grand scheme of medicine, biology, pharmacology, and toxicology. Exp Biol Med (Maywood) 2017; 242:1559–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuberi A, Lutz C. Mouse models for drug discovery. Can new tools and technology improve translational power? ILAR J 2016; 57:178–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tagle DA. The NIH microphysiological systems program: developing in vitro tools for safety and efficacy in drug development. Curr Opin Pharmacol 2019; 48:146–54 [DOI] [PubMed] [Google Scholar]
- 13.Ingber DE. Is it time for reviewer 3 to request human organ chip experiments instead of animal validation studies? Adv Sci (Weinh) 2020; 7:2002030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahadian S, Civitarese R, Bannerman D, Mohammadi MH, Lu R, Wang E, Davenport-Huyer L, Lai B, Zhang B, Zhao Y, Mandla S, Korolj A, Radisic M. Organ-on-a-chip platforms: a convergence of advanced materials, cells, and microscale technologies. Adv Healthc Mater 2018; 7:1700506. [DOI] [PubMed] [Google Scholar]
- 15.Zhang YS, Arneri A, Bersini S, Shin SR, Zhu K, Goli-Malekabadi Z, Aleman J, Colosi C, Busignani F, Dell'Erba V, Bishop C, Shupe T, Demarchi D, Moretti M, Rasponi M, Dokmeci MR, Atala A, Khademhosseini A. Bioprinting 3D microfibrous scaffolds for engineering endothelialized myocardium and heart-on-a-chip. Biomaterials 2016; 110:45–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li X, George SM, Vernetti L, Gough AH, Taylor DL. A glass-based, continuously zonated and vascularized human liver acinus microphysiological system (vLAMPS) designed for experimental modeling of diseases and ADME/TOX. Lab Chip 2018; 18:2614–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol 2014; 32:760–72 [DOI] [PubMed] [Google Scholar]
- 18.Polini A, Prodanov L, Bhise NS, Manoharan V, Dokmeci MR, Khademhosseini A. Organs-on-a-chip: a new tool for drug discovery. Expert Opin Drug Discov 2014; 9:335–52 [DOI] [PubMed] [Google Scholar]
- 19.Tanataweethum N, Zelaya A, Yang F, Cohen RN, Brey EM, Bhushan A. Establishment and characterization of a primary murine adipose tissue-chip. Biotechnol Bioeng 2018; 115:1979–87 [DOI] [PubMed] [Google Scholar]
- 20.Ronaldson-Bouchard K, Vunjak-Novakovic G. Organs-on-a-chip: a fast track for engineered human tissues in drug development. Cell Stem Cell 2018; 22:310–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kratz SRA, Holl G, Schuller P, Ertl P, Rothbauer M. Latest trends in biosensing for microphysiological organs-on-a-chip and body-on-a-chip systems. Biosensors (Basel) 2019; 9:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peel S, Corrigan AM, Ehrhardt B, Jang KJ, Caetano-Pinto P, Boeckeler M, Rubins JE, Kodella K, Petropolis DB, Ronxhi J, Kulkarni G, Foster AJ, Williams D, Hamilton GA, Ewart L. Introducing an automated high content confocal imaging approach for organs-on-chips. Lab Chip 2019; 19:410–21 [DOI] [PubMed] [Google Scholar]
- 23.Kim JH, Park JY, Jin S, Yoon S, Kwak JY, Jeong YH. A microfluidic chip embracing a nanofiber scaffold for 3D cell culture and real-time monitoring. Nanomaterials (Basel) 2019; 9:588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curzer HJ, Perry G, Wallace MC, Perry D. The three Rs of animal research: what they mean for the Institutional Animal Care and Use Committee and why. Sci Eng Ethics 2016; 22:549–65 [DOI] [PubMed] [Google Scholar]
- 25.Ekert JE, Deakyne J, Pribul-Allen P, Terry R, Schofield C, Jeong CG, Storey J, Mohamet L, Francis J, Naidoo A, Amador A, Klein JL, Rowan W. Recommended guidelines for developing, qualifying, and implementing complex in vitro models (CIVMs) for drug discovery. SLAS Discov 2020; 25:1174–90 [DOI] [PubMed] [Google Scholar]
- 26.Low LA, Mummery C, Berridge BR, Austin CP, Tagle DA. Organs-on-chips: into the next decade. Nat Rev Drug Discov 2020. doi: 10.1038/s41573-020-0079-3. [DOI] [PubMed] [Google Scholar]
- 27.Apati A, Varga N, Berecz T, Erdei Z, Homolya L, Sarkadi B. Application of human pluripotent stem cells and pluripotent stem cell-derived cellular models for assessing drug toxicity. Expert Opin Drug Metab Toxicol 2019; 15:61–75 [DOI] [PubMed] [Google Scholar]
- 28.Rudmann DG. The emergence of microphysiological systems (organs-on-chips) as paradigm-changing tools for toxicologic pathology. Toxicol Pathol 2019; 47:4–10 [DOI] [PubMed] [Google Scholar]
- 29.Heringa MB, Park M, Kienhuis AS, Vandebriel RJ. The value of organs-on-chip for regulatory safety assessment. ALTEX 2020; 37:208–22 [DOI] [PubMed] [Google Scholar]
- 30.Atchison L, Zhang H, Cao K, Truskey GA. A tissue engineered blood vessel model of Hutchinson-Gilford progeria syndrome using human iPSC-derived smooth muscle cells. Sci Rep 2017; 7:8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ribas J, Zhang YS, Pitrez PR, Leijten J, Miscuglio M, Rouwkema J, Dokmeci MR, Nissan X, Ferreira L, Khademhosseini A. Biomechanical strain exacerbates inflammation on a progeria-on-a-chip model. Small 2017; 13. doi: 10.1002/smll.201603737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G, McCain ML, Yang L, He A, Pasqualini FS, Agarwal A, Yuan H, Jiang D, Zhang D, Zangi L, Geva J, Roberts AE, Ma Q, Ding J, Chen J, Wang DZ, Li K, Wang J, Wanders RJ, Kulik W, Vaz FM, Laflamme MA, Murry CE, Chien KR, Kelley RI, Church GM, Parker KK, Pu WT. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med 2014; 20:616–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Meer AD, Orlova VV, ten Dijke P, van den Berg A, Mummery CL. Three-dimensional co-cultures of human endothelial cells and embryonic stem cell-derived pericytes inside a microfluidic device. Lab Chip 2013; 13:3562–8 [DOI] [PubMed] [Google Scholar]
- 34.van den Berg A, Mummery CL, Passier R, van der Meer AD. Personalised organs-on-chips: functional testing for precision medicine. Lab Chip 2019; 19:198–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang SY, Weber EJ, Sidorenko VS, Chapron A, Yeung CK, Gao C, Mao Q, Shen D, Wang J, Rosenquist TA, Dickman KG, Neumann T, Grollman AP, Kelly EJ, Himmelfarb J, Eaton DL. Human liver-kidney model elucidates the mechanisms of aristolochic acid nephrotoxicity. JCI Insight 2017; 2:e95978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Truskey GA. Human microphysiological systems and organoids as in vitro models for toxicological studies. Front Public Health 2018; 6:185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu L, Koo Y, Akwitti C, Russell T, Gay E, Laskowitz DT, Yun Y. Three-dimensional (3D) brain microphysiological system for organophosphates and neurochemical agent toxicity screening. PLoS One 2019; 14:e0224657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang JW, Shen YC, Lin KC, Cheng SJ, Chen SL, Chen CY, Kumar PV, Lin SF, Lu HE, Chen GY. Organ-on-a-chip: opportunities for assessing the toxicity of particulate matter. Front Bioeng Biotechnol 2020; 8:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang H, Abouleila Y, Si L, Ortega-Prieto AM, Mummery CL, Ingber DE, Mashaghi A. Human organs-on-chips for virology. Trends Microbiol 2020; 28:934–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hawkins KG, Casolaro C, Brown JA, Edwards DA, Wikswo JP. The microbiome and the gut-liver-brain axis for central nervous system clinical pharmacology: challenges in specifying and integrating in vitro and in silico models. Clin Pharmacol Ther 2020; 108:929–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ribeiro AJS, Yang X, Patel V, Madabushi R, Strauss DG. Liver microphysiological systems for predicting and evaluating drug effects. Clin Pharmacol Ther 2019; 106:139–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paek J, Park SE, Lu Q, Park KT, Cho M, Oh JM, Kwon KW, Yi YS, Song JW, Edelstein HI, Ishibashi J, Yang W, Myerson JW, Kiseleva RY, Aprelev P, Hood ED, Stambolian D, Seale P, Muzykantov VR, Huh D. Microphysiological engineering of self-assembled and perfusable microvascular beds for the production of vascularized three-dimensional human microtissues. ACS Nano 2019; 13:7627–43 [DOI] [PubMed] [Google Scholar]
- 43.Herron LA, Hansen CS, Abaci HE. Engineering tissue-specific blood vessels. Bioeng Transl Med 2019; 4:e10139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown JA, Faley SL, Shi Y, Hillgren KM, Sawada GA, Baker TK, Wikswo JP, Lippmann ES. Advances in blood-brain barrier modeling in microphysiological systems highlight critical differences in opioid transport due to cortisol exposure. Fluids Barriers CNS 2020; 17:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee CS, Leong KW. Advances in microphysiological blood-brain barrier (BBB) models towards drug delivery. Curr Opin Biotechnol 2020; 66:78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ahn SI, Sei YJ, Park HJ, Kim J, Ryu Y, Choi JJ, Sung HJ, MacDonald TJ, Levey AI, Kim Y. Microengineered human blood-brain barrier platform for understanding nanoparticle transport mechanisms. Nat Commun 2020; 11:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davis BN, Yen R, Prasad V, Truskey GA. Oxygen consumption in human, tissue-engineered myobundles during basal and electrical stimulation conditions. APL Bioeng 2019; 3:026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Truskey GA. Development and application of human skeletal muscle microphysiological systems. Lab Chip 2018; 18:3061–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang X, Hong S, Yen R, Kondash M, Fernandez CE, Truskey GA. A system to monitor statin-induced myopathy in individual engineered skeletal muscle myobundles. Lab Chip 2018; 18:2787–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lind JU, Busbee TA, Valentine AD, Pasqualini FS, Yuan H, Yadid M, Park SJ, Kotikian A, Nesmith AP, Campbell PH, Vlassak JJ, Lewis JA, Parker KK. Instrumented cardiac microphysiological devices via multimaterial three-dimensional printing. Nat Mater 2017; 16:30308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lind JU, Yadid M, Perkins I, O'Connor BB, Eweje F, Chantre CO, Hemphill MA, Yuan H, Campbell PH, Vlassak JJ, Parker KK. Cardiac microphysiological devices with flexible thin-film sensors for higher-throughput drug screening. Lab Chip 2017; 17:3692–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sidorov VY, Samson PC, Sidorova TN, Davidson JM, Lim CC, Wikswo JP. I-Wire heart-on-a-chip I: three-dimensional cardiac tissue constructs for physiology and pharmacology. Acta Biomater 2017; 48:68–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Rafatian N, Feric NT, Cox BJ, Aschar-Sobbi R, Wang EY, Aggarwal P, Zhang B, Conant G, Ronaldson-Bouchard K, Pahnke A, Protze S, Lee JH, Davenport Huyer L, Jekic D, Wickeler A, Naguib HE, Keller GM, Vunjak-Novakovic G, Broeckel U, Backx PH, Radisic M. A platform for generation of chamber-specific cardiac tissues and disease modeling. Cell 2019; 176:913–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Conant G, Lai BFL, Lu RXZ, Korolj A, Wang EY, Radisic M. High-content assessment of cardiac function using heart-on-a-chip devices as drug screening model. Stem Cell Rev Rep 2017; 13:335–46 [DOI] [PubMed] [Google Scholar]
- 55.Huebsch N, Loskill P, Deveshwar N, Spencer CI, Judge LM, Mandegar MA, Fox CB, Mohamed TM, Ma Z, Mathur A, Sheehan AM, Truong A, Saxton M, Yoo J, Srivastava D, Desai TA, So PL, Healy KE, Conklin BR. Miniaturized iPS-cell-derived cardiac muscles for physiologically relevant drug response analyses. Sci Rep 2016; 6:24726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mathur A, Loskill P, Shao K, Huebsch N, Hong S, Marcus SG, Marks N, Mandegar M, Conklin BR, Lee LP, Healy KE. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep 2015; 5:8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schroer A, Pardon G, Castillo E, Blair C, Pruitt B. Engineering hiPSC cardiomyocyte in vitro model systems for functional and structural assessment. Prog Biophys Mol Biol 2019; 144:3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maass C, Sorensen NB, Himmelfarb J, Kelly EJ, Stokes CL, Cirit M. Translational assessment of drug-induced proximal tubule injury using a kidney microphysiological system. CPT Pharmacometrics Syst Pharmacol 2019; 8:316–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakolish C, Chen Z, Dalaijamts C, Mitra K, Liu Y, Fulton T, Wade TL, Kelly EJ, Rusyn I, Chiu WA. Predicting tubular reabsorption with a human kidney proximal tubule tissue-on-a-chip and physiologically-based modeling. Toxicol in Vitro 2020; 63:104752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chapron A, Chapron BD, Hailey DW, Chang SY, Imaoka T, Thummel KE, Kelly E, Himmelfarb J, Shen D, Yeung CK. An improved vascularized, dual-channel microphysiological system facilitates modeling of proximal tubular solute secretion. ACS Pharmacol Transl Sci 2020; 3:496–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phillips JA, Grandhi TSP, Davis M, Gautier JC, Hariparsad N, Keller D, Sura R, Van Vleet TR. A pharmaceutical industry perspective on microphysiological kidney systems for evaluation of safety for new therapies. Lab Chip 2020; 20:468–76 [DOI] [PubMed] [Google Scholar]
- 62.Xiao S, Coppeta JR, Rogers HB, Isenberg BC, Zhu J, Olalekan SA, McKinnon KE, Dokic D, Rashedi AS, Haisenleder DJ, Malpani SS, Arnold-Murray CA, Chen K, Jiang M, Bai L, Nguyen CT, Zhang J, Laronda MM, Hope TJ, Maniar KP, Pavone ME, Avram MJ, Sefton EC, Getsios S, Burdette JE, Kim JJ, Borenstein JT, Woodruff TK. A microfluidic culture model of the human reproductive tract and 28-day menstrual cycle. Nat Commun 2017; 8:14584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Komeya M, Kimura H, Nakamura H, Yokonishi T, Sato T, Kojima K, Hayashi K, Katagiri K, Yamanaka H, Sanjo H, Yao M, Kamimura S, Inoue K, Ogonuki N, Ogura A, Fujii T, Ogawa T. Long-term ex vivo maintenance of testis tissues producing fertile sperm in a microfluidic device. Sci Rep 2016; 6:21472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pamies D, Barreras P, Block K, Makri G, Kumar A, Wiersma D, Smirnova L, Zang C, Bressler J, Christian KM, Harris G, Ming GL, Berlinicke CJ, Kyro K, Song H, Pardo CA, Hartung T, Hogberg HT. A human brain microphysiological system derived from induced pluripotent stem cells to study neurological diseases and toxicity. ALTEX 2017; 34:362–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kwak BS, Jin SP, Kim SJ, Kim EJ, Chung JH, Sung JH. Microfluidic skin chip with vasculature for recapitulating the immune response of the skin tissue. Biotechnol Bioeng 2020; 117:1853–63 [DOI] [PubMed] [Google Scholar]
- 66.Hardwick RN, Betts CJ, Whritenour J, Sura R, Thamsen M, Kaufman EH, Fabre K. Drug-induced skin toxicity: gaps in preclinical testing Cascade as opportunities for complex in vitro models and assays. Lab Chip 2020; 20:199–214 [DOI] [PubMed] [Google Scholar]
- 67.Kim K, Jeon HM, Choi KC, Sung GY. Testing the effectiveness of curcuma longa leaf extract on a skin equivalent using a pumpless skin-on-a-chip model. Int J Mol Sci 2020; 21:3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Romero-Lopez M, Li Z, Rhee C, Maruyama M, Pajarinen J, O'Donnell B, Lin TH, Lo CW, Hanlon J, Dubowitz R, Yao Z, Bunnell Ba Lin H, Tuan RS, Goodman SB. Macrophage effects on mesenchymal stem cell osteogenesis in a three-dimensional in vitro bone model. Tissue Eng Part A 2020. doi: 10.1089/ten.TEA.2020.0041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Achberger K, Probst C, Haderspeck J, Bolz S, Rogal J, Chuchuy J, Nikolova M, Cora V, Antkowiak L, Haq W, Shen N, Schenke-Layland K, Ueffing M, Liebau S, Loskill P. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform. Elife 2019; 8:e46188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wright CB, Becker SM, Low LA, Tagle DA, Sieving PA. Improved ocular tissue models and eye-on-a-chip technologies will facilitate ophthalmic drug development. J Ocul Pharmacol Ther 2020; 36:25–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci Rep 2017; 7:42296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Novak R, Ingram M, Marquez S, Das D, Delahanty A, Herland A, Maoz BM, Jeanty SSF, Somayaji MR, Burt M, Calamari E, Chalkiadaki A, Cho A, Choe Y, Chou DB, Cronce M, Dauth S, Divic T, Fernandez-Alcon J, Ferrante T, Ferrier J, FitzGerald EA, Fleming R, Jalili-Firoozinezhad S, Grevesse T, Goss JA, Hamkins-Indik T, Henry O, Hinojosa C, Huffstater T, Jang KJ, Kujala V, Leng L, Mannix R, Milton Y, Nawroth J, Nestor BA, Ng CF, O'Connor B, Park TE, Sanchez H, Sliz J, Sontheimer-Phelps A, Swenor B, Thompson G, 2nd Touloumes GJ, Tranchemontagne Z, Wen N, Yadid M, Bahinski A, Hamilton GA, Levner D, Levy O, Przekwas A, Prantil-Baun R, Parker KK, Ingber DE. Robotic fluidic coupling and interrogation of multiple vascularized organ chips. Nat Biomed Eng 2020; 4:407–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Edington CD, Chen WLK, Geishecker E, Kassis T, Soenksen LR, Bhushan BM, Freake D, Kirschner J, Maass C, Tsamandouras N, Valdez J, Cook CD, Parent T, Snyder S, Yu J, Suter E, Shockley M, Velazquez J, Velazquez JJ, Stockdale L, Papps JP, Lee I, Vann N, Gamboa M, LaBarge ME, Zhong Z, Wang X, Boyer LA, Lauffenburger DA, Carrier RL, Communal C, Tannenbaum SR, Stokes CL, Hughes DJ, Rohatgi G, Trumper DL, Cirit M, Griffith LG. Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci Rep 2018; 8:4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Miyoshi T, Hiratsuka K, Saiz EG, Morizane R. Kidney organoids in translational medicine: disease modeling and regenerative medicine. Dev Dyn 2020; 249:34–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Weng KC, Kurokawa YK, Hajek BS, Paladin JA, Shirure VS, George SC. Human induced pluripotent stem-cardiac-endothelial-tumor-on-a-chip to assess anticancer efficacy and cardiotoxicity. Tissue Eng Part C Methods 2020; 26:44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin Z, Li Z, Li EN, Li X, Del DC, Shen H, Hao T, O'Donnell B, Bunnell BA, Goodman SB, Alexander PG, Tuan RS., Lin H. Osteochondral tissue chip derived from iPSCs: modeling OA pathologies and testing drugs. Front Bioeng Biotechnol 2019; 7:411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sharma A, Sances S, Workman MJ, Svendsen CN. Multi-lineage human iPSC-derived platforms for disease modeling and drug discovery. Cell Stem Cell 2020; 26:309–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang Q, Zhang X, Truskey GA. Vascular microphysiological systems to model diseases. Cell Gene Ther Insights 2020; 6:93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chramiec A, Teles D, Yeager K, Marturano-Kruik A, Pak J, Chen T, Hao L, Wang M, Lock R, Tavakol DN, Lee MB, Kim J, Ronaldson-Bouchard K, Vunjak-Novakovic G. Integrated human organ-on-a-chip model for predictive studies of anti-tumor drug efficacy and cardiac safety. Lab Chip 2020. doi: 10.1039/d0lc00424c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sakolish C, Weber EJ, Kelly EJ, Himmelfarb J, Mouneimne R, Grimm FA, House JS, Wade T, Han A, Chiu WA, Rusyn I. Technology transfer of the microphysiological systems: a case study of the human proximal tubule tissue chip. Sci Rep 2018; 8:14882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Y, Sakolish C, Chen Z, Phan DTT, Bender RHF, Hughes CCW, Rusyn I. Human in vitro vascularized micro-organ and micro-tumor models are reproducible organ-on-a-chip platforms for studies of anticancer drugs. Toxicology 2020; 445:152601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sakolish C, House JS, Chramiec A, Liu Y, Chen Z, Halligan SP, Vunjak-Novakovic G, Rusyn I. Tissue-engineered bone tumor as a reproducible human in vitro model for studies of anticancer drugs. Toxicol Sci 2020; 173:65–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gough A, Vernetti L, Bergenthal L, Shun TY, Taylor DL. The microphysiology systems database for analyzing and modeling compound interactions with human and animal organ models. Appl in Vitro Toxicol 2016; 2:103–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schurdak M, Vernetti L, Bergenthal L, Wolter QK, Shun TY, Karcher S, Taylor DL, Gough A. Applications of the microphysiology systems database for experimental ADME-Tox and disease models. Lab Chip 2020; 20:1472–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Vernetti L, Gough A, Baetz N, Blutt S, Broughman JR, Brown JA, Foulke-Abel J, Hasan N, In J, Kelly E, Kovbasnjuk O, Repper J, Senutovitch N, Stabb J, Yeung C, Zachos NC, Donowitz M, Estes M, Himmelfarb J, Truskey G, Wikswo JP, Taylor DL. Corrigendum: functional coupling of human microphysiology systems: intestine, liver, kidney proximal tubule, blood-brain barrier and skeletal muscle. Sci Rep 2017; 7:44517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alexander PG, Clark KL, Tuan RS. Prenatal exposure to environmental factors and congenital limb defects. Birth Defects Res C Embryo Today 2016; 108:243–73 [DOI] [PubMed] [Google Scholar]
- 87.Low LA, Tagle DA. Organs-on-chips: progress, challenges, and future directions. Exp Biol Med (Maywood) 2017; 242:1573–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ewart L, Fabre K, Chakilam A, Dragan Y, Duignan DB, Eswaraka J, Gan J, Guzzie-Peck P, Otieno M, Jeong CG, Keller DA, de Morais SM, Phillips JA, Proctor W, Sura R, Van Vleet T, Watson D, Will Y, Tagle D, Berridge B. Navigating tissue chips from development to dissemination: a pharmaceutical industry perspective. Exp Biol Med (Maywood) 2017; 242:1579–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jang KJ, Otieno MA, Ronxhi J, Lim HK, Ewart L, Kodella KR, Petropolis DB, Kulkarni G, Rubins JE, Conegliano D, Nawroth J, Simic D, Lam W, Singer M, Barale E, Singh B, Sonee M, Streeter AJ, Manthey C, Jones B, Srivastava A, Andersson LC, Williams D, Park H, Barrile R, Sliz J, Herland A, Haney S, Karalis K, Ingber DE, Hamilton GA. Reproducing human and cross-species drug toxicities using a liver-chip. Sci Transl Med 2019; 11:eeax516. [DOI] [PubMed] [Google Scholar]
- 90.Fowler S, Chen WLK, Duignan DB, Gupta A, Hariparsad N, Kenny JR, Lai WG, Liras J, Phillips JA, Gan J. Microphysiological systems for ADME-related applications: current status and recommendations for system development and characterization. Lab Chip 2020; 20:446–67 [DOI] [PubMed] [Google Scholar]
- 91.Fabre K, Berridge B, Proctor WR, Ralston S, Will Y, Baran SW, Yoder G, Van Vleet TR. Introduction to a manuscript series on the characterization and use of microphysiological systems (MPS) in pharmaceutical safety and ADME applications. Lab Chip 2020; 20:1049–57 [DOI] [PubMed] [Google Scholar]
- 92.Fabre KM, Delsing L, Hicks R, Colclough N, Crowther DC, Ewart L. Utilizing microphysiological systems and induced pluripotent stem cells for disease modeling: a case study for blood brain barrier research in a pharmaceutical setting. Adv Drug Deliv Rev 2019; 140:129–35 [DOI] [PubMed] [Google Scholar]
- 93.Ainslie GR, Davis M, Ewart L, Lieberman LA, Rowlands DJ, Thorley AJ, Yoder G, Ryan AM. Microphysiological lung models to evaluate the safety of new pharmaceutical modalities: a biopharmaceutical perspective. Lab Chip 2019; 19:3152–61 [DOI] [PubMed] [Google Scholar]
- 94.Baudy AR, Otieno MA, Hewitt P, Gan J, Roth A, Keller D, Sura R, Van Vleet TR, Proctor WR. Liver microphysiological systems development guidelines for safety risk assessment in the pharmaceutical industry. Lab Chip 2020; 20:215–25 [DOI] [PubMed] [Google Scholar]
- 95.Peterson NC, Mahalingaiah PK, Fullerton A, Di Piazza M. Application of microphysiological systems in biopharmaceutical research and development. Lab Chip 2020; 20:697–708 [DOI] [PubMed] [Google Scholar]
- 96.Peters MF, Choy AL, Pin C, Leishman DJ, Moisan A, Ewart L, Guzzie-Peck PJ, Sura R, Keller DA, Scott CW, Kolaja KL. Developing in vitro assays to transform gastrointestinal safety assessment: potential for microphysiological systems. Lab Chip 2020; 20:1177–90 [DOI] [PubMed] [Google Scholar]
- 97.Eckstrum K, Striz A, Ferguson M, Zhao Y, Welch B, Solomotis N, Olejnik N, Sprando R. Utilization of a model hepatotoxic compound, diglycolic acid, to evaluate liver organ-chip performance and in vitro to in vivo concordance. Food Chem Toxicol 2020; 146:111850. [DOI] [PubMed] [Google Scholar]
- 98.Marx U, Akabane T, Andersson TB, Baker E, Beilmann M, Beken S, Brendler-Schwaab S, Cirit M, David R, Dehne EM, Durieux I, Ewart L, Fitzpatrick SC, Frey O, Fuchs F, Griffith LG, Hamilton GA, Hartung T, Hoeng J, Hogberg H, Hughes DJ, Ingber DE, Iskandar A, Kanamori T, Kojima H, Kuehnl J, Leist M, Li B, Loskill P, Mendrick DL, Neumann T, Pallocca G, Rusyn I, Smirnova L, Steger-Hartmann T, Tagle DA, Tonevitsky A, Tsyb S, Trapecar M, Van de Water B, Van den Eijnden-van Raaij J, Vulto P, Watanabe K, Wolf A, Zhou X, Roth A. Biology-inspired microphysiological systems to advance patient benefit and animal welfare in drug development. ALTEX 2020; 37:364–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006; 126:663–76 [DOI] [PubMed] [Google Scholar]
- 100.Kwist K, Bridges WC, Burg KJ. The effect of cell passage number on osteogenic and adipogenic characteristics of D1 cells. Cytotechnology 2016; 68:1661–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Briske-Anderson MJ, Finley JW, Newman SM. The influence of culture time and passage number on the morphological and physiological development of Caco-2 cells. Proc Soc Exp Biol Med 1997; 214:248–57 [DOI] [PubMed] [Google Scholar]
- 102.Esquenet M, Swinnen JV, Heyns W, Verhoeven G. LNCaP prostatic adenocarcinoma cells derived from low and high passage numbers display divergent responses not only to androgens but also to retinoids. J Steroid Biochem Mol Biol 1997; 62:391–9 [DOI] [PubMed] [Google Scholar]
- 103.Yu H, Cook TJ, Sinko PJ. Evidence for diminished functional expression of intestinal transporters in Caco-2 cell monolayers at high passages. Pharm Res 1997; 14:757–62 [DOI] [PubMed] [Google Scholar]
- 104.Takahashi K, Okita K, Nakagawa M, Yamanaka S. Induction of pluripotent stem cells from fibroblast cultures. Nat Protoc 2007; 2:3081–9 [DOI] [PubMed] [Google Scholar]
- 105.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007; 131:861–72 [DOI] [PubMed] [Google Scholar]
- 106.Liang P, Sallam K, Wu H, Li Y, Itzhaki I, Garg P, Zhang Y, Vermglinchan V, Lan F, Gu M, Gong T, Zhuge Y, He C, Ebert AD, Sanchez-Freire V, Churko J, Hu S, Sharma A, Lam CK, Scheinman MM, Bers DM, Wu JC. Patient-specific and genome-edited induced pluripotent stem cell-derived cardiomyocytes elucidate single-cell phenotype of Brugada syndrome. J Am Coll Cardiol 2016; 68:2086–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Burridge PW, Li YF, Matsa E, Wu H, Ong SG, Sharma A, Holmstrom A, Chang AC, Coronado MJ, Ebert AD, Knowles JW, Telli ML, Witteles RM, Blau HM, Bernstein D, Altman RB, Wu JC. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med 2016; 22:547–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nishizawa M, Chonabayashi K, Nomura M, Tanaka A, Nakamura M, Inagaki A, Nishikawa M, Takei I, Oishi A, Tanabe K, Ohnuki M, Yokota H, Koyanagi-Aoi M, Okita K, Watanabe A, Takaori-Kondo A, Yamanaka S, Yoshida Y. Epigenetic variation between human induced pluripotent stem cell lines is an indicator of differentiation capacity. Cell Stem Cell 2016; 19:341–54 [DOI] [PubMed] [Google Scholar]
- 109.Kim K, Doi A, Wen B, Ng K, Zhao R, Cahan P, Kim J, Aryee MJ, Ji H, Ehrlich LI, Yabuuchi A, Takeuchi A, Cunniff KC, Hongguang H, McKinney-Freeman S, Naveiras O, Yoon TJ, Irizarry RA, Jung N, Seita J, Hanna J, Murakami P, Jaenisch R, Weissleder R, Orkin SH, Weissman IL, Feinberg AP, Daley GQ. Epigenetic memory in induced pluripotent stem cells. Nature 2010; 467:285–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rajamohan D, Matsa E, Kalra S, Crutchley J, Patel A, George V, Denning C. Current status of drug screening and disease modelling in human pluripotent stem cells. Bioessays 2013; 35:281–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Doss MX, Sachinidis A. Current challenges of iPSC-based disease modeling and therapeutic implications. Cells 2019; 8:403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Christoffersson J, Mandenius CF. Fabrication of a microfluidic cell culture device using photolithographic and soft lithographic techniques. Methods Mol Biol 2019; 1994:227–33 [DOI] [PubMed] [Google Scholar]
- 113.Duffy DC, McDonald JC, Schueller OJ, Whitesides GM. Rapid prototyping of microfluidic systems in poly(dimethylsiloxane). Anal Chem 1998; 70:4974–84 [DOI] [PubMed] [Google Scholar]
- 114.Bhattacharjee N, Urrios A, Kang S, Folch A. The upcoming 3D-printing revolution in microfluidics. Lab Chip 2016; 16:1720–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang YS, Yue K, Aleman J, Moghaddam KM, Bakht SM, Yang J, Jia W, Dell'Erba V, Assawes P, Shin SR, Dokmeci MR, Oklu R, Khademhosseini A. 3D bioprinting for tissue and organ fabrication. Ann Biomed Eng 2017; 45:148–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Quiros-Solano WF, Gaio N, Stassen O, Arik YB, Silvestri C, Van Engeland NCA, Van der Meer A, Passier R, Sahlgren CM, Bouten CVC, van den Berg A, Dekker R, Sarro PM. Microfabricated tuneable and transferable porous PDMS membranes for organs-on-chips. Sci Rep 2018; 8:13524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van Meer BJ, de Vries H, Firth KSA, van Weerd J, Tertoolen LGJ, Karperien HBJ, Jonkheijm P, Denning C, Ijzerman AP, Mummery CL. Small molecule absorption by PDMS in the context of drug response bioassays. Biochem Biophys Res Commun 2017; 482:323–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Shirure VS, George SC. Design considerations to minimize the impact of drug absorption in polymer-based organ-on-a-chip platforms. Lab Chip 2017; 17:681–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sano E, Mori C, Matsuoka N, Ozaki Y, Yagi K, Wada A, Tashima K, Yamasaki S, Tanabe K, Yano K, Torisawa YS. Tetrafluoroethylene-propylene elastomer for fabrication of microfluidic organs-on-chips resistant to drug absorption. Micromachines (Basel ) 2019; 10:793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Domansky K, Leslie DC, McKinney J, Fraser JP, Sliz JD, Hamkins-Indik T, Hamilton GA, Bahinski A, Ingber DE. Clear castable polyurethane elastomer for fabrication of microfluidic devices. Lab Chip 2013; 13:3956–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rogers HB, Zhou LT, Kusuhara A, Zaniker E, Shafaie S, Owen BC, Duncan FE, Woodruff TK. Dental resins used in 3D printing technologies release ovo-toxic leachates. Chemosphere 2020; 270:129003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li Y, Yang Y, Xiong A, Wu X, Xie J, Han S, Zhao S. Comparative gene expression analysis of lymphocytes treated with exosomes derived from ovarian cancer and ovarian cysts. Front Immunol 2017; 8:607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yin P, Lv H, Li Y, Deng Y, Zhang L, Tang P. Exosome-mediated genetic information transfer, a missing piece of osteoblast-osteoclast communication puzzle. Front Endocrinol (Lausanne) 2017; 8:336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wikswo JP, Curtis EL, Eagleton ZE, Evans BC, Kole A, Hofmeister LH, Matloff WJ. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip 2013; 13:3496–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moraes C, Labuz JM, Leung BM, Inoue M, Chun TH, Takayama S. On being the right size: scaling effects in designing a human-on-a-chip. Integr Biol (Camb) 2013; 5:1149–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Park D, Lee J, Chung JJ, Jung Y, Kim SH. Integrating organs-on-chips: multiplexing, scaling, vascularization, and innervation. Trends Biotechnol 2020; 38:99–112 [DOI] [PubMed] [Google Scholar]
- 127.Sin A, Chin KC, Jamil MF, Kostov Y, Rao G, Shuler ML. The design and fabrication of three-chamber microscale cell culture analog devices with integrated dissolved oxygen sensors. Biotechnol Prog 2004; 20:338–45 [DOI] [PubMed] [Google Scholar]
- 128.Herland A, Maoz BM, Das D, Somayaji MR, Prantil-Baun R, Novak R, Cronce M, Huffstater T, Jeanty SSF, Ingram M, Chalkiadaki A, Benson Chou D, Marquez S, Delahanty A, Jalili-Firoozinezhad S, Milton Y, Sontheimer-Phelps A, Swenor B, Levy O, Parker KK, Przekwas A, Ingber DE. Quantitative prediction of human pharmacokinetic responses to drugs via fluidically coupled vascularized organ chips. Nat Biomed Eng 2020; 4:421–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Abaci HE, Shuler ML. Human-on-a-chip design strategies and principles for physiologically based pharmacokinetics/pharmacodynamics modeling. Integr Biol (Camb) 2015; 7:383–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Polini A, Del Mercato LL, Barra A, Zhang YS, Calabi F, Gigli G. Towards the development of human immune-system-on-a-chip platforms. Drug Discov Today 2019; 24:517–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci U S A 2016; 113:E7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang B, Radisic M. Organ-on-a-chip devices advance to market. Lab Chip 2017; 17:2395–420 [DOI] [PubMed] [Google Scholar]
- 133.Tourovskaia A, Fauver M, Kramer G, Simonson S, Neumann T. Tissue-engineered microenvironment systems for modeling human vasculature. Exp Biol Med (Maywood) 2014; 239:1264–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Weber EJ, Chapron A, Chapron BD, Voellinger JL, Lidberg KA, Yeung CK, Wang Z, Yamaura Y, Hailey DW, Neumann T, Shen DD, Thummel KE, Muczynski KA, Himmelfarb J, Kelly EJ. Development of a microphysiological model of human kidney proximal tubule function. Kidney Int 2016; 90:627–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Vunjak-Novakovic G, Bhatia S, Chen C, Hirschi K. HeLiVa platform: integrated heart-liver-vascular systems for drug testing in human health and disease. Stem Cell Res Ther 2013; 4 Suppl 1:S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Qu Y, Feric N, Pallotta I, Singh R, Sobbi R, Vargas HM. Inotropic assessment in engineered 3D cardiac tissues using human induced pluripotent stem cell-derived cardiomyocytes in the Biowire(TM) II platform. J Pharmacol Toxicol Methods 2020; 105:106886. [DOI] [PubMed] [Google Scholar]
- 137.Sutherland JJ, Jolly RA, Goldstein KM, Stevens JL. Assessing concordance of drug-induced transcriptional response in rodent liver and cultured hepatocytes. PLoS Comput Biol 2016; 12:e1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Low LA, Tagle DA. Tissue chips - innovative tools for drug development and disease modeling. Lab Chip 2017; 17:3026–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yeung CK, Koenig P, Countryman S, Thummel KE, Himmelfarb J, Kelly EJ. Tissue chips in space-challenges and opportunities. Clin Transl Sci 2020; 13:8–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Takebe T, Zhang B, Radisic M. Synergistic engineering: organoids meet organs-on-a-chip. Cell Stem Cell 2017; 21:297–300 [DOI] [PubMed] [Google Scholar]