Abstract
Cytokinesis, the final step of mitosis, is critical for maintaining the ploidy level of cells. Cytokinesis is a complex, highly regulated process and its failure can lead to genetic instability and apoptosis, contributing to the development of cancer. Human hepatocellular carcinoma is often accompanied by a high frequency of aneuploidy and the DNA ploidy pattern observed in human hepatocellular carcinoma results mostly from impairments in cytokinesis. Many key regulators of cytokinesis are abnormally expressed in human hepatocellular carcinoma, and their expression levels are often correlated with patient prognosis. Moreover, preclinical studies have demonstrated that the inhibition of key cytokinesis regulators can suppress the growth of human hepatocellular carcinoma. Here, we provide an overview of the current understanding of the signaling networks regulating cytokinesis, the key cytokinesis regulators involved in the initiation and development of human hepatocellular carcinoma, and their applications as potential diagnostic and therapeutic biomarkers.
Keywords: Hepatocellular carcinoma, cytokinesis, therapeutic targets
Impact statement
Cytokinesis is a complex, highly regulated process, and its failure can lead to genetic instability, apoptosis, and cancer. Abnormal expression of cytokinesis regulators has been widely detected in cancers, including HCC, indicating crucial roles for cytokinesis regulators in HCC diagnosis and therapy. Moreover, our laboratory recently reported that cytokinesis regulators, such as RACGAP1, ECT2, and PRC1, can act as oncogenic drivers for HCC early recurrence post-surgery. However, cytokinesis is a short and dynamic stage during mitosis, and cytokinesis regulators often exhibit versatile functions in multiple oncogenic signaling network. Therefore, we still know little regarding how to target cytokinesis regulators for HCC treatment. Here, we summarize the updates on the roles and small-molecule inhibitors of cytokinesis regulators in HCC, aiming to accelerate both basic and translational studies and focus more attention on this topic.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary liver cancer with over half a million cases diagnosed annually worldwide.1,2 The major risk factors for HCC are cirrhosis, chronic infection with hepatitis B virus (HBV) and hepatitis C virus (HCV), alcoholic liver disease, and nonalcoholic fatty liver disease.3,4
For early HCC, multiple treatment options, such as liver transplantation, surgical resection, radiofrequency ablation, transcatheter arterial chemoembolization (TACE), and systemic targeted chemotherapy, are available. 5 Unfortunately, the clinical symptoms of early HCC are atypical, and as a consequence, most HCC patients are diagnosed at an intermediate or advanced stage. Due to the scarcity of organ donors and the high rates of recurrence following resection, the prognosis of advanced HCC is dismal. Although sorafenib, a kinase inhibitor, is widely offered as the standard first-line therapy for advanced HCC, its efficacy remains unsatisfactory as a monotherapy, and novel combination therapies are being explored. 6 Hence, the outlook remains bleak for patients with advanced-stage HCC, with a median survival time <10 months and a 5-year survival rate of <5%. 7 There remains an immediate need for the development of novel therapeutic strategies to improve the outcome of advanced-stage HCC patients. Uncontrolled cell division is the most prominent feature of cancer cells, and this review focuses on cytokinesis, the final step of mitotic cell division, as a feasible avenue to identify potential novel targets to tackle HCC.
Cytokinesis: Concept and molecular mechanisms
The concept of cytokinesis
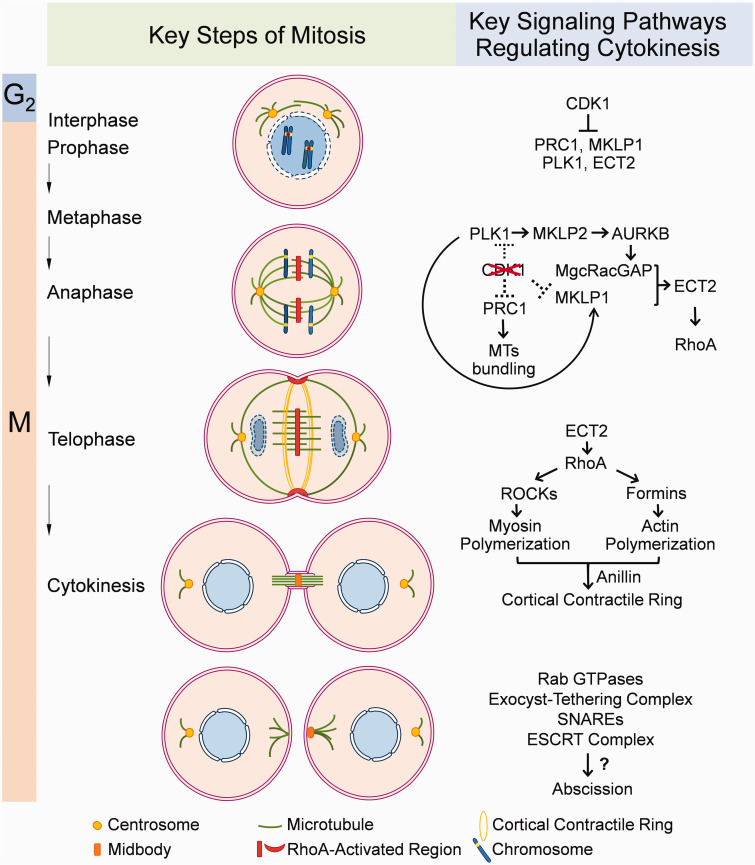
Cell division is the process by which a mother cell divides into two or more daughter cells. For eukaryotic somatic cells, this process is referred to as mitosis, which consists of nuclear division and cytoplasmic division. 8 Nuclear division includes five phases, namely interphase, prophase, metaphase, anaphase, and telophase, during which the replicated chromosomes are evenly distributed to the two poles of the spindle apparatus and enveloped by nuclear membranes to form two daughter nuclei. 9 Cytoplasmic division, which is referred to as cytokinesis, generally initiates at late stages of nuclear division and finishes shortly after the telophase. During cytokinesis, the cleavage furrow is formed along the cell division plane under the synergistic action of both the central spindle and the actomyosin contraction ring. The cleavage furrow is further deepened to shrink the overlapping microtubules (MTs) of the central spindle into a tight bundle, thus forming a narrow intracellular bridge connecting the two daughter cells. Eventually, the intracellular bridge is abscised, and the two daughter cells become completely detached from each other 10 (Figure 1).
Figure 1.
An overview of critical biological processes of cytokinesis and key signaling pathways involved in each process. (A color version of this figure is available in the online journal.)
In the human liver, approximately 30% of hepatocytes are polyploid due to endoreplication, cytokinesis failure, and cell fusion. 11 The biological importance of liver polyploidization remains unclear. Previously, scientists hypothesized that excess genetic material would provide a material basis for higher transcription and translation activities on a per cell basis. 12 However, microarray comparison on gene expression profiles of fluorescence-activated cell sorting-isolated diploid, tetraploid and octoploid hepatocytes revealed that the difference in RNA transcription was subtle. 13 In contrast, a study focused on stochastic production of mRNA from transcription sites (transcriptional bursts) demonstrated that liver polyploidy could dampen the intrinsic variability associated with transcriptional bursts, leading to more controlled gene expression. 14 More recent research on Cdk1-knockout livers, which produced a large proportion of the mononucleate polyploid hepatocytes following partial hepatectomy, showed that polyploidy promotes the anaerobic energy production by decreasing the expression of mitochondrial and de novo lipid biosynthesis genes and increasing the expression of glycolytic genes. 15 The altered metabolic profile and genetic instability due to excess genetic material should contribute to higher resistance to stresses such as chemical irritants, inflammation, and imbalanced metabolic pathways. Recently, in vivo lineage tracing in mouse models showed that polyploid hepatocytes readily formed liver tumors via frequent ploidy reduction. 16 Comprehensive analysis of the ploidy spectra in HCC specimens demonstrated that highly polyploid tumors are associated with a poor prognosis. 17 Considering the high frequency of polyploidy in the liver and its importance in cell proliferation, a thorough understanding of cytokinesis regulation might inspire novel diagnostic and therapeutic strategies for HCC.
Key regulators of cytokinesis
Cytokinesis consists of the following three critical steps: the initial formation of the cleavage furrow, deepening of the cleavage furrow, and abscission of the intracellular bridge. 18 All of these processes are precisely controlled by specific groups of kinases (Figure 1).
The initial formation of the cleavage furrow
The timing and position of cleavage furrow formation during cytokinesis are closely coupled with chromosome segregation during nuclear division, and both processes are highly dependent on the mitotic spindle. During the metaphase-to-anaphase transition, CDK1 (cyclin-dependent kinase 1) activity dramatically decreases, relieving the inhibition of the CDK1 substrates PRC1 (protein regulating cytokinesis 1), MKLP1 (mitotic kinesin-like protein 1), PLK1 (Polo-like kinase 1), and ECT2 (epithelial cell transforming 2)19,20 PRC1 can then promote microtubule bundling at the spindle midzone to form the central spindle, where MKLP1 and MgcRacGAP (also known as RACGAP1, Rac GTPase Activating Protein 1) assemble to form a heterotetrameric complex called centralspindlin. 21 PRC1 also recruits PLK1 (Polo-like kinase 1), which in turn activates MgcRacGAP for the recruitment of ECT2 to the central spindle. 22
The chromosomal passenger complex (CPC) also translocates from the centromeres to the central spindle during anaphase with the help of MKLP2 (Mitotic Kinesin-Like Protein 2), a plus-end-directed kinesin activated by PLK1. 23 The CPC is formed by the kinase module AURKB (Aurora B Kinase), a localization module consisting of the scaffolding protein INCENP (Inner Centromere Protein), Survivin, Borealin, and the guanine exchange factor TD-60 (Telophase Disk Protein of 60 KDa), which is not stably associated with the CPC. 24 The activity of AURKB is indispensable for the function of the central spindle and the centralspindlin complex since KIF2A (Kinesin Family Member 2 A), KIF4A (Kinesin Family Member 4 A), MKLP1, and MgcRacGAP are all substrates of AURKB. 25 The phosphorylation gradient created by AURKB along the midzone MTs provides a critical spatiotemporal cue for furrow positioning during anaphase. 26
Decreased CDK1 activity also leads to the removal of inhibitory phosphorylation on the myosin regulatory light chain. 27 Moreover, the phosphorylation of the centralspindlin complex by AURKB and PLK1 generates a docking site for ECT2 at the central spindle. 22 ECT2 then activates RhoA (Ras Homolog Family Member A), eventually leading to the assembly and recruitment of myosin II around the cell equator. 28 Myosin II interacts with actin filaments to promote node condensation to form the contractile ring. 29 On the other hand, myosin II accumulation is suppressed at regions of high astral MT density around both poles, thus generating a region of relatively low contractility around the poles. 30 This spatial and temporal regulation of Actomyosin contractility initiates the formation of the cleavage furrow in the equatorial region.
The deepening of the cleavage furrow
During telophase, the membrane-bound pool of centralspindlin recruits ECT2, which promotes RhoA activation at the equatorial plasma membrane. 30 Activated RhoA induces the activation of Rho-associated protein kinases (ROCKs) and formins which promote the polymerization of myosin and actin to form filaments. 31 Meanwhile, the scaffold protein anillin recruits F-actin, septins, myosin II, and more ECT2 to the cortex in a Rho-dependent manner. 32 Such a positive feedback loop leads to the fast assembly of the cortical contractile ring. Myosin II forms bipolar filaments to exert forces on actin filaments. Activated ROCKs increase the phosphorylation of the myosin light chains and eventually enhance myosin II contraction.33,34 As a result of myosin II contraction and ring component disassembly in the around-the-ring direction, the cortical contractile ring constricts. 35 Since the contractile ring is bound to the cell membrane with connecting proteins such as anillin, its constriction leads to deepening of the cleavage furrow until a narrow intracellular bridge separating the two daughter cells is formed. 36
The abscission of the intracellular bridge
After telophase, nuclear envelopes have formed within the daughter cells, but the cells are still connected via the intracellular bridge. The intracellular bridge contains extremely densely packed MTs and a structure named the midbody. The connection between the midbody and the plasma membrane is mainly mediated by an interaction between the C1 domain of the centralspindlin subunit MgcRacGAP and polyanionic phosphoinositide lipids within the plasma membrane. 37 The intracellular bridge is abscised nonsymmetrically on either side of the midbody marking the completion of cytokinesis. The detailed mechanisms underlying bridge abscission are still not fully understood. However, recent evidence suggests that vesicle trafficking and membrane fusion mediated by Rab GTPases, the exocyst-tethering complex, SNAREs (Syntaxin-2 and endobrevin), and the ESCRT (endosomal sorting) complex are critical for this process. 10
Cytokinesis regulators as therapeutic targets in HCC
Cytokinesis failure generates tetraploid cells with four copies of each chromosome (4 N) and two centrosomes. 38 Most tetraploid cells undergo apoptosis, but some tetraploid cells are able to escape intrinsic regulation through either the loss of critical tumor suppressors, such as p53 and p21, or overexpression of oncogenes, such as Bcl-2.39–41 Since their genetic information is highly redundant, tetraploid cells are more tolerant to genetic damage, accumulating mutations, insertions, and deletions in their genome.42,43 Additionally, redundant centrosomes interfere with spindle formation during the next round of cell division, thus resulting in the generation of aneuploid progeny.44,45 Therefore, cytokinesis failure is a double-edged sword for cancer. First, cytokinesis failure can induce apoptosis in cancer cells, suggesting that manual intervention in cytokinesis could be a potential strategy to kill cancer cells. Second, a small fraction of the tetraploid intermediate could be the source of chromatin instability (CIN). 46 Occasionally some surviving tetraploid cells and their aneuploid progeny gain a survival advantage from their genetic variations, eventually leading to malignant transformation.38,47
According to the NCI Cancer Genome Anatomy Project, approximately 80% of HCC tissues are aneuploid, with dysregulation of many key proteins involved in cytokinesis. 48 For example, a whole-genome and whole-exome sequencing study demonstrated that the HCC-C1 subtype could be identified due to mitotic checkpoint defects associated with mutations in PLK1 and ECT2. 49 Therefore, key regulators of cytokinesis could be exploited as potential diagnostic biomarkers and therapeutic targets in HCC, which are topics discussed later in this review (Table 1).
Table 1.
A summary of the expression status and representative inhibitors of key regulators for cytokinesis as therapeutic targets of HCC.
| Protein name | Expression status in HCC | Representative inhibitors | Ref. |
|---|---|---|---|
| CDK1 | Up-regulated | JNJ-7706621, RO3306 | 50–57 |
| PRC1 | Up-regulated | N.A. | 58–61 |
| KIF4A | Up-regulated | N.A. | 25 , 62 , 63 |
| KIF4B | Up-regulated | N.A. | 62 , 64 |
| MKLP1 (KIF23) | Up-regulated | N.A. | 64–66 |
| MKLP2 (KIF20A) | Up-regulated | Paprotrain, BKS0349 | 67–71 |
| PLK1 | Up-regulated | DAP-81, BI 2536, BI 6727, Ro3280, TAK-960, NMS-P937, Poloxin, Poloxipan, Purpurogallin | 72–76 |
| PLK4 | LOH occurrs at the PLK4 locus in 50% HCC; protein level of PLK4 is significantly higher in HCC | CFI-400945, Centrinone/centrinone B, YLT-11 | 77–81 |
| AURKB | Up-regulated | VE-465, AZD1152-HQPA, AZD115229, MK0457, Deguelin | 82–90 |
| Survivin | Up-regulated | YM155, WM-127, Etoposide | 91–99 |
| ECT2 | Up-regulated | N.A. | 100–102 |
| RhoA | Up-regulated | L07, Y16, Zoledronic acid, CCG-203971, and CCG-1423 | 103–114 |
| MgcRacGAP | Up-regulated | MINC1 | 115–118 |
CDK1
CDK1 is one of the most important and widely studied regulators of nuclear division and cytokinesis in HCC. 119 High CDK1 activity is required for proper assembly of the mitotic apparatus and the alignment of chromosomes, while CDK1 inhibition is a prerequisite for the initiation of cytokinesis. 120 Therefore, inhibition of CDK1 can normalize both critical processes of mitosis. It has been demonstrated that CDK1 is highly expressed in HCC tissues compared to normal tissues at both the RNA and protein levels, which contributes to more active nuclear division but less active cytokinesis.50–52,121 Significant upregulation of the CDK1 mRNA level can be observed in very early HCC tissues, compared to cirrhotic liver tissues, thus making CDK1 a potential diagnostic marker. 53 It belongs to the serine/threonine kinase family, and therefore contains a catalytic kinase subunit suitable for specific targeting with drug-like small molecules. 54
CDK1 inhibitors have been tested for HCC treatment due to the overexpression status of CDK1 in HCC. The therapeutic effects of these inhibitors are mainly attributed to the induction of mitosis failure, the suppression of kinase activity, and the normalization of cytokinesis. JNJ-7706621 is a pan-inhibitor of CDKs and aurora kinases. Danhier et al. reported that the combination of JNJ-7706621 and paclitaxel could synergically suppress the growth of transplantable liver cancer in mice. 55 ATP-competitive RO3306 is a specific inhibitor of CDK1. Wu et al. reported that combining RO3306 with sorafenib could potently suppress the growth of patient-derived HCC xenografts by reducing the stemness of liver cancer stem cells via inhibition of the CDK1/PDK1/β-Catenin signaling pathway. 121 We previously demonstrated that CDK1 could phosphorylate B-cell CLL/lymphoma 9 (BCL9) at Thr 172 to promote mitotic Wnt signaling activity and the growth of HCC cells. 56 Moreover, a CDK1 siRNA interference study demonstrated that the inhibition of cell proliferation resulted in the apoptosis of HCC cells, suggesting that CDK1 could be a promising therapeutic target in HCC 57 ,.57,58
PRC1
As previously described, PRC1 plays a vital role in the bundling of MTs during cytokinesis, and its overexpression in HCC has been documented by several independent research groups.59–61 Significant upregulation of the PRC1 mRNA level can be observed in very early HCC tissues, compared to cirrhotic liver tissues, thus making it a potential diagnostic marker. 53 Higher PRC1 expression is significantly correlated with worse tumor staging and a worse prognosis. 60 HCC cells overexpressing PRC1 exhibit strong resistance against conventional chemotherapeutic reagents such as 5-Fu and Taxol. 59 In experimental models of HCC, knockdown of PRC1 by an adenovirus could remarkably sensitize HCC cells to Taxol. 61 Since PRC1 regulates the cell cycle through protein-protein interactions and not by kinase activity, its microtubule-binding domain could provide an ideal target for the development of small-molecule inhibitors. Currently there is no commercially available small-molecule drug targeting PRC1, and its inhibition is generally achieved by experimental genetic tools, such as shRNA and microRNA.60,122
The kinesin superfamily of proteins
Kinesin superfamily motor proteins facilitate the transport of mRNAs, protein complexes, and organelles along microtubules in an ATP-dependent manner. 123 Functional screening studies with esiRNA libraries indicated that at least four kinesins are involved in cytokinesis, including KIF4A, KIF4B, MKLP1, and MKLP2. 69
Meta-analysis of the Oncomine database suggested that KIF4A expression was upregulated in HCC, and that a higher KIF4A level was correlated with poorer overall survival and disease-free survival. Overexpression of KIF4A led to faster proliferation of HCC cells, while depletion of KIF4A resulted in abnormal chromosome segregation followed by apoptosis. 63 On the other hand, a higher KIF4B RNA level was also observed in HCC tissues compared to normal liver tissues but this difference was not significantly correlated with the prognosis of HCC patients. 64
The kinesin-6 family motor protein MKLP1, also known as KIF23, is a key regulator of cytokinesis. 65 RT-PCR analysis showed that KIF23 was frequently expressed in HCC tissues but not in normal liver tissues. KIF23 has two splicing isoforms, namely KIF23 V1, which is longer and localized in the nucleus, and KIF23 V2, which is shorter and distributed in the cytoplasm. Interestingly, KIF23 V1 was detected in 57.6% of HCC specimens, while KIF23 V2 was detected in 94.4% of HCC specimens. Prognostic analysis suggested that elevated KIF23 V1 expression was correlated with longer five-year survival, while KIF23 V2 was not significantly associated with three- or five-year survival. 66 In a more recent bioinformatic study in which a 14-gene signature was developed to predict the prognosis of HCC patients based on data from a cohort in The Cancer Genome Atlas (TCGA), a higher KIF23 RNA level contributed to a higher risk score, which was correlated with a poorer prognosis. 67 These conflicting observations suggest that detailed mechanistic analysis is urgently needed to understand the functions of KIF23 and its two splicing variants.
MKLP2 (also known as KIF20A), which is also a kinesin-6 family motor protein, mediates the recruitment of AURKB to the equatorial cortex to promote furrow ingression during cytokinesis. 68 MKLP2 mRNA is undetectable in normal human hepatocytes, but it accumulates in a large proportion of human HCC cells, with the highest expression observed in tumors with genomic instability. 69 An analysis performed with the TCGA database suggested that higher MKLP2 expression was correlated with poorer overall survival and relapse-free survival for HCC patients. 70
A cell-permeable acrylonitrile compound named Paprotrain has been developed to inhibit the ATPase activity of MKLP2. It demonstrated antitumor activity against ovarian clear cell carcinoma cells. 71 More recently, BKS0349, a 10-fold more potent analog of Paprotrain, was reported to be able to reduce the number and size of endometriotic lesions in an experimental mouse model of ovarian endometriosis. 124 However, the therapeutic effects of Paprotrain and BKS0349 on HCC have not been evaluated.
Polo-like kinases
The appropriate spatial-temporal regulation of PLK1 activity is critical for cytokinesis. Both overexpression and loss of expression can contribute to malignant transformation, depending on stage of disease and the genetic background of the tissue. For example, Gray et al. reported that most pancreatic cancer specimens showed increased PLK1 expression, and that PLK1 knockdown induced G2/M cell cycle arrest and a drastic reduction in proliferation rates in pancreatic cancer cells. 125 Similar observations suggesting PLK1 as an oncoprotein have been reported for most types of solid tumors, such as lung cancer, breast cancer, and colorectal cancer.126–129 On the other hand, PLK1 overexpression prevented the development of Kras-induced and Her2-induced mammary gland tumors via CIN-induced apoptosis in mouse models, and PLK1 overexpression correlated with improved survival in ER-negative and HER2-positive breast cancer subtypes in a TCGA cohort. 130 Moreover, PLK1 inhibition promoted the development of adenomatous polyps in two independent ApcMin/+ mouse models, suggesting its tumor-suppressive potential in APC-truncated colon cancer cells. 72
PLK1 has been reported to be significantly overexpressed in HCC tissues compared to corresponding normal liver tissues in a number of different cohorts and could be used as an independent marker for predicting prognosis.73–75 Many natural and synthetic compounds have been identified to inhibit the kinase activity of PLK1, including ATP competitors, such as DAP-81, BI 2536, BI 6727(volasertib), Ro3280, TAK-960 and NMS-P937, as well as inhibitors of POLO-Box Domain, such as Poloxin, Poloxipan, and Purpurogallin, and these compounds exert a good antitumor effect by inducing apoptosis in vitro. 76 In a randomized phase 2 trial (NCT00804856), the combination of BI 6727 and low-dose cytarabine (LDAC) significantly improved the response rate of acute myelocytic leukemia patients unsuitable for intensive induction chemotherapy compared to LDAC alone. 131 However, the effect of PLK1 inhibitors has not been systematically evaluated in HCC.
Polo-like kinase 4 (PLK4) is another serine/threonine kinase mainly localized at the centriole, spindle midzone, and midbody, and it is critical for centriole duplication and the ECT2 mediated activation of RhoA during cytokinesis. 132 A genetic study showed that mice with PLK4 haploinsufficiency exhibited a significantly increased incidence of spontaneous liver and lung cancers. 77 A later analysis of clinical specimens demonstrated that loss of heterozygosity (LOH) occurred at the PLK4 locus in 50% of HCC cases, resulting in reduced PLK4 mRNA expression. 78 These studies suggest that PLK4 protects hepatoma from malignant transformation. Paradoxically, a more recent study showed that the protein level of PLK4 was significantly higher in HCC tissues than in healthy liver tissues, and that knockdown of PLK4 remarkably reduced the growth of HCC cells in vitro and in vivo. 79 In another study analyzing SNPs of the PLK4 gene locus, the functional SNP rs3811741 (G/A) was associated with a higher risk of HCC. This SNP, located on the enhancer of PLK4, was strongly modified by histone H3K4Me1 and H3K27Ac. This SNP positively regulates PLK4 transcription, thus promoting centrosome amplification and cell proliferation. 80 These studies indicate that PLK4 is an oncoprotein. Such contradictions in the data on the roles of PLK4 in HCC among studies might be caused by the different observation indexes utilized in different studies. Since the degradation of PLK4 is promoted by autophosphorylation, the protein level of PLK4 might not be linearly dependent on the copy number of the gene locus or mRNA transcript. 81 Nevertheless, a comprehensive analysis to understand the role of PLK4 in the development and treatment of HCC is still pending.
Several PLK4 inhibitors have been developed as potential therapeutic agents for cancer, such as CFI-400945, centrinone/centrinone B and YLT-11. 133 In vitro drug sensitivity tests showed that HCC cell lines with higher PLK4 expression, such as Huh7 and BEL‐7402, were more sensitive to CFI-400945 than cell lines with lower PLK expression, such as MHCC-97L and MHCC-97H. 96 However, considering that both a decrease and an increase in PLK4 activity might contribute to the development of HCC, the therapeutic effects and safety profiles of PLK4 inhibitors need careful evaluation in vivo.
The CPC (AURKB and survivin)
The CPC plays important roles during both nuclear division and cytoplasmic division, including the regulation of the mitotic checkpoint, the assembly of spindle MTs, and the recruitment and activation of key proteins involved in cytokinesis. 25 Perturbation of the CPC leads to chromosome segregation errors and cytokinesis failure. 134 Hence, analysis of CPC components could provide further insights into the role of cytokinesis dysregulation in HCC.
The kinase module of CPC is the serine/threonine kinase AURKB, which is localized in the centromeres during early mitosis and then at the spindle midzone after anaphase.82,135 Several studies have shown that AURKB expression is significantly higher in HCC tissues than in noncancerous tissues, and that its expression level is associated with the tumor grade and prognosis of cancer patients, indicating that AURKB could be an independent prognostic marker for HCC.83–86
Many selective inhibitors have been developed targeting AURKB, such as VE-465, AZD1152-HQPA, AZD115229, MK0457, and Deguelin.87–90 Lin et al. showed that AZD1152-HQPA induced proliferation blockade, histone H3 dephosphorylation, cell cycle disturbance, and apoptosis in HCC cell lines in vitro. 84 Benten et al. also reported that PHA-739358, an aurora A/B/C kinase inhibitor (currently undergoing phase II clinical trials), suppressed the growth of HCC cells. 136 These preclinical studies strongly suggest that AURKB inhibitors can act as promising therapeutic agents for HCC.
In CPC, Survivin is an important mediator of the centromere and midbody docking of AURKB. 137 In addition to its role in mitosis, Survivin plays multiple regulatory roles in critical biological processes involved in malignant transformation, such as apoptosis, autophagy, epithelial-to-mesenchymal transition, and angiogenesis.91,138–140 Normal hepatocytes express very low levels of Survivin, while Survivin mRNA and protein are frequently detected in HCC specimens. 92 Overexpression of Survivin promotes cell proliferation and drug resistance in HCC.92,93 In contrast, a microRNA miR-203 can suppress the expression of Survivin, thus leading to reduced proliferation of HCC cells. 94 These observations suggest that Survivin is a promising therapeutic target in HCC.
The best-studied Survivin suppressor is YM155. Instead of directly binding with Survivin, YM155 disrupts the ILF3/p54 complex, which is necessary for the transcription of Survivin. 95 The safety profile of YM155 has been evaluated in a phase I safety and pharmacokinetic study in patients with EGFR TKI-refractory advanced non-small cell lung cancer (NSCLC). 96 Xia et al. reported that YM155 exerted significantly better therapeutic effect than sorafenib in an orthotopic mouse model established with HCC cells exhibiting elevated Survivin and p-Survivin expression. 93 YM155 has been shown to sensitize HCC cells to the BCL2 family inhibitor ABT-263. 97 In addition to YM155, WM-127 (a matrine derivative) was identified to suppress the expression of Survivin in a Survivin-targeted drug screening platform, and it inhibited the growth of HCC cells both in vitro and in vivo. 98 Moreover, etoposide was identified as a compound blocking the Survivin-Borealin interaction in a high-throughput screen based on bimolecular fluorescence complementation (BiFC) technology. 99 Etoposide has been widely used as a chemotherapeutic agent to inhibit topoisomerase II activity, and it is appealing to explore new applications for this drug related to regulating cytokinesis. 100
ECT2
ECT2 is one of the most important guanine nucleotide exchange factors involved in cytokinesis. ECT2 is significantly overexpressed in HCC and correlated with early recurrence of HCC 125 . Knockdown of ECT2 results in reduced HCC cell division and migration in vitro, and reduced xenograft growth in vivo. 101 ECT2 can be negatively regulated by miR-490-5p in HCC cells. 102 Specific inhibitors targeting pleckstrin homology (PH) domain of ECT2 have been developed and preliminarily tested in non-small cell lung cancer for their anticancer activity, as reported by a conference abstract. 141 However, neither intensive evaluation nor mechanistic study has been conducted for these compounds yet. There remains an urgent need for specific inhibitors of ECT2.
RhoA
RhoA is a small GTPase protein that shuttles between an inactive GDP-bound state and an active GTP-bound state and exhibits intrinsic GTPase activities. 142 RhoA-GTP activates its effector proteins, such as ROCKs and Formins, by displacing their autoinhibitory domains, thus leading to the phosphorylation of their substrates. 143 Therefore, RhoA plays indispensable roles in actin organization, myosin contractility, cell cycle maintenance, and cellular morphological polarization. 103 Malignant HCC tissues frequently express high RhoA mRNA and protein levels, whereas benign liver tissues have a minimal level, and a high level of RhoA is significantly associated with a poor prognosis.104–107 Knockdown of RhoA can sensitize HCC cells to TNF-α-induced apoptosis. 108
Several small-molecule compounds have been developed to inhibit RhoA activity, such as HL07, Y16, zoledronic acid, CCG-203971, and CCG-1423.109–112 Zoledronic acid was shown to delay disease progression of HCC bone metastases in a small-scale clinical trial.113,114 It was also reported that an advanced HCC patient with bone metastasis showed a complete response after sorafenib therapy plus zoledronic acid in a case report. 115 The applications of RhoA inhibitors for the treatment of HCC remain to be further explored.
MgcRacGAP
As the critical component of centralspindlin, MgcRacGAP is highly expressed in HCC tissues compared to healthy liver tissues, and its expression level correlates with early recurrence in HCC patients. 116 Silencing MgcRacGAP inhibits cell migration, invasion, and proliferation.116,117 MiR-15-5p suppresses the expression of MgcRacGAP. 118 In a high-throughput screen of 342,046 compounds, MINC1 was identified as a selective inhibitor of MgcRacGAP, and cell experiments showed that MINC1 treatment caused cytokinesis failure and multinucleation. 144 The antitumor effects and safety profile of MINC1 await further comprehensive evaluations.
Conclusions and perspective
Cytokinesis is critical for cell division and requires precise spatiotemporal regulation. Although some studies have verified the new roles of cytokinesis regulators in signaling regulation, the complete picture of the functions of cytokinesis regulators in every unique cell cycle stage has still not been elucidated. More precise cellular and molecular mechanistic studies should be performed to dissect several key points:
What are the novel roles of cytokinesis regulators in cellular signaling regulation during interphase in HCC cells? For example, it is known that Survivin plays a vital role in the suppression of apoptotic signaling, 145 and that PRC1 reinforces Wnt signalling. 60 The fact that these molecules have critical functions in both resting cells and proliferating cells increases the success rate of targeting them for HCC therapy.
What is the potential regulatory mechanism of the redistribution of cytokinesis regulators from key mitotic machines to the nucleus during the cytokinesis process? Protein functions are closely related to subcellular localization. It is fascinating how cytokinesis regulators translocate to different compartments during the progression of the cell cycle. More studies in this field could lead us to a deeper understanding of the precise regulatory network during mitosis and cytokinesis.
Are there any factors with nonclassic cancer-specific roles in cytokinesis regulation? Such an Achilles' heel might be the ideal therapeutic target for precise targeting of HCC cells.
Do the expression level and kinase activity of cytokinesis regulators change after conventional HCC therapies? Are these changes related to resistance to conventional therapies? For example, in vitro empirical evidence has demonstrated that PRC1, Survivin, and ECT2 contribute to the development of chemoresistance, even though mechanistic studies have indicated signaling pathways other than cytokinesis dysregulation.59,92,93,146 One possible explanation may be that the drug treatment schemes and durations used in such in vitro experiments using HCC cell lines are obviously different from the real clinical condition. Therefore, these short-term in vitro studies are suitable for the evaluation of direct cytotoxicity but impropriate for analyzing the development of drug resistance which involves changes in genetic materials due to cytokinesis dysregulation and the selection of clones with a survival advantage. Systematic comparisons performed with clinical HCC specimens or animal models are still urgently needed to validate the roles of these cytokinesis regulators in chemoresistance
What are the potential risks of targeting cytokinesis regulators for HCC therapy? Most of the key cytokinesis regulators are highly expressed and associated with a poor prognosis, as proven by a retrospective study of pathological samples of HCC. Although inhibition of cytokinesis regulators suppresses the proliferation of HCC cells in preclinical models, till now none of these specific inhibitors targeting cytokinesis regulators have been approved for HCC treatment. The clinical benefits and potential risks associated with long-term inhibition of these therapeutic targets have not been systematically evaluated in HCC patients. Recently, SP600125 (a c-Jun N-terminal kinase (JNK) inhibitor) was reported to suppress CDK1 activity, but it led to endoreplication in cells in the G2 phase in a CDK2-dependent manner, suggesting the potential risk of increasing polyploidy. 147 Compared to other organs, the liver has a higher tolerance for tetraploidy, so caution must be taken when applying information obtained from other organs to HCC.
Overall, numerous preclinical studies have demonstrated cytokinesis regulators to be promising prognostic biomarkers and therapeutic targets in HCC, but much more effort must be dedicated to this field to comprehensively understand the altered signaling network and identify the Achilles' heel during cytokinesis for the treatment of HCC.
Footnotes
AUTHORS’ CONTRIBUTIONS: All authors participated in the design, literature review and preparation of the manuscript. KMH, and JC supervised the whole process and edited the manuscript. YQ and YP wrote the manuscript. ML and MR prepared the table and figure.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: This research was supported by grants from the Zhejiang Provincial Natural Science Foundation of China for Distinguished Young Scholars (LR21H160001), Start-up Grant of HZNU (4125C5021820470), National Natural Science Foundation of China (81802338, 82072646) (to JC); National Natural Science Foundation of China (81903143) (to YQ).
ORCID iD: Kam M Hui https://orcid.org/0000-0003-1820-1399
References
- 1.Mittal S, El-Serag HB. Epidemiology of hepatocellular carcinoma: consider the population. J Clin Gastroenterol 2013; 47: S2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68:394–424 [DOI] [PubMed] [Google Scholar]
- 3.Park JW, Chen M, Colombo M, Roberts LR, Schwartz M, Chen PJ, Kudo M, Johnson P, Wagner S, Orsini LS, Sherman M. Global patterns of hepatocellular carcinoma management from diagnosis to death: the BRIDGE study. Liver Int 2015; 35:2155–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Massoud O, Charlton M. Nonalcoholic fatty liver disease/nonalcoholic steatohepatitis and hepatocellular carcinoma. Clin Liver Dis 2018; 22:201–11 [DOI] [PubMed] [Google Scholar]
- 5.Llovet JM. Updated treatment approach to hepatocellular carcinoma. J Gastroenterol 2005; 40:225–35 [DOI] [PubMed] [Google Scholar]
- 6.Ikeda M, Morizane C, Ueno M, Okusaka T, Ishii H, Furuse J. Chemotherapy for hepatocellular carcinoma: current status and future perspectives. Jpn J Clin Oncol 2018; 48:103–14 [DOI] [PubMed] [Google Scholar]
- 7.Sarveazad A, Agah S, Babahajian A, Amini N, Bahardoust M. Predictors of 5 year survival rate in hepatocellular carcinoma patients. J Res Med Sci 2019; 24:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burke DJ, Church D. Protein synthesis requirements for nuclear division, cytokinesis, and cell separation in Saccharomyces cerevisiae. Mol Cell Biol 1991; 11:3691–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pickett-Heaps JD, Northcote DH. Organization of microtubules and endoplasmic reticulum during mitosis and cytokinesis in wheat meristems. J Cell Sci 1966; 1:109–20 [DOI] [PubMed] [Google Scholar]
- 10.Barr FA, Gruneberg U. Cytokinesis: placing and making the final cut. Cell 2007; 131:847–60 [DOI] [PubMed] [Google Scholar]
- 11.Donne R, Saroul-Aïnama M, Cordier P, Celton-Morizur S, Desdouets C. Polyploidy in liver development, homeostasis and disease. Nat Rev Gastroenterol Hepatol 2020; 17:391–405 [DOI] [PubMed] [Google Scholar]
- 12.Schoenfelder KP, Fox DT. The expanding implications of polyploidy. J Cell Biol 2015; 209:485–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu P, Prost S, Caldwell H, Tugwood JD, Betton GR, Harrison DJ. Microarray analysis of gene expression of mouse hepatocytes of different ploidy. Mamm Genome 2007; 18:617–26 [DOI] [PubMed] [Google Scholar]
- 14.Bahar Halpern K, Tanami S, Landen S, Chapal M, Szlak L, Hutzler A, Nizhberg A, Itzkovitz S. Bursty gene expression in the intact mammalian liver. Mol Cell 2015; 58:147–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miettinen TP, Pessa HK, Caldez MJ, Fuhrer T, Diril MK, Sauer U, Kaldis P, Björklund M. Identification of transcriptional and metabolic programs related to mammalian cell size. Curr Biol 2014; 24:598–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto T, Wakefield L, Peters A, Peto M, Spellman P, Grompe M. Proliferative polyploid cells give rise to tumors via ploidy reduction. Nat Commun 2021; 12:646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bou-Nader M, Caruso S, Donne R, Celton-Morizur S, Calderaro J, Gentric G, Cadoux M, L’Hermitte A, Klein C, Guilbert T, Albuquerque M, Couchy G, Paradis V, Couty J-P, Zucman-Rossi J, Desdouets C. Polyploidy spectrum: a new marker in HCC classification. Gut 2020; 69:355–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verma V, Mogilner A. Classical and emerging regulatory mechanisms of cytokinesis in animal cells. Biology 2019; 8:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fededa JP, Gerlich DW. Molecular control of animal cell cytokinesis. Nat Cell Biol 2012; 14:440–47 [DOI] [PubMed] [Google Scholar]
- 20.Niiya F, Tatsumoto T, Lee KS, Miki T. Phosphorylation of the cytokinesis regulator ECT2 at G2/M phase stimulates association of the mitotic kinase Plk1 and accumulation of GTP-bound RhoA. Oncogene 2006; 25:827–37 [DOI] [PubMed] [Google Scholar]
- 21.Lee K-Y, Esmaeili B, Zealley B, Mishima M. Direct interaction between centralspindlin and PRC1 reinforces mechanical resilience of the central spindle. Nat Commun 2015; 6:7290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim H, Guo F, Brahma S, Xing Y, Burkard ME. Centralspindlin assembly and 2 phosphorylations on MgcRacGAP by polo-like kinase 1 initiate Ect2 binding in early cytokinesis. Cell Cycle 2014; 13:2952–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neef Rd Preisinger C, Sutcliffe J, Kopajtich R, Nigg EA, Mayer TU, Barr FA. Phosphorylation of mitotic kinesin-like protein 2 by polo-like kinase 1 is required for cytokinesis. J Cell Biol 2003; 162:863–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carmena M, Wheelock M, Funabiki H, Earnshaw WC. The chromosomal passenger complex (CPC): from easy rider to the godfather of mitosis. Nat Rev Mol Cell Biol 2012; 13:789–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kitagawa M, Lee SH. The chromosomal passenger complex (CPC) as a key orchestrator of orderly mitotic exit and cytokinesis. Front Cell Dev Biol 2015; 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zaytsev AV, Segura-Peña D, Godzi M, Calderon A, Ballister ER, Stamatov R, Mayo AM, Peterson L, Black BE, Ataullakhanov FI, Lampson MA, Grishchuk EL. Bistability of a coupled Aurora B kinase-phosphatase system in cell division. Elife 2016; 5:e10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wheatley SP, Hinchcliffe EH, Glotzer M, Hyman AA, Sluder G., Wang Yl. CDK1 inactivation regulates anaphase spindle dynamics and cytokinesis in vivo. J Cell Biol 1997; 138:385–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Basant A, Glotzer M. Spatiotemporal regulation of RhoA during cytokinesis. Curr Biol 2018; 28:R570–R80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laporte D, Zhao R, Wu J-Q. Mechanisms of contractile-ring assembly in fission yeast and beyond. Semin Cell Dev Biol 2010; 21:892–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werner M, Munro E, Glotzer M. Astral signals spatially bias cortical myosin recruitment to break symmetry and promote cytokinesis. Curr Biol 2007; 17:1286–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dekraker C, Boucher E, Mandato CA. Regulation and assembly of actomyosin contractile rings in cytokinesis and cell repair. Anat Rec 2018; 301:2051–66 [DOI] [PubMed] [Google Scholar]
- 32.Hickson GRX, O'Farrell PH. Anillin: a pivotal organizer of the cytokinetic machinery. Biochem Soc Trans 2008; 36:439–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mishra M, Kashiwazaki J, Takagi T, Srinivasan R, Huang Y, Balasubramanian MK, Mabuchi I. In vitro contraction of cytokinetic ring depends on myosin II but not on actin dynamics. Nat Cell Biol 2013; 15:853–59 [DOI] [PubMed] [Google Scholar]
- 34.Matsumura F. Regulation of myosin II during cytokinesis in higher eukaryotes. Trends Cell Biol 2005; 15:371–77 [DOI] [PubMed] [Google Scholar]
- 35.Khaliullin RN, Green RA, Shi LZ, Gomez-Cavazos JS, Berns MW, Desai A. A positive-feedback-based mechanism for constriction rate acceleration during cytokinesis in Caenorhabditis elegans. Elife 2018; 7:e36073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun L, Guan R, Lee IJ, Liu Y, Chen M, Wang J, Wu J-Q, Chen Z. Mechanistic insights into the anchorage of the contractile ring by anillin and Mid1. Dev Cell 2015; 33:413–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lekomtsev S, Su KC, Pye VE, Blight K, Sundaramoorthy S, Takaki T, Collinson LM, Cherepanov P, Divecha N, Petronczki M. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature 2012; 492:276–9 [DOI] [PubMed] [Google Scholar]
- 38.Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005; 437:1043–7 [DOI] [PubMed] [Google Scholar]
- 39.Cross SM, Sanchez CA, Morgan CA, Schimke MK, Ramel S, Idzerda RL, Raskind WH, Reid BJ. A p53-dependent mouse spindle checkpoint. Science 1995; 267:1353–6 [DOI] [PubMed] [Google Scholar]
- 40.Castedo M, Coquelle A, Vivet S, Vitale I, Kauffmann A, Dessen P, Pequignot MO, Casares N, Valent A, Mouhamad S, Schmitt E, Modjtahedi N, Vainchenker W, Zitvogel L, Lazar V, Garrido C, Kroemer G. Apoptosis regulation in tetraploid cancer cells. EMBO J 2006; 25:2584–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shen Y-J, DeLong CJ, Tercé F, Kute T, Willingham MC, Pettenati MJ, Cui Z. Polyploid formation via chromosome duplication induced by CTP:Phosphocholine cytidylyltransferase deficiency and bcl-2 overexpression: identification of two novel endogenous factors. J Histochem Cytochem 2005; 53:725–33 [DOI] [PubMed] [Google Scholar]
- 42.Castedo M, Coquelle A, Vitale I, Vivet S, Mouhamad S, Viaud S, Zitvogel L, Kroemer G. Selective resistance of tetraploid cancer cells against DNA damage-induced apoptosis. Ann N Y Acad Sci 2006; 1090:35–49 [DOI] [PubMed] [Google Scholar]
- 43.Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Grönroos E, Endesfelder D, Joshi T, Mouradov D, Gibbs P, Ward RL, Hawkins NJ, Szallasi Z, Sieber OM, Swanton C. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov 2014; 4:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gönczy P. Centrosomes and cancer: revisiting a long-standing relationship. Nat Rev Cancer 2015; 15:639–52 [DOI] [PubMed] [Google Scholar]
- 45.Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev 2009; 28:85–98 [DOI] [PubMed] [Google Scholar]
- 46.Storchova Z, Pellman D. From polyploidy to aneuploidy, genome instability and cancer. Nat Rev Mol Cell Biol 2004; 5:45–54 [DOI] [PubMed] [Google Scholar]
- 47.Castedo M, Senovilla L, Vitale I, Kroemer G. Tetraploid cancer cell precursors in ovarian carcinoma. Cell Cycle 2012; 11:3157–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duijf PHG, Schultz N, Benezra R. Cancer cells preferentially lose small chromosomes. Int J Cancer 2013; 132:2316–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaisaingmongkol J, Budhu A, Dang H, Rabibhadana S, Pupacdi B, Kwon SM, Forgues M, Pomyen Y, Bhudhisawasdi V, Lertprasertsuke N, Chotirosniramit A, Pairojkul C, Auewarakul CU, Sricharunrat T, Phornphutkul K, Sangrajrang S, Cam M, He P, Hewitt SM, Ylaya K, Wu X, Andersen JB, Thorgeirsson SS, Waterfall JJ, Zhu YJ, Walling J, Stevenson HS, Edelman D, Meltzer PS, Loffredo CA, Hama N, Shibata T, Wiltrout RH, Harris CC, Mahidol C, Ruchirawat M, Wang XW. Common molecular subtypes among Asian hepatocellular carcinoma and cholangiocarcinoma. Cancer Cell 2017; 32:57–70.e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jin J, Xu H, Li W, Xu X, Liu H, Wei F. LINC00346 acts as a competing endogenous RNA regulating development of hepatocellular carcinoma via modulating CDK1/CCNB1 axis. Front Bioeng Biotechnol 2020; 8:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang X, Wang L, Yan Y. Identification of potential key genes and pathways in hepatitis B virus-associated hepatocellular carcinoma by bioinformatics analyses. Oncol Lett 2020; 19:3477–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang WX, Pan YY, You CG. CDK1, CCNB1, CDC20, BUB1, MAD2L1, MCM3, BUB1B, MCM2, and RFC4 may be potential therapeutic targets for hepatocellular carcinoma using integrated bioinformatic analysis. Biomed Res Int 2019; 2019:1245072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu M, Liu Z, Li X, Zhang A, Lin D, Li N. Analysis of potential key genes in very early hepatocellular carcinoma. World J Surg Oncol 2019; 17:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Brown NR, Korolchuk S, Martin MP, Stanley WA, Moukhametzianov R, Noble MEM, Endicott JA. CDK1 structures reveal conserved and unique features of the essential cell cycle CDK. Nat Commun 2015; 6:6769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Danhier F, Ucakar B, Magotteaux N, Brewster ME, Préat V. Active and passive tumor targeting of a novel poorly soluble cyclin dependent kinase inhibitor, JNJ-7706621. Int J Pharm 2010; 392:20–8 [DOI] [PubMed] [Google Scholar]
- 56.Chen J, Rajasekaran M, Xia H, Kong SN, Deivasigamani A, Sekar K, Gao H, Swa HL, Gunaratne J, Ooi LL, Xie T, Hong W, Hui KM. CDK1-mediated BCL9 phosphorylation inhibits clathrin to promote mitotic wnt signalling. EMBO J 2018; 37:e99395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li L, Huang K, Zhao H, Chen B, Ye Q, Yue J. CDK1-PLK1/SGOL2/ANLN pathway mediating abnormal cell division in cell cycle may be a critical process in hepatocellular carcinoma. Cell Cycle 2020; 19:1236–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shao YY, Li YS, Hsu HW, Lin H, Wang HY, Wo RR, Cheng AL, Hsu CH. Potent activity of composite cyclin dependent kinase inhibition against hepatocellular carcinoma. Cancers 2019; 11:1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y, Shi F, Xing GH, Xie P, Zhao N, Yin YF, Sun SY, He J, Wang Y, Xuan SY. Protein regulator of cytokinesis PRC1 confers chemoresistance and predicts an unfavorable postoperative survival of hepatocellular carcinoma patients. J Cancer 2017; 8:801–08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BK, Ooi LL, Hong W, Hui KM. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the wnt/β-catenin signalling pathway. Gut 2016; 65:1522–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu X, Li Y, Meng L, Liu XY, Peng A, Chen Y, Liu C, Chen H, Sun S, Miao X, Zhang Y, Zheng L, Huang K. Reducing protein regulator of cytokinesis 1 as a prospective therapy for hepatocellular carcinoma. Cell Death Dis 2018; 9:534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu C, Zhao J, Bibikova M, Leverson JD, Bossy-Wetzel E, Fan J-B, Abraham RT, Jiang W. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol Biol Cell 2005; 16:3187–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang Y, Wang H, Lian Y, Wu X, Zhou L, Wang J, Deng M, Huang Y. Upregulation of kinesin family member 4A enhanced cell proliferation via activation of akt signaling and predicted a poor prognosis in hepatocellular carcinoma. Cell Death Dis 2018; 9:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen J, Li S, Zhou S, Cao S, Lou Y, Shen H, Yin J, Li G. Kinesin superfamily protein expression and its association with progression and prognosis in hepatocellular carcinoma. J Cancer Res Ther 2017; 13:651–59 [DOI] [PubMed] [Google Scholar]
- 65.Glotzer M. The molecular requirements for cytokinesis. Science 2005; 307:1735–9 [DOI] [PubMed] [Google Scholar]
- 66.Sun X, Jin Z, Song X, Wang J, Li Y, Qian X, Zhang Y, Yin Y. Evaluation of KIF23 variant 1 expression and relevance as a novel prognostic factor in patients with hepatocellular carcinoma. BMC Cancer 2015; 15:961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang BH, Yang J, Jiang L, Lyu T, Kong LX, Tan YF, Li B, Zhu YF, Xi AY, Xu X, Yan LN, Yang JY. Development and validation of a 14-gene signature for prognosis prediction in hepatocellular carcinoma. Genomics 2020; 112:2763–71 [DOI] [PubMed] [Google Scholar]
- 68.Kitagawa M, Fung SY, Onishi N, Saya H, Lee SH. Targeting Aurora B to the equatorial cortex by MKlp2 is required for cytokinesis. PLoS One 2013; 8:e64826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gasnereau I, Boissan M, Margall-Ducos G, Couchy G, Wendum D, Bourgain-Guglielmetti F, Desdouets C, Lacombe ML, Zucman-Rossi J, Sobczak-Thépot J. KIF20A mRNA and its product MKlp2 are increased during hepatocyte proliferation and hepatocarcinogenesis. Am J Pathol 2012; 180:131–40 [DOI] [PubMed] [Google Scholar]
- 70.Lu M, Huang X, Chen Y, Fu Y, Xu C, Xiang W, Li C, Zhang S, Yu C. Aberrant KIF20A expression might independently predict poor overall survival and recurrence-free survival of hepatocellular carcinoma. IUBMB Life 2018; 70:328–35 [DOI] [PubMed] [Google Scholar]
- 71.Kawai Y, Shibata K, Sakata J, Suzuki S, Utsumi F, Niimi K, Sekiya R, Senga T, Kikkawa F, Kajiyama H. KIF20A expression as a prognostic indicator and its possible involvement in the proliferation of ovarian clear‑cell carcinoma cells. Oncol Rep 2018; 40:195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raab M, Sanhaji M, Matthess Y, Hörlin A, Lorenz I, Dötsch C, Habbe N, Waidmann O, Kurunci-Csacsko E, Firestein R, Becker S, Strebhardt K. PLK1 has tumor-suppressive potential in APC-truncated colon cancer cells. Nat Commun 2018; 9:1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.He ZL, Zheng H, Lin H, Miao XY, Zhong DW. Overexpression of polo-like kinase1 predicts a poor prognosis in hepatocellular carcinoma patients. World J Gastroenterol 2009; 15:4177–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sun W, Su Q, Cao X, Shang B, Chen A, Yin H, Liu B. High expression of polo-like kinase 1 is associated with early development of hepatocellular carcinoma. Int J Genom 2014; 2014:312130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang XQ, Zhu YQ, Lui KS, Cai Q, Lu P, Poon RT. Aberrant polo-like kinase 1-Cdc25A pathway in metastatic hepatocellular carcinoma. Clin Cancer Res 2008; 14:6813–20 [DOI] [PubMed] [Google Scholar]
- 76.Kumar S, Kim J. PLK-1 targeted inhibitors and their potential against tumorigenesis. BioMed Res Int 2015; 2015:705745–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ko MA, Rosario CO, Hudson JW, Kulkarni S, Pollett A, Dennis JW, Swallow CJ. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet 2005; 37:883–8 [DOI] [PubMed] [Google Scholar]
- 78.Rosario CO, Ko MA, Haffani YZ, Gladdy RA, Paderova J, Pollett A, Squire JA, Dennis JW, Swallow CJ. Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci U S A 2010; 107:6888–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bao J, Yu Y, Chen J, He Y, Chen X, Ren Z, Xue C, Liu L, Hu Q, Li J, Cui G, Sun R. MiR-126 negatively regulates PLK-4 to impact the development of hepatocellular carcinoma via ATR/CHEK1 pathway. Cell Death Dis 2018; 9:1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Meng L, Zhou Y, Ju S, Han J, Song C, Kong J, Wu Y, Lu S, Xu J, Yuan W, Zhang E, Wang C, Hu Z. A cis-eQTL genetic variant in PLK4 confers high risk of hepatocellular carcinoma. Cancer Med 2019; 8:6476–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J Cell Biol 2010; 188:191–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Portella G, Passaro C, Chieffi P. Aurora B: a new prognostic marker and therapeutic target in cancer. Curr Med Chem 2011; 18:482–96 [DOI] [PubMed] [Google Scholar]
- 83.Tovuu LO, Utsunomiya T, Imura S, Morine Y, Ikemoto T, Arakawa Y, Mori H, Hanaoka J, Kanamoto M, Sugimoto K, Saito Y, Yamada S, Asanoma M, Shimada M. The role of Aurora B expression in non-tumor liver tissues of patients with hepatocellular carcinoma. Int J Clin Oncol 2014; 19:622–8 [DOI] [PubMed] [Google Scholar]
- 84.Lin ZZ, Jeng YM, Hu FC, Pan HW, Tsao HW, Lai PL, Lee PH, Cheng AL, Hsu HC. Significance of Aurora B overexpression in hepatocellular carcinoma. Aurora B overexpression in HCC. BMC Cancer 2010; 10:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sistayanarain A, Tsuneyama K, Zheng H, Takahashi H, Nomoto K, Cheng C, Murai Y, Tanaka A, Takano Y. Expression of Aurora-B kinase and phosphorylated histone H3 in hepatocellular carcinoma. Anticancer Res 2006; 26:3585–93 [PubMed] [Google Scholar]
- 86.Tanaka S, Arii S, Yasen M, Mogushi K, Su NT, Zhao C, Imoto I, Eishi Y, Inazawa J, Miki Y, Tanaka H. Aurora kinase B is a predictive factor for the aggressive recurrence of hepatocellular carcinoma after curative hepatectomy. Br J Surg 2008; 95:611–9 [DOI] [PubMed] [Google Scholar]
- 87.Fu S, Li Y, Huang J, Liu T, Hong Z, Chen A, Bast RC, Kavanagh JJ, Gershenson DM, Sood AK, Hu W. Aurora kinase inhibitor VE 465 synergistically enhances cytotoxicity of carboplatin in ovarian cancer cells through induction of apoptosis and downregulation of histone 3. Cancer Biol Ther 2012; 13:1034–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mortlock AA, Foote KM, Heron NM, Jung FH, Pasquet G, Lohmann JJ, Warin N, Renaud F, De Savi C, Roberts NJ, Johnson T, Dousson CB, Hill GB, Perkins D, Hatter G, Wilkinson RW, Wedge SR, Heaton SP, Odedra R, Keen NJ, Crafter C, Brown E, Thompson K, Brightwell S, Khatri L, Brady MC, Kearney S, McKillop D, Rhead S, Parry T, Green S. Discovery, synthesis, and in vivo activity of a new class of pyrazoloquinazolines as selective inhibitors of Aurora B kinase. J Med Chem 2007; 50:2213–24 [DOI] [PubMed] [Google Scholar]
- 89.Giles FJ, Swords RT, Nagler A, Hochhaus A, Ottmann OG, Rizzieri DA, Talpaz M, Clark J, Watson P, Xiao A, Zhao B, Bergstrom D, Le Coutre PD, Freedman SJ, Cortes JE. MK-0457, an Aurora kinase and BCR-ABL inhibitor, is active in patients with BCR-ABL T315I leukemia. Leukemia 2013; 27:113–7 [DOI] [PubMed] [Google Scholar]
- 90.Yu X, Liang Q, Liu W, Zhou L, Li W, Liu H. Deguelin, an Aurora B kinase inhibitor, exhibits potent anti-Tumor effect in human esophageal squamous cell carcinoma. EBioMedicine 2017; 26:100–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li Z, Ren W, Zeng Q, Chen S, Zhang M, Zhao Y, Cheng J, Wang X. Effects of survivin on angiogenesis in vivo and in vitro. Am J Transl Res 2016; 8:270–83 [PMC free article] [PubMed] [Google Scholar]
- 92.Ito T, Shiraki K, Sugimoto K, Yamanaka T, Fujikawa K, Ito M, Takase K, Moriyama M, Kawano H, Hayashida M, Nakano T, Suzuki A. Survivin promotes cell proliferation in human hepatocellular carcinoma. Hepatology 2000; 31:1080–5 [DOI] [PubMed] [Google Scholar]
- 93.Xia H, Chen J, Shi M, Deivasigamani A, Ooi LL, Hui KM. The over-expression of survivin enhances the chemotherapeutic efficacy of YM155 in human hepatocellular carcinoma. Oncotarget 2015; 6:5990–6000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei W, Wanjun L, Hui S, Dongyue C, Xinjun Y, Jisheng Z. miR-203 inhibits proliferation of HCC cells by targeting survivin. Cell Biochem Funct 2013; 31:82–5 [DOI] [PubMed] [Google Scholar]
- 95.Yamauchi T, Nakamura N, Hiramoto M, Yuri M, Yokota H, Naitou M, Takeuchi M, Yamanaka K, Kita A, Nakahara T, Kinoyama I, Matsuhisa A, Kaneko N, Koutoku H, Sasamata M, Kobori M, Katou M, Tawara S, Kawabata S, Furuichi K. Sepantronium bromide (YM155) induces disruption of the ILF3/p54(nrb) complex, which is required for survivin expression. Biochem Biophys Res Commun 2012; 425:711–6 [DOI] [PubMed] [Google Scholar]
- 96.Shimizu T, Nishio K, Sakai K, Okamoto I, Okamoto K, Takeda M, Morishita M, Nakagawa K. Phase I safety and pharmacokinetic study of YM155, a potent selective survivin inhibitor, in combination with erlotinib in patients with EGFR TKI refractory advanced non-small cell lung cancer. Cancer Chemother Pharmacol 2020; 86:211–19 [DOI] [PubMed] [Google Scholar]
- 97.Zhao X, Ogunwobi OO, Liu C. Survivin inhibition is critical for bcl-2 inhibitor-induced apoptosis in hepatocellular carcinoma cells. PLoS One 2011; 6:e21980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yin H, Que R, Liu C, Ji W, Sun B, Lin X, Zhang Q, Zhao X, Peng Z, Zhang X, Qian H, Chen L, Yao Y, Su C. Survivin-targeted drug screening platform identifies a matrine derivative WM-127 as a potential therapeutics against hepatocellular carcinoma. Cancer Lett 2018; 425:54–64 [DOI] [PubMed] [Google Scholar]
- 99.Yue L, Li L, Li D, Yang Z, Han S, Chen M, Lan S, Xu X, Hui L. High-throughput screening for survivin and borealin interaction inhibitors in hepatocellular carcinoma. Biochem Biophys Res Commun 2017; 484:642–47 [DOI] [PubMed] [Google Scholar]
- 100.Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr Med Chem Anticancer Agents 2005; 5:363–72 [DOI] [PubMed] [Google Scholar]
- 101.Chen J, Xia H, Zhang X, Karthik S, Pratap SV, Ooi LL, Hong W, Hui KM. ECT2 regulates the rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. J Hepatol 2015; 62:1287–95 [DOI] [PubMed] [Google Scholar]
- 102.Yu Y, Cai O, Wu P, Tan S. MiR-490-5p inhibits the stemness of hepatocellular carcinoma cells by targeting ECT2 . J Cell Biochem 2019; 120:967–76 [DOI] [PubMed] [Google Scholar]
- 103.Gallo G. RhoA-kinase coordinates F-actin organization and myosin II activity during semaphorin-3A-induced axon retraction. J Cell Sci 2006; 119:3413–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang D, Dou K, Xiang H, Song Z, Zhao Q, Chen Y, Li Y. Involvement of RhoA in progression of human hepatocellular carcinoma. J Gastroenterol Hepatol 2007; 22:1916–20 [DOI] [PubMed] [Google Scholar]
- 105.Li XR, Ji F, Ouyang J, Wu W, Qian LY, Yang KY. Overexpression of RhoA is associated with poor prognosis in hepatocellular carcinoma. Eur J Surg Oncol 2006; 32:1130–4 [DOI] [PubMed] [Google Scholar]
- 106.Bai Y, Xie F, Miao F, Long J, Huang S, Huang H, Lin J, Wang D, Yang X, Bian J, Mao J, Wang X, Mao Y, Sang X, Zhao H. The diagnostic and prognostic role of RhoA in hepatocellular carcinoma. Aging 2019; 11:5158–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang F, Xu X, Zhang N, Chen Z. Identification and integrated analysis of hepatocellular carcinoma-related circular RNA signature. Ann Transl Med 2020; 8:294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yin K, Zhao G, Huang X, Gao G, Sun H, Wei Q, Liu Q, Li M, Xu C, Zhu S, Ba Z, Yan G. Inhibition of RhoA expression by adenovirus-mediated siRNA combined with TNF-α induced apoptosis of hepatocarcinoma cells. Bio Med Mater Eng 2015; 26:S2055–67 [DOI] [PubMed] [Google Scholar]
- 109.Zhang Y, Deng J, Ma S, Xue L, Zhu J, Zhu WL, Jiang HL, Li J, Miao LY. The effect of first-in-class small molecule RhoA inhibitor, HL07, on the phenylephrine-induced artery contraction. Curr Pharm Des 2012; 18:4258–64 [DOI] [PubMed] [Google Scholar]
- 110.Shang X, Marchioni F, Evelyn CR, Sipes N, Zhou X, Seibel W, Wortman M, Zheng Y. Small-molecule inhibitors targeting G-protein-coupled rho guanine nucleotide exchange factors. Proc Natl Acad Sci U S A 2013; 110:3155–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pietrovito L, Comito G, Parri M, Giannoni E, Chiarugi P, Taddei ML. Zoledronic acid inhibits the RhoA-mediated amoeboid motility of prostate cancer cells. Curr Cancer Drug Targets 2019; 19:807–16 [DOI] [PubMed] [Google Scholar]
- 112.Evelyn CR, Wade SM, Wang Q, Wu M, Iñiguez-Lluhí JA, Merajver SD, Neubig RR. CCG-1423: a small-molecule inhibitor of RhoA transcriptional signaling. Mol Cancer Ther 2007; 6:2249–60 [DOI] [PubMed] [Google Scholar]
- 113.Katamura Y, Aikata H, Hashimoto Y, Kimura Y, Kawaoka T, Takaki S, Waki K, Hiramatsu A, Kawakami Y, Takahashi S, Kenjo M, Chayama K. Zoledronic acid delays disease progression of bone metastases from hepatocellular carcinoma. Hepatol Res 2010; 40:1195–203 [DOI] [PubMed] [Google Scholar]
- 114.Honda Y, Aikata H, Honda F, Nakano N, Nakamura Y, Hatooka M, Morio K, Kobayashi T, Fukuhara T, Nagaoki Y, Kawaoka T, Hiramatsu A, Imamura M, Kawakami Y, Chayama K. Clinical outcome and prognostic factors in hepatocellular carcinoma patients with bone metastases medicated with zoledronic acid. Hepatol Res 2017; 47:1053–60 [DOI] [PubMed] [Google Scholar]
- 115.Natori T, Yamaguchi M. [ Complete response after sorafenib therapy plus zoledronic acid for advanced hepatocellular carcinoma with bone metastasis – a case report. Gan to Kagaku Ryoho Cancer Ryoho 2013; 40:635–7 [PubMed] [Google Scholar]
- 116.Wang SM, Ooi LL, Hui KM. Upregulation of rac GTPase-activating protein 1 is significantly associated with the early recurrence of human hepatocellular carcinoma. Clin Cancer Res 2011; 17:6040–51 [DOI] [PubMed] [Google Scholar]
- 117.Yang XM, Cao XY, He P, Li J, Feng MX, Zhang YL, Zhang XL, Wang YH, Yang Q, Zhu L, Nie HZ, Jiang SH, Tian GA, Zhang XX, Liu Q, Ji J, Zhu X, Xia Q, Zhang ZG. Overexpression of rac GTPase activating protein 1 contributes to proliferation of cancer cells by reducing hippo signaling to promote cytokinesis. Gastroenterology 2018; 155:1233–49e22 [DOI] [PubMed] [Google Scholar]
- 118.Wang MY, Chen DP, Qi B, Li MY, Zhu YY, Yin WJ, He L, Yu Y, Li ZY, Lin L, Yang F, Lin ZR, Liu JQ. Pseudogene RACGAP1P activates RACGAP1/rho/ERK signalling axis as a competing endogenous RNA to promote hepatocellular carcinoma early recurrence. Cell Death Dis 2019; 10:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Santamaría D, Barrière C, Cerqueira A, Hunt S, Tardy C, Newton K, Cáceres JF, Dubus P, Malumbres M, Barbacid M. Cdk1 is sufficient to drive the mammalian cell cycle. Nature 2007; 448:811–5 [DOI] [PubMed] [Google Scholar]
- 120.Potapova TA, Daum JR, Byrd KS, Gorbsky GJ. Fine tuning the cell cycle: activation of the Cdk1 inhibitory phosphorylation pathway during mitotic exit. Mol Biol Cell 2009; 20:1737–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu CX, Wang XQ, Chok SH, Man K, Tsang SHY, Chan ACY, Ma KW, Xia W, Cheung TT. Blocking CDK1/PDK1/β-Catenin signaling by CDK1 inhibitor RO3306 increased the efficacy of sorafenib treatment by targeting cancer stem cells in a preclinical model of hepatocellular carcinoma. Theranostics 2018; 8:3737–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang H, Zhao H, Yu ZY, Feng X, Fu BS, Qiu CH, Zhang JW. MicroRNA-194 inhibits cell invasion and migration in hepatocellular carcinoma through PRC1-mediated inhibition of wnt/β-catenin signaling pathway. Dig Liver Dis 2019; 51:1314–22 [DOI] [PubMed] [Google Scholar]
- 123.Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol 2009; 10:682–96 [DOI] [PubMed] [Google Scholar]
- 124.Ferrero H, Corachán A, Quiñonero A, Bougeret C, Pouletty P, Pellicer A, Domínguez F. Inhibition of KIF20A by BKS0349 reduces endometriotic lesions in a xenograft mouse model. Mol Hum Reprod 2019; 25:562–71 [DOI] [PubMed] [Google Scholar]
- 125.Gray PJ, Jr., Bearss DJ, Han H, Nagle R, Tsao MS, Dean N, Von Hoff DD. Identification of human polo-like kinase 1 as a potential therapeutic target in pancreatic cancer. Mol Cancer Ther 2004; 3:641–6 [PubMed] [Google Scholar]
- 126.Wolf G, Elez R, Doermer A, Holtrich U, Ackermann H, Stutte HJ, Altmannsberger HM, Rübsamen-Waigmann H, Strebhardt K. Prognostic significance of polo-like kinase (PLK) expression in non-small cell lung cancer. Oncogene 1997; 14:543–9 [DOI] [PubMed] [Google Scholar]
- 127.King SI, Purdie CA, Bray SE, Quinlan PR, Jordan LB, Thompson AM, Meek DW. Immunohistochemical detection of polo-like kinase-1 (PLK1) in primary breast cancer is associated with TP53 mutation and poor clinical outcome. Breast Cancer Res 2012; 14:R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Weichert W, Kristiansen G, Schmidt M, Gekeler V, Noske A, Niesporek S, Dietel M, Denkert C. Polo-like kinase 1 expression is a prognostic factor in human colon cancer. World J Gastroenterol 2005; 11:5644–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu Z, Sun Q, Wang X. PLK1, a potential target for cancer therapy. Transl Oncol 2017; 10:22–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.de Cárcer G, Venkateswaran SV. Plk1 overexpression induces chromosomal instability and suppresses tumor development. Nat Commun 2018; 9:3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Döhner H, Lübbert M, Fiedler W, Fouillard L, Haaland A, Brandwein JM, Lepretre S, Reman O, Turlure P, Ottmann OG, Müller-Tidow C, Krämer A, Raffoux E, Döhner K, Schlenk RF, Voss F, Taube T, Fritsch H, Maertens J. Randomized, phase 2 trial of low-dose cytarabine with or without volasertib in AML patients not suitable for induction therapy. Blood 2014; 124:1426–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Press MF, Xie B, Davenport S, Zhou Y, Guzman R, Nolan GP, O'Brien N, Palazzolo M, Mak TW, Brugge JS, Slamon DJ. Role for polo-like kinase 4 in mediation of cytokinesis. Proc Natl Acad Sci U S A 2019; 116:11309–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhao Y, Wang X. PLK4: a promising target for cancer therapy. J Cancer Res Clin Oncol 2019; 145:2413–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hindriksen S, Meppelink A, Lens SM. Functionality of the chromosomal passenger complex in cancer. Biochem Soc Trans 2015; 43:23–32 [DOI] [PubMed] [Google Scholar]
- 135.Krenn V, Musacchio A. The Aurora B kinase in chromosome bi-orientation and spindle checkpoint signaling. Front Oncol 2015; 5:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Benten D, Keller G, Quaas A, Schrader J, Gontarewicz A, Balabanov S, Braig M, Wege H, Moll J, Lohse AW, Brummendorf TH. Aurora kinase inhibitor PHA-739358 suppresses growth of hepatocellular carcinoma in vitro and in a xenograft mouse model. Neoplasia 2009; 11:934–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vader G, Kauw JJ, Medema RH, Lens SM. Survivin mediates targeting of the chromosomal passenger complex to the centromere and midbody. EMBO Rep 2006; 7:85–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wheatley SP, Altieri DC. Survivin at a glance. J Cell Sci 2019; 132:jcs223826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chang YJ, Li LT, Chen HA, Hung CS, Wei PL. Silencing survivin activates autophagy as an alternative survival pathway in HCC cells. Tumour Biol 2014; 35:9957–66 [DOI] [PubMed] [Google Scholar]
- 140.Liu F, Sun Y, Liu B, Lu J, Li H, Zhu H, Gao H, Zhou X, Chang H. Insulin-like growth factor-1 induces epithelial-mesenchymal transition in hepatocellular carcinoma by activating survivin. Oncol Rep 2018; 40:952–58 [DOI] [PubMed] [Google Scholar]
- 141.Beyer TE, Smith RM, Song Z, Gokhale V, Moses SA, Meuillet EJ. Abstract 2835: discovery of novel inhibitors for ECT2 as a novel therapeutic strategy for lung cancer. Cancer Res 2012; 72:2835 [Google Scholar]
- 142.Sit S-T, Manser E. Rho GTPases and their role in organizing the actin cytoskeleton. J Cell Sci 2011; 124:679–83 [DOI] [PubMed] [Google Scholar]
- 143.Kühn S, Geyer M. Formins as effector proteins of rho GTPases. Small GTPases 2014; 5:e29513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.van Adrichem AJ, Fagerholm A, Turunen L, Lehto A, Saarela J, Koskinen A, Repasky GA, Wennerberg K. Discovery of MINC1, a GTPase-activating protein small molecule inhibitor, targeting MgcRacGAP. Comb Chem High Throughput Screen 2015; 18:3–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Garg H, Suri P, Gupta JC, Talwar GP, Dubey S. Survivin: a unique target for tumor therapy. Cancer Cell Int 2016; 16:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Xu D, Wang Y, Wu J, Zhang Z, Chen J, Xie M, Tang R, Chen C, Chen L, Lin S, Luo X, Zheng J. ECT2 overexpression promotes the polarization of tumor-associated macrophages in hepatocellular carcinoma via the ECT2/PLK1/PTEN pathway. Cell Death Dis 2021; 12:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Kim JA, Lee J, Margolis RL, Fotedar R. SP600125 suppresses Cdk1 and induces endoreplication directly from G2 phase, independent of JNK inhibition. Oncogene 2010; 29:1702–16 [DOI] [PMC free article] [PubMed] [Google Scholar]