Abstract
Triple-negative breast cancer (TNBC) is one of the most common malignances worldwide. Concurrently, the incidence of TNBC has continued to rise in recent years. It is reported that long non-coding RNAs (lncRNAs) are involved in biological processes in numerous cancers including TNBC. Small nucleolar RNA host gene 11 (SNHG11) has already been studied and reported in some cancers. However, the role of SNHG11 in TNBC remains unknown. RT-qPCR was used to measure gene expression in the current study. CCK-8, colony formation, flow cytometry, Transwell and western blotting experiments were also performed to determine the biological function of SNHG11 in TNBC cells. Luciferase reporter and RIP assays were performed to measure relationship between genes. In the present study, the results indicated SNHG11 was highly expressed in TNBC tissues and cell lines. Moreover, SNHG11 aggravated cell proliferation and migration, and whereas it attenuated cell apoptosis in TNBC. Furthermore, SNHG11 sponged microRNA 2355-5p (miR-2355-5p) in TNBC. Silencing SNHG11 increased miR-2355-5p expression. In addition, chromobox 5 (CBX5) was identified to be targeted by miR-2355-5p in TNBC. It was also suggested that CBX5 silencing suppressed cell proliferation and migration. Furthermore, overexpressed CBX5 recovered the inhibitive influence of SNHG11 silencing on proliferative and migrative abilities of TNBC cells. Overall, SNHG11 acted as a tumor promoter in TNBC and regulated TNBC cell growth by modulating the miR-2355-5p/CBX5 axis, which indicated that it may be used as a biomarker for TNBC treatment.
Keywords: TNBC, SNHG11, miR-2355-5p, CBX5
Introduction
As reported, triple-negative breast cancer (TNBC) is one of the most common malignant tumors and the main cause of cancer-related mortality in women (1,2). TNBC is a breast cancer subtype renowned for its capacity to affect younger women, metastasize early despite optimal adjuvant treatment and carry a poor prognosis (3,4). Despite significant progress in the diagnosis and treatment of TNBC, the long-term survival rate of TNBC patients remains unsatisfactory (5). Thus, it is of great significance to make more efforts to reduce the mortality of TNBC patients. There is an urgent need to further probe potential regulatory patterns in TNBC development.
Long non-coding RNAs (lncRNAs) are non-coding RNAs with a length greater than 200 nucleotides (6,7). Studies have revealed that lncRNAs play important roles in numerous aspects such as dose compensation effect, epigenetic regulation, cell cycle regulation and cell differentiation regulation (8-10). It is often reported that lncRNAs serve as key regulators via sponging microRNAs (miRNAs) and targeting messenger RNAs (mRNAs) (6), and ultimately participate in regulation of cancers. Previous studies have reported that lncRNAs are identified as potential biomarkers in a plurality of human cancers (11). For example, lncRNA NNT-AS1 was revealed to promote proliferative ability in glioma via sponging miR-494-3p (12). ST8SIA6-AS1 facilitated proliferative and migrative abilities of hepatocellular carcinoma via sponging miR-5195-3p (13). Moreover, UCA1 accelerated endometrial cancer progression via interacting with KLF5(14). Hence, in the present study the aim was to explore the detailed mechanism by which lncRNAs regulate TNBC proliferation and migration. Previously, it has been revealed that small nucleolar RNA host gene 11 (SNHG11) acted as a tumor promoter in lung (15), prostate (16) and liver cancer (17), but it has not been studied in TNBC. Therefore, the present study mainly investigated the role and regulatory mode of SNHG11 in TNBC.
miRNAs are another subgroup of non-coding RNAs with a length of 18-25 nucleotides (18-20). miRNAs can specifically bind with the 3'-untranslated region (UTR) of mRNAs, thereby modulating gene expression post-transcriptionally and taking part in various cancer type progressions. For example, miR-124-3p directly targeted SARM1 to contribute to prostate cancer progression (21). As a tumor suppressor in lung cancer, miR-93 downregulated the expression of TBP-2(22). In recent years, microRNA 2355-5p (miR-2355-5p) has received a lot of attention in some cancers. As reported, miR-2355-5p has been revealed to play a role in the suppression of cancer development in a variety of cancers, including chondrosarcoma (23) and bladder cancer (24). However, the interaction between SNHG11 and miR-2355-5p in TNBC has not been elucidated and requires further investigation.
In summary, the present study was designed to explore the biological role and regulatory network of SNHG11 in TNBC. The findings suggested that SNHG11 could be a potential biomarker for TNBC treatment.
Materials and methods
Tissue samples
A total of 42 pairs of TNBC tissues and adjacent noncancerous tissues were obtained from TNBC patients who were clinically and pathologically diagnosed with TNBC at the Jiangsu Taizhou People's Hospital from July 2018 to July 2019. Written informed consent was obtained from all participants and study was approved by the Ethics Committee of Jiangsu Taizhou People's Hospital (Taizhou, China). The age range of patients with TNBC was 26-67 years, with a mean age of 47.2±5.1 years. The distance between the tumor tissue edge and the adjacent tissue from which samples were obtained was >5 cm. The collected tissues were rapidly frozen in liquid nitrogen and stored at -80˚C for the following research. All patients had not received anticancer treatment.
Cell lines
TNBC cell lines BT-20, BT-549, MDA-MB-231, Hs 578T and an immortalized human epithelial cell line MCF10A were offered by the American Type Culture Collection (ATCC). In brief, cells were cultured in Roswell Park Memorial Institute 1640 (RPMI-1640) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine, 100 U/ml penicillin (Sigma-Aldrich; Merck KGaA) and 100 µg/ml streptomycin (Sigma-Aldrich; Merck KGaA). The cells were maintained in a humid atmosphere at 37˚C in the presence of 5% CO2.
Cell transfection
For downregulating SNHG11 in BT-20 and BT-549 cells, two short hairpin RNAs (sh-RNA#1 and sh-RNA#2; 3 µg) specifically targeting SNHG11 with sh-NC (cat. no. C02003) as the negative control were designed and synthesized by Shanghai GenePharma Co., Ltd. The sequences of sh-SNHG11#1/2 are as follows: sh-SNHG11#1: 5'-CCTTGGGTCTGGAAACTGTTA-3'; sh-SNHG11#2: 5'-CCGTGTGTGTTATATCATGAT-3'. The full length of CBX5 (mRNA) was synthesized and subcloned into pcDNA3.1 (Invitrogen; Thermo Fisher Scientific, Inc.) plasmid to produce pcDNA3.1/CBX5. For miR-2355-5p overexpression, BT-20 and BT-549 cells were transfected with miR-2355-5p mimics or (negative control) NC mimics. miR-2355-5p mimics and NC mimics (cat. no. B01001) were both obtained from Shanghai GenePharma Co., Ltd. BT-20 and BT-549 cells (1x106 cells/well) were seeded in 24-well plates and 500 µl DMEM was added to each well. When the cells reached 40-60% confluence, the aforementioned vectors were transfected into cells at a final concentration of 50 nM using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C with 5% CO2 according to the manufacturer's instructions. Cells were harvested 48 h after transfection.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNAs were extracted from tissues and cells with TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.) reagent kits as recommended. Total RNAs were reverse transcribed into complementary DNA (cDNA) using PrimeScript RT reagent kits (cat. no. RRO36A; Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. A SYBR® Premix Ex Taq™ II reagent kit (cat. no. RR820A; Takara) was utilized to perform RT-qPCR with an ABI7500 real-time qPCR system (ABI; Thermo Fisher Scientific, Inc.). RNU6 (U6) was used as the internal reference for miR-2355-5p. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was applied as an internal reference for SNHG11 and CBX5. The relative quantification was calculated with the 2-∆∆Cq method (25). The following primers were used: SNHG11 forward, 5'-TGGGAGTTGTCATGTTGGGA-3' and reverse, 5'-ACTCGTCACTCTTGGTCTGT-3'; miR-2355-5p forward, 5'-ATTGTCCTTGCTGTTTGGAGAT-3' and reverse, 5'-GCGAGCACAGAATTAATACGAC-3'; CBX5 forward, 5'-GCAGACGTTAGCGTGAGTG-3' and reverse, 5'-GCGGAATTCGGATCCCTCGAGTT-3'; U6 forward, 5'-CTCGCTTCGGCAGCACATA-3' and reverse, 5'-CGCTTCACGAATTTGCGTG-3'; GAPDH forward, 5'-GATCATCAGCAATGCCTCCT-3' and reverse, 5'-TGAGTCCTTCCACGATACCA-3'. The following thermocycling conditions were used: Initial denaturation at 95˚C for 3 min, followed by 40 cycles at 95˚C for 5 sec, 60˚C for 30 sec and at 72˚C for 45 sec, before final extension at 72˚C for 3 min.
Cell Counting Kit-8 (CCK-8) assay
Cell proliferation in response to SNHG11 knockdown was measured with CCK-8 (Dojindo Molecular Technologies, Inc.) following the manufacturer's instructions. The cells were seeded into 96-well plates (1x103 cells/well) in triplicate and at 24, 48 and 72 h. Then, CCK-8 reagent was added, and the cells were maintained for a further 2 h at 37˚C. The chromogenic reaction was allowed in a culture incubator for 4 h. The absorption was recorded at 450 nm by Tecan Infinite F500 microplate reader (Tecan Group, Ltd.).
Colony formation assay
The colony formation assay was utilized to assess the cell proliferation. Transfected cells were diluted and seeded onto 6-well plates with a density of ~1x103 cells/well. The cells were then incubated at 37˚C for 14 days and fixed with paraformaldehyde at room temperature for 15 min and stained with 0.1% crystal violet (Beyotime Institute of Biotechnology) at room temperature for 30 min when colonies (≥50 cells) were visible. After 48 h, the plates were photographed under a light microscope (DM1000; Leica Microsystems GmbH), and the number of colonies were counted (magnification, x100).
Flow cytometric analysis
Briefly, transfected cells were collected and resuspended with phosphate-buffered saline (PBS). Then, the density of cells was adjusted to 1x106 cells/ml. Transfected cells were double stained by propidium iodide and Annexin V-fluorescein isothiocyanate (20 mg/ml; BD Pharmingen; BD Biosciences) at room temperature for 10 min in line with the manufacturer's instructions. Finally, cell apoptosis was detected via flow cytometry using a FACScan® (BD Biosciences) equipped with CellQuest Pro software 5.1 (BD Biosciences).
Transwell assay
The capacity for cell migration was assessed using Transwell chambers (8 µm diameter; Corning, Inc.) without Matrigel. After being cultured for 48 h, the cells were harvested, washed with PBS, and resuspended in RPMI-1640 medium without FBS. In total, 100 µl of suspension containing 5x104 cells was added into the upper chambers. Transwell chambers were placed into a 24-well plate that had already been covered with 500 µl of RPMI-1640 medium containing 10% of FBS. At 24 h after inoculation, nonmigratory cells were gently removed, and the migratory cells were fixed with 100% methanol at room temperature for 15 min, stained with 0.5% crystal violet at room temperature for 20 min, washed with PBS, and imaged using an inverted microscope (magnification, x200; Olympus Corporation). The Transwell invasion assay was similar to the migration assay except that the chamber was precoated with Matrigel (BD Biosciences) overnight at room temperature. The migration and invasion abilities were measured by respectively counting the migrated and invasive cells.
Western blotting
Total proteins in cells were extracted with Radio Immunoprecipitation Assay (RIPA) lysis buffer containing protease and phosphatase inhibitors [Roche Diagnostics (Shanghai) Co., Ltd.]. Protein concentration was detected by a bicinchoninic acid kit. Then the proteins (5 µg/sample) were separated using 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride (PVDF) membrane. The membranes were blocked with 5% skim milk powder at room temperature for 1 h and incubated with the primary antibodies (Abcam) of antibodies to E-cadherin (cat. no. ab1416; 1:50) and N-cadherin (cat. no. ab98952; 1:500) and GAPDH (cat. no. ab8245; 1:500) at 4˚C overnight. Then the membranes were washed with Tris-buffered saline 20% Tween-20 and further incubated with the horseradish peroxidase-conjugated goat anti-rabbit antibody (cat. no. sc-2357; 1:5,000; Santa Cruz Biotechnology, Inc.) at room temperature for 1 h. Proteins on the membrane were visualized by an enhanced chemiluminescence detection kit (BB-3501; Amersham Pharmacia Biotech; Cytiva) and Bio-Rad image analysis system (Bio-Rad Laboratories, Inc.).
Bioinformatics analysis
The downstream gene of SNHG11 and the downstream gene of miR-2355-5p were predicted using the starBase database (http://starbase.sysu.edu.cn/). The interaction between SNHG11 miR-2355-5p was also predicted using the starBase database. The binding fragment of miR-2355-5p on the 3'UTR of CBX5 was predicted using TargetScan (http://www.targetscan.org/vert_72/).
Luciferase reporter assay
To detect the binding ability between SNHG11 (CBX5) and miR-2355-5p, wild-type sequences of SNHG11 (CBX5 3'UTR) and the mutant SNHG11 (CBX5 3'UTR) were cloned into the firefly luciferase gene reporter vector pmiRGLO (Promega Corporation). The plasmid was synthesized by Invitrogen; Thermo Fisher Scientific, Inc. The pmirGLO-SNHG11 (CBX5 3'UTR)-Wt or pmirGLO-SNHG11(CBX5 3'UTR)-Mut was co-transfected with miR-2355-5p mimics or NC mimics (Guangzhou RiboBio Co., Ltd.) into BT-20 and BT-549 cells. Then, 48 h after transfection using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) at 37˚C with 5% CO2. The luciferase assay was performed using the dual-luciferase reporter assay system kit (Promega Corporation) according to the manufacturer's instructions. The luciferase activity was analyzed using a Modulus single-tube multimode reader (Promega Corporation) in comparison with Renilla luciferase activity.
RNA pull down
Biotinylated SNHG11 (SNHG11 probe-biotin) and negative control (SNHG11 probe-no biotin) purchased from Sangon Biotech Co., Ltd., were transfected into BT-20 and BT-549 cells using Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h at room temperature. The cells were lysed (Roche Diagnostics) and collected after transfection. A total of 20 µl biotin-labeled RNA and 40 µl streptavidin Dynabeads (Thermo Fisher Scientific, Inc.) were added to 1 ml of BT-20 and BT-549 cell supernatant lysate for 6 h at 25°C. Beads were isolated from the supernatant (centrifugation at 2,500 x g for 5 min at 4°C) and washed with wash buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 2 M NaCl and 0.1% Tween-20) followed by centrifugation (2,500 x g for 5 min at 4˚C). Isolated beads were mixed with 20 µl SDS loading buffer (Beyotime Institute of Biotechnology) and boiled for 10 min at 100˚C. The bound RNAs were subjected to RT-qPCR for quantification and analysis.
RNA immunoprecipitation (RIP) assay
RIP assay was performed under the instructions of the EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit (EMD Millipore). Cells in different groups were lysed in lysis buffer (5 mM HEPES; pH 7.4; 85 mM KCl; 0.5% NP40; Thermo Fisher Scientific, Inc.) which contained protease inhibitor cocktail and RNase inhibitor. The cells were incubated with RIP buffer containing magnetic beads coated with Ago2 antibodies (cat. no. MABE56; EMD Millipore). IgG (cat. no. 12-370; EMD Millipore) group was applied as a negative control group. After incubation at 4˚C for 2 h, the coprecipitated RNA was eluted from the beads and measured by PCR analysis.
Statistical analysis
The data were obtained and presented as the mean ± standard deviation. SPSS 20.0 software (IBM Corp.) and Graphpad Prism 7.0 software (GraphPad Software, Inc.) were used for data evaluation and charting. One-way analysis of variance (ANOVA) followed by a Tukey's post hoc test or Student's t-test was performed for comparison between groups. A log rank test was performed for Kaplan-Meier survival curves. In addition, the relationship among genes was determined by Spearman correlation analysis. All experiments were repeated three times. P<0.05 was considered to indicate a statistically significant difference.
Results
SNHG11 upregulation is significantly reflected in TNBC and represents an unsatisfactory prognosis
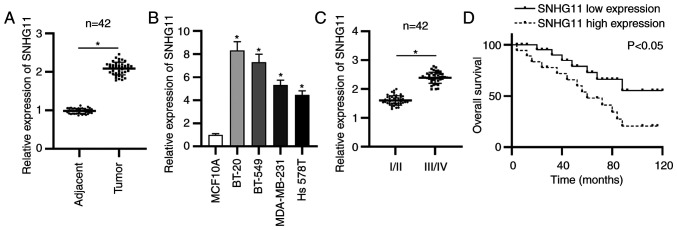
To probe role of lncRNA SNHG11 in TNBC, the SNHG11 expression in TNBC was first detected via RT-qPCR analysis. The data revealed that SNHG11 expression was significantly upregulated in TNBC tissues (Fig. 1A). SNHG11 overexpression was also revealed in 4 human TNBC cells compared with that of normal mammary epithelial cells (MCF10A) (Fig. 1B). In addition, a marked increase of SNHG11 expression was presented at advanced stages of patients with TNBC (Fig. 1C). Moreover, Kaplan-Meier analysis revealed that TNBC patients with a high level of SNHG11 exhibited poorer overall survival compared to patients with a low expression level of SNHG11 (Fig. 1D). Collectively, SNHG11 was highly expressed in TNBC and high SNHG11 expression was related to a poor prognosis.
Figure 1.
SNHG11 upregulation is revealed in TNBC. (A) RT-qPCR revealed SNHG11 expression in TNBC tissues and adjacent ones. (B) SNHG11 expression was examined via RT-qPCR in TNBC cells. (C) RT-qPCR assessed SNHG11 expression in patients at early or advanced stages. (D) Kaplan-Meier analysis assessed the overall survival rate of TNBC patients. *P<0.05. TNBC, triple-negative breast cancer; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; SNHG11, small nucleolar RNA host gene 11.
SNHG11 aggravates cell proliferative and migrative capabilities of TNBC cells
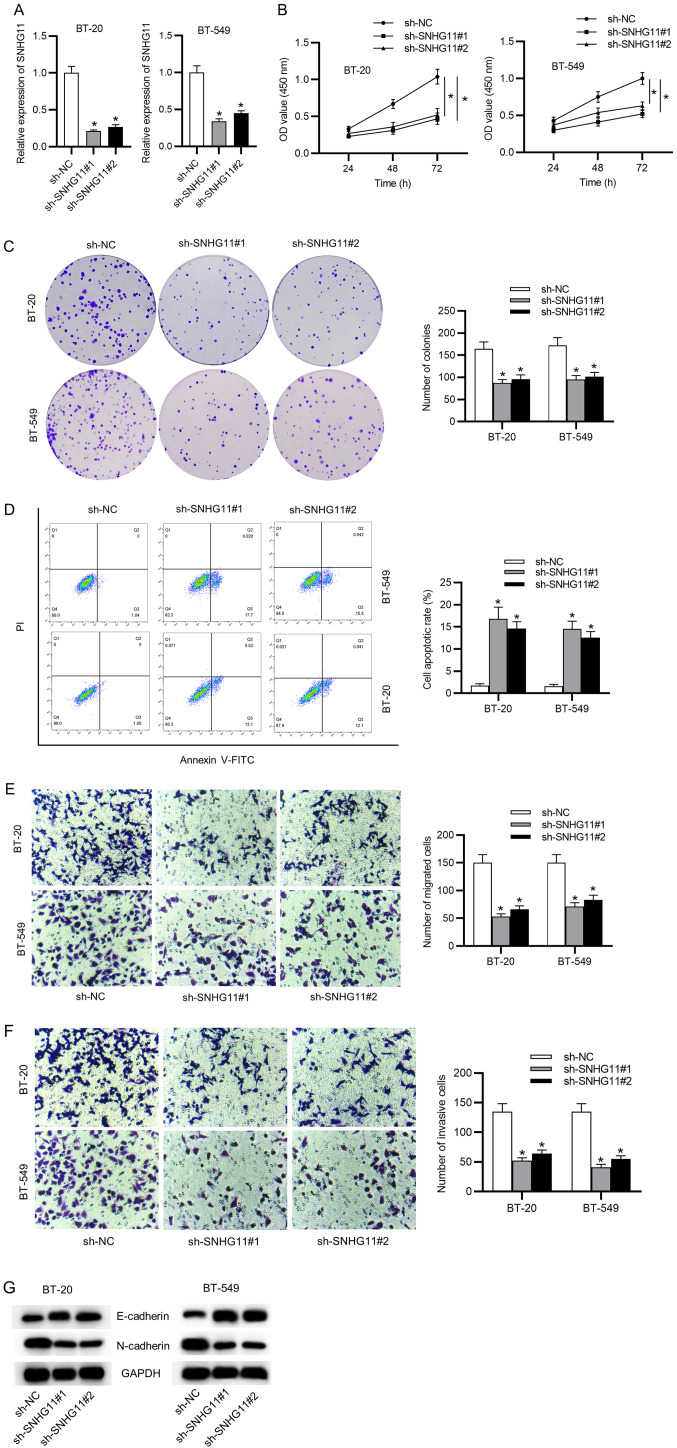
To explore possible biological roles of SNHG11 in tumorigenesis and development of TNBC, SNHG11 expression was first knocked down using sh-SNHG11#1 or sh-SNHG11#2. In succession, silencing efficacy of sh-SNHG11 in BT-20 and BT-549 cells was confirmed with RT-qPCR analysis (Fig. 2A). Then, it was determined that SNHG11 knockdown inhibited cell viability through CCK-8 results (Fig. 2B). It had an inhibitory effect on colony formation (Fig. 2C), while it induced cell apoptosis in TNBC cells (Fig. 2D). In addition, Transwell experiments revealed that SNHG11 knockdown significantly hampered cell migration and invasion capacities in TNBC (Fig. 2E and F). Western blot analysis examined the levels of epithelial-mesenchymal transition (EMT) markers (E-cadherin and N-cadherin). As presented in Fig. 2G, the expression of the epithelial marker E-cadherin was markedly upregulated, and the mesenchymal marker N-cadherin expression was obviously reduced by silencing of SNHG11 in TNBC. Collectively, SNHG11 accelerated the cell growth and migration in TNBC.
Figure 2.
SNHG11 promotes cell proliferative and migratory abilities in TNBC. (A) RT-qPCR measured knockdown efficacy of sh-SNHG11#1/2. (B and C) Cell viability and proliferative ability after silencing of SNHG11 were respectively assessed via CCK-8 and colony formation assays. (D) The apoptotic rate after silencing of SNHG11 was assessed with flow cytometric analysis. (E and F) Transwell assays validated the migrative and invasive abilities of cells under SNHG11 knockdown. (G) Western blot analysis verified the levels of EMT markers in each group. *P<0.05. SNHG11, small nucleolar RNA host gene 11; TNBC, triple-negative breast cancer; sh-, short hairpin; CCK-8, Cell Counting Kit-8; EMT, epithelial-mesenchymal transition; NC, negative control.
SNHG11 sponges miR-2355-5p in TNBC
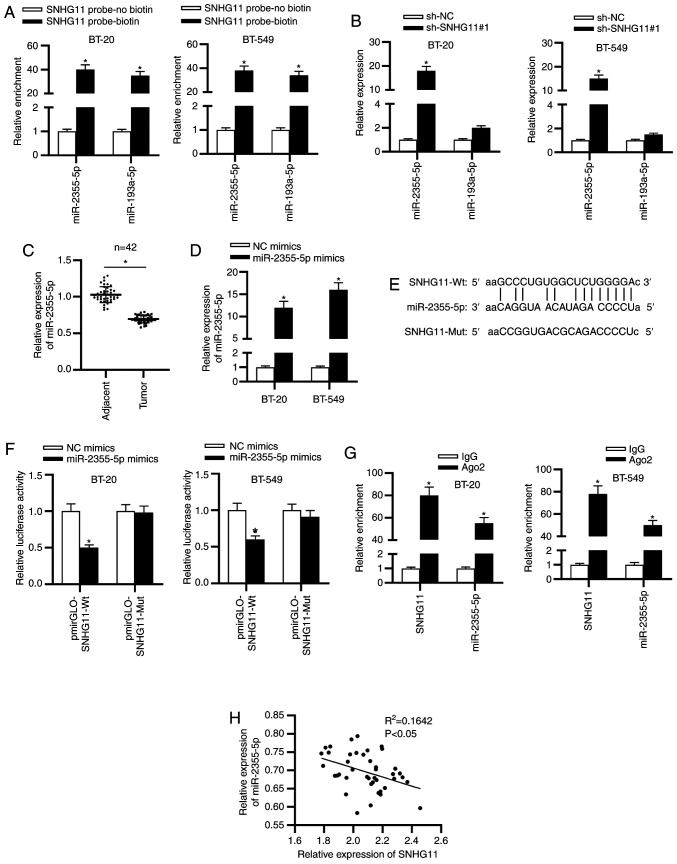
To probe the underlying mechanism of SNHG11 in TNBC, starBase v2.0 was used and 2 miRNAs (miR-2355-5p and miR-193a-5p) (condition: Pan-cancer: 8 cancer types) which may bind with SNHG11 were identified. To select the qualified miRNA, a pull down assay was performed. As revealed in Fig. 3A, both miR-2355-5p and miR-193a-5p were enriched in the SNHG11-probe biotin group compared with the SNHG11-probe no biotin group. Next, it was revealed that only miR-2355-5p could be upregulated with sh-SNHG11#1 (Fig. 3B). Thus, miR-2355-5p was identified to be the target of SNHG11 and selected for the following experiments. It was revealed from RT-qPCR analysis that miR-2355-5p expression exhibited a significant decrease in TNBC tissues (Fig. 3C). The overexpression efficacy of miR-2355-5p was examined as revealed in Fig. 3D. The binding sequence of miR-2355-5p with SNHG11 was predicted by starBase (Fig. 3E). To verify interaction between SNHG11 and miR-2355-5p, luciferase reporter and RIP assays were conducted. The results indicated that the luciferase activity of SNHG11-Wt reporters was significantly decreased by miR-2355-5p mimics, whereas SNHG11-Mut reporters presented no marked change (Fig. 3F). Based on the results from the RIP assay, it was revealed that SNHG11 and miR-2355-5p were significantly enriched in Ago2 group (Fig. 3G). Additionally, a negative correlation was revealed between SNHG11 and miR-2355-5p in TNBC tissues (Fig. 3H). Hence, it was concluded that miR-2355-5p was sponged by SNHG11 in TNBC.
Figure 3.
SNHG11 is a sponge of miR-2355-5p in TNBC. (A) RNA-pull down assays assessed the potential binding between SNHG11 and the two predicated miRNAs. (B) The effect of SNHG11 silencing on the expression levels of two candidate miRNAs was analyzed with RT-qPCR. (C) RT-qPCR assessed miR-2355-5p expression in TNBC tissues and adjacent ones. (D) RT-qPCR confirmed overexpression efficiency of miR-2355-5p in BT-20 and BT-549 cells. (E) Putative binding fragment of SNHG11 and miR-2355-5p was predicted by starBase. (F) The interaction between SNHG11 and miR-2355-5p was confirmed via luciferase reporter experiments. (G) RIP assay validated the binding of SNHG11 and miR-2355-5p. (H) Spearman's correlation analysis verified the expression correlation between SNHG11 and miR-2355-5p in TNBC tissues. *P<0.05. SNHG11, small nucleolar RNA host gene 11; miR-2355-5p, microRNA-2355-5p; TNBC, triple-negative breast cancer; miRNAs, microRNAs; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; RIP, RNA immunoprecipitation; sh-, short hairpin; NC, negative control; Wt, wild-type; Mut mutant.
CBX5 was a downstream target of miR-2355-5p
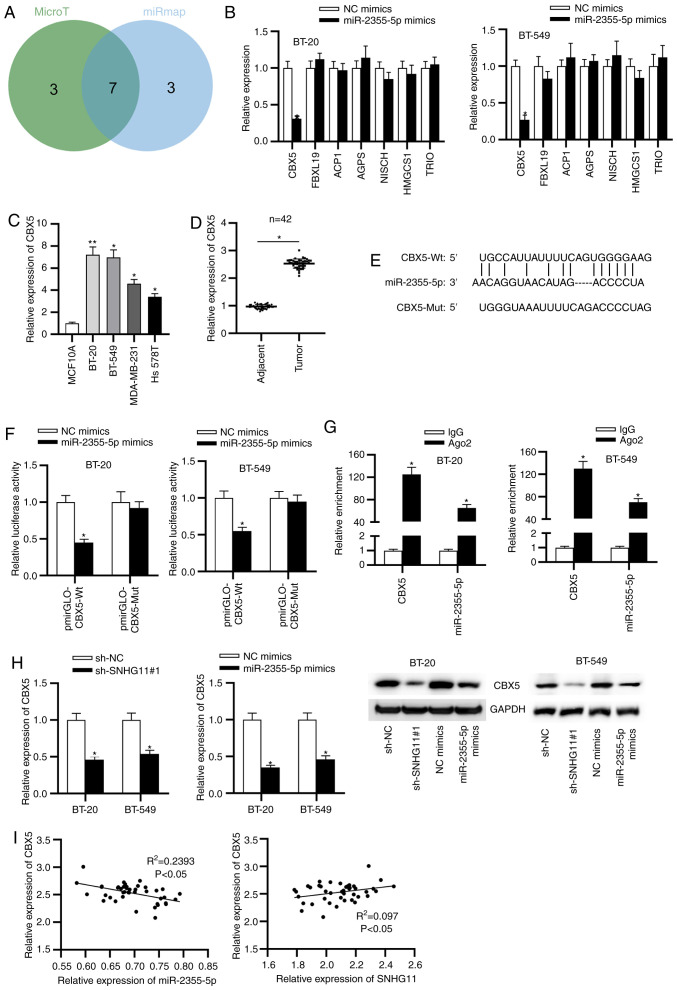
To determine the target of miR-2355-5p, starBase v2.0 was used to obtain potential downstream targets of miR-2355-5p. There were 7 potential mRNAs that were identified as follows: CBX5, FBXL19, ACP1, AGPS, NISCH, HMGCS1 and TRIO (Fig. 4A). After miR-2355-5p upregulation, the expression levels of the aforementioned mRNAs were examined, and the results revealed that only CBX5 was downregulated upon miR-2355-5p overexpression (Fig. 4B). Therefore, CBX5 was demonstrated to be targeted by miR-2355-5p and selected for the subsequent experiments. In addition, it was revealed that CBX5 expression was upregulated in TNBC tissues and cells (Fig. 4C and D). The binding fragment of miR-2355-5p on 3'UTR of CBX5 was predicted by TargetScan (Fig. 4E). Additionally, luciferase activity of CBX5-Wt was decreased with miR-2355-5p upregulation (Fig. 4F). Concurrently, both CBX5 and miR-2355-5p were enriched in Ago2-conjugated beads compared with normal IgG (Fig. 4G). In addition, SNHG11 silencing or miR-2355-5p upregulation suppressed the expression of CBX5 (Fig. 4H). Furthermore, CBX5 expression in TNBC was negatively correlated with miR-2355-5p expression but positively correlated to SNHG11 expression in TNBC tissues (Fig. 4I). To sum up, CBX5 was directly targeted by miR-2355-5p, and SNHG11 upregulated CBX5 by sponging miR-2355-5p.
Figure 4.
CBX5 is a downstream of miR-2355-5p. (A) Seven potential target genes of miR-2355-5p through the starBase database are presented in the Venn diagram. (B) RT-qPCR examined the expression levels of candidate mRNAs after miR-2355-5p upregulation. (C) CBX5 expression was assessed with RT-qPCR in TNBC cells. (D) RT-qPCR determined CBX5 expression in TNBC tissues and adjacent ones. (E) Predicted sequence between CBX5 and miR-2355-5p was obtained from TargetScan. (F) The combination of CBX5 with miR-2355-5p was evaluated via luciferase reporter assay. (G) The binding ability of CBX5 and miR-2355-5p was verified through enrichment in Ago2-conjugated beads compared to in normal IgG via RIP assay. (H) The effect of SNHG11 knockdown or miR-2355-5p overexpression on CBX5 expression was confirmed by RT-qPCR and western blot analysis in TNBC cells. (I) Correlation between CBX5 and miR-2355-5p (SNHG11) expression in TNBC tissues was c with Spearman's correlation analysis. *P<0.05 and **P<0.01. CBX5, chromobox 5; miR-2355-5p, microRNA-2355-5p; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; TNBC, triple-negative breast cancer; RIP, RNA immunoprecipitation; SNHG11, small nucleolar RNA host gene 11; NC, negative control; sh-, short hairpin.
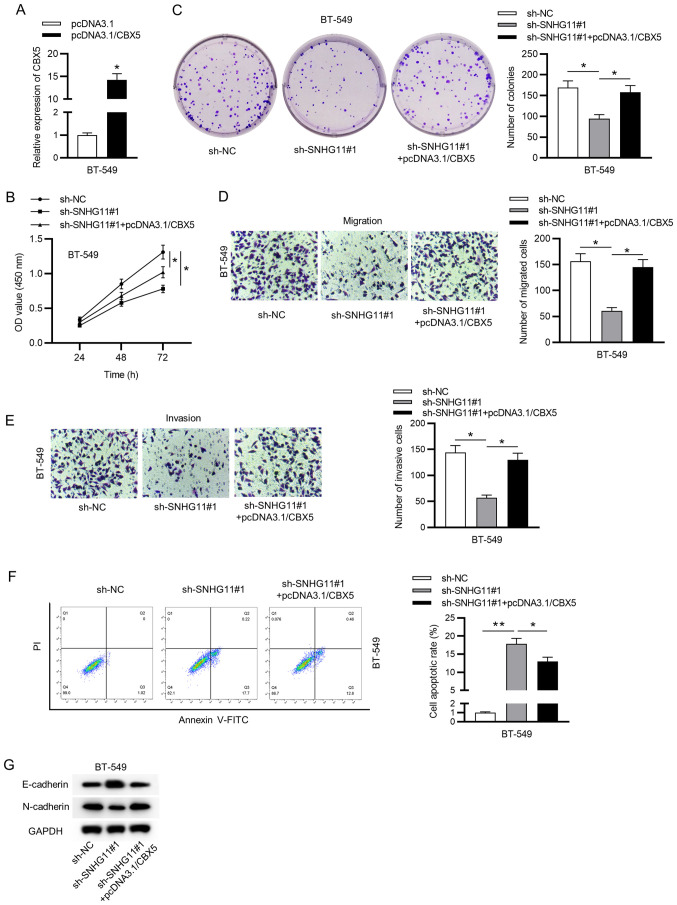
SNHG11 aggravates TNBC proliferation and migration by sponging miR-2355-5p and regulating CBX5
Finally, to determine the impact of the SNHG11/miR-2355-5p/CBX5 axis on TNBC, rescue experiments were implemented in BT-549 cells. Firstly, the overexpressed efficiency of CBX5 in BT-549 cells was validated by RT-qPCR analysis (Fig. 5A). Furthermore, upregulated CBX5 reversed the inhibitive influence of SNHG11 silencing on cell viability and proliferation in TNBC (Fig. 5B and C). Additionally, overexpressed CBX5 rescued the migrative and invasive abilities suppressed under SNHG11 downregulation in TNBC (Fig. 5D and E). Flow cytometric analysis indicated that knockdown of SNHG11 promoted cell apoptosis, but this effect could be reversed by overexpressing CBX5 (Fig. 5F). Moreover, upregulated CBX5 counteracted SNHG11 silencing-mediated attenuation of the EMT process (Fig. 5G). Collectively, our data revealed that SNHG11 accelerated the cell growth of TNBC via modulating the miR-2355-5p/CBX5 axis.
Figure 5.
SNHG11 induces proliferation and migration in TNBC through the miR-2355-5p/CBX5 axis. (A) RT-qPCR examined the overexpression efficacy of CBX5. (B and C) CCK-8 and colony formation assays assessed the viability and proliferative ability of BT-549 cells in each group. (D and E) Transwell assays assessed the migration and invasion of cells under indicated transfection. (F) The apoptosis of BT-549 cells after indicated transfection was evaluated through flow cytometric assay. (G) Western blot analysis measured the protein levels of EMT markers in each group. *P<0.05 and **P<0.01. SNHG11, small nucleolar RNA host gene 11; TNBC, triple-negative breast cancer; miR-2355-5p, microRNA-2355-5p; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; CCK-8, Cell Counting Kit-8; EMT, epithelial-mesenchymal transition; sh-, short hairpin; NC, negative control.
Discussion
Recently, numerous studies have revealed that SNHG11 functions as an oncogene in lung (12), prostate (13) and liver (14). In the present study, initial evidence was provided of the high expression of SNHG11 in TNBC tissues and cells. Then loss-of-function assays were conducted to determine the function of SNHG11 in TNBC. It was revealed that SNHG11 knockdown inhibited TNBC cell viability, proliferation, migration, invasion and the EMT process, whereas it contributed to TNBC cell apoptosis. Therefore, the results also indicated that SNHG11 acted as a tumor promoter in the cell growth of TNBC.
As is well known, lncRNAs regulate the behaviors of cancer cells via sponging miRNAs (26). LncRNAs can reduce the regulatory effect of miRNAs (27). The target gene of SNHG11 was predicted and it was determined that miR-2355-5p was a putative target gene of SNHG11. Previous studies have verified that miR-2355-5p has a suppressive effect in various cancers, including chondrosarcoma (23) and bladder cancer (24). In our present study, it was determined that miR-2355-5p expression was downregulated in TNBC. As reported, lncRNAs contain structural domains that can sense and bind other RNAs via complementary base pairing (28). Hence, further mechanism experiments were performed and it was demonstrated that there was a direct binding sequence between SNHG11 and miR-2355-5p, and that there was a negative expression correlation between these two genes. In brief, binding of SNHG11 and miR-2355-5p could be observed in TNBC.
A previous study has revealed that functional interactions in competing endogenous RNA (ceRNA) networks aid in coordinating a number of biologic processes and, when perturbed, contribute to disease pathogenesis (29). Therefore, to complete the ceRNA mode, starBase 2.0 was utilized to predict the downstream target genes. Among mRNA candidates, CBX5 has previously been reported to aggravate gastric cancer progression (30). Moreover, CBX5 acted as an oncogene and could be positively regulated by BRD4 in ovarian cancer (31). In the present study, it was confirmed that miR-2355-5p could bind to CBX5, and their expression levels were negatively correlated in TNBC tissues. Such ceRNAs thus alter their abundance by competing for binding to miRNAs (32). In addition, as reported, lncRNAs play vital roles in the regulation of cancers via modulating mRNA or the epigenetic landscape of their target genes (33). Hence, subsequent rescue assays verified that overexpression of CBX5 could rescue the inhibitive influence of SNHG11 silencing on TNBC. In summary, lncRNA SNHG11 regulated CBX5 expression via sponging miR-2355-5p in TNBC.
To sum up, the present data initially demonstrated the upregulated expression of SNHG11 in TNBC. SNHG11 upregulated the expression of CBX5 to play a tumor-promoting role in TNBC via sponging miR-2355-5p, which indicated a novel regulatory axis in TNBC. These findings revealed that SNHG11 may be a potential biomarker for TNBC treatment.
Acknowledgements
Not applicable.
Funding Statement
Funding: The current work was supported by Jiangsu Taizhou People's Hospital 2019 Hospital-level Scientific Research Fund Project (grant no. ZL201917).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
LY and WZ conceived and designed the experiments. LY, WZ, PW, QZ, AC, XY and KS carried out the experiments. LY, WZ and KS analyzed the data. LY, WZ and KS drafted the manuscript. All authors agreed to be accountable for all aspects of the work. LY, WZ and KS confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved from the Ethics Committee of Jiangsu Taizhou People's Hospital (Taizhou, China) and informed consent was obtained from the participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.DeSantis CE, Miller KD, Goding Sauer A, Jemal A, Siegel RL. Cancer statistics for African Americans, 2019. CA Cancer J Clin. 2019;69:211–233. doi: 10.3322/caac.21555. [DOI] [PubMed] [Google Scholar]
- 2.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 3.Bergin ART, Loi S. Triple-negative breast cancer: Recent treatment advances. F1000Res. 2019;8(1342) doi: 10.12688/f1000research.18888.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lyons TG, Traina TA. Emerging novel therapeutics in triple-negative breast cancer. Adv Exp Med Biol. 2019;1152:377–399. doi: 10.1007/978-3-030-20301-6_20. [DOI] [PubMed] [Google Scholar]
- 5.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends-an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 6.Hombach S, Kretz M. Non-coding RNAs: Classification, biology and functioning. Adv Exp Med Biol. 2016;937:3–17. doi: 10.1007/978-3-319-42059-2_1. [DOI] [PubMed] [Google Scholar]
- 7.Riva P, Ratti A, Venturin M. The long non-coding RNAs in neurodegenerative diseases: Novel mechanisms of pathogenesis. Curr Alzheimer Res. 2016;13:1219–1231. doi: 10.2174/1567205013666160622112234. [DOI] [PubMed] [Google Scholar]
- 8.Chu C, Quinn J, Chang HY. Chromatin isolation by RNA purification (ChIRP) J Vis Exp. 2012;61(e3912) doi: 10.3791/3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer TR, Mattick JS. Structure and function of long noncoding RNAs in epigenetic regulation. Nat Struct Mol Biol. 2013;20:300–307. doi: 10.1038/nsmb.2480. [DOI] [PubMed] [Google Scholar]
- 10.Ju C, Liu R, Zhang YW, Zhang Y, Zhou R, Sun J, Lv XB, Zhang Z. Mesenchymal stem cell-associated lncRNA in osteogenic differentiation. Biomed Pharmacother. 2019;115(108912) doi: 10.1016/j.biopha.2019.108912. [DOI] [PubMed] [Google Scholar]
- 11.Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017;77:3965–3981. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng D, Chen D, Lin F, Wang X, Lu L, Luo S, Chen J, Xu X. LncRNA NNT-AS1 promote glioma cell proliferation and metastases through miR-494-3p/PRMT1 axis. Cell Cycle. 2020;19:1621–1631. doi: 10.1080/15384101.2020.1762037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Y, Jiang A. ST8SIA6-AS1 promotes hepatocellular carcinoma by absorbing miR-5195-3p to regulate HOXB6. Cancer Biol Ther. 2020;21:647–655. doi: 10.1080/15384047.2020.1743150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T, Wang X, Zhai J, Wang Q, Zhang B. doi: 10.1089/cbr.2019.3278. Long noncoding RNA UCA1 facilitates endometrial cancer development by regulating KLF5 and RXFP1 gene expressions. Cancer Biother Radiopharm: May 15, 2020 (Epub ahead of print). [DOI] [PubMed] [Google Scholar]
- 15.Liu S, Yang N, Wang L, Wei B, Chen J, Gao Y. lncRNA SNHG11 promotes lung cancer cell proliferation and migration via activation of Wnt/β-catenin signaling pathway. J Cell Physiol. 2020;235:7541–7553. doi: 10.1002/jcp.29656. [DOI] [PubMed] [Google Scholar]
- 16.Bicak M, Wang X, Gao X, Xu X, Väänänen RM, Taimen P, Lilja H, Pettersson K, Klein RJ. Prostate cancer risk SNP rs10993994 is a trans-eQTL for SNHG11 mediated through MSMB. Hum Mol Genet. 2020;29:1581–1591. doi: 10.1093/hmg/ddaa026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang W, Huang F, Lei Z, Luo H. LncRNA SNHG11 promotes proliferation, migration, apoptosis, and autophagy by regulating hsa-miR-184/AGO2 in HCC. Onco Targets Ther. 2020;13:413–421. doi: 10.2147/OTT.S237161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simonson B, Das S. MicroRNA therapeutics: The next magic bullet? Mini Rev Med Chem. 2015;15:467–474. doi: 10.2174/1389557515666150324123208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morales S, Monzo M, Navarro A. Epigenetic regulation mechanisms of microRNA expression. Biomol Concepts. 2017;8:203–212. doi: 10.1515/bmc-2017-0024. [DOI] [PubMed] [Google Scholar]
- 20.Jámbor I, Szabó K, Zeher M, Papp G. The importance of microRNAs in the development of systemic autoimmune disorders. Orv Hetil. 2019;160:563–572. doi: 10.1556/650.2019.31349. (In Hu) [DOI] [PubMed] [Google Scholar]
- 21.Yan K, Hou L, Liu T, Jiao W, Ma Q, Fang Z, Zhang S, Song D, Liu J, Gao X, Fan Y. lncRNA OGFRP1 functions as a ceRNA to promote the progression of prostate cancer by regulating SARM1 level via miR-124-3p. Aging (Albany NY) 2020;12:8880–8892. doi: 10.18632/aging.103007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, Liang M, Zhang Y, Yuan B, Gao W, Shi Z, Bai J. miR-93, miR-373, and miR-17-5p negatively regulate the expression of TBP2 in lung cancer. Front Oncol. 2020;10(526) doi: 10.3389/fonc.2020.00526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheng C, Zhang Z, Cheng F, Shao Z. Exosomal lncRNA RAMP2-AS1 derived from chondrosarcoma cells promotes angiogenesis through miR-2355-5p/VEGFR2 axis. Onco Targets Ther. 2020;13:3291–3301. doi: 10.2147/OTT.S244652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D, Chen J, Gao J, Zhang Y, Ma Y, Wei W, Wei Y. LncRNA DDX11-AS1 promotes bladder cancer occurrence via protecting LAMB3 from downregulation by sponging miR-2355-5p. Cancer Biother Radiopharm. 2020;35:319–328. doi: 10.1089/cbr.2019.3021. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Soudyab M, Iranpour M, Ghafouri-Fard S. The role of long non-coding RNAs in breast cancer. Arch Iran Med. 2016;19:508–517. [PubMed] [Google Scholar]
- 27.Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing miRNA-lncRNA interactions. Methods Mol Biol. 2016;1402:271–286. doi: 10.1007/978-1-4939-3378-5_21. [DOI] [PubMed] [Google Scholar]
- 28.Guil S, Esteller M. RNA-RNA interactions in gene regulation: The coding and noncoding players. Trends Biochem Sci. 2015;40:248–256. doi: 10.1016/j.tibs.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 29.Karreth FA, Pandolfi PP. ceRNA cross-talk in cancer: When ce-bling rivalries go awry. Cancer Discov. 2013;3:1113–1121. doi: 10.1158/2159-8290.CD-13-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo J, Zhang Z, Pan L, Zhou Y. Identification of miR-758-3p as potential modulator of CBX5 expression in gastric cancer. Technol Cancer Res Treat. 2018;17(1533033818816061) doi: 10.1177/1533033818816061. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Pongas G, Kim MK, Min DJ, House CD, Jordan E, Caplen N, Chakka S, Ohiri J, Kruhlak MJ, Annunziata CM. BRD4 facilitates DNA damage response and represses CBX5/Heterochromatin protein 1 (HP1) Oncotarget. 2017;8:51402–51415. doi: 10.18632/oncotarget.17572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smillie CL, Sirey T, Ponting CP. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit Rev Biochem Mol Biol. 2018;53:231–245. doi: 10.1080/10409238.2018.1447542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romero-Barrios N, Legascue MF, Benhamed M, Ariel F, Crespi M. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018;46:2169–2184. doi: 10.1093/nar/gky095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.