Abstract
Background
Current algorithm for Congenital Chagas Disease (cCD) diagnosis is unsatisfactory due to low sensitivity of the parasitological methods. Moreover, loss to follow-up precludes final serodiagnosis after nine months of life in many cases. A duplex TaqMan qPCR kit for Trypanosoma cruzi DNA amplification was prospectively evaluated in umbilical cord (UCB) and peripheral venous blood (PVB) of infants born to CD mothers at endemic and non-endemic sites of Argentina.
Methods
We enrolled and followed-up 370 infants; qPCR was compared to gold-standard cCD diagnosis following studies of diagnostic accuracy guidelines.
Findings
Fourteen infants (3·78%) had cCD. The qPCR sensitivity and specificity were higher in PVB (72·73%, 99·15% respectively) than in UCB (66·67%, 96·3%). Positive and negative predictive values were 80 and 98·73% and 50 and 98·11% for PVB and UCB, respectively. The Areas under the Curve (AUC) of ROC analysis for qPCR and micromethod (MM) were 0·81 and 0·67 in UCB and 0·86 and 0·68 in PVB, respectively. Parasitic loads ranged from 37·5 to 23,709 parasite equivalents/mL. Discrete typing Unit Tc V was identified in five cCD patients and in six other cCD cases no distinction among Tc II, Tc V or Tc VI was achieved.
Interpretation
This first prospective field study demonstrated that qPCR was more sensitive than MM for early cCD detection and more accurate in PVB than in UCB. Its use, as an auxiliary diagnostic tool to MM will provide more accurate records on cCD incidence.
Funding
FITS SALUD 001-CHAGAS (FONARSEC, MINCyT, Argentina) to the Public-Private Consortium (INGEBI-CONICET, INP-ANLIS MALBRAN and Wiener Laboratories); ERANET-LAC-HD 328 to AGS and PICT 2015-0074 (FONCYT, MinCyT) to AGS and FA.
Keywords: Congenital Chagas disease, Early diagnosis, Real Time PCR, Trypanosoma cruzi, Discrete typing unit, Parasitic load
Research in context.
Evidence before this study
Successful control of vectorial and transfusional transmission has exposed public health importance of Congenital Chagas disease (cCD), which nowadays is the main route of Chagas disease (CD) urbanization in non-endemic areas. In 2010, the WHO recommended governments to launch systems of early detection of new infections and congenital infections in newborns. It was estimated that new cases of cCD in endemic regions represented 22% of all new cases of CD. Six years later, the Panamerican Health Organization incorporated cCD in the Elimination of mother-to-child transmission (EMTCT) -Plus initiative to boost control of vertical infections in the region, together with HIV, Syphilis and Hepatitis B.
Current diagnosis of cCD is based on positive microscopic tests performed shortly after birth and/or by serological reactivity after 9 months of age. However, this diagnostic algorithm is unsatisfactory due to the low sensitivity and operator-dependence of parasitological methods and loss to follow-up once the neonate leaves maternity, a frequent event in rural endemic areas, where population is more vulnerable.
Polymerase chain reaction (PCR) methodology has been proposed as a higher sensitive and specific laboratory tool compared to current diagnostics algorithm for early diagnosis of cCD, but no prospective field evaluations have been done.
Added value of this study
This is the first prospective field evaluation of Real Time PCR (qPCR) accuracy for early diagnosis of cCD in endemic and non-endemic sites for CD, in comparison with current diagnostics algorithm following Standards for Reporting of Diagnostic Accuracy guidelines. The study compared the performance of a qPCR assay using as starting clinical material, umbilical cord and peripheral blood samples of newborns and infants born to CD women. Furthermore, parasitic loads were estimated and discrete typing units of the infecting parasite populations were genotyped using molecular amplification strategies. The qPCR assay accuracy was higher in peripheral venous blood than in umbilical cord blood samples and its clinical sensitivity was almost twice than that of the current parasitological method.
Implications of all the available evidence
The implementation of this qPCR assay will improve early detection of cases, provide more accurate records on the number of infants born to CD mothers in endemic and non-endemic countries and allow estimating the likely number of cases missed in places where only traditional parasitological procedures are still employed.
Furthermore, early qPCR diagnosis will contribute to maximize prompt treatment of infected neonates, with high impact in public health.
Alt-text: Unlabelled box
1. Introduction
Successful control of vectorial and transfusional transmission has exposed public health importance of congenital Chagas disease (cCD).
Due to migrations from endemic areas to vector-free suburban and urban centers, cCD is mainly accountable for Chagas disease (CD) urbanization [1]. The parasite may be transmitted from mother-to-child during pregnancy or at delivery and may infect twins and siblings in successive generations [2,3].
Current diagnosis of cCD is based on positive parasitological tests shortly after birth and/or serological reactivity after nine months of age. Parasitological techniques detect motile parasites in umbilical cord blood (UCB) or peripheral venous blood (PVB), concentrate parasites by centrifugation using capillary (microhematocrite) or microcentrifuge tubes (microMethod (MM)) followed by microscopic examination of the buffy coat. If the test is negative at birth, it can be repeated at one month of age, when the peak of parasitemia is usually observed. These parasitological tests require prompt processing of the sample to allow visualization of motile parasites, experienced laboratory operators and at least half an hour of microscopic observation per sample for adequate sensitivity. However, all these conditions are seldom achieved in public health laboratories. Other parasitological strategies, such as hemoculture, are not routinely used for diagnosis of cCD. The sensitivity of parasitological methods is disappointing, and in more than 50% of neonates, final diagnosis can only be done by means of serological analysis. A main concern is that 75-80% of infants do not go back to the health centers for diagnosis confirmation, precluding their opportunity to treatment, which is highly successful when implemented during the first year of life [4,5].
Home-brewed and Real Time PCR (qPCR) studies have encouraged the use of molecular diagnostics for early detection of cCD [4], [5], [6], [7]. The incorporation of nucleic acid amplification methods to the current diagnostics algorithm would allow early detection of a higher proportion of infected cases that are not detected by parasitological methods, preventing their loss to follow-up and allowing their prompt treatment. Indeed, the efficacy of treatment with Nifurtimox or Benznidazole increases closer to birth with higher opportunity to demonstrate cure, while if left untreated the infection progresses to chronic CD with a fall in the cure rate [8]. Access to treatment before the first year of age has shown 99% of efficacy and is the main strategy to interrupt congenital transmission to future generations.
T. cruzi qPCR tests have recently become commercially available in Europe [9]. However, no evaluation of standardized qPCR kits has been done so far in the context of prospective blind-based studies [[4], [5], [6], [7],9]. Our central hypothesis proposes that a qPCR standardized method will depict better accuracy than that of current parasitological tests to detect cCD at birth and/or within the first months of age in infants born to seropositive mothers. Our aim was to evaluate the sensitivity and specificity of a duplex TaqMan Real Time qPCR kit designed for qualitative detection of T. cruzi DNA in UCB and PVB samples. The performance of this index test was compared to that of current parasitological diagnosis in prospective clinical samples from neonates born to infected mothers that attended public health centers in endemic and non-endemic sites of Argentina. Furthermore, to characterize T. cruzi infection in cCD infants, parasitic loads and discrete typing units (DTUs) were determined directly from their qPCR positive blood samples [10,11].
2. Methods
2.1. Study design
This study followed the endorsements for “studies of diagnostic accuracy” (STARD). It aimed to estimate, on a blind basis, the accuracy (sensitivity and specificity) of a qPCR prototype kit as index test, using as gold standard the current diagnostics algorithm for cCD: MM at birth and between four to eight weeks of age and serological analysis after nine months of age [11,12].
The evaluation was performed at five public health care centers (HC) in Argentina: the Instituto Nacional de Parasitología Dr. M. Fatala Chabén - ANLIS Dr. C. G. Malbrán (INP), in Buenos Aires city (HC1), and the remainder in endemic Provinces: Instituto de Maternidad y Ginecología “Nuestra Señora de las Mercedes”, San Miguel de Tucumán, Tucumán (HC2), Hospital “Dr. Julio C. Perrando” in Resistencia, Chaco (HC3); Hospital Regional “Dr. Ramón Carrillo”, Santiago del Estero city and Centro Integral de Salud La Banda, Santiago del Estero (HC4 and HC5, respectively). The personnel involved in the field study together with the researchers responsible for the project design, execution and analysis formed the Congenital Chagas Disease Study Group (Supplemental File).
2.2. Ethical statement
Protocols were approved by the Bioethics committees of the participant centers, after first approval of the Institutional review board (IRB) at “Centro de Educación Médica e Investigaciones Clínicas Norberto Quirno” (CEMIC), Buenos Aires.
2.3. Participants
T. cruzi-infected women older than 18 years old diagnosed by routine serological analysis following the Guidelines of the National Ministry of Health [13] and residing within 50 km from the corresponding HC, were eligible for enrollment and invited to participate. Those living outside the catchment area were excluded. Mother-newborns sample size was estimated using EpiData Software version 3.1 (http://www.epidata.dk). Assuming an expected sensitivity of 90% for qPCR and 50% for MM and a 4% mean rate of vertical transmission (5·55% in 2011; 5·08% in 2012; 3·08% in 2013) [14]; at least 483 mothers were planned to be enrolled, expecting 5% of loss to follow-up. Eligible women signed a written informed consent before enrollment. Only live births occurring between 8.00 am to 8.00 pm were enrolled at endemic provinces. Health center 1, without a Maternity service, enrolled outpatients aged between one and 22 weeks, weighing more than 3 kilograms.
Clinical and socio demographic characteristics relevant to T. cruzi infection and potential risk factors for cCD transmission were documented: residence in rural area, presence of triatomine bugs inside patients’ houses, maternal HIV co-infection, mothers stating having received trypanocidal treatment, previous gestations, older siblings infected by T. cruzi, vaginal delivery, breastfeeding at four to eight weeks visit and ten months visit, and cracked nipples with bleeding.
Complete cases were children from whom at least one early sample was collected for qPCR and MM analysis and a second one around ten months of age for serological analysis.
2.4. Sample collection for laboratory tests
Five mL of UCB was drawn after birth, as reported [7]. The UCB was collected in two tubes, one containing EDTA (4 mL) to perform the index test and the other one containing heparin (1 mL) to perform the reference test. Heparin-treated samples were stored for a maximum of 12 hours at 4°C and prior to microscopic observation the tubes were stored 30 minutes at room temperature.
Peripheral venous blood was obtained at birth if UCB was not available, or at four to 16 weeks of life in 1·5 mL EDTA-treated tubes for qPCR and 1 mL of heparinized-blood tubes for MM. Finally, PVB was drawn at ten months of age in Vacutainer tubes for serological examination. An aliquot was centrifuged for serum collection, kept in buffered glycerin and sent to INP laboratory for serology analysis. A volume of 1·2 mL of EDTA-PVB was collected in 2 mL microtubes for genotyping of T. cruzi DTUs.
All EDTA-blood samples were homogenized with a Guanidinium-based stabilizing reagent (DNAgard, Biomatrica, USA) in the proportion 1:4 (vol: vol) of stabilizer: blood and stored at 4°C for qPCR.
At each period of sample collection, samples were codified and kept in a double-blind form for all participants in the study.
2.5. Index test
A TaqMan based duplex qPCR assay targeting T. cruzi satellite DNA sequence and an internal amplification control (IAC) was developed and standardized. The satellite DNA based primers and probe sequences are primer cruzi forward: 5`GGGAGTCAGAGRCACTCTC3`, primer cruzi reverse: 5`AATTCCTCCAAGMAGCGGATA3` and cruzi probe: FAM-CACACACTGGACACCAAACAACCC-BHQ1. They were designed after alignment of database T. cruzi satellite DNA sequences (TriTrypDB, RRID:SCR_007043) to have a relatively well-conserved sequence recognized in all DTUs. The primers and probe sequences for IAC were modified from those published by Duffy and colleagues [10]. They are primer IAC forward: 5`CGTCATGGAACAGCACGTAC3`, primer IAC reverse: 5`ACCAAGACGAAAGCTAAAACACC3` and IAC probe HEX-TGACTGGATTTGGAGCATCTGTTC-BHQ1 (underlined Cs are locked nucleic acid residues).
The assay has been developed into a commercial product manufactured by Wiener Laboratorios, S.A.I.C and released to the market as “T. cruzi DNA test” for in-vitro diagnostics use (IVD), after approval by the “Administración Nacional de Medicamentos, Alimentos y Tecnología Médica”, ANMAT, Argentina, under the ID PM 1102-173 (Supplemental Data). The index test was carried out at LaBMECh, INGEBI in collaboration with Wiener Laboratorios, S.A.I.C., where the batch of the kit for the field study was produced. Nucleic Acids were extracted from 250 µL of stabilized blood, adding 5 µL of linear IAC (40 pg/µL) prior extraction with the High Pure PCR Template Preparation kit (Roche Diagnostics, Germany) [10]. No more than 12 blood samples plus a negative control (seronegative blood) were purified in the same experiment. The qPCR was performed in duplicates from each DNA extract. The kit included Uracil DNA Glycosylase to avoid carry-over contamination from previously amplified products. The reaction contained 5 µL of DNA sample in a final volume of 20 µL. The qPCR plates were run in an ABI 7500 thermocycler (Applied Biosystems, California, USA). Cycling conditions were one cycle of 2 min at 50°C, 10 min at 95°C and 45 cycles at 95°C, 15 sec and 58°C, 1 min.
2.6. External Quality assurance of qPCR performance
An external quality assurance of qPCR performance was built-up at the Research & Development Department of INP to evaluate the methodology implemented. Three proficiency testing panels made of seronegative human blood samples spiked with 1·5 (P102 sample), 15 (P103 sample) and 150 (P104 sample) parasite equivalents/mL of T. cruzi (a Tc V strain isolated from a cCD case [4]) and a negative blood control without parasites (P101 sample) were evaluated in three laboratories using different thermocyclers: ABI 7500 (Applied Biosystem, California, USA), Light Cycler 96 (Roche Diagnostic Rotkreuz, Switzerland), CFX96 Touch Real Time PCR Detection System (Bio Rad laboratories, California, USA). Each operator ran the assays from each panel in duplicates and in two consecutive days. The Cts were registered and qualitative results were expressed as non-detectable or positive. The reports were sent to the reference laboratory for statistical analysis. Intra- and inter-laboratory qPCR results from the proficiency testing panels were analyzed by SPSS Statistics software. The Cohen kappa coefficient was used to analyze the closeness of the agreement and the differences between qualitative qPCR results obtained from the samples tested at the three participating laboratories. The Analyse-It software for Microsoft Excel 5.30.2 Build 7069-17990 was used for this analysis. The performances of each participating Laboratory were compared in terms of the Z score, in accordance with ISO 13528:2015(E) [11]. The Z-score was negative or positive when data points were below or above the mean, respectively.
2.7. Reference standards
The MM was done in each HC on duplicates using 500µL of heparinized blood collected into Vacutainer tubes and loaded into 2mL microtubes, as reported [12]. Serological assays were performed at ten to 12 months of age using Chagatest ELISA recombinante v.3.0 and Chagatest HAI assays (Wiener Laboratories, Argentina) at the INP, using the cut-off values recommended by the manufacturers. Two positive serological tests were necessary for cCD diagnosis; in case of discordance, an indirect immunofluorescence (IFI) test was performed, following the recommendations of the manufacturer [15].
2.8. Definition and rationale for test positivity
2.8.1. Index test
Pre-analytical issues such as incorrect sample volume, coagulated sample, viscous sample hard to pipette, detected upon sample reception at LabMECh in INGEBI, determined that an UCB or PVB sample had to be excluded for analysis of qPCR accuracy. Analytical issues such as getting outlier (cycle threshold) Ct values of IAC, which were estimated for each qPCR run using the Tukey´s criteria (Ct > 75th percentile + 1.5× interquartile distance of median Ct) [10] determined that the qPCR result was taken as not valid for analysis of accuracy.
A sample was defined as qPCR positive only if both DNA duplicates were qPCR positive with (quantification cycle) Cq values<39, using a threshold value of 0·04 (ABI 7500 thermocycler) and was reported as non-detectable when the Cq of both duplicates were ≥39 or were non-detectable. Samples showing only one qPCR positive replicate (Cq <39) and the other one negative were considered as inconclusive and excluded for analysis of qPCR accuracy.
2.8.2. Reference tests
After 15 minutes of microscopic observation (40x) of each MM duplicate, the motility of trypomastigotes was indicative of a positive sample [12]. Serological positivity was determined according to the guidelines of the manufacturers for each test.
Clinical information and reference standard results were not available to the performers/readers of the index test. The index test results were not available to the health personnel responsible for the analyses of the reference tests. Only a positive MM or reactivity of two serological tests confirmed cCD and were the criterions to derive the patient for treatment, as indicated by national guidelines.
2.9. qPCR analysis of clinical samples
Standard operative procedures for sample collection, conservation, transportation, DNA extraction, qPCR amplification and interpretation of qPCR results were redacted. Estimation of qPCR sensitivity and specificity was done only in valid samples. Sensitivity was calculated as the proportion of samples with the target condition (cCD) which tested positive using the index test. Its specificity was calculated as the proportion of samples without cCD, which tested negative using the index test. Missing data on the index or reference tests precluded analysis of the case. In this study, qPCR positive results were not used for taking clinical decisions.
2.10. Estimation of parasitic loads
The kit was designed for qualitative amplification of T. cruzi DNA. However, parasitic loads were quantified to further characterize those qPCR positive samples. Thus, a standard quantitative curve was constructed as follows: a pellet of epimastigotes (CL-Brener stock) quantitated using a Neubauer chamber was suspended in EDTA-Blood and mixed with DNAgard to a final concentration of 105 parasites/mL. DNA extraction was performed as described above and diluted serially (1/10) in DNA matrix negative for T. cruzi, which was extracted from a seronegative PVB sample mixed with DNAgard. Quantification curves ranged from 10,000 to 0·5 par.eq/mL and were amplified in each plate together with samples and controls. The qPCR conditions were the same as those for clinical samples and the parasitic loads were automatically calculated by the thermocycler software and expressed in par.eq/mL.
2.11. Identification of T. cruzi DTU groups
The qPCR positive samples with parasitic loads higher than 5 par.eq/mL were genotyped using two different genotyping algorithms: 1) a Multiplex TaqMan Real Time qPCR algorithm for distinction of the six DTUs [16], and 2) Hemi-nested PCRs targeting the spliced Leader-Intergenic Regions I and II to identify T. cruzi I (350bp) and T. cruzi II/V/VI (300bp) groups, respectively [17]. The latter strategy has higher sensitivity than qPCR but cannot distinguish among individual T. cruzi II, T. cruzi V or T. cruzi VI genotypes [17].
2.12. Statistical Analysis
For data analysis we used the STATA software, version 11 and Epidat software, version 3.1. A descriptive analysis was done over all included cases. Absolute and relative frequencies of variables concerning characteristics of mothers, pregnancy and delivery of the index case were calculated. For proportions, 95% confidence intervals (CI) were obtained. The same analysis was performed in complete followed-up cases with final diagnosis. Comparison of proportions between cCD and not-cCD cases was performed using Chi2 or Fischer exact tests, and comparison of means was done using Student's or Wilcoxon rank-sum tests, as appropriate. For analysis of qPCR accuracy, receiver operating characteristic (ROC) curves and the corresponding areas under the curves (AUC) were carried out. A statistical comparison of the areas under two ROC curves derived from the same set of patients` samples was done by taking in account the correlation between the areas that is induced by the paired nature of the data, according to Hanley and Mc Neil [18] using a non-parametric approach [19]. A statistically significant level of p<0·05 was assumed. Non-significant p values were expressed as p:ns.
2.13. Role of the funding source
The study sponsors did not have any role in study design, collection, analysis, and interpretation of data, writing of the report and in the decision to submit the paper for publication.
3. Results
3.1. Participants and samples
Out of 622 screened seropositive pregnant women, 559 were enrolled (Fig. 1). Study cases were classified into two groups according to the period during which the first sample was collected. Group 1 clustered cases from which the first sample was obtained before the neonate abandoned the maternity service (within the first 72 hours of age) and Group 2 included cases which first sample was collected when the outpatient had his first control between four days and 22 weeks after birth.
Fig. 1.
Flowchart of Prospective Field study to evaluate qPCR kit prototype for diagnosis of cCD. PVB: Peripheral Venous Blood; UCB: Umbilical Cord Blood; MM: MicroMethod. *In four enrolled babies no sample could be withdrawn. No visit: the patient did not attend the appointment for collection of the follow-up sample.
In Group 1, 256 mother-newborn binomials were enrolled. In 95 newborns, UCB samples were obtained and PVB in 161. Out of them, 85 cases had a second PVB collected between two and 28 weeks and 147 completed follow-up around 10 months of age (68 with an UCB and 79 with a PVB first sample) (Fig. 1).
In Group 2, 303 binomials were enrolled. The first PVB sample was obtained from 259 neonates at a median age of 33 days (min four days, max 22 weeks). The remaining cases were not included because 44 women who had given their informed consent did not return to the corresponding HC (14·52% loss to follow-up); 223/259 tested infants were followed-up at around 10 months of age for serodiagnosis.
In sum, 370 cases were considered as complete followed-up cases (Fig. 1). The number and type of samples from them are detailed in Supplementary Table 1. Out of 370 infants with complete follow-up, 132 were born to CD mothers residing in an urban non-endemic area (Buenos Aires city and surroundings), 75 in Tucumán, 56 in Chaco and 107 in Santiago del Estero endemic provinces. Fourteen infants (3·78%) were diagnosed as cCD based on the gold-standard diagnostic algorithm. The rates of congenital transmission, the number of UCB and PVB samples and the number of complete followed-up cases distributed per HC are shown in Table 1.
Table 2.
Positivity by MicroMethod and qPCR in valid samples obtained from complete followed-up cases
| Sample Type (cCD / tested samples) | Diagnostic accuracy study | MicroMethod (positive/ tested samples)* | qPCR (positive/ tested samples)* |
|---|---|---|---|
| UCBa (3/57) | (1/57) | (2/57) | |
| Sensitivity % (95% CI) | 33.33 (0.00-100.00) | 66.67 (0.00-100.00) | |
| Specificity % (95% CI) | 100.00 (99.07-100.00) | 96.30 (90.33-100.00) | |
| PPV % (95% CI) | 50.00 (50.00-100.00) | 50.00 (50.00-100.00) | |
| NPV % (95% CI) | 96.43 (90.68-100.00) | 98.11 (93.51-100.00) | |
| PVBb (11/246) | (4/246) | (8/246) | |
| Sensitivity % (95% CI) | 36.36 (3.39-64.34) | 72.73 (41.86-100.00) | |
| Specificity % (95% CI) | 100.00 (99.79-100.00) | 99.15 (97.76-100.00) | |
| PPV % (95% CI) | 97.11 (94.79-100.00) | 80.00 (50.21-100.00) | |
| NPV % (95% CI) | 97.18 (94.91-99.44) | 98.73 (97.09-100.00) | |
| All samplesc (14/350) | (6/350) | (10/350) | |
| Sensitivity % (95% CI) | 40.00 (11.87-68.13) | 66.67 (39.48-93.86) | |
| Specificity % (95% CI) | 100.00 (99.85-100.00) | 98.81 (97.49-100.00) | |
| PPV % (95% CI) | 100.00 (91.67-100.00) | 71.43 (44.19-98.66) | |
| NPV % (95% CI) | 97.38 (95.55-99.22) | 98.51 (97.11-99.96) | |
| UCB or PVBd (2/46) | (0/46) | (2/46) | |
| Sensitivity % (95% CI) | 100.00 (75.00-100.00) | ||
| Specificity % (95% CI) | 97.73 (92.19-100.00) | ||
| PPV % (95% CI) | 66.67 (0.00-100.00) | ||
| NPV % (95% CI) | 100.00 (98.84-100.00) |
cCD: Congenital Chagas Disease; a- UCB: Umbilical Cord Blood collected at birth; b- PVB: Peripheral Venous Blood collected after 72 hours, till five months of life (day 4-133); c- UCB and PVB collected from day 0-133; d- Cases with two blood samples, one collected at birth and the other one in the second visit.
*Numbers in bold between parentheses indicate the rate of positive results out of the tested samples using the MicroMethod or qPCR. CI: Confidence Interval; PPV: Positive Predictive Value; NPV: Negative Predictive Value.
3.2. Diagnostic accuracy of qPCR
The qPCR assay was carried out in 421 samples from 370 babies. Out of them, 350 (57 UCB and 293 PVB) were included for analysis of qPCR accuracy (Table 2 and Supplementary Table 2). The remaining cases were excluded because their samples were not valid at pre-analytical or analytical stages, as mentioned in Methods. The proportion of not valid samples was similar between UCB (3/67; 4·47%) and PVB samples (26/349; 7·44%) (p:ns). Excluded qPCR results due to outlier values of IAC or to discordance between duplicates was also similar between UCB and PVB valid samples (5/64 (7·8%) and 29/323 (8·9%), respectively (p: ns, Supplementary Table 2). The qPCR accuracy was estimated taking in account the source of the sample (UCB or PVB) and the moment at which it was withdrawn (before or after leaving the Maternity Service, around 72 hs after birth) (Table 2). The highest level of qPCR accuracy (72·73% of sensitivity and 99·15% of specificity) was achieved in 246 PVB samples obtained between four and 133 days of life (median: 34·5 days), whereas qPCR in UCB samples (N=57) had 66·67% of sensitivity and 96·3% of specificity (Table 2). The AUC of ROC analysis for qPCR was 0·8594 and 0·8148 for PVB and UCB samples, respectively while the AUC for MM was 0·6818 and 0·6667 (Fig. 2). No significant differences were observed between the AUCs of ROC curves for paired MM and qPCR data obtained in PVB and UCB samples (p=0·08).
Table 3.
a: MicroMethod, qPCR, serological and Discrete Typing Unit results in congenital Chagas Disease patients.
| ID | Site | Umbilical Cord blood |
Peripheral Venous Blood |
Serodiagnosis at 10 months |
Discrete Typing Units |
||||||||||
| MM | qPCR | Par Load | MM | qPCR | Par Load | Age (days) | Diagnosis | ELISA | HAI | IFI | SL IR II | SL-IR I | TaqMan PCR | ||
| 1001 | HC1 | NA | NA | x | Neg | Neg | x | 64 | Reactive | 0·759 Ɛ | 64 ¤ | 32 ø | Tc II/V/VI§ | Neg | Neg |
| 1006 | HC1 | NA | NA | x | Neg | Neg | x | 118 | Reactive | 4·442§ | 256 ¤ | ND | ND | ND | ND |
| 1039 | HC1 | NA | NA | x | Neg | Pos | 78·8 | 33 | Discordant*◊ | 0·015 ß | 128 ¤ | NR | Tc II/V/VI | Neg | Neg |
| 1044 | HC1 | NA | NA | x | Pos | Pos | 3684 | 87 | Non Reactive* | 0·015 ß | NR ¤ | ND | Tc II/V/VI | Neg | Tc V |
| 1083 | HC1 | NA | NA | x | Neg | Pos | 356 | 36 | Reactive | 2·985 ß | 256 ¤ | ND | Tc II/V/VI | Neg | Neg |
| 1111 | HC1 | NA | NA | x | Pos | Pos | 641 | 10 | Non Reactive* | 0·036 ¥ | NR ¤ | ND | Tc II/V/VI | Neg | Tc V |
| 1167 | HC1 | NA | NA | x | Pos | Pos | 37·5 | 49 | Reactive | 2·501 ß | 256 ¤ | ND | Tc II/V/VI | Neg | Neg |
| 2037 | HC2 | NA | NA | x | Pos | Pos | 0 | 9 | Non Reactive* | 0·018 § | NR ¤ | ND | ND | ND | ND |
| 2047 | HC2 | NA | NA | x | Pos | Neg | x | 11 | Reactive | 2·624 § | 256 ¤ | ND | Tc II/V/VI§ | Neg | Tc V |
| 4045 | HC3 | Neg | Pos | 1418 | Neg | Pos | 23709 | 54 | Reactive | 2·624 ¥ | 256 ¤ | ND | Tc II/V/VI | Neg | Tc V |
| 4055 | HC3 | Neg | Neg | x | Neg | Pos | 53·4 | 41 | Reactive | 2·531 ß | 128 ¤ | ND | Tc II/V/VI | Neg | Neg |
| 4057 | HC3 | NA | NA | x | Neg | Pos | 82·0 | 45 | Reactive | 1·418 ¥ | 256 ¤ | ND | Tc II/V/VI | Neg | Neg |
| 4058 | HC3 | Pos | Pos | 1370 | Neg | Neg | x | NA | Non Reactive* | 0·059 ¥ | NR ¤ | ND | Tc II/V/VI | Neg | Tc V |
| 6004 | HC4 | NA | NA | x | Pos | Neg | x | 1 | Non Reactive* | 0·017 § | NR ¤ | ND | ND | ND | ND |
| b: MicroMethod, qPCR, serological and Discrete Typing Unit results in non congenital Chagas Disease patients. | |||||||||||||||
| 1110 | HC1 | NA | NA | x | Neg | Pos | 0·44 | 12 | Non Reactive | NR | NR ¤ | ND | ND | ND | ND |
| 4044 | HC3 | NA | NA | x | Neg | Pos | 3·64 | 69 | Non Reactive | NR | NR ¤ | ND | ND | ND | ND |
| 4047 | HC3 | Neg | Pos | 39 | NA | NA | x | x | Non Reactive | NR | NR ¤ | ND | Tc II/V/VI | Tc I | Neg |
| 5069 | HC5 | Neg | Pos | 0·45 | NA | NA | x | x | Non Reactive | NR | NR ¤ | ND | ND | ND | ND |
MM: MicroMethod; Par load: Parasitic load in par.eq/mL * Treated patient: NA: Not available sample; ND: Not done; Pos: Positive; Neg: Negative; x: Not tested/No data; NR: Non Reactive
Ɛ cut-off: 0·152; § cut-off: 0·318; ß cut-off: 0·316; ¥ cut-off: 0·322; ¤ cut-off: dilution factor 16; ø cut-off: dilution factor 32; ◊ Case diagnosed by a positive MM on a second
routine control at 3 months of age and treated. § DTUs were identified in qPCR positive samples obtained at ten month of age.
Fig. 2.
Analysis of ROC curves to compare the accuracy of qPCR (right panels) and MicroMethod (left panels) for diagnosis of cCD in UCB (a) and PVB (b) samples in infants born to seropositive women. AUC: Area Under the Curve. Std-Error: Standard-Error; CI: Confidence Interval.
Forty six neonates were tested in two consecutive samples, one withdrawn at birth and the other one between four to 133 days of life (Table 2). Two were MM negative cCD patients diagnosed by serology at 10 months of age. The qPCR was positive in both, in one of them (4045, Table 3) from the UCB sample (1,418 par.eq/mL) and from the PVB collected at 54 days of life (23,709 par.eq/mL). In the other one (4055, Table 3) qPCR was positive in the PVB obtained 41 days after birth (54 par.eq/mL) but not in the UCB sample.
Table 4.
Operational parameters of the qPCR assay in complete followed-up infants, using as criteria of positivity Cq values in one or both DNA replicates.
| Sample Type | Rate of cCD by Gold Standard | qPCR results | Rate of detection by qPCR | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|
| % (95% CI) | % (95% CI) | % (95% CI) | % (95% CI) | ||||
| UCB | 3/57 | 1 rep· Cq<37 | 2/57 | 66·67 (0·00-100·00) | 96·30 (90·33-100·00) | 50·00 (50·00-100·00) | 98·11 (93·51-100·00) |
| 3/62 | 1 rep· Cq<39 | 2/62 | 66·67 (0·00-100·00) | 88·14 (79·04-97·23) | 22·22 (0·00-54·94) | 98·11 (93·51-100·00) | |
| 3/57 | 2 rep· Cq<39 | 2/57 | 66·67 (0·00-100·00) | 96·30 (90·33-100·00) | 50·00 (50·00-100·00) | 98·11 (93·51-100·00) | |
| PVB | 11/248 | 1 rep· Cq<37 | 8/248 | 72·73 (41·86-100·00) | 98·31 (96·46-100·00) | 66·67 (35·83-97·51) | 98·73 (97·09-100·00) |
| 13/264 | 1 rep· Cq<39 | 10/264 | 76·92 (50·17-100·00) | 92·83 (89·44-96·22) | 35·71 (16·18-55·25) | 98·73 (97·09-100·00) | |
| 11/246 | 2 rep· Cq<39 | 8/246 | 72·73 (41·86-100·00) | 99·15 (97·76-100·00) | 80·00 (50·21-100·00) | 98·73 (97·09-100·00) |
UCB: Umbilical Cord Blood; PVB: Peripheral Venous Blood; Rep: replicate; Ct: threshold cycle; CI: confidence interval; PPV: Positive Predictive Value; NPV: Negative Predictive Value.
Five qPCR positive samples from cCD were MM negative. Their diagnosis was assessed by serological analysis, except in case 1039 which presented discordant serological findings and was diagnosed based on a positive MM performed at three months of age in a sample not available for qPCR analysis (Table 3). Four cCD cases had false negative qPCR results in PVB. Out of them, two cases were also MM negative (1001 and 1006) and two MM positive (2047 and 6004). Other five cCD cases had MM negative and qPCR positive results:1039 (78·8 par.eq/mL), 1083 (356 par.eq/mL), 4045 (1418 par.eq/mL at birth and 23,709 par.eq/mL at 54 days of age), 4055 (53·4 par.eq/mL) and 4057 (82 par.eq/mL) (Table 3). Besides, case 4058 was diagnosed by means of MM positivity in UCB, in agreement with the qPCR-based result. Based on the MM result, this cCD patient received trypanocidal treatment and during follow-up, the PVB sample was negative by MM and qPCR, suggesting parasitological response to treatment.
3.3. Genotyping of T. cruzi discrete typing units from cCD patients
After decoding samples and identifying those belonging to cCD patients, T. cruzi DTUs were identified directly from the qPCR positive DNA extracts (Fig. 3). The multiplex qPCR algorithm allowed identification of Tc V in five patients (Supplementary Figure 1) and SL-IR I-II PCRs allowed identification of Tc II/V/VI group in six additional cCD cases that were non-detectable by qPCR genotyping (Fig. 3). Samples from cCD 1001 and 2047 cases were qPCR negative at their first visit but qPCR positive at 10 months of age (labeled §, Table 3) and so, DTU typing was carried out from the latter ones. A qPCR positive UCB sample from the non-cCD case 4047 showed a mixture of Tc I and Tc II/V/VI (Fig. 3a and b, 4047). Discrete typing units Tc III or Tc IV were not detected.
Fig. 3.
Genotyping of Discrete typing Units in qPCR positive samples from cCD patients. a. 1•5% Agarose gel electrophoresis showing identification of Tc II/V/VI samples using SL-IR II Heminested PCR. PCR positive samples 1001 and 2047 were obtained from the corresponding infants at ten months of age. b. 1•5% Agarose gel electrophoresis showing identification of Tc I samples using SL-IR I Heminested PCR. The arrows indicate the specific amplicon. SCN: Seronegative blood sample; NTC: Non Template control; Tc I control: Silvio X-10 stock; Tc V control: MNCl2 stock; Mk 1 kb+, 1 kilobase plus DNA ladder molecular weight marker. Numbers above wells in red font indicate UCB samples, numbers in green font indicate PVB withdrawn at first follow-up time point and numbers in blue font indicate PVB collected at ten months of age.
3.4. External Quality assurance (EQC) of qPCR performance
Qualitative PCR results were informed by the three participant laboratories. All laboratory units informed as non-detectable qPCR all negative P101 control samples and as qPCR positive all samples spiked with T. cruzi parasites in the range between 1·5 and 150 par.eq/mL (P102, P103 and P104 samples). Supplementary Table 3 shows the descriptive statistics of the study. A Kappa coefficient = 1 was obtained among the results reported by each laboratory, indicating perfect agreement. In this data set, 99% of values gave a Z-score between -3 and 3, which means that values lied within three standard deviations. Supplementary Table 3b shows that all the EQC results obtained by the participating laboratories dropped within the accepted limits.
4. Discussion
The 63d World Health Assembly (2010) advised governments to launch systems of early detection, in particular for diagnosis of new infections and congenital infections in newborns. It was estimated that all new cases of cCD represented 22% (8,668/38,593) of the new cases of CD [20]. A report of CD in Argentina at that time estimated between 200,000 and 376,000 seropositive women giving birth to between 23,000 and 43,000 children, out of whom between 1,140 to 2,145 would be infected [21,22]. In 2016, the Panamerican Health Organization (PAHO) incorporated cCD in the framework for elimination of mother-to-child transmission (EMTCT Plus) to boost control of vertical infections in the region, together with HIV, Syphilis and Hepatitis B [23].
Home-brewed conventional PCR and qPCR based studies have encouraged the use of molecular diagnostics for early detection of cCD [4,10]. Once the method is standardized and personnel is trained, molecular methods exhibit high sensitivity and specificity and the possibility of being monitored through the implementation of internal and external quality controls [24]. In the last years, T. cruzi qPCR tests became commercially available in Europe [8]. However, no evaluations have been made in the context of prospective field studies, precluding assess their accuracy in both endemic and non-endemic scenarios.
In the present study, the offspring of CD women resident in localities of high endemicity of Northern Argentina (HC2 to HC5, Table 1) as well as of women living in Buenos Aires city, not endemic for CD (HC1, Table 1), were tested following STARD guidelines. The overall rate of cCD was 3·78% (14/370 cases), similar to what was estimated in other studies in Argentina [30,31]. The highest percentage of cCD children was detected in the province of Chaco (HC3, 7·1%) followed by those detected in Buenos Aires (HC1, 5·3%). Current parasitological assays require laboratory conditions that are difficult to fulfill in general laboratories, such as timely observation of the sample and availability of skilled personnel [25,26,27]. Moreover, quality controls for parasitological observation are not currently carried out in health centers. The MM has a detection limit of around 50 parasites/mL. In contrast, the qPCR assay herein performed in samples from newborns/neonates, with an analytical sensitivity below 1 parasite/mL10, would early detect a proportion of MM negative cases, allowing their prompt treatment and reducing their loss to follow-up after nine months of life, when infants should be tested using serological methods [4,28,29]. Regarding serological methods for cCD diagnosis, the possibility of immune tolerance to parasite antigens in cCD might explain some false-negative serological results observed in infected infants. On the other hand, the transmission of maternal antibodies to newborns which results in false-positive serological diagnosis can be overcome using molecular tools. Indeed, qPCR positive findings are indicative of the presence of the parasite in the tested sample independently of the host`s immune status.
In the present study, the rate of early diagnosis determined by MM was 1·89% (7/370 cCD cases), whereas qPCR detected 2·70% (10/370 cases), out of which five were also MM positive. The five remaining cases were MM negative, so these infants were diagnosed as cCD only at ten months of age. On the other hand, two patients, whose samples were collected at one (6004) and 11 days (2047) of age, were MM positive but qPCR negative. This finding was not expected, given the high analytical sensitivity of the qPCR. Inhibition of qPCR was not observed in these samples as indicated by correct IAC amplification, and the sample collected from case 2047 at ten months of age was qPCR positive and could be genotyped (Table 3 and Fig. 3). Technical problems such as DNA degradation during collection, storage and/or transportation of the blood sample from the endemic locality to the qPCR laboratory might have been possible causes for these false negative qPCR findings.
Four non-infected cases gave false positive qPCR findings, two in UCB (4047: 39 par.eq/mL and 5069: 0·45 par.eq/mL, Table 3). Sample from case 4047 was genotyped and gave a mixed TcI plus TcII/V/VI population, indicating not only the presence of satellite DNA but also of spliced-leader intergenic nuclear sequences (Table 3 and Fig. 3). A parasitic load of 39 par.eq/mL seems too high to be a false positive result due to laboratory cross-contamination with another infected sample. Besides, amplicon carry-over contamination is unlikely because the kit incorporates an Uracil DNA Glycosylase system. Thus, it might be speculated that this UCB sample could contain maternal parasite traces despite the care and umbilical cord decontamination step done during collection of the sample (see Methods). Otherwise, spontaneous cure, which is the clearance of the infection without the need of treatment, could be a plausible explanation for this observation, as previously reported [32], [33], [34]. The other two false positive samples were processed simultaneously to true positive ones: PVB 4044 (3·64 par.eq/mL) was processed together with cCD 4045 (23,709 par.eq/mL) and PVB 1110 (0·44 par.eq/mL) was processed together with cCD 1111 (641 par.eq/mL).The high parasitic loads of the above-mentioned samples suggest that cross-contamination during sample collection, transportation and/or DNA extraction might have occurred (Table 3). Indeed, samples collected in HC4 exhibited a higher proportion of inconclusive results (13·33%, Table 1) because of discordant qPCR duplicates, which in part could have arisen by cross-contamination between consecutive samples.
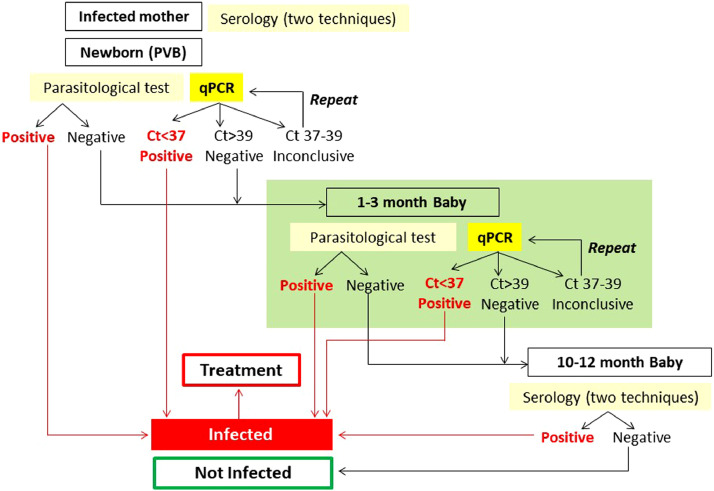
Maximum qPCR accuracy (sensitivity 72·73%, specificity 91·9%) was obtained in PVB collected around one month of age (34·5 days). It has been reported that a proportion of samples collected from cCD patients close to delivery may present low parasitic loads. Indeed, cCD case 4045 showed higher parasitic loads in the PVB collected at day 54 than in the UCB sample, and case 4055 was qPCR positive at 41 days of age and negative in the UCB sample (Table 3). ROC curve analysis suggested higher accuracy for qPCR in PVB than in UCB samples, although the difference did not reach significance levels (p=0·08). Previous studies observed that diagnosis has higher sensitivity around the first month of age and at that time, the likelihood of false positive results derived from passage of maternal T. cruzi DNA to the newborn is discarded [4,5,34,35]. The study designed as criterion of qPCR positivity or negativity the concordance of results between duplicates, which led to the detection to 7·8% and 8·9% of discordant findings in UCB and PVB, respectively. To diminish the proportion of discordant results, additional analyses were performed. The first one considered as qPCR positive a sample with at least one qPCR replicate detectable at Cq<39. In PVB samples, this criterion allowed a higher qPCR sensitivity (76·92%) but lower specificity (92·83%) and in UCB, sensitivity was 66·67% and specificity only 88·14%. Indeed, this criterion gave rise to 18 false positive qPCR results among 264 tested PVB and 7 false positive qPCR results among 62 tested UCB samples. To obtain higher qPCR accuracy when testing only one replicate, the maximum Cq value for qPCR positivity was lowered below 37. In this case, sensitivity was 72·73% and specificity 98·31% for PVB and 66·67% and 96·30% for UCB, respectively (Table 4). These findings allowed us to propose an algorithm for reliable diagnosis of cCD, employing qPCR and MM at delivery and around one month of life in PVB samples (Fig. 4). A Cq <37 in at least one replicate determines a qPCR positive result. If the Cq is >37 and <39 in only one replicate, the result should be considered inconclusive and therefore DNA extraction and qPCR amplification should be repeated. Finally, a Cq >39 is interpreted as a non-detectable qPCR result (Fig. 4).
Table 1.
Distribution of cases and samples for congenital Chagas Disease diagnosis per each recruiting Health Center
| Prospective Field Study Cases | Enrolled Cases | HC1 | HC2 | HC3 | HC4 | HC5 | TOTAL |
|---|---|---|---|---|---|---|---|
| 151 | 85 | 114 | 122 | 87 | 559 | ||
| Complete Followed-up Cases | 132 | 75 | 56 | 51 | 56 | 370 | |
| Congenital Chagas Disease Cases (%) | 7 (5·3) | 2 (2·7) | 4 (7·1) | 1 (2·0) | 0 (0·0) | 14 (3·8) | |
| Numbers of UCB samples for qPCR analysis from cases with complete follow-up and diagnosis | UCB collected samples | 0 | 25 | 31 | 0 | 11 | 67 |
| Included for analysis of qPCR accuracy° | 0 | 21 | 29 | 0 | 7 | 57 | |
| Without parasitological result a | 0 | 2 | 0 | 0 | 0 | 2 | |
| Not valid samples for qPCR b | 0 | 0 | 0 | 0 | 3 | 3 | |
| Inconclusive qPCR results c | 0 | 2 | 2 | 0 | 1 | 5 | |
| Numbers of PVB samples for qPCR analysis from cases with complete follow-up and diagnosis | PVB collected samples | 129 | 77 | 52 | 45 | 51 | 354 |
| Included for qPCR accuracy | 122 | 68 | 42 | 35 | 26 | 293 | |
| Outpatient >5 months of age | 1 | 2 | 1 | 0 | 1 | 5 | |
| Without parasitological result a | 0 | 0 | 1 | 0 | 0 | 1 | |
| Not valid samples for qPCR b | 0 | 0 | 0 | 4 | 22 | 26 | |
| Inconclusive qPCR results c | 6 | 7 | 8 | 6 | 2 | 29 | |
| Numbers of UCB and PVB samples for qPCR analysis from cases with complete follow-up and diagnosis | Total samples | 129 | 102 | 83 | 45 | 62 | 421 |
| Included for qPCR accuracy | 122 | 89 | 71 | 35 | 33 | 350 | |
| Outpatient >5 months of age | 1 | 2 | 1 | 0 | 1 | 5 | |
| Without parasitological result a | 0 | 2 | 1 | 0 | 0 | 3 | |
| Not valid samples for qPCR b | 0 | 0 | 0 | 4 | 25 | 29 | |
| Inconclusive qPCR results c | 6 | 9 | 10 | 6 | 3 | 34 |
HC: Health Center; UCB: Umbilical Cord Blood; PVB: Peripheral Venous Blood.
(%) Percentage of congenital Chagas disease infants diagnosed out of complete followed-up cases per Health Center. Analysis of qPCR accuracy was done only from cases whose early samples were collected before 5 months of age, a-the sample was obtained but the MM test was not done; b- Not valid samples were those with incorrect volume, detection of coagulation or difficulty in pipetting due to viscosity. c- Inconclusive qPCR results due to discordance between duplicates and/or outlier Ct values of IAC.
Fig. 4.
Proposed algorithm for diagnosis of cCD. Detectable MM and/or qPCR findings of a neonate´s PVB-based DNA lysate are reported as cCD. P: Positive, N: Negative, *If one of the serological techniques is not reactive, a third technique should be performed.
Establishment of EQC assurance [37] is relevant for accompanying PCR-based field studies for monitoring the quality of the qPCR procedures carried out. Accordingly, EQC was implemented in three different laboratories that used different thermocyclers and were operated by different technicians. Statistical analysis showed concordant qualitative qPCR results (Kappa coefficient=1). Moreover, all samples spiked with 1·5 to 150 par.eq/mL, as well as the positive DNA controls provided by the kit gave amplification signals, whereas non-spiked seronegative blood gave non-detectable qPCR findings, demonstrating the high inter-laboratory reproducibility of the assay.
Genotyping allowed detection of Tc V in those cCD samples harboring 641-23,709 par.eq/mL. Samples with lower parasitic loads could be only genotyped by means of conventional SL-IR based heminested PCRs, resulting in Tc II/V/VI. Indeed, the latter strategy is more sensitive than qPCR, although it cannot provide a distinction among TcII, TcV and TcVI DTUs [16,17]. The predominance of Tc V in cCD has been reported in the southern cone [36], in agreement with its predominance in the general population, suggesting no associations between certain DTUs and cCD [1,17,38]. The only sample exhibiting Tc I plus Tc II/V/VI mixed populations was an UCB from a non-infected newborn. It is tempting to speculate that Tc I strains circulate in bloodstream at lower parasitic loads and accordingly are more difficult to be detected. A recent case report of a cCD child who tested negative by microscopic observation and PCR done at 20 days and 6 months of age, identified a Tc I isolate obtained by hemoculture at seven months of age [39].
We did not find clinical and sociodemographic characteristics associated to the likelihood of cCD transmission, except for a higher frequency of previously infected brothers and sisters of the cCD cases confirming previous evidence of family clustering of cCD [3]. Previous works suggested an association between infants’ parasitic loads and the severity of clinical manifestations in cCD 26Ç [26], [27]. In our cohort, two cases (1044 and 4045, Table 3) depicted high parasitic loads. The former case was born at week 36th (2·5 kgr) without any complication except hospitalization due to bronchiolitis and the latter was born at week 36th (2·78 kgr) without complications except one admission to hospital at one month of age due to an ovarian surgery not related to cCD.
Previous international qPCR validation studies were done in adult blood samples and hence PCR clinical sensitivity was estimated from 10 or 5 mL of blood treated with equal volumes of Guanidinium Hidrochloride 6M, EDTA 0.2 M, pH 8·00 (GE) as stabilizing agent, volume that is feasible to obtain from UCB but problematic to collect from neonates´PVB [10,40]. The present study employed only 1·2 mL of neonatal blood plus 0·3 mL of a commercial stabilizer. The combination of DNA extraction and amplification steps allows results in just over three hours, in a straightforward way with standardized reagents and quality controls.
Nevertheless, the commercial stabilizer herein used resulted not optimal for blood storage. Many samples became viscous and difficult to manipulate, so they could not be processed (not valid samples, Table 1). It is expected that further prospective studies using EDTA or GE blood [41,42] will improve the kit performance.
During the study, 189 babies (33·8%) could not be followed-up. This occurred particularly in the three endemic sites with higher predominance of rural populations (Table 1, HC3, 4 and 5), while HC2 is located in an endemic area with certification of vector-borne transmission control. Two cases from HC4, without complete follow-up (unpublished results), had positive qPCR findings with high parasitic loads, which suggested cCD. Therefore, these cases were reported to the site for diagnosis confirmation and treatment.
National guidelines have mentioned qPCR as a potential laboratory tool for early cCD diagnosis. However, no field prospective works had been performed so far [6,29]. Also, current guidelines do not consider performing tests around one month of age, which we found optimal for qPCR and parasitological studies, and accordingly we have included it in the proposed algorithm. Consequently, this study may contribute to implement qPCR for IVD use, aiming to improve current early diagnosis of cCD in those countries that only perform parasitological methods. The sensitivity of this qPCR kit was almost twice than that of the MM. Messenger and coworkers [5] detected 68·6% versus 16·7% of sensitivity in UCB for qPCR and MM, respectively, reaching a cumulative sensitivity of 84·2% and 34·2% when a second analysis was performed in the same patients at one month of age.
Future evaluation of the qPCR kit performance in molecular biology laboratories located at endemic areas using EDTA or GE-treated samples, would reduce the problems related to sample transportation observed in this study. Its use, in combination with the MM, should improve early detection of cases and provide more accurate records on the number of cCD infants in endemic and non-endemic countries. Such data would be useful in estimating the likely number of cases missed in places where only traditional parasitological procedures are still employed.[43] Moreover, incorporation of standard curves for quantification of parasitic loads may provide a useful tool to monitoring patients under treatment and to study associations between parasitic burden and cCD clinical severity. Indeed, qPCR-based early diagnosis will contribute to maximize prompt treatment to infected neonates with high impact in public health [22,42,43].
Description of this data
The file FITS SALUD CHAGAS 001 Study_public data base.cvs contains all the variables used in the analysis of the manuscript “Multicenter Field Evaluation of Real time PCR Kit prototype for Early Diagnosis of Congenital Chagas disease”. The data set is structured with one record per each screened T. cruzi infected mother-newborn binomial. Each of the 622 screened binomials was assigned with an ID, which is the first variable included in the data set, and subsequently variables on binomials' characteristics of interest and laboratory results for the 559 recruited binomials have been incorporated. Variables were obtained from: i) study forms to collect information from clinical records or participants´ answers, ii) specific data forms for laboratory results, and iii) from previous variables. In the file FonarsecStudy_DataDictionary.doc is detailed the information for each variable.
The analysis of this data set was done with Stata version 11.2, and some complementary analysis was performed using EpiDat version 3.1.
Contributors
AGS, SSE and FR designed the trial with input from JB, ME, MLC and FA. Trial coordination, management and clinical data collection was performed by SAB, ED, SL and MLC with the input of GA, LL, SS, ELA, RHL and the Congenital Chagas Disease Working Group. Samples were processed by AFB, JCR, SB, MB, SAB, CLA and KS. PCR data were generated by AFB and SAB and EQC data were generated by JB and CLA with the input of MR and LI. Underlying data were verified by AGS, SL, SSE, APP and MLC. Data analysis was performed by ED, AFB, AGS with the input of FA and MLC. AGS, ED, AFB and SAB wrote the first draft of the manuscript and all coauthors contributed to subsequent drafts and revisions. All authors read and approved the final manuscript.
Data Sharing Statement
Individual participant data that underlie the results reported in this article have been de-identified and deposited in the database of the Congenital Chagas disease Working Group, which is available at https://data.mendeley.com/datasets/c6882kknhd/1.
Funding Resources
This work has received financial support from the following grants: FITS SALUD 001-CHAGAS (FONARSEC Ministery of Science, Technology and Innovation from Argentina) to the Public-Private Consortium (INGEBI-CONICET, INP-ANLIS MALBRAN and Wiener Laboratories S.R.L), from ERANET-LAC-HD 328 to AGS and PICT 2015-0074 (FONCYT, MinCyT) to AGS and FA.
Declaration of Competing Interest
None of the authors received payment or service from a third part at any time. None of the authors have any patents relevant to the work.
Acknowledgments
The study is part of the project entitled FITS SALUD CHAGAS 001 (FONARSEC, MINCYT, Argentina) designed and executed by a Public-Private Consortium formed by Instituto de Investigaciones en Ingeniería Genética y Biología Molecular “Dr Héctor Torres” (INGEBI) - Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET); Instituto Nacional de Parasitología “Dr. Mario Fatala Chaben” (INP), dependent of ANLIS MALBRAN – MINSAL and Centro de Investigación y Biotecnología (CIBIO) de Wiener Laboratorios S.A.I.C.
AGS, SSS, JB, SAL and FA are members of the “Carrera del Investigador Científico”, from the Consejo Nacional de Investigaciones Científicas y Técnicas de Argentina (CONICET). AGS, SAL and SSE are members of the NHEPACHA Network (Red Iberoamericana “Nuevas Herramientas para el Diagnóstico y la Evaluación del Paciente con Enfermedad de Chagas”).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103450.
Appendix. Supplementary materials
References
- 1.Bisio M, Seidenstein ME, Burgos JM. Urbanization of congenital transmission of Trypanosoma cruzi: prospective polymerase chain reaction study in pregnancy. Trans R Soc Trop Med Hyg. 2011;105(10):543–549. doi: 10.1016/j.trstmh.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 2.Bittencourt AL. Possible risk factors for vertical transmission of Chagas' disease. Rev Inst Med Trop Sao Paulo. 1992;34(5):403–408. doi: 10.1590/s0036-46651992000500006. Review. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez Negrete O, Mora MC, Basombrio MA. High prevalence of congenital Trypanosoma cruzi infection and family clustering in Salta, Argentina. Pediatrics. 2005;115:e668–e672. doi: 10.1542/peds.2004-1732. [DOI] [PubMed] [Google Scholar]
- 4.Bua J, Volta BJ, Perrone AE. How to improve the early diagnosis of Trypanosoma cruzi infection: relationship between validated conventional diagnosis and quantitative DNA amplification in congenitally infected children. PLoS Negl Trop Dis. 2013;7(10):e2476. doi: 10.1371/journal.pntd.0002476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Messenger LA, Gilman RH, Verastegui M, et al. Toward Improving Early Diagnosis of Congenital Chagas Disease in an Endemic Setting. Clin Infect Dis. 2017;65(2):268-275. [DOI] [PMC free article] [PubMed]
- 6.Velázquez EB, Rivero R, De Rissio AM. Predictive role of polymerase chain reaction in the early diagnosis of congenital Trypanosoma cruzi infection. Acta Trop. 2014;137:195–200. doi: 10.1016/j.actatropica.2014.05.016. [DOI] [PubMed] [Google Scholar]
- 7.Buekens P, Cafferata ML, Alger J. Congenital Transmission of Trypanosoma cruzi in Argentina, Honduras, and Mexico: An Observational Prospective Study. Am J Trop Med Hyg. 2018;98(2):478–485. doi: 10.4269/ajtmh.17-0516. FebEpub 2017 Nov 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sosa-Estani S, Colantonio L, Segura EL. Therapy of Chagas disease: implications for levels of prevention. J Trop Med. 2012;2012 doi: 10.1155/2012/292138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abras A, Ballart C, Llovet T, Roig C, Gutiérrez C, Tebar S, Berenguer P, Pinazo MJ, Posada E, Gascón J, Schijman AG, Gállego M, Muñoz C. Introducing automation to the molecular diagnosis of Trypanosoma cruzi infection: a comparative study of sample treatments, DNA extraction methods and Real Time PCR assays. PLoS One. 2018;13(4) doi: 10.1371/journal.pone.0195738. Apr 17doi: 10.1371/journal.pone.0195738. eCollection 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duffy T, Cura CI, Ramirez JC. Analytical performance of a multiplex Real Time PCR assay using TaqMan probes for quantification of Trypanosoma cruzi satellite DNA in blood samples. PLoS Negl Trop Dis. 2013;7(1):e2000. doi: 10.1371/journal.pntd.0002000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ISO 13528:2015 (E) Second edition 2015-08-01 Statistical Methods for use in Proficiency testing by interlaboratory comparison
- 12.De Rissio AM, Riarte AR, Garcia MM, Esteva MI, Quaglino M, Ruiz AM. Congenital Trypanosoma cruzi infection. Efficacy of its monitoring in an urban reference health center in a non-endemic area of Argentina. Am J Trop Med Hyg. 2010;82(5):838–845. doi: 10.4269/ajtmh.2010.08-0383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ministerio de Salud de la Nación . Subsecretaría de Promoción y Prevención, Dirección de Enfermedades Transmisibles por Vectores, Coordinación Nacional de Control de Vectores, Programa Nacional de Chagas. Ministerio de Salud de la Nación; 2012. Guía para la Atención al Paciente Infectado con T. cruzi (Enfermedad de Chagas). ANLIS-Malbrán. CeNDIE - INP Secretaría de Programas Sanitarios. [Google Scholar]
- 14.Iniciativa del Cono Sur para controlar e eliminar la enfermedad de Chagas (INCOSUR); https://www.argentina.gob.ar/salud/epidemiologia/boletines2012)
- 15.Salud Administración Nacional de Laboratorios e Institutos de. Manual de Laboratorio. Primera Edición; 1999. Dr. Carlos G. Malbran”. Instituto Nacional de Parasitología “Dr. Mario Fatala Chaben”. Diagnóstico en Parasitosis. [Google Scholar]
- 16.Cura CI, Duffy T, Lucero RH. Multiplex Real Time PCR Assay Using TaqMan Probes for the Identification of Trypanosoma cruzi DTUs in Biological and Clinical Samples. PLoS Negl Trop Dis. 2015;9(5) doi: 10.1371/journal.pntd.0003765. May 19e0003765eCollection 2015 May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burgos JM, Altcheh J, Bisio M. Direct molecular profiling of minicircle signatures and lineages of Trypanosoma cruzi bloodstream populations causing congenital Chagas disease. Int J Parasitol. 2007;37(12):1319–1327. doi: 10.1016/j.ijpara.2007.04.015. OctEpub 2007 May 10. [DOI] [PubMed] [Google Scholar]
- 18.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148(3):839–843. doi: 10.1148/radiology.148.3.6878708. Sep. [DOI] [PubMed] [Google Scholar]
- 19.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. Sep. [PubMed] [Google Scholar]
- 20.World Health Organization. Chagas disease in Latin America: an epidemiological update based on 2010 estimates. Weekly Epidemiol Rec. 2015;90:33–44. [PubMed] [Google Scholar]
- 21.Howard EJ, Xiong X, Carlier Y, Sosa-Estani S, Buekens P. Frequency of the congenital transmission of Trypanosoma cruzi: a systematic review and meta-analysis. BJOG. 2014;121(1):22–33. doi: 10.1111/1471-0528.12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Picado A, Cruz I, Redard-Jacot M. The burden of congenital Chagas disease and implementation of molecular diagnostic tools in Latin America. BMJ Glob Heal. 2018 doi: 10.1136/bmjgh-2018-001069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan American Health Organization. EMTCT Plus. Framework for elimination of mother-to-child transmission of HIV, Syphilis, Hepatitis B, and Chagas. PAHO/CHA/17-009, 2017.(https://www.paho.org/hq/dmdocuments/2017/2017-cha-etmi-plus-marco-vih-hep-chagas.pdf).
- 24.Ramírez JC, Parrado R, Sulleiro E, de la Barra A, Rodríguez M, Villarroel S, Irazu L, Alonso-Vega C, Alves F, Curto MA, García L, Ortiz L, Torrico F, Gascón J, Flevaud L, Molina I, Ribeiro I, Schijman AG. First external quality assurance program for bloodstream Real Time PCR monitoring of treatment response in clinical trials of Chagas disease. PLoS One. 2017 Nov 27;12(11) doi: 10.1371/journal.pone.0188550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Freilij H, Muller L, Gonzalez Cappa SM. Direct micromethod for diagnosis of acute and congenital Chagas’ disease. J. Clin. Microbiol. 1983;18(2):327–330. doi: 10.1128/jcm.18.2.327-330.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torrico MC, Solano M, Guzman JM. Estimation of the parasitemia in Trypanosoma cruzi human infection: high parasitemias are associated with severe and fatal congenital Chagas disease. Rev Soc Bras Med Trop. 2005;38(Suppl 2):58–61. [PubMed] [Google Scholar]
- 27.Russomando G, de Tomassone MM, de Guillen I. Treatment of congenital Chagas' disease diagnosed and followed up by the polymerase chain reaction. Am J Trop Med Hyg. 1998;59(3):487–491. doi: 10.4269/ajtmh.1998.59.487. [DOI] [PubMed] [Google Scholar]
- 28.Cura CI, Ramírez JC, Rodríguez M. Comparative Study and Analytical Verification of PCR Methods for the Diagnosis of Congenital Chagas Disease. J Mol Diagn. 2017;19(5):673–681. doi: 10.1016/j.jmoldx.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Mora MC, Sanchez Negrette O, Marco D. J. Parasitol. 2005;91(6):1468–1473. doi: 10.1645/GE-549R.1. [DOI] [PubMed] [Google Scholar]
- 30.Fabbro DL, Danesi E, Olivera V. Trypanocide treatment of women infected with Trypanosoma cruzi and its effect on preventing congenital Chagas. PLoS Negl Trop Dis. 2014;8:e3312. doi: 10.1371/journal.pntd.0003312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeledón R, Dias JCP, Brilla-Salazar A. Does a spontaneous cure for Chagas' disease exist? Revista da Sociedade Brasileira de Medicina Tropical. 1988;21:15–20. doi: 10.1590/s0037-86821988000100003. [DOI] [PubMed] [Google Scholar]
- 32.Dias JC, Dias E, Martins-Filho OA. Further evidence of spontaneous cure in human Chagas disease. Rev Soc Bras Med Trop. 2008;41(5):505–506. doi: 10.1590/s0037-86822008000500014. Sep-Oct. [DOI] [PubMed] [Google Scholar]
- 33.Carlier Y, Truyens C. Congenital Chagas disease as an ecological model of interactions between Trypanosoma cruzi parasites, pregnant women, placenta and fetuses. Acta Trop. 2015;151:103–115. doi: 10.1016/j.actatropica.2015.07.016. Nov. [DOI] [PubMed] [Google Scholar]
- 34.Bua J, Volta BJ, Velazquez EB, et al. . Vertical transmission of Trypanosoma cruzi infection: quantification of parasite burden in mothers and their children by parasite DNA amplification. Trans R Soc Trop Med Hyg. 2012 Oct;106(10):623-8. [DOI] [PubMed]
- 35.Sosa-Estani S. Congenital transmission of Trypanosoma cruzi infection in Argentina. Rev Soc Bras Med Trop. 2005;38(Suppl 2):29–32. Spanish. [PubMed] [Google Scholar]
- 36.Virreira M, Alonso-Vega C, Solano M. Congenital Chagas disease in Bolivia is not associated with DNA polymorphism of Trypanosoma cruzi. Am J Trop Med Hyg. 2006;75(5):871–879. Nov. [PubMed] [Google Scholar]
- 37.Volta BJ, Perrone AE, Rivero R, Scollo K, Bustos PL, Bua J. Vol. 141. Pediatrics; 2018. pp. S451–S455. (Some Limitations for Early Diagnosis of Congenital Chagas Infection by PCR). Apr. [DOI] [PubMed] [Google Scholar]
- 38.Parrado R, Ramirez JC, de la Barra A. Real Time PCR for the Evaluation of Treatment Response in Clinical Trials of Adult Chronic Chagas Disease: Usefulness of Serial Blood Sampling and qPCR Replicates. Antimicrob Agents Chemother. 2018 doi: 10.1128/AAC.01191-18. Dec 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schijman AG, Bisio M, Orellana L, et al. International study to evaluate PCR methods for detection of Trypanosoma cruzi DNA in blood samples from Chagas disease patients. PLoS Negl Trop Dis. 2011 Jan 11;5(1):e931. doi: 10.1371/journal.pntd.0000931. [DOI] [PMC free article] [PubMed]
- 40.Ramírez JC, Cura CI, da Cruz Moreira O, et al. Analytical Validation of Quantitative Real Time PCR Methods for Quantification of Trypanosoma cruzi DNA in Blood Samples from Chagas Disease Patients. J Mol Diagn. 2015 Sep;17(5):605-15. doi: 10.1016/j.jmoldx.2015.04.010. [DOI] [PMC free article] [PubMed]
- 41.Alonso-Vega C, Billot C, Torrico F. Achievements and challenges upon the implementation of a program for national control of congenital Chagas in Bolivia: results 2004-2009. PLoS Negl Trop Dis. 2013;7(7):e2304. doi: 10.1371/journal.pntd.0002304. Jul 11Print 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porrás AI, Yadon ZE, Altcheh J. Target Product Profile (TPP) for Chagas Disease Point-of-Care Diagnosis and Assessment of Response to Treatment. PLoS Negl Trop Dis. 2015;9(6) doi: 10.1371/journal.pntd.0003697. eCollection 2015 Jun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alonso-Padilla J, Gallego M, Schijman AG, Gascon J. Vol. 17. Expert Rev Mol Diagn; 2017. pp. 699–710. (Molecular diagnostics for Chagas disease: up to date and novel methodologies). JulReview. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.