Abstract
Peroxidized phosphatidylethanolamine (PEox) species have been identified by liquid chromatography mass spectrometry (LC-MS) as predictive biomarkers of ferroptosis, a new program of regulated cell death. However, the presence and subcellular distribution of PEox in specific cell types and tissues have not been directly detected by imaging protocols. By applying gas cluster ion beam secondary ion mass spectrometry (GCIB-SIMS) imaging with a 70 keV (H2O)n+ (n > 28000) cluster ion beam, we were able to map PEox with 1.2 μm spatial resolution at the single cell/subcellular level in ferroptotic H9c2 cardiomyocytes and cortical/hippocampal neurons after traumatic brain injury. Application of this protocol affords visualization of physiologically relevant levels of very low abundance (20 pmolμmol−1 lipid) peroxidized lipids in subcellular compartments and their accumulation in disease conditions.
Keywords: cell death, imaging, lipid, mass spectrometry, oxidation
Lipid reprograming has been recognized as a common response to a number of physiological and pathological stimuli.[1] Contemporary liquid chromatography-mass spectrometry (LC-MS) based lipidomics has identified and structurally characterized thousands of individual molecular species of the lipidome.[2] However, even the most advanced LC-MS protocols from bulk extracts provide no information on the intracellular distribution of lipid molecules. Several MS imaging techniques: matrix-assisted laser desorption/ionization (MALDI), liquid extraction surface analysis (LESA), desorption electrospray ionization (DESI) and secondary ion mass spectrometry (SIMS), have been developed to explore the distribution of lipids albeit with a limited spatial resolution or chemical sensitivity.[3] The latest modifications of MALDI and a specific type of SIMS using gas cluster ion beams (GCIB-SIMS) have proved to be excellent for mapping the most abundant intact lipids of biomembranes in cells.[4] Furthermore, imaging high molecular weight phospholipids (PLs) in subcellular structures at ca. 1 μm resolution using GCIB-SIMS has become possible as the Winograd group introduced a ca. 70 keV CO2 cluster ion beam.[5] However, the signaling phospholipid molecules, which are present in cells at very low steady-state concentrations, still remained undetectable by conventional GCIB-SIMS. The latest success has been attained with the employment of a H2O cluster ion beam, which results in a 100-fold signal amplification due to the enhanced protonation.[6] The combination of the H2O cluster with high kinetic energy, ca. 70 keV, has led to both increased sensitivity and spatial resolution.[7] This has provided us with an advantage in attempts to image very low abundance oxidized phospholipid species acting as signaling molecules at the single cell level.
We chose to apply the newly developed protocol for the intracellular imaging of phospholipid death signals in ferroptosis, a novel death program.[8] Our previous work using LC-MS has identified a subset of oxidized phosphatidylethanolamine PE (PEox) species, namely PEox containing arachidonoyl- and adrenoyl acyls, which play a major role in ferroptosis[9] (Scheme 1). Given that ferroptosis has been associated with close to 100 different disease conditions and injuries, the PEox species and their metabolic pathways are important targets for therapeutic interventions.[10] Here we present the first successful results describing the application of H2O cluster GCIB-SIMS imaging in studies of PEox in ferroptosis in cardiomyocytes and neurons in frozen-hydrated samples at the single cell/subcellular level. Unlike chemical fixation, frozen-hydration guarantees that the chemistry of the biological system will be preserved as it has been shown previously.[7,11] For example, Newell et al. observed the improved ion detection of biomolecules in a variety of plant, animal and human tissues.[11d] Tian et al. showed the enhancement of various lipids (for example, cardiolipin and PE) and detected compartmentalized metabolites in frozen-hydrated tissue and HeLa cells.[7,11b]
Scheme 1.
Enzymatic oxidation of phosphatidylethanolamine (PE) leads to production of hydroperoxy lipid death signals.
A glutathione peroxidase 4 (GPX4) inhibitor, RSL3 (500 nM), induced death in 70% of H9c2 cardiomyocytes and this effect was fully prevented by an anti-ferroptosis agent, Ferrostatin-1 (Fer-1) (Figure 1 and single channel images in the Supporting Information, Figure S2). GCIB-SIMS imaging employing a H2O cluster ion beam revealed three major species of hydroperoxy-PE (PE-OOH): PEp(36:4)-OH/OOH, PE(38:4)-OOH and PE(40:4)-OOH along with their non-oxidized precursor ions in both control and treated cells (Figure 1B). However, LC-MS revealed that the amounts of PEox signals were increased 2–3 times in RSL3-treated cells and their content was returned to control levels by Fer-1 treatment (Figure 1C,D). This trend, increasing PEox signal upon RSL3 treatment and return to control levels with the addition of Fer-1, was also seen during GCIB-SIMS imaging (Supporting Information, Figure S2). Our current imaging study indicates that the PEox species in ferroptotic H9c2 cells are non-uniformly distributed. This is supported by our previous studies which indicated that these species occur in localized compartments (endoplasmic reticulum, mitochondria and mitochondria associated membranes [MAMs]) and may exist as individual molecules, clusters or protein adducts that eventually compromise the plasma membrane.[9,12] The PEox species were distributed throughout the cell at the 2.5 h RSL3 time point (Figure 1B) and approximately 15%, 30% and 25% of the PEp(36:4)-OH/OOH, PE(38:4)-OOH and PE(40:4)-OOH species were co-localized with their non-oxidized precursor PEp(36:4), PE(38:4) and PE(40:4) species, respectively [Figure 1E]. PEox species were confirmed by LC-MS and MS/MS analysis including calibration curves with PEox standards (Supporting Information, Figures S3–S6). A GPX4 co-factor, GSH (glutathione, reduced form) detected at a mass-to-charge ratio (m/z) of 306.08 decreased significantly upon ferroptosis induction in RSL3-treated H9c2 cells. These levels were partially preserved by treatment with Fer-1 as determined by GCIB-SIMS signal intensities (Supporting Information, Table S1).
Figure 1.
Detection of peroxidized PE species in RSL3-treated H9c2 cells by GCIB-SIMS imaging. 1A: Assessment of LDH release from RSL3-treated ferroptotically dying H9c2 cells; 1B: GCIB-SIMS imaging of RSL3 treated H9c2 cells provides the total ion signal (red) to identify cellular outlines, non-oxidized (blue) and hydroperoxy (green) PE species as given to the left, Scalar intensity multiplication was performed on the non-oxidized and oxidized PE species for clarity, images are 1.2 μm pixel size, the zoomed inset is a direct 3x magnification. Scale bar corresponds to 100 microns (20 microns in zoomed images); 1C: LC-MS analysis of peroxidized PE species in cell extracts; 1D: Heat map of peroxidized PE species in control, RSL3-treated and RSL3+Ferrostatin-1 treated H9c2 cell extracts; 1E: Percentage of PEox species colocalized with their precursor PE species as detected by GCIB-SIMS imaging.
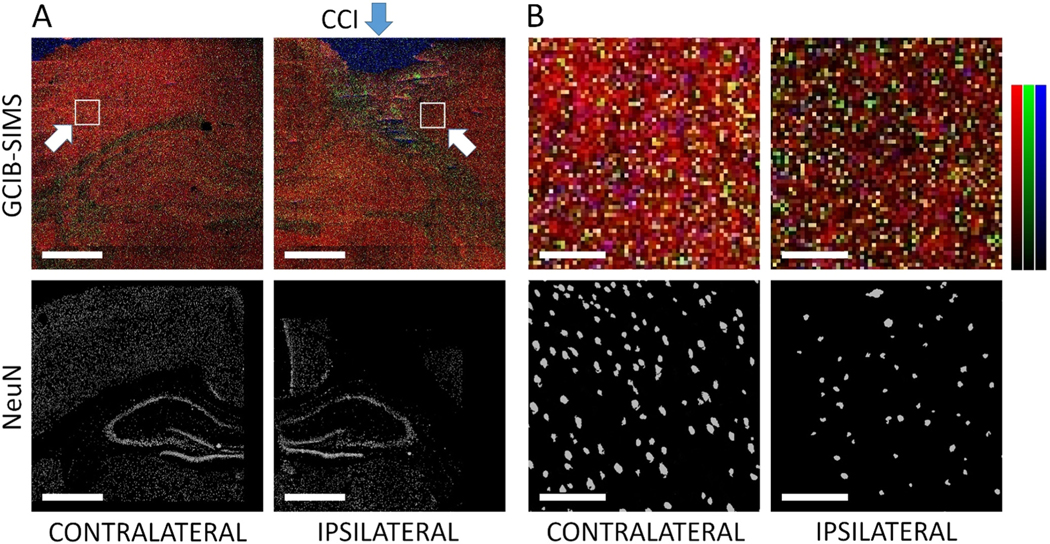
To explore the applicability of the GCIB-SIMS protocol to tissues, we next imaged and assessed peroxidized lipid signals in rat brain cortex. Given that traumatic brain injury (TBI) is associated with ferroptotic death,[13] we performed these assessments in each of two distinct areas of the brain: the ipsilateral pericontusional cortex and corresponding contra-lateral (non-contusional) cortex (Figure 2 and the grayscale image in the Supporting Information, Figure S7). GCIB-SIMS imaging identified two species of oxidized lipids: (PE(38:4)-OH, m/z 782.5, blue) and an oxidized form of arachidonic acid (AA(20:4)-OH, m/z 319.23, green) (Figure 2B). Both the PE(38:4)-OH and the AA(20:4)-OH were significantly increased in the ipsilateral region (Supporting Information, Figures S8–S10) and their structures were confirmed by MS/MS analysis (Supporting Information, Figure S11). Orthogonal partial least square-discriminant analysis (OPLS-DA) displayed two specific groupings of neurons with respect to major lipid signals (Figure 3). Neurons in the ipsilateral region were statistically significantly different and were distinguishable from those in a corresponding contralateral region and clustered differently along the component axes. The ipsilateral neurons displayed decreased ratios of phospholipid signals (ipsilateral to contralateral), in line with our previous MALDI imaging results.[14] In order to determine which lipid species contributed the most towards distinguishing the ipsilateral from the contralateral neurons, we applied a plot of values for the variable importance for the prediction (VIP) vs. their rank, which indicated that free fatty acids (FFAs) were most discriminatory in defining differences among the neuronal groupings (Supporting Information, Figure S12). The presence of FFAs in the ipsilateral region could be a result of phospholipase A2 (PLA2) cleavage of phospholipids forming FFA and lyso-PLs, as well as oxidation of PUFA-containing PLs with subsequent cleavage by specialized Ca2+-independent PLA2 (iPLA2) to form lysoPLs and FFAoxs.[15] Some level of in-source fragmentation of precursor peroxidized molecules cannot be completely excluded. In fact, our assessments based on the decomposition of HOO-AA-PE, HO-AA-PE and AA-PE showed that approximately 25 mol% of FFA-OH (relative to the precursor ion) can be generated in the course of GCIB-SIMS imaging (Supporting Information, Figure S6D). Selection of the imaging conditions minimizing this undesirable fragmentation is the goal of current efforts.
Figure 2.
Comparison of GCIB-SIMS ion images with IHC confocal microscopy differentiates pericontusional and non-contusional neurons in controlled cortical impact (CCI) brain. A) top right: GCIB-SIMS ion intensity map showing loss of PE(38:4) [M–H]- (m/z 766.5) in the contusional and pericontusional regions, ipsilateral hemisphere. Top left: Corresponding region from the contralateral hemisphere. Bottom right: Binary image from NeuN (neuronal) IHC staining identifies locations of neuron loss (contusional region) and the immediately surrounding cortical region (pericontusional). (bottom left) Corresponding NeuN image from the contralateral hemisphere. Scale bar=1000 μm. B) Ipsilateral pericontusional and corresponding contralateral non-contusional regions (white arrows, (A) top). The binary layers of the NeuN IHC images (bottom images) were used to identify individual neurons from the GCIB-SIMS pericontusional/ipsilateral (top right) and non-contusional/contralateral (top left) cortical regions. Red: m/z 766.5 PE(38:4), Green: m/z 319.2 FFA(20:4)OH, Blue: m/z 782.5 PE(38:4)OH. Intensity color bars are relative to the maximum intensity of each ion: 0% to 10% for the green and blue channels, 0% to 35% for the red channel. Scale bar=100 μm.
Figure 3.
OPLS-DA comparison of 20 ipsilateral pericontusional neurons and 20 contralateral non-contusional neurons. GCIB-SIMS ion intensity signals from areas corresponding to each of 20 neurons from the pericontusional region (top right) along with 20 corresponding non-contusional neurons from the contralateral hemisphere (left) were used for OPLS-DA analysis. This showed differential clustering of pericontusional (ipsilateral, red in plot) and non-contusonal (contralateral, green in plot) neurons. Each neuron is one point on the OPLS-DA score plot.
GCIB-SIMS imaging with H2O clusters on frozen-hydrated tissue and cells has allowed us, for the first time, to simultaneously visualize low abundance PEox and FFAox species and significant changes in metabolites such as GSH that occur as a result of RSL3-induced ferroptotic death in cardiomyocytes and in the neuronal cell death in response to TBI. Furthermore, frozen-hydration closely preserves chemical gradients to their natural state. These critical characteristics are most likely responsible for its ability to visualize low abundance PEox and FFAox species in cardiomyocytes and neuronal cells, due to less interference from highly abundant lipids. This advancement in GCIB-SIMS technology will lead to its multiple applications in studies of signaling roles of different phospholipids in the context of their production in different intracellular compartments and intercellular communications.[16]
Supplementary Material
Acknowledgements
This research was funded in part by the following grants from the National Institutes of Health: R01NS076511, R01NS061817, U01AI156924, P01HL114453, R01CA165065, R01AI145406, U01AI156923, R01CA243142, R01GM134715, R01GM113908, R01HL151078, R01NS117000, U54GM103529-08; Natural Science Foundation of China (81873209, 81703696) and 111 Project of Chinese MoE (B13038).
Footnotes
Supporting information and the ORCID identification number(s) for the author(s) of this article can be found under: https://doi.org/10.1002/anie.202102001.
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Louis J. Sparvero, Department of Environmental and Occupational Health and Center for Free Radical and Antioxidant Health University of Pittsburgh, Pittsburgh, PA 15261 (USA); Department of Critical Care Medicine and Children’s Neuroscience Institute UPMC Children’s Hospital of Pittsburgh Pittsburgh, PA 15261 (USA).
Hua Tian, Department of Chemistry, Pennsylvania State University University Park, State College, PA 16802 (USA).
Andrew A. Amoscato, Department of Environmental and Occupational Health and Center for Free Radical and Antioxidant Health University of Pittsburgh, Pittsburgh, PA 15261 (USA)
Wan-Yang Sun, College of Pharmacy, Jinan University Guangzhou, Guangdong 510632 (China).
Tamil S. Anthonymuthu, Department of Environmental and Occupational Health and Center for Free Radical and Antioxidant Health University of Pittsburgh, Pittsburgh, PA 15261 (USA) Department of Critical Care Medicine and Children’s Neuroscience Institute UPMC Children’s Hospital of Pittsburgh Pittsburgh, PA 15261 (USA).
Yulia Y. Tyurina, Department of Environmental and Occupational Health and Center for Free Radical and Antioxidant Health University of Pittsburgh, Pittsburgh, PA 15261 (USA)
Oleksandr Kapralov, Department of Environmental and Occupational Health and Center for Free Radical and Antioxidant Health University of Pittsburgh, Pittsburgh, PA 15261 (USA).
Sabzali Javadov, Department of Physiology, School of Medicine University of Puerto Rico, San Juan, PR 00936-5067 (USA).
Rong-Rong He, College of Pharmacy and School of Traditional Chinese Medicine Jinan University, Guangzhou, Guangdong 510632 (China).
Simon C. Watkins, Department of Cell Biology and Center for Biological Imaging University of Pittsburgh, Pittsburgh, PA 15261 (USA)
Nicholas Winograd, Department of Chemistry, Pennsylvania State University University Park, State College, PA 16802 (USA).
Valerian E. Kagan, Department of Environmental and Occupational Health and Center for Free Radical and Antioxidant Health University of Pittsburgh, Pittsburgh, PA 15261 (USA); Department of Critical Care Medicine and Children’s Neuroscience Institute UPMC Children’s Hospital of Pittsburgh Pittsburgh, PA 15261 (USA).
Hülya Bayır, Department of Environmental and Occupational Health and Center for Free Radical and Antioxidant Health University of Pittsburgh, Pittsburgh, PA 15261 (USA); Department of Critical Care Medicine and Children’s Neuroscience Institute UPMC Children’s Hospital of Pittsburgh Pittsburgh, PA 15261 (USA).
References
- [1].a) Kapralov AA, Yang Q, Dar HH, Tyurina YY, Anthonymuthu TS, Kim R, St Croix CM, Mikulska-Ruminska K, Liu B, Shrivastava IH, Tyurin VA, Ting HC, Wu YL, Gao Y, Shurin GV, Artyukhova MA, Ponomareva LA, Timashev PS, Domingues RM, Stoyanovsky DA, Greenberger JS, Mallampalli RK, Bahar I, Gabrilovich DI, Bayır H, Kagan VE, Nat. Chem. Biol 2020, 16, 278–290; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Tyurina YY, Tyurin VA, Anthonymuthu T, Amoscato AA, Sparvero LJ, Nesterova AM, Baynard ML, Sun W, He R, Khaitovich P, Vladimirov YA, Gabrilovich DI, Bayır H, Kagan VE, Chem. Phys. Lipids 2019, 221, 93–107; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Conrad M, Kagan VE, Bayir H, Pagnussat GC, Head B, Traber MG, Stockwell BR, Genes Dev. 2018, 32, 602–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].a) Pulfer M, Murphy RC, Mass Spectrom. Rev 2003, 22, 332–364; [DOI] [PubMed] [Google Scholar]; b) Hayakawa J, Okabayashi Y, J. Pharm. Biomed. Anal 2004, 35, 583–592; [DOI] [PubMed] [Google Scholar]; c) Ikeda K, Oike Y, Shimizu T, Taguchi R, Chromatogr J. B 2009, 877, 2639–2647; [DOI] [PubMed] [Google Scholar]; d) Tyurina YY, Polimova AM, Maciel E, Tyurin VA, Kapralova VI, Winnica DE, Vikulina AS, Domingues MR, McCoy J, Sanders LH, Bayır H, Greenamyre JT, Kagan VE, Free Radical Res. 2015, 49, 681–691; [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Nakanishi H, Ogiso H, Taguchi R, Methods Mol. Biol 2009, 579, 287–313; [DOI] [PubMed] [Google Scholar]; f) Lydic TA, Goo YH, Clin. Transl. Med 2018, 7, 4; [DOI] [PMC free article] [PubMed] [Google Scholar]; g) Holčapek M, Liebisch G, Ekroos K, Anal. Chem 2018, 90, 4249–4257. [DOI] [PubMed] [Google Scholar]
- [3].Vickerman JC, Analyst 2011, 136, 2199–2217. [DOI] [PubMed] [Google Scholar]
- [4].a) Angerer TB, Blenkinsopp P, Fletcher JS, Int. J. Mass Spectrom 2015, 377, 591–598; [Google Scholar]; b) Toyoda N, Matsuo J, Aoki T, Yamada I, Fenner DB, Nucl. Instrum. Methods Phys. Res. Sect. B 2002, 190, 860–864; [Google Scholar]; c) Niehaus M, Soltwisch J, Belov ME, Dreisewerd K, Nat. Methods 2019, 16, 925–931; [DOI] [PubMed] [Google Scholar]; d) Kompauer M, Heiles S, Spengler B, Nat. Methods 2017, 14, 90–96. [DOI] [PubMed] [Google Scholar]
- [5].Tian H, Sparvero LJ, Blenkinsopp P, Amoscato AA, Watkins SC, Bayır H, Kagan VE, Winograd N, Angew. Chem. Int. Ed 2019, 58, 3156–3161; Angew. Chem. 2019, 131, 3188–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].a) Sheraz nee Rabbani S, Razo IB, Kohn T, Lockyer NP, Vickerman JC, Anal. Chem 2015, 87, 2367–2374; [DOI] [PubMed] [Google Scholar]; b) Sheraz nee Rabbani S, Barber A, Fletcher JS, Lockyer NP, Vickerman JC, Anal. Chem 2013, 85, 5654–5658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Sheraz S, Tian H, Vickerman JC, Blenkinsopp P, Winograd N, Cumpson P, Anal. Chem 2019, 91, 9058–9068. [DOI] [PubMed] [Google Scholar]
- [8].Stockwell BR, Free Radical Biol. Med 2018, 120, S7. [Google Scholar]
- [9].Kagan VE, Mao G, Qu F, Angeli JP, Doll S, Croix CS, Dar HH, Liu B, Tyurin VA, Ritov VB, Kapralov AA, Amoscato AA, Jiang J, Anthonymuthu T, Mohammadyani D, Yang Q, Proneth B, Klein-Seetharaman J, Watkins S, Bahar I, Greenberger J, Mallampalli RK, Stockwell BR, Tyurina YY, Conrad M, Bayır H, Nat. Chem. Biol 2017, 13, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Stockwell BR, Jiang X, Gu W, Trends Cell Biol. 2020, 30, 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Piwowar AM, Fletcher JS, Kordys J, Lockyer NP, Winograd N, Vickerman JC, Anal. Chem 2010, 82, 8291–8299; [DOI] [PubMed] [Google Scholar]; b) Pareek V, Tian H, Winograd N, Benkovic SJ, Science 2020, 368, 283–290; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Yoon S, Lee TG, Nano Convergence 2018, 5, 24; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Newell CL, Vorng JL, MacRae JI, Gilmore IS, Gould AP, Angew. Chem. Int. Ed 2020, 59, 18194–18200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Maguire JJ, Tyurina YY, Mohammadyani D, Kapralov AA, Anthonymuthu TS, Qu F, Amoscato AA, Sparvero LJ, Tyurin VA, Planas-Iglesias J, He R-R, Klein-Seetharaman J, Bayır H, Kagan VE, Biochim. Biophys. Acta 2017, 1862, 8–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kenny EM, Fidan E, Yang Q, Anthonymuthu TS, New LA, Meyer EA, Wang H, Kochanek PM, Dixon CE, Kagan VE, Bayır H, Crit. Care Med 2019, 47, 410–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sparvero LJ, Amoscato AA, Fink AB, Anthonymuthu T, New LA, Kochanek PM, Watkins S, Kagan VE, Bayır H, J. Neurochem 2016, 139, 659–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].a) Anthonymuthu TS, Kenny EM, Amoscato AA, Lewis J, Kochanek PM, Kagan VE, Bayır H, Biochim. Biophys. Acta. Mol. Basis Dis 2017, 1863, 2601–2613; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Sun WY, Tyurin VA, Mikulska-Ruminska K, Shrivastava IH, Anthonymuthu TS, Zhai YJ, Pan MH, Gong HB, Lu DH, Sun J, Duan WJ, Korolev S, Abramov AY, Angelova PR, Miller I, Beharier O, Mao GW, Dar HH, Kapralov AA, Amoscato AA, Hastings TG, Greenamyre TJ, Chu CT, Sadovsky Y, Bahar I, Bayır H, Tyurina YY, He RR, Kagan VE, Nat. Chem. Biol 2021, 10.1038/s41589-020-00734-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Manuscript data including GCIB-SIMS imaging data sets is publicly available in Mendeley Data at: DOI: 10.17632/c3k5w84792.1 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.