Fig. 3.
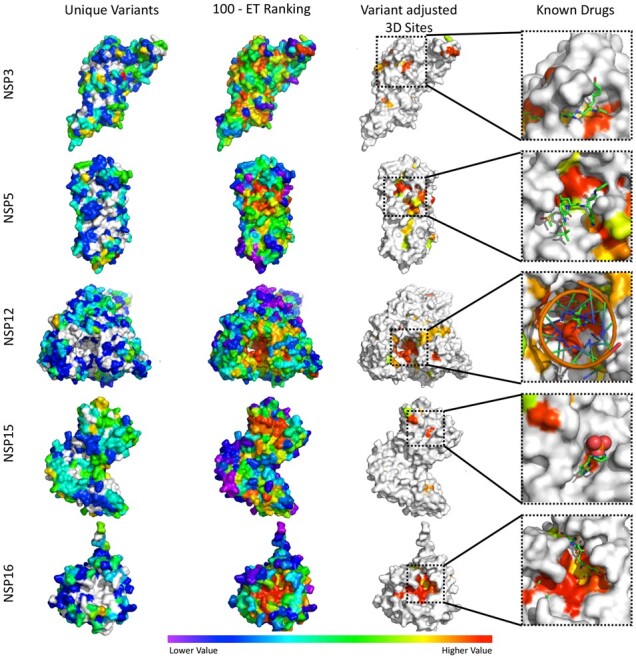
Identification of variant adjusted 3D sites (5 Å) and their colocalization with known drug binding sites. Variant adjusted 3D sites for NSP3 (6w9c), NSP5 (6yb7), NSP12 (7bv1), NSP15 (6wlc) and NSP16 (6w4h) were identified as clusters of surface residues with low ET ranks and a lack of mutations in the current outbreak. In the known drugs panels, variant adjusted 3D sites were identified using apo form structures, then mapped to the co-structures of NSP3 with peptide inhibitor vir251 (PDB:6wx), NSP5 with potential drug 13 b (PDB:6y2f), NSP12 with drug remdesivir (7bv2), NSP15 in complex with potential drug tipiracil (PDB:6wxc) and NSP16 with sinefungin (PDB:6wkq). For structures in the ‘Unique Variants’ column ‘Lower Values’ in the color scale correspond to fewer variants, while ‘Higher Values’ correspond to more variants and white residues have no reported variants in the analyzed SARS-CoV-2 strains. For the ‘100—ET Ranking’, ‘Variant Adjusted 3D Sites’ and ‘Known Drugs’ columns, ‘Lower Values’ correspond to less phylogenetic conservation while ‘Higher Values’ correspond to more phylogenetic conservation
