Abstract
Lipid bilayer membranes are a central structural feature of living cells, providing a wide range of functions including partitioning of organelles, mediating cell interaction with the environment, and modulating intracellular signaling processes. By capturing the fluidity of the natural membranes in a reductionist in vitro model, substrate supported lipid bilayers have emerged as a compelling model system for these structures. Furthermore, the ability to control the composition and mobility of this system at micro- and nano-scales inspired several new routes of biological and biotechnological investigation. Here, we describe key methods used to create multicomponent lipid bilayers, discuss design considerations important to making these systems, and demonstrate this process in the specific context of understanding juxtacrine cell signaling. Different fabrication techniques were combined to first pattern a surface with barriers to lipid diffusion and then spatially control the exposure of this surface to lipid vesicles, leading to local formation of bilayers of different composition. This multicomponent system was used as a platform for to mimic the natural organization of a T Cells and Antigen Presenting Cells by presenting ligands to the T Cell Receptor and LFA-1 that are tethered to separate, closely juxtaposed regions of bilayer. Other technologies like using photochemical polymerization of lipids to pattern bilayers has also been discussed. The information gathered from evaluating membrane interactions in patterned lipid bilayers may lead to the development of membrane based biomedical devices for conducting novel cell-based assays and potentially high throughput drug screens targeting membranes or membrane associated components.
Keywords: lipid bilayers, patterning, microfluidics, microfabrication, lithography, microcontact printing
Introduction:
Supported lipid bilayers provide a powerful model of natural cell membranes and have found widespread use across a diverse spectrum of research (Brian & McConnell, 1984; Sackmann, 1996). The basic system consists of a phospholipid bilayer in close proximity to an appropriate solid support (Boxer, 2000). A thin layer of water separates the bilayer from the underlying substrate, allowing lateral mobility of membrane molecules and thus capturing the fluidity of the cellular counterpart (Singer & Nicolson, 1972) in a reductionist format well suited for microscopy and other experimental manipulations. A major application of this platform has been in the study of juxtacrine signaling, with the supported lipid bilayer used in place of one of the interacting cells (Manz & Groves, 2010). In this approach, engineered biomolecules based on cell adhesion proteins are tethered to the bilayer, allowing their spatial reorganization under the influence of an adherent cell. This model has been particularly successful in understanding the immune synapse, a small (~ 70 μm2) area of contact between a lymphocyte and Antigen Presenting Cell (APC) which focuses their communication (Chan, Lawrence, Dustin, Ferguson, Golan & Springer, 1991; Dean et al, 2003; Grakoui et al, 1999; Groves & Dustin, 2003). Initial studies examined the interaction of T cells with supported lipid bilayers presenting peptide-loaded major histocompatibility complex (pMHC) and Intercellular adhesion molecule −1 (ICAM-1) proteins, which are normally on the APC surface (Grakoui et al, 1999). Live-cell microscopy revealed that microclusters containing pMHC bound to T Cell Receptor (TCR) are actively transported towards the center of this artificial synapse, while ICAM-1 bound to its receptor Lymphocyte function-associated antigen-1 (LFA-1) are localized to the peripheral regions, revealing the complex dynamics behind formation of the archetypal “bullseye” pattern indicative of the mature synapse (Monks, Freiberg, Kupfer, Sciaky & Kupfer, 1998). Importantly, these rearrangements would not be observed in a system where the protein ligands are immobilized to the substrate. Since those initial observations, this system has been developed extensively to understand the impact of ligand mobility on cell function, including studies of additional receptor-ligand pairs and juxtacrine signaling in other systems (Manz & Groves, 2010). In addition, the highly controllable, reductionist nature of the supported lipid bilayer model has gained it much attention as a general platform for studying cell adhesion, fundamental membrane physiology, and biosensor/biotechnology applications.
The ability to locally control membrane mobility and composition provides new opportunities and routes of investigation in virtually all of these areas. This is perhaps counter-intuitive as the distinctive feature of the supported lipid bilayer model is long-range lateral mobility, which would serve to erase any local gradient in membrane composition. Moreover, the main approaches used to form supported lipid bilayers – fusion of lipid vesicles to a surface and layer-by-layer deposition in a Langmuir trough – are designed to create membranes of uniform composition across large surfaces. However, cellular physiology is replete with examples of local membrane order and separation, from the presence of diffusional barriers important to epithelial cell polarity or axon compartmentalization to phenomena at smaller scales such as membrane microdomains. On an experimental front, supported lipid bilayers are attractive for studying cell interaction with laterally mobile ligands, but this basic configuration cannot simultaneously support spreading required for normal function, as these attachments provide insufficient resistance to the forces applied by adherent cells. Lastly, the ability to create arrays of supported lipid bilayer of different compositions on a single surface greatly increases the utility of this system in screening and biotechnology applications, an approach demonstrated in current generations of chip-based gene and protein analysis.
The earliest approaches to create precise, microscale patterns of supported lipid bilayers were based on a peculiar aspect of the underlying substrate (Groves, Ulman & Boxer, 1997). Specifically, these approaches used the aggregation and fusion of lipid vesicles, applied in an aqueous solution to the substrate, to form the extended lipid bilayer (Figure 1A). This process was observed to occur on only a limited number of surfaces, specifically those that are predominantly silicon oxide in composition, such as quartz, glass, or oxide layer present on crystalline silicon. Other materials, including plastic (photoresist), aluminum oxide, or metals, do not readily support vesicle fusion into an extended bilayer. Materials such as conductive metal surfaces of gold (Au), titanium (Ti), and silver (Ag) are of major interest in biosensor technologies and other applications (Parthasarathy & Groves, 2004; Yoon et al, 2006), particularly with the attachment of biological molecules and larger assemblies to the material surface (Figure 1B). Additional steps have been developed to allow formation of supported lipid bilayers on these materials. In one of approach, siloxane-terminated alkane-thiols are attached to the metal, which mimics the glass surface and allows facile rupture and fusion of lipid vesicles (Taylor, 2007) (Figure 1C). Other tethering approached have been successfully applied to get bilayer on these difficult surfaces (Sackmann & Tanaka, 2000). However, by patterning these materials onto surface using fabrication technologies developed in the microelectronics industry, it is possible to create microscale, and later nano-scale, barriers that locally disrupt the lipid bilayer and restrain membrane mobility (Figure 1B). In another variation on this theme, microfluidic lithography can be used to generate patterns of two different self-assembled monolayers on the surface followed by the formation of bilayers in the lipid adhered regions (Figure 1C) (Dutta, Pulsipher & Yousaf, 2010). In this study, bio-orthogonal quinone-oxyamine chemistry was used to form electroactive fluid lipid bilayers on conductive surfaces. Apart from providing a platform to study various bio-specific ligand-receptor interactions, because the immobilization of various ligands results in the formation of electroactive oxime bond, ligand density and extent of reaction can be quantified and monitored by cyclic voltammetry. Subsequent development of techniques for making patterns of proteins (microcontact printing, in particular) or even non-fluid regions of lipid bilayer (Morigaki, Baumgart, Offenhäusser & Knoll, 2001) allowed the use of biologically important molecules into these systems. Intriguingly, the ability of proteins with biological activity to serve as features within a fluid supported lipid bilayer inspired the development of surfaces combining multiple functionalities into a single system. In particular, we used this approach to promote spreading of anchorage-dependent cells across supported lipid bilayers by patterning arrays of cell-adhesive stepping stones of extracellular matrix proteins, interspersed amongst the artificial membrane (Kam & Boxer, 2001; Perez, Nelson, Boxer & Kam, 2005). This arrangement allowed cells to attain a well-understood morphology in culture while interacting with proteins or other biomolecules tethered to the lipid bilayer.
Figure 1.
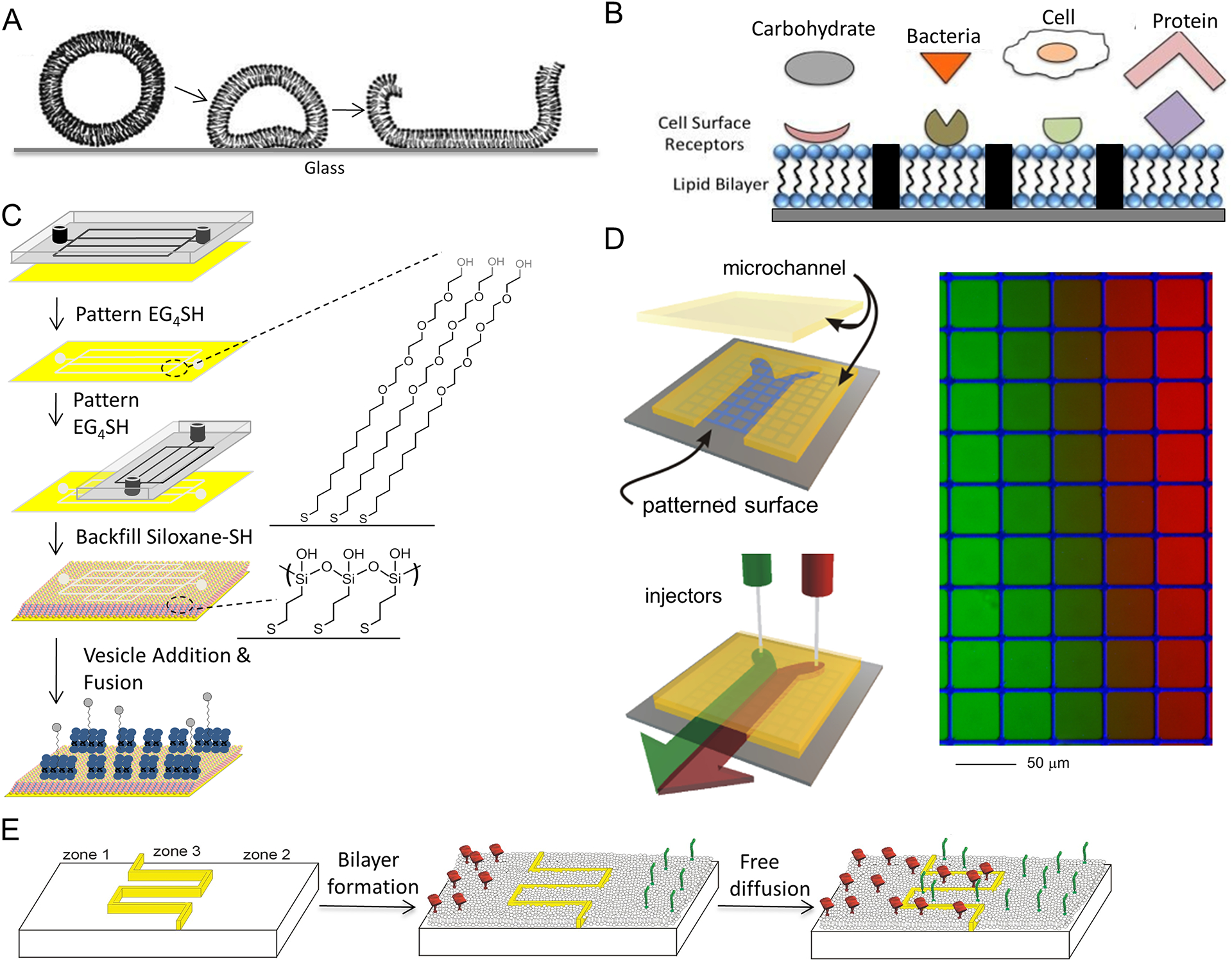
(A) Schematics of how lipid vesicles come down to the surface, rupture and fuse spontaneously to form lipid bilayer structure. (B) Schematics of how solid supported lipid bilayers can form platforms to study different biomolecular interactions. (C) Schematic for the generation of patterned fluid lipid bilayers using a microfluidic cassette: A PDMS microfluidic cassette was reversibly sealed to a bare gold substrate, and a solution of tetra(ethylene) glycol-terminated alkanethiol (EG4SH) in EtOH was flowed through the channels; forming a patterned EG4SH SAM by microfluidic lithography (μFL); the same cassette was then placed perpendicular to the newly patterned EG4SH SAM, and μFL was performed again; the substrate was then immersed in a solution of HO3SiSH in EtOH for ~16h, in order to backfill the remaining un-patterned gold with a SAM; a solution of lipid vesicles containing H2Q in Tris buffer was then added on the surface to undergo spontaneous fusion and form bilayers, only in regions containing HO3SiSH. EG4SH and HO3SiSH prepared for use as described in (Dutta, Pulsipher & Yousaf, 2010). Reprinted with permission from (Dutta, Pulsipher & Yousaf, 2010). Copyright 2010 American Chemical Society. (D) Microfluidic cassettes consisting of converging channels offer a different opportunity to create complex supported lipid bilayers. Reprinted with permission from (Kam & Boxer, 2000). Copyright 2000 American Chemical Society. (E) Schematic illustrating self-aligning patterns of multiple SLBs. Reprinted with permission from (Shen, Tsai, Shi & Kam, 2009). Copyright 2009 American Chemical Society.
While prepatterning a surface with diffusion barriers allowed control over membrane mobility, the supported bilayers in these systems were otherwise typically uniform in composition from location to location. This was also related to method in which bilayers are formed, typically the application of a solution containing vesicles of a single membrane composition to the surface. One approach to gain local control over membrane composition is to expose different types of lipid vesicles to each area of the substrate. This is difficult with typical, macroscopic laboratory bench tools, as the substrate and vesicles must all remain submerged for this process, and diffusion within a solution acts to erase any gradients in vesicle composition. What emerged was the use of microfluidic systems to create convergent streams of vesicle solutions that impinge on the target surface (Figure 1D) (Kam & Boxer, 2000; Kam & Boxer, 2003). The laminar nature of flow within these chambers minimizes mixing between these streams, allowing local control over solution composition and spatially resolved manipulation of a supported lipid bilayer. It is noted that following membrane formation using such a configuration, a supported lipid bilayer on a uniform surface would undergo mixing within the plane of this structure, erasing any changes in composition from place to place. For this reason, microfluidics was combined with surface prepatterning to limit and control the extent of this mixing, leading to formation of such entities as concentration gradients of lipid composition across a single surface (Figure 1D).
Returning to the desire to capture the local changes in membrane composition observed at cell-cell interfaces and across the cell membrane, we describe here the further evolution of this approach to create surfaces containing multiple patches of lipid bilayer that are of different composition and with geometry that can be controlled at sub-micrometer scales (Shen, Tsai, Shi & Kam, 2009). These scales are difficult to achieve using microfluidics directly, as limited mixing does occur between streams in these microfluidic cassettes, typically on the order of several to tens of micrometers. To achieve this higher resolution, we took advantage of the 2D fluidity of the lipid bilayers, creating inlets and canals that connected small features within a target area to large reservoirs of composition that are defined far from this region (Figure 1E). The coarse resolution afforded by microfluidics was used to define the composition of these reservoirs, and the surface-bound canals were used to introduce these contiguous lipid membranes into the target area. Using contemporary nanofabrication technologies, it is conceptually possible to control the local composition of a supported lipid bilayer system with resolution on the order of tens of nanometers. We present in the following sections the implementation of this approach, describing key materials and devices used to create multicomponent, micropatterned lipid bilayer systems. In addition, we describe additional technologies that in combination with or in place of these techniques provide a robust toolbox for the next generation of lipid bilayers.
MATERIALS:
Silicon wafers (5”, low doping, (100) orientation, prime grade; UniversityWafer, Boston, MA)
Glass coverslip
Poly(methylmethacrylate) (25 kDa and 950 kDA) (cat# M230006; MicroChem, Newton, MA) in 1:1 Methyl isobutyl ketone and Iso propyl alcohol (solvents from Sigma-Aldrich, St. Louis, MO)
Aquasave (formulation 53ZA; Mitsubishi Rayon Co., New York, NY)
3:1 Iso Propyl Alcohol-water mixture (solvents from Sigma-Aldrich, St. Louis, MO)
Acetone (Sigma-Aldrich)
SU-8 2025 (MicroChem)
Dimethylchlorosilane (Sigma-Aldrich)
Sylgard 184 polydimethoxysiloxane elastomer (PDMS; Dow Corning, Midland, MI).
Acrylic sheets (3 mM thick), custom cut and drilled
Lipids: Texas Red-DHPE (cat# T1395MP; Life Technologies, Grand Island, NY), NBD-PE (cat# 810143; Avanti Polar Lipids, Alabaster, AL), DiD (cat# D7757; Life Technologies), DOPC (cat# 850375; Avanti), Biotinyl-Cap-PE (cat# 870277; Avanti), DOGS-NTA (cat# 790404; Avanti)
Milli Q water
Bovine Serum Albumin (cat# A7030, Sigma-Aldrich)
Alexa-488 conjugated streptavidin (cat# S32354, Life Technologies)
Phosphate Buffer Saline (pH 7.4)
Biotinylated-OKT3 (cat# 13-0037; Affymetrix-ebioscience, Santa Clara, CA)
Cy5-labeled ICAM-1-6His (cat# 10346-H03H-5; Life Technologies)
RPMI-1640 medium (cat# 11875119; Life Technologies)
L-glutamine (2mM)
10% Fetal Bovine Serum (FBS), Characterized (cat# SH30071; HyClone, Thermo Scientific)
HEPES (10 mM) (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
Penicillin (100 U/mL) in Phosphate Buffer Solution
Streptomycin (100 μg/mL) in Phosphate Buffer Solution
2-mercaptoethanol (50 μM) in Phosphate Buffer Solution
EQUIPMENT:
Plasma cleaner (Basic Plasma Cleaner; Harrick Plasma, Ithaca, NY)
Spin coater
Süss MicroTec MJB3 Mask Aligner
FEI XL-30 scanning electron microscope
Semicore electron beam evaporator
Vacuum desiccator (cat# 08-594-15B; Thermo Scientific)
Sonicator (Branson 1510; Thermo Scientific)
Membrane filters and lipid extruder (cat# 610000 with 50 nm pore-diameter; Avanti)
Fluorescence microscope (IX81; Olympus)
METHOD:
A. Prepatterning of diffusion barriers onto array surface
The fabrication techniques required for creating barriers of metal, photoresist, or other materials are well established but for the most part require access to clean room facilities, each of which will have a specific array of equipment for use. As such, we highlight here the aspects of patterning that are important for defining barriers to lipid membrane diffusion. Specifically, the following considerations are key to developing a good barrier patterning process:
The working substrate is typically glass or quartz. The chemistry of these surfaces is well suited for supported lipid bilayers while their transparency is needed for subsequent microscopy and cell culture. Silicon wafers, which are opaque but more common for use in clean room processes, are occasionally used.
The resist material (light- or radiation-sensitive layer that use used to define and process a given pattern) must be easy to remove completely from the glass surface. Positive resists, which become easier to dissolve following exposure to light or radiation, are thus typically chosen for this specific application. The resist layer is on the order of 200 nm to a few micrometers in thickness. It must be able to resist all cleaning steps applied to the coverslip.
The material to be patterned can be photoresist, metal (chrome, titanium, gold, etc.), or organic molecule such as protein or poly(ethylene glycol).
The feature dimensions required for processing depend on the end application. The micrometer-scale resolution readily achievable by photolithography is well suited for many subcellular studies of cell function. Finer, nanoscale resolution is suited for the study of membrane biophysics or phenomena involving molecular assemblies, and typically requires electron beam patterning.
Photolithography
This well-established approach is the workhorse of the microelectronics industry. It involves coating a substrate with a photoresist layer, exposing this layer to light (typically UV or shorter wavelength) which chemically modified the resist layer chemistry, followed by development of the resist layer to expose designated regions of the surface. Typically, metal or other material is deposited on the substrate in patterns defined by the resist exposure, and these layers can be of very thin dimensions (several nanometers). However, the earliest papers of lipid bilayer patterning by Groves, Ulman, and Boxer (Groves, Ulman & Boxer, 1997) demonstrated that photoresist itself could be used as the diffusion barrier, without the need for further development.
The choice of photoresist material for successful bilayer patterning reflects a balance between the needs of the lipid system and processes available in the clean room. Glass surfaces are typically exposed to harsh acid/base solutions, high temperature (425°C), or other aggressive treatment to produce the clean surfaces required for bilayer formation. As such, resists that are difficult to completely remove often pose challenges to lipid bilayer formation. For this reason, positive resists, which become easier to dissolve upon exposure to light, are typically chosen for this purpose. The cleanroom facilities used in our processes have well defined processes for the Shipley S1805 positive resist, driving our choice of this material for processes. However, it is noted that if the resist itself is used as a barrier, a negative photoresist, which becomes more solid and difficult to remove upon exposure, can also be used. For this purpose, the SU8 series of resists, which are formulated for microfluidics and other high-aspect-ratio applications, is very attractive.
Another major consideration in photolithography is the type of photomask that is used to define barriers. This typically involves the use of a contact aligner (the Süss MicroTec MJB3 Mask Aligner indicated in Equipment) or projection system, which are well suited for resolution on the order of a micrometer. To reach these resolutions, the photomasks must be of high quality, typically a metal-on-glass construct. These are readily available, but comparatively expensive. If the barriers are of 10 μm or larger dimension, it is possible to use high-resolution (10,000 dpi) transparency masks, which are often employed for microfluidics fabrication. Such masks are available through specialty printers CAD/Art services, Bandon, OR.
Standard photolithography approaches were used to create the barrier system in Figure 3A–C below.
Figure 3.
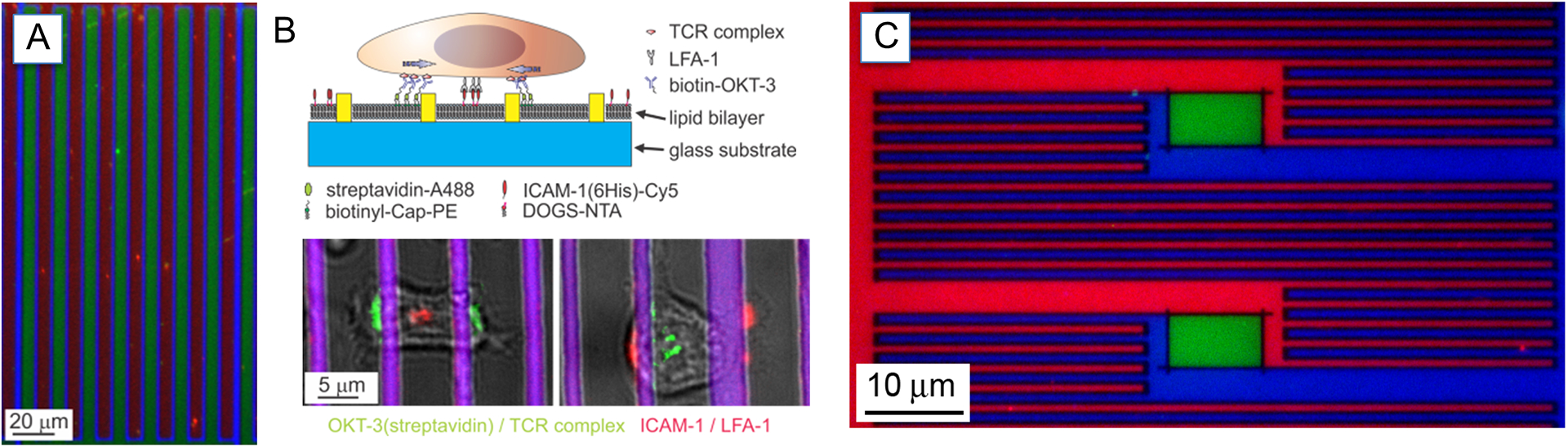
(A) Example of interdigitating lipid bilayers. A serpentine barrier of photoresist (blue) separated bilayers containing a small amount of TR (red) or NBD (green) labeled lipids. (B) Schematic of the use of these segregated bilayers to spatially control the distribution of membrane-tethered anti-CD3 (OKT3, green) and ICAM-1 (red). (C) A finer three-component pattern with submicrometer resolution was achieved using e-beam lithography. A more complicated barrier geometry allowed patterning of an increasing number of proteins. This surface was exposed to lipid vesicles containing small amounts of lipid labeled with TR (red), NBD (green), or DiD (blue). Reprinted with permission from (Shen, Tsai, Shi & Kam, 2009). Copyright 2009 American Chemical Society.
Electron-beam lithography
Finer barriers (< 100 nm dimension) can be created using electron beam lithography; as the converse the higher resolution offered by electron vs. light microscopy, defining a pattern by a stream of electrons offers the ability to define barriers to lipid diffusion on the order of tens of nanometers (Tsai, Sun, Gao, Hone & Kam, 2008).
In this case, a major concern is the use of a glass substrate as the working surface. Electron beam lithography typically required a conductive substrate to drain off accumulated charges, which would otherwise distort and deflect the electron beam. Glass, as an insulator, does not inherently allow this process. To address this issue, a charge dispersion layer of conductive material can be applied to the surface prior to writing. In the material list above, this purpose is served by Aquasav, a conductive, water-soluble polymer that is applied to the resist layer prior to writing.
In selection of resists, it is recognized that an electron-beam sensitive layer, rather than light-sensitive, must be used in this form of lithography. In the approach described here, we use poly(methyl methacrylate) (PMMA, MicroChem, Newton, MA) as the resist, as it provides acceptable resolution but is readily and cleanly removed from the working surface, for the same reasons stated above for using a positive photoresist. As described by Tsai and colleagues (Tsai, Sun, Gao, Hone & Kam, 2008), two layers of PMMA, each of different molecular weight and development characteristics, improves metal patterning (in a process termed lift-off). Please see that manuscript for additional details.
This electron-beam lithography approach was used to create the finer barriers shown in Figure 3D below.
Lastly, while these two general approaches have found widespread use for patterning of supported lipid bilayers, there are many additional technologies available, many of which offer advanced capabilities such as the ability to turn barriers on off during the experiment or allow directed coupling of biomolecules to the bilayer. Please see the discussion section for key examples of these methods.
B. Fabrication and assembly of three-stream laminar flow chamber:
To create the closely apposed lipid bilayer regions indicated schematically in Figure 1E the lithography techniques described in the last section were used to create a serpentine barrier within a working region of a glass slide. This was then fitted with a three-channel flow chamber, shown conceptually in Figure 2A. These chambers were designed in AutoCAD (Autodesk, San Rafael, CA) and printed on transparency sheet at 10,000 dpi (CAD/Art services, Bandon, OR). The master for the flow chamber was fabricated on a 2-inch wafer using a 25 μm thick layer of SU-8 2025 (MicroChem), then vapor-primed with dimethylchlorosilane (Sigma, St. Louis, MO) in a vacuum desiccator for 30 min. Channels were made by replica molding of Sylgard 184 polydimethoxysiloxane elastomer (PDMS, Dow Corning, Midland, MI). The PDMS channels were drilled using standard syringes to provide flow inlets and outlets, and aligned by eye onto the coverslip with interdigitating patterns (without plasma treatment, thus preventing permanent bonding). The whole structure was then sandwiched and pressed between two 3 mm-thick acrylic sheets, and connected with polyethylene tubings (Figure 2B). Gravity flow was established by placing the inlet tubings in sample solution and the opening of outlet tubings 30~50 cm lower than the inlets. To prevent the layers from separating during flow, pressure was applied across the acrylic sheets using binder clips (Figure 2B).
Figure 2.
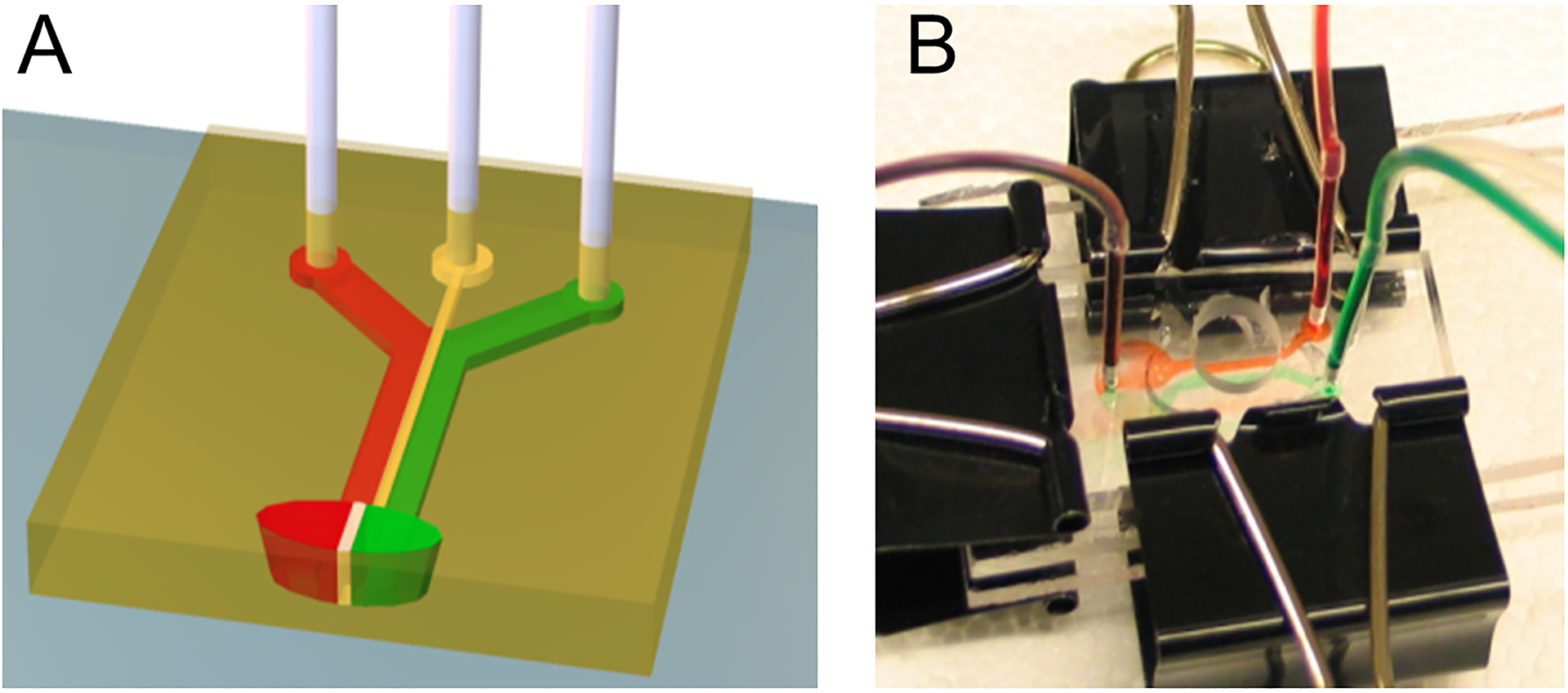
(A) Schematic diagram and (B) benchtop-implementation of a three-stream, converging laminar flow configuration used to define patterns of bilayer formation on micropatterned regions.
C. Formation of multicomponent bilayer systems defined at subcellular scales.
Preparation of small unilamellar vesicles and supported lipid bilayer:
Lipid components were mixed in chloroform at 1 mol % and dried in round-bottom flask under N2 flow, and desiccated for 2 hours with house vacuum. The mixtures were reconstituted by bath sonication in deionized water at 2.5 mg/ml, and extruded through 50-nm pore membranes. The solutions were diluted 1:1 in PBS (pH 7.4) before being applied to the surface in the three-stream laminar flow chamber for the bilayer formation process. Vesicle solutions were allowed to interact with the surface for 2 minutes, and then the entire chamber was washed exclusively with deionized water.
Protein capture in lipid bilayer:
DOPC lipids supplemented with either 0.02 mol % Biotinyl-Cap-PE or 6 mol % of DOGS-NTA were used to form supported lipid bilayers. After overnight diffusion, the surface was blocked for 1 hour with 1mg/ml BSA and incubated 20 minutes with 2 μg/ml Alexa-488 conjugated streptavidin. The surface was then washed three times with PBS (pH 7.4), and incubated 20 minutes with 5 μg/ml monobiotinylated-OKT3 and 2 μg/ml Cy5-labeled ICAM-1–6His before being washed again for three times with PBS.
Cell culture:
Day-8 human CD4 + T cell blasts frozen in liquid nitrogen were thawed and allowed to recover overnight in RPMI-1640 medium supplemented with 2 mL L-glutamine, 10% FBS, 10 mM HEPES, 100 U/mL penicillin, 100 μg/mL streptomycin and 50 μM 2-mercaptoethanol. Before being seeded onto bilayers, cells were pelleted and washed twice in extracellular buffer (ECB; 130 mM NaCl, 5 mM KCl, 1.5 mM CaCl2, 1mM MgCl2, 25 mM HEPES, 1 mg/mL BSA, and 5 mM glucose, pH 7.4). The flow chamber containing interdigitated lipid bilayers was equilibrated with ECB and warmed up to 37°C, and the resuspended cells in pre-warmed (37°C) ECB were then injected and live-imaged on a fluorescence microscope equipped with temperature controlled unit (Bioptechs, Butler, PA).
Results
Two different uses of multicomponent supported lipid bilayer surfaces are illustrated in Figure 3. In the first example (Figure 3A–C), an interdigitating barrier is used to create two apposed lipid bilayer regions of different composition together at sub-cellular scales. Figure 3A illustrates this general approach with two bilayers containing a small amount each of different fluorescent dyes, while in Figure 3B, one region contains a biotinylated lipid while the other a poly-His modified counterpart. These moieties were used to bind anti-CD3 (OKT3) and ICAM-1 onto the respective areas of the bilayer. These surfaces were effective in keeping the two ligands separate within the interface of T cells with this artificial system, while maintaining their lateral mobility. The second example (Figure 3C) illustrates a more complex geometry creating including in the barrier pattern isolated regions that were not connected with the adjoining regions (i.e., corresponding to the green region of Figure 3C, surrounded by the dark metal barriers). These isolated regions were aligned with the central stream of the three-flow device shown in Figure 2; in this case, the bilayers were formed by introducing vesicles containing small amounts of lipids labeled with Texas Red (TR, red), Nitro-2-1,3-BenzoxaDiazol-4-yl (NBD, green), and Cyanine dye (DiD, blue) into the left, central, and right streams respectively. Following bilayer formation, the TR and DiD lipids diffused into the central region of the field of view, confined to their respective bilayer compartments. Conversely, NBD in these outer-connected regions diffused out of the central region. In the central, isolated region, there is no exchange of lipids, so no TR or DiD diffused into this region, and no loss of NBD was observed. The final concentrations of NBD in the isolated and outer-connected regions are defined by the relative areas of those outer-connected regions that are exposed to different vesicles. In Figure 3B, the outer-connected regions are defined by the relative areas of those outer-connected regions that are exposed to the different vesicles. In Figure 3D, the outer-connected regions provide extensive reservoirs for diffusion, and NBD was not detectable in those areas, representing at least a 100 fold difference in relative concentration.
Discussion:
The protocol described here introduces a method to form regions of supported bilayer membranes of different compositions and scales at micrometers and smaller aligned on a solid surface. The platform mimics the natural organization of T-cell receptors and antigen-presenting cells by presenting ligands to the T-cell receptor and LFA-1 in separate, closely aligned regions of lipid bilayers.
Highlights:
Fluid lipid bilayer can be formed on a range of substrates- glass, mica or silicon oxide
Multiple bilayer regions can be formed on a single surface
Spatial resolution to the scales of tens of nanometers is determined by the barrier in zone 3.
Any known barrier materials including metals, photoresists or proteins can be used, thus providing few restrictions to the fabrication technique
Only a single bilayer deposition step is required to form multiple bilayer components of different concentrations
Alternative methods of forming lipid bilayer arrays:
Several other methods exist for the patterning of lipid bilayers by the modification of the substrate or by microcontact printing. We focus on two specific methods in this section. First, the Knoll lab has described a method to imprint a pattern within the lipid bilayer by photochemical polymerization of the photoactive lipid diacetylene phosphocholine (Morigaki, Baumgart, Offenhäusser & Knoll, 2001). Method: Bilayers of diacetylene phosphocholine was deposited onto glass substrates from the air/water interface by the Langmuir-Blodgett (LB) and Langmuir-Schaefer (LS) methods (surface pressure Π = 35 mNm−1). Prior to the photopolymerization, oxygen was removed from the aqueous solution by an argon gas purge. Bilayer was polymerized by UV light, where a pattern can be imposed using a mask. After the patterned polymerization, monomeric diacetylene phosphocholine was removed by immersing the sample in ethanol and subsequent rinsing it extensively with Milli-Q water.
Second, the Yousaf lab has demonstrated the use of microfluidic lithography (μFL) to pattern two different self-assembled monolayers (SAMs) on gold followed by the subsequent addition of electroactive lipid vesicles for the generation of fluid lipid bilayers capable of immobilizing a variety of ligands and biomolecules (Dutta, Pulsipher & Yousaf, 2010). To support a fluid lipid bilayer on gold, a SAM of siloxane-terminated alkanethiol (HO3SiOH) was formed on the surface. Lipid vesicles were prepared with a hydroquinone containing alkane (H2Q), which upon mixing became spontaneously embedded into the vesicle. This solution was then ruptured onto the HO3SiOH SAM to generate a fluid bilayer on the substrate. After bilayer formation, H2Q was electrochemically oxidized to quinone (Q) to enable the selective coupling of soluble oxyamine containing (RONH2) ligands, obtained from the bilayer. Rhodamine-oxyamine (Rho-ONH2) and glucose-oxyamine (Glu-ONH2) were immobilized to microarray patterns of electroactive bilayers and then characterized by electrochemistry and fluorescence microscopy. Method: A polydimethylsilane (PDMS) micro- fluidic cassette was reversibly sealed to a bare gold surface. To pattern a SAM by microfluidic lithography (μFL), a 1 mM solution of tetra(ethylene) glycol-terminated alkanethiol (EG4SH) in ethanol was allowed to flow through the channels for 60 s. Without the cassette being removed, ethanol flowed through the channels, which were suction evacuated to clean the surface. The same cassette was then placed perpendicular to the newly patterned EG4SH SAM, and μFL was performed under the same conditions for a second time, as previously described. The substrate was then rinsed with ethanol, dried, and immersed in a 1 mM solution of HO3SiSH in EtOH for 16 h in order to backfill the remaining gold with a SAM. To generate bilayer patterns, gold surfaces with pre-patterned EG4SH and HO3SiSH SAMs were immersed into a 3 mM solution of egg-POPC/H2Q (9:1) in Tris buffer for 1 h. Substrates were then rinsed with PBS, dried in air, and oxidized electrochemically to present Q-terminated patterned bilayers. Rhod-ONH2 was reacted with the Q-terminated bilayers (7 mM in MeOH, rt, 4 h). Similarly, glucose oxyamine (glc-ONH2) was also immobilized onto micro- arrays of Q-terminated bilayers (20 mM in MeOH, rt, 4 h), and TRITC-conjuagted Concanavalin A (ConA) was added (1 mg/mL in DMSO, rt, 2 h). The resultant microarrays displaying Rhod and ConA were imaged by fluorescence microscopy.
Acknowledgements:
This work was supported by the National Institutes of Health, R01AI088377 and PN2EY016586.
References:
- Boxer SG (2000) Molecular transport and organization in supported lipid membranes. Current Opinion in Chemical Biology ;, 4, 704–709. [DOI] [PubMed] [Google Scholar]
- Brian AA & McConnell HM (1984) Allogeneic stimulation of cytotoxic T cells by supported planar membranes. Proceedings of the National Academy of Sciences of the United States of America, 81, 6159–6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PY, Lawrence MB, Dustin ML, Ferguson LM, Golan DE & Springer TA (1991) Influence of Receptor Lateral Mobility On Adhesion Strengthening Between Membranes Containing Lfa-3 and Cd2. Journal of Cell Biology ;, 115, 245–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean C et al. (2003) Neurexin mediates the assembly of presynaptic terminals. Nature neuroscience, 6, 708–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta D, Pulsipher A & Yousaf MN (2010) Selective Tethering of Ligands and Proteins to a Microfluidically Patterned Electroactive Fluid Lipid Bilayer Array. Langmuir, 26, 9835–9841. [DOI] [PubMed] [Google Scholar]
- Grakoui A et al. (1999) The immunological synapse: a molecular machine controlling T cell activation. Science, 285, 221–227. [DOI] [PubMed] [Google Scholar]
- Groves JT & Dustin ML (2003) Supported planar bilayers in studies on immune cell adhesion and communication. Journal of Immunological Methods, 278, 19–32. [DOI] [PubMed] [Google Scholar]
- Groves JT, Ulman N & Boxer SG (1997) Micropatterning of Fluid Lipid Bilayers on Solid Supports. Science, 275, 651–653. [DOI] [PubMed] [Google Scholar]
- Kam L & Boxer SG (2000) Formation of supported lipid bilayer composition arrays by controlled mixing and surface capture. Journal of the American Chemical Society, 122, 12901–12902. [Google Scholar]
- Kam L & Boxer SG (2001) Cell adhesion to protein-micropatterned-supported lipid bilayer membranes. Journal of Biomedical Materials Research, 55, 487–495. [DOI] [PubMed] [Google Scholar]
- Kam L & Boxer SG (2003) Spatially selective manipulation of supported lipid bilayers by laminar flow: Steps toward biomembrane microfluidics. Langmuir, 19, 1624–1631. [Google Scholar]
- Manz BN & Groves JT (2010) Spatial organization and signal transduction at intercellular junctions. Nat Rev Mol Cell Biol, 11, 342–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monks CR, Freiberg BA, Kupfer H, Sciaky N & Kupfer A (1998) Three-dimensional segregation of supramolecular activation clusters in T cells. Nature, 395, 82–86. [DOI] [PubMed] [Google Scholar]
- Morigaki K, Baumgart T, Offenhäusser A & Knoll W (2001) Patterning Solid-Supported Lipid Bilayer Membranes by Lithographic Polymerization of a Diacetylene Lipid. Angewandte Chemie International Edition, 40, 172–174. [DOI] [PubMed] [Google Scholar]
- Parthasarathy R & Groves JT (2004) Protein patterns at lipid bilayer junctions. Proc Natl Acad Sci U S A, 101, 12798–12803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez TD, Nelson WJ, Boxer SG & Kam L (2005) E-Cadherin Tethered to Micropatterned Supported Lipid Bilayers as a Model for Cell Adhesion. Langmuir, 21, 11963–11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sackmann E (1996) Supported membranes: scientific and practical applications. Science, 271, 43–48. [DOI] [PubMed] [Google Scholar]
- Sackmann E & Tanaka M (2000) Supported membranes on soft polymer cushions: fabrication, characterization and applications. [Review] [29 refs]. Trends In Biotechnology, 18, 58–64. [DOI] [PubMed] [Google Scholar]
- Shen K, Tsai J, Shi P & Kam LC (2009) Self-aligned supported lipid bilayers for patterning the cell-substrate interface. J Am Chem Soc, 131, 13204–13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SJ & Nicolson GL (1972) The Fluid Mosaic Model of the Structure of Cell Membranes. Science, 175, 720–731. [DOI] [PubMed] [Google Scholar]
- Taylor JD, Phillips KS, and Cheng Q (2007) Microfluidic fabrication of addressable tethered lipid bilayer arrays and optimization using SPR with silane-derivatized nanoglassy substrates. Lab on a Chip, 7, 927–930. [DOI] [PubMed] [Google Scholar]
- Tsai J, Sun E, Gao Y, Hone JC & Kam LC (2008) Non-Brownian diffusion of membrane molecules in nanopatterned supported lipid bilayers. Nano Letters, 8, 425–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon TY et al. (2006) Topographic control of lipid-raft reconstitution in model membranes. Nature Materials, 5, 281–285. [DOI] [PubMed] [Google Scholar]


